Significance
Mistakes in chromosome attachment activate the spindle assembly checkpoint (SAC) to delay anaphase onset. However, it is poorly understood how this checkpoint is silenced to initiate anaphase once all chromosomes are attached properly. Our research uncovers the SAC silencing network (SSN) in budding yeast. Chromosome bipolar attachment applies tension on chromosomes. The SSN prevents SAC silencing prior to tension generation but triggers SAC silencing once chromosomes are under tension, thereby ensuring that cells enter anaphase only after bipolar attachment. Our data indicate that the coordination of SAC silencing with chromosome attachment is achieved through the modulation of the phosphorylation of a kinetochore protein.
Abstract
Improper kinetochore attachments activate the spindle assembly checkpoint (SAC) to prevent anaphase onset, but it is poorly understood how this checkpoint is silenced to allow anaphase onset. Chromosome bipolar attachment applies tension on sister kinetochores, and the lack of tension delays anaphase onset. In budding yeast, the delay induced by tension defects depends on the intact SAC as well as increase in ploidy (Ipl1)/Aurora kinase and a centromere-associated protein ShuGOshin (Sgo1). Here we provide evidence indicating that Ipl1-dependent phosphorylation of the kinetochore protein Duo1 and Mps1 interacting (Dam1) prevents SAC silencing when tension is absent. The nonphosphorylatable dam1 mutant cells, as well as sgo1 mutant cells, are competent in SAC activation but unable to prevent SAC silencing in response to tension defects. We further found that phosphomimetic dam1 mutants exhibited delayed anaphase onset mainly due to the failure in SAC silencing, but destabilized kinetochore attachment likely plays a minor role in this delay. Because the tension resulting from bipolar attachment triggers the dephosphorylation of Dam1 by protein phosphatase 1, this dephosphorylation likely coordinates SAC silencing with chromosome bipolar attachment. Therefore, Sgo1, Ipl1 kinase, Dam1, and protein phosphatase 1 comprise the SAC silencing network that ensures the correct timing for anaphase onset.
The attachment of sister kinetochores to microtubules emanating from opposite spindle poles establishes bipolar attachment, a process essential for sister chromatid segregation. Mistakes in this process activate the spindle assembly checkpoint (SAC) to delay anaphase onset. The SAC components include mitotic arrest-deficient 1 (Mad1), Mad2, Mad3, budding uninhibited by benzimidazole 1 (Bub1), Bub3 and monopolar spindle (Mps1), which are well conserved in all eukaryotes (1–3). Unattached kinetochores recruit some checkpoint proteins, such as Mad1 and Bub1, generating an inhibitory signal to delay anaphase entry (4, 5). The current view is that some active SAC components sequester cell division cycle 20 (Cdc20), the activator of anaphase promoting complex (APC), thereby preventing APCCdc20 activation. Because APCCdc20 mediates the degradation of securin precocious dissociation of sisters 1 (Pds1), the anaphase inhibitor, active SAC delays anaphase onset by stabilizing Pds1 (6). Once all chromosomes have achieved bipolar attachment, the SAC must be silenced for the initiation of anaphase, in which sister chromatids segregate. However, the link between chromosome attachment and SAC silencing is still missing.
Bipolar attachment generates tension on chromosomes, but chromosomes become tension-defective when sister-chromatid cohesion is eliminated or when two sister kinetochores are attached by microtubules emanating from the same spindle pole (syntelic attachment). The SAC is required for anaphase entry delay in response to tension defects. Interestingly, in budding yeast, increase in ploidy 1 (Ipl1) kinase (the Aurora B homolog) and a centromere-localized protein ShuGOshin (Sgo1) are also required to prevent anaphase entry in cells lacking tension; nevertheless, they are dispensable for the cell cycle arrest induced by unattached kinetochores (7–9). One explanation is that Ipl1 kinase destabilizes tension-defective attachments to generate unattached kinetochores, which not only facilitates error-correction but also activates the SAC (10, 11). Because Sgo1 does not play a role in destabilizing attachments, it is also possible that Ipl1 and Sgo1 regulate SAC activity through an uncharacterized mechanism.
Recent studies indicate that several mechanisms may contribute to SAC silencing, including the dissociation of SAC components from Cdc20 (12), and the stripping of Mad1 and Mad2 from the kinetochore by a dynein motor (13). However, these mechanisms fail to establish the link between SAC silencing and chromosome attachment. Recent evidence indicates the role of protein phosphatase 1 (PP1) in SAC silencing, because high levels of PP1, which antagonizes Ipl1 kinase, promote SAC silencing in yeast cells (14, 15). However, the relevant substrate(s) of Ipl1/PP1 for SAC silencing remains unidentified.
Here, we present evidence for the existence of the SAC silencing network (SSN) in budding yeast, which includes Sgo1, Ipl1 kinase, PP1, and a kinetochore protein Dam1. Our data suggest that Ipl1 phosphorylates Dam1 to prevent SAC silencing before tension generation; nevertheless, tension-induced Dam1 dephosphorylation triggers SAC silencing. Therefore, the SSN coordinates SAC silencing with chromosome bipolar attachment through the modulation of Dam1 phosphorylation, ensuring that anaphase onset occurs at the right time.
Results
Ipl1 and Sgo1 Are Part of the SSN.
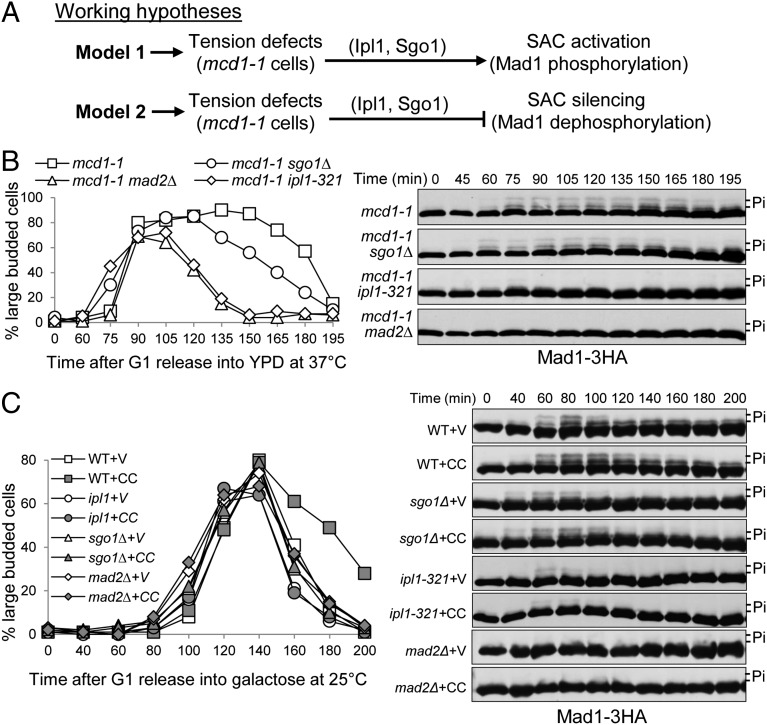
Ipl1 and Sgo1 are required to delay anaphase onset in response to tension defects. One possibility is that Ipl1 and Sgo1 are part of the SAC and required for SAC activation in the absence of tension, or they prevent SAC silencing in cells lacking tension (Fig. 1A). To distinguish these possibilities, we assessed the SAC activation and silencing processes in ipl1 and sgo1 mutants in the absence of tension by examining the phosphorylation of a SAC protein Mad1, which indicates SAC activation (16, 17). Because mitotic chromosome determinant 1 (Mcd1) is one of the cohesin subunits and mcd1-1 mutant cells fail to generate tension on sister chromatids when incubated at 37 °C (7, 8), we analyzed the kinetics of Mad1 phosphorylation in mcd1-1 MAD1–3HA cells with dysfunctional Ipl1, Sgo1, or a SAC component Mad2.
Fig. 1.
ipl1 and sgo1 mutants exhibit premature SAC silencing. (A) Two working models for the checkpoint function of Ipl1 and Sgo1 in response to tension defects. (B) Mad1 phosphorylation kinetics in checkpoint mutant cells lacking functional cohesin Mcd1. MAD1–3HA cells with the indicated genotypes were synchronized in G1 phase with α-factor at 25 °C, and then released into 37 °C yeast peptone dextrose (YPD) medium. α-factor was added back to block rebudding. Mad1 protein was detected after Western blotting with anti-HA antibody. The budding index is shown on the Left panel, and Mad1 protein modification during the cell cycle is shown on the Right. The slow migrating bands represent phosphorylated Mad1. All of the time course experiments in this project were repeated at least twice. (C) Mad1 phosphorylation kinetics in checkpoint mutants in the presence of CIK1-CC–induced syntelic attachments. A vector (V) or a PGALCIK1-CC (CC) plasmid was introduced into cells with the indicated genotypes. The transformants were grown in raffinose medium to midlog phase and then synchronized in G1 phase. The cells were released into galactose medium to induce CIK1-CC overexpression. α-factor was restored after budding to block the second round of cell cycle. The budding index and Mad1 protein level are shown.
The cells were synchronized in G1-phase and then released into the cell cycle at 37 °C to inactivate Mcd1. The slow migrating bands of Mad1 were observed after release for 60 min in mcd1-1 cells, indicating Mad1 phosphorylation and SAC activation. We noticed the reduction in band-shift at the later time points (180 and 195 min), which is consistent with the decrease of large-budded cells (Fig. 1B). In clear contrast, no Mad1 phosphovariants were detected in mcd1-1 mad2Δ mutant cells during the cell cycle, indicating complete abolishment of SAC activity (16, 17). In mcd1-1 sgo1Δ cells, obvious Mad1 phosphorylation was detected starting from 60 min, but the slow migrating forms disappeared much sooner compared with mcd-1 single mutants. For the mcd1-1 ipl1-321 cells, weak Mad1 phosphovariants were noticed at 75 min and then disappeared (Fig. 1B). Quantitative analysis using ImageJ indicates the percentage of phosphorylated Mad1 in these cells during the cell cycle (Fig. S1A). Thus, unlike the SAC mutant mad2Δ, the SAC can be activated to some degree in ipl1 and sgo1 mutants in response to tension defects. In agreement with the kinetics of Mad1 phosphorylation, mcd1-1 cells arrested as large-budded cells after release into 37 °C medium until 180 min. The accumulation of large-budded cells was abolished by mad2Δ and ipl1-321 mutants, but a partial suppression was observed in mcd1-1 sgo1Δ mutants (Fig. 1B).
We recently found that the loss of function of the chromosome instability and karyogamy 1 (Cik1)/karyogamy 3 (Kar3) motor complex increases the frequency of syntelic attachment, which also results in tension-defective chromosomes. Moreover, overexpression of the coiled-coil domain of CIK1 (CIK1-CC) disrupts Cik1–Kar3 interaction and induces an anaphase entry delay that depends on both Ipl1 and Sgo1 (9, 18). Therefore, we also compared the Mad1 phosphorylation kinetics in wild-type (WT), ipl1, and sgo1 mutant cells overexpressing CIK1-CC. G1-synchronized cells carrying a PGALCIK1-CC plasmid were released into 25 °C medium containing galactose to induce CIK1-CC overexpression. WT cells overexpressing CIK1-CC exhibit more persistent Mad1 phosphorylation compared with the vector control (Fig. 1C). The delayed disappearance of Mad1 phosphovariants induced by CIK1-CC was partially suppressed by sgo1Δ and completely suppressed by ipl1–321. As a control, no Mad1 phosphorylation was detected in mad2Δ mutants. Consistently, mad2Δ, ipl1–321, and sgo1Δ mutants also abolished the delayed transition from large-budded cells to single cells induced by CIK1-CC overexpression (Fig. 1C). The result supports a unique concept that Ipl1 and Sgo1 prevents SAC silencing in response to tension defects by inhibiting Mad1 dephosphorylation, but this does not exclude the possibility that Ipl1 is also involved in SAC activation.
The Phosphorylation of Dam1 by Ipl1 Is Required to Prevent Anaphase Entry in Response to Tension Defects.
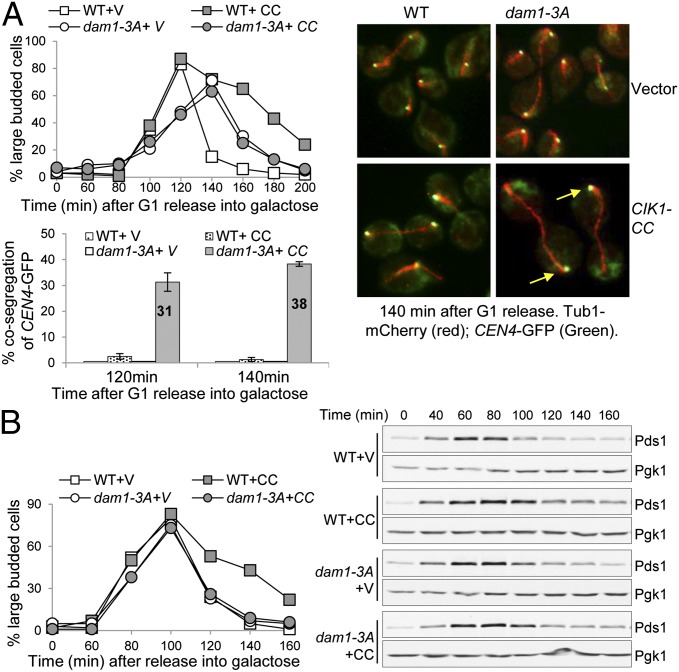
We have shown that ipl1–321 mutants are sensitive to syntelic attachments induced by CIK1-CC overexpression (9). We reason that an Ipl1-dependent phosphorylation event is essential for the viability in cells with syntelic attachments. Kinetochore protein Dam1 is one of the substrates of Ipl1 kinase, and replacement of three of the four Ipl1 kinase consensus sites (S257, S265, and S292) with alanine generates a viable dam1–3A mutant (19). We introduced a PGALCIK1-CC plasmid into WT and dam1–3A mutant cells. dam1–3A mutant cells harboring a PGALCIK1-CC plasmid were unable to grow on galactose plates that induce CIK1-CC overexpression. Moreover, 85% of dam1–3A cells lost viability after CIK1-CC overexpression for 6 h, compared with 16% viability loss for WT cells (Fig. S1B).
To determine the cause of the lethality of dam1–3A cells overexpressing CIK1-CC, we analyzed the cell cycle progression and chromosome segregation. To this end, we introduced the PGALCIK1-CC plasmid into WT and dam1–3A cells with a GFP-marked centromere of chromosome IV (CEN4–GFP) and mCherry-labeled microtubules (Tub1-mCherry). After G1 release into galactose medium, overexpression of CIK1-CC delayed cell cycle progression in WT cells as more cells remained as large-budded presumably due to the induction of syntelic attachment, but this delay was not observed in dam1–3A cells (Fig. 2A). We examined CEN4–GFP segregation in WT and dam1–3A mutant cells with an elongated spindle at 120 and 140 min. Overexpression of CIK1-CC only induced cosegregation of two CEN4–GFP dots with one spindle pole in 3% of WT cells, but the frequency of cosegregation increased to more than 30% in dam1–3A mutant cells (Fig. 2A), likely causing viability loss.
Fig. 2.
The nonphosphorylatable dam1–3A mutants are sensitive to the induction of syntelic attachments. (A) CIK1-CC overexpression causes chromosome missegregation in dam1–3A mutant cells. A vector (V) or a PGALCIK1-CC (CC) plasmid was introduced into WT and dam1–3A cells with CEN4–GFP TUB1–mCherry. The transformants were first arrested in G1 phase in raffinose medium and then released into galactose medium at 30 °C. Cells were collected at the indicated time points for the examination of fluorescence signals. The budding index is shown in the Left panel. The Right panel shows the distribution of CEN4–GFP and spindle morphology in some representative cells. The arrows indicate cells with cosegregated CEN4–GFP. The percentage of cells with an elongated spindle as well as cosegregated CEN4–GFP dots at 120 and 140 min is shown in the Lower panel (n > 100). The percentage is the average from three independent experiments. (B) dam1–3A mutation suppresses the delayed Pds1 degradation induced by CIK1-CC overexpression. G1-arrested PDS1–18myc and dam1–3A PDS1–18myc cells with a vector or a PGALCIK1-CC plasmid were released into 30 °C galactose medium. α-factor was restored after budding. The Pds1 protein levels were determined after Western blotting. Pgk1 protein levels are used as a loading control. The budding index is shown in the Left panel, and Pds1 levels are shown in the Right panel.
To further assess the checkpoint function in dam1–3A cells in response to tension defects, we examined Pds1 protein levels, as its degradation marks anaphase onset. G1-arrested PDS1-18myc (c-Myc) and dam1-3A PDS1-18myc cells carrying either a vector or a PGALCIK1-CC plasmid were released into galactose medium. An obvious delay in Pds1 degradation was observed in WT cells overexpressing CIK1-CC, but this delay was abolished in dam1–3A mutant cells (Fig. 2B). Therefore, dam1–3A mutants are unable to delay anaphase entry in response to syntelic attachments. We also examined the checkpoint competency of dam1–3A cells in response to tension defects induced by Mcd1 inactivation. Pds1 protein was stabilized in mcd1-1 cells incubated at 37 °C, but this stabilization was abolished in mad1Δ, sgo1Δ, and dam1–3A mutant strains (Fig. S2A). Therefore, tension defects induced by syntelic attachments or cohesin inactivation fail to delay anaphase onset in nonphosphorylatable dam1–3A cells, supporting the conclusion that Dam1 phosphorylation by Ipl1 is necessary for the checkpoint response to tension defects.
As ipl1 and sgo1Δ mutants show intact SAC function when treated with spindle poison nocodazole (7, 8), we compared the cell cycle progression in WT and dam1–3A cells in the presence of nocodazole. After G1 release into the medium containing 20 µg/mL of nocodazole, both WT and dam1–3A cells arrested as large-budded cells with stabilized Pds1 (Fig. S2B), suggesting that the SAC is functional in dam1–3A cells. Taken together, nonphosphorylatable dam1–3A mutants behave like ipl1 and sgo1 mutants, which show intact SAC function but fail to arrest the cell cycle in response to tension defects.
dam1–3A Mutants Show Premature SAC Silencing in Response to Tension Defects.
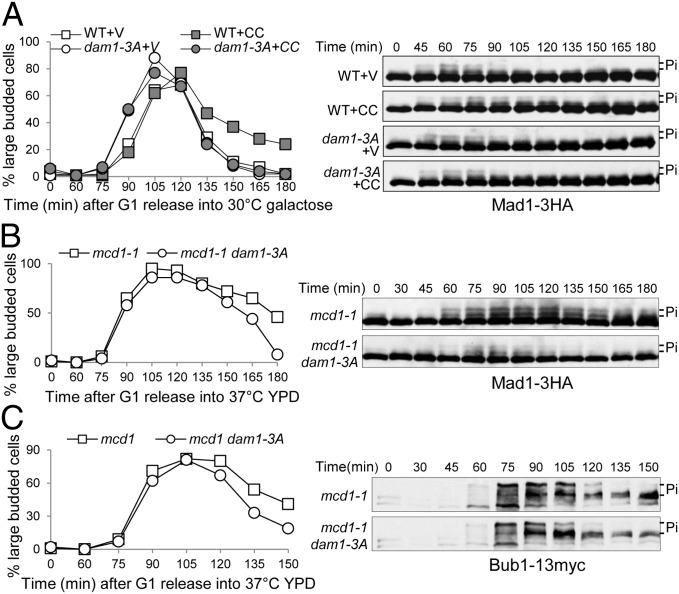
Because our data indicate premature SAC silencing in ipl1 and sgo1Δ mutants in response to tension defects, we also analyzed the SAC silencing process in dam1–3A cells. G1-arrested MAD1–3HA and dam1–3A MAD1-3HA cells carrying either a vector or PGALCIK1-CC plasmid were released into galactose medium at 30 °C. The delayed Mad1 dephosphorylation induced by CIK1-CC overexpression was eliminated in dam1–3A mutant cells (Fig. 3A). We also examined Mad1 phosphorylation kinetics in synchronized mcd1-1 and mcd1-1 dam1–3A cells incubated at 37 °C. Clearly, dam1–3A cells were unable to maintain the phosphorylation status of Mad1 in the absence of tension (Fig. 3B), which could be a result of increased Mad1 dephosphorylation or impaired Mad1 phosphorylation.
Fig. 3.
dam1–3A mutants exhibit premature SAC silencing. (A) dam1–3A cells show premature Mad1 dephosphorylation when CIK1-CC is overexpressed. G1-arrested MAD1–3HA and dam1–3A MAD1–3HA cells harboring a vector (V) or a PGALCIK1-CC (CC) plasmid were released into 30 °C galactose medium. α-factor was added back after budding. Mad1 protein was detected after Western blotting. The budding index and Mad1 protein level are shown. (B) dam1–3A cells show premature Mad1 dephosphorylation when cohesin Mcd1 is inactivated. mcd1-1 MAD1–3HA and mcd1-1 dam1–3A MAD1–3HA cells growing at 25 °C were synchronized in G1 and then released into 37 °C YPD medium. Cell lysates were prepared at the indicated times for Western blotting with anti-HA antibody. The budding index and Mad1 modification are shown. (C) dam1–3A cells show premature Bub1 dephosphorylation in the absence of cohesion. mcd1-1 BUB1–13myc and mcd1-1 dam1–3A BUB1–13myc cells growing at 25 °C were synchronized in G1 phase and then released into 37 °C YPD medium. Cell lysates were prepared every 15 min, and Bub1 modification was analyzed after Western blotting. Shown here are the budding index and Bub1 protein levels.
In addition to Mad1, other SAC proteins may also become dephosphorylated before SAC silencing. Another SAC component, Bub1, is a phosphoprotein (20, 21); thus, we analyzed its phosphorylation kinetics in synchronized mcd1-1 and mcd1-1 dam1–3A mutant cells incubated at 37 °C. These two strains exhibited similar Bub1 phosphorylation levels at 75 and 90 min after G1 release, but mcd1-1 dam1–3A cells showed obvious premature dephosphorylation (Fig. 3C). One explanation is that Ipl1-dependent Dam1 phosphorylation blocks Bub1 dephosphorylation to prevent SAC silencing.
The Phosphomimetic dam1–3D Cells Show SAC-Dependent Delay in Anaphase Entry.
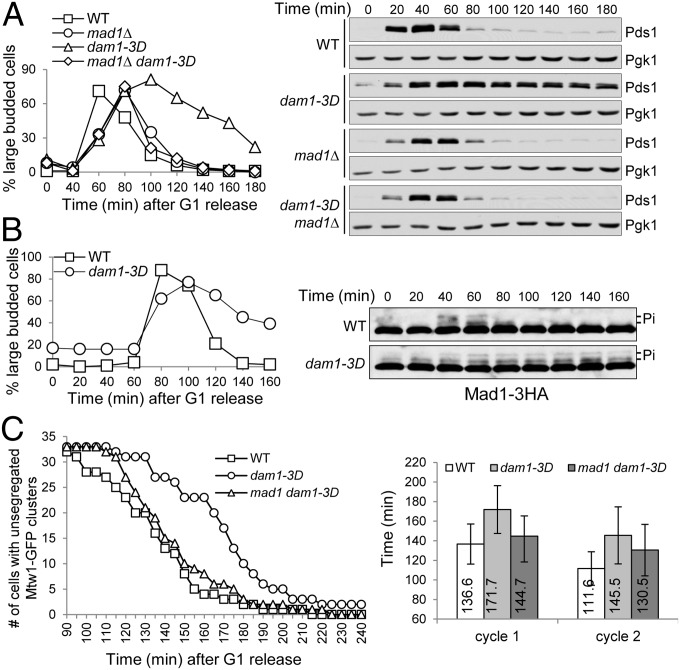
If Dam1 phosphorylation prevents SAC silencing, phosphomimetic dam1 mutants are expected to show impaired SAC silencing. Replacement of three of the four Ipl1 consensus Ser (S) residues in Dam1 with phosphomimetic residue Asp (D) generates viable dam1–3D (S257D S265D S292D) mutants that show a slow growth phenotype (19). This phenotype could be a result of SAC activation. To test this possibility, we crossed dam1–3D to SAC mutants mad1Δ and mad2Δ. Interestingly, we obtained viable dam1–3D mad1Δ and dam1–3D mad2Δ double mutants, although the double mutants showed similar sick growth as dam1–3D single mutant cells. Using dam1–3D mad1Δ mutants, we assessed if dam1–3D mutants show an SAC-dependent anaphase entry delay by examining Pds1 protein levels in synchronized cells. After G1 release into cell cycle, dam1–3D mutant cells showed stabilized Pds1 protein levels, indicating delayed anaphase entry. Consistently, the mutant cells also exhibited a significant delay in the transition from large-budded to unbudded G1 cells. Deletion of the SAC checkpoint MAD1, however, abolished Pds1 stabilization and delayed M to G1 transition (Fig. 4A).
Fig. 4.
dam1–3D mutants show SAC silencing defects. (A) dam1–3D cells exhibit SAC-dependent anaphase entry delay. G1-arrested PDS1–18myc cells with the indicated genotypes were released into YPD medium at 30 °C. α-factor was added back after budding to block further cell cycle. Cell lysates were prepared every 20 min, and the Pds1 protein levels were determined after Western blotting. The budding index and Pds1 protein levels are shown. Pgk1 protein levels are used as a loading control. (B) dam1–3D cells exhibit persistent Mad1 phosphorylation. G1-arrested MAD1–3HA and dam1–3D MAD1–3HA cells were released into YPD medium at 30 °C. α-factor was restored after budding. Mad1 protein was detected after Western blotting. Shown here are the budding index and Mad1 protein levels. (C) Live-cell imaging of chromosome segregation in dam1–3D and dam1–3D mad1Δ cells with GFP-tagged kinetochore protein Mtw1. Cells with the indicated genotypes were first arrested in G1 and then loaded onto the pad containing complete synthetic medium. The images were acquired every 5 min for 6 h. The Left panel shows the cell numbers with unsegregated Mtw1–GFP clusters over time. The cell numbers for the three strains used in this experiment are WT, 32; dam1–3D, 33; dam1–3D mad1Δ, 33. The average doubling time for the three strains during the first and second cell cycle is shown on the Right panel. The doubling time for the first cell cycle starts from G1 release to the first time point showing the segregation of two Mtw1–GFP clusters into two daughter cells. The doubling time for the second cell cycle is defined as the time difference between two Mtw1–GFP cluster segregation points in the sequential cell cycles.
To further analyze the SAC activation status in dam1–3D mutant cells, we examined Mad1 and Bub1 phosphorylation kinetics during the cell cycle in dam1–3D cells. Strikingly, the mutant cells showed substantially delayed dephosphorylation of these two SAC components, indicating persistent SAC activation (Fig. 4B and Fig. S3A). Therefore, phosphomimetic dam1–3D cells have difficulty entering anaphase due to active SAC.
dam1–3D Mutant Cells Show Defects in SAC Silencing.
Next, we asked what causes the anaphase entry delay in dam1–3D cells. Because previous data indicate the role of Ipl1 kinase in destabilizing tension-defective kinetochore attachments (10, 11), one possibility is that unattached kinetochores in phosphomimetic dam1–3D cells activate the SAC to delay anaphase onset. If that is the case, dysfunctional SAC will allow dam1–3D cells to enter anaphase with unattached kinetochores and result in chromosome missegregation. Using the established colony sectoring assay (22), we determined the chromosome loss rate in dam1–3D strains with or without functional SAC. Both WT and dam1–3D single mutants showed a low chromosome loss rate (0.1% and 0.21%, respectively). The rate for dam1–3D mad2Δ mutants (0.53%) is similar to that of mad2Δ (0.48%), indicating that the kinetochore attachment defect in dam1–3D cells is not significant (Fig. S3B). The fact that dam1–3D cells are viable in the absence of MAD1 or MAD2 also supports this notion. Therefore, the destabilized kinetochore attachment cannot fully explain the dramatic anaphase entry delay in dam1–3D cells.
To further clarify if kinetochore detachment is responsible for the delayed anaphase onset in dam1–3D mutant cells, we performed live-cell imaging to visualize the chromosome segregation process in two sequential cell cycles. We speculate that a haploid yeast cell lacking a whole chromosome is unable to accomplish mitosis. Thus, the successful chromosome segregation in both daughter cells indicates that no chromosome missegregation occurs during the preceding cell cycle. WT, dam1–3D, and dam1–3D mad1Δ cells were first arrested in G1 phase and then spotted onto the surface of a slide with agarose medium pad. For each strain, about 30 cells were visualized for the segregation of kinetochore clusters [mis twelve-like 1 (Mtw1)-GFP] for two sequential cell divisions. dam1–3D cells showed a clear delay in kinetochore cluster segregation, and 1 of the 33 dam1–3D cells never finished the first segregation, but this delay was suppressed by mad1Δ (Fig. 4C and Fig. S4). Among all of the daughters of the 33 dam1–3D mad1Δ cells, only two of them failed to segregate chromosomes during the second round of cell cycle, suggesting that 31 of the 33 dam1–3D mad1Δ cells had faithful chromosome segregation during the first round of mitosis. Therefore, the failure of SAC silencing, but not the destabilized kinetochore attachment, may play a major role in the delayed anaphase entry in dam1–3D cells.
Chromosome bipolar attachment applies tension on chromosomes that separates sister kinetochores/centromeres before anaphase entry; thus, we also assessed the bipolar attachment process in dam1–3D cells by examining the kinetics of sister centromere separation (CEN4–GFP) in synchronized cdc13-1 and cdc13-1 dam1–3D cells that arrest in preanaphase at high temperature due to the activation of the DNA damage checkpoint (23). After G1 release into 34 °C medium, cdc13-1 and cdc13-1 dam1–3D cells showed similar kinetics for the separation of CEN4–GFP dots (Fig. S5A), indicating that the defect in bipolar attachment is not obvious in dam1–3D cells.
If the SAC silencing process is impaired in dam1–3D cells, the mutant cells will show more dramatic delay in anaphase entry after SAC activation by nocodazole treatment. As dam1–3D cells show significant anaphase entry delay, we first arrested the cells in preanaphase using cdc13-1 in the presence or absence of nocodazole, and then analyzed the recovery process. G1-arrested cdc13-1 and cdc13-1 dam1–3D cells were released into 32 °C medium with or without nocodazole. After nocodazole washout, the cells were released into 25 °C medium. cdc13-1 and cdc13-1 dam1–3D mutants without nocodazole treatment showed similar kinetics for the transition from large-budded cells to single cells. Our explanation is that cdc13-1–induced arrest allowed enough time for SAC silencing even in cdc13-1 dam1–3D mutants. Once exposed to nocodazole, however, cdc13-1 dam1–3D mutants exhibited a significant delay in this transition compared with cdc13-1 cells, which could be attributed by impaired SAC silencing (Fig. S5B).
Ipl1 Kinase and PP1 Control SAC Silencing Through Dam1.
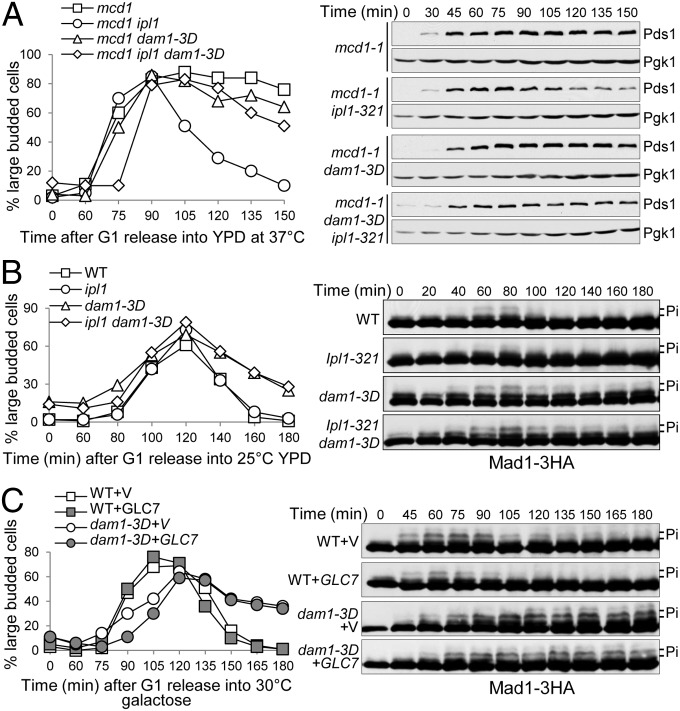
If the failure of Dam1 phosphorylation by Ipl1 contributes to the premature anaphase onset in ipl1 mutant cells lacking tension, phosphomimetic dam1 mutant will suppress this mutant phenotype. To test this idea, we first compared the cell cycle progression in synchronized mcd1-1, mcd1-1 ipl1–321, and mcd1-1 ipl1–321 dam1–3D cells incubated at 37 °C. As shown previously, mcd1-1 cells exhibited stabilized Pds1 protein, but this stabilization was abolished in mcd1-1 ipl1–321 mutant cells. However, dam1–3D mutant suppressed the premature Pds1 degradation in mcd1-1 ipl1–321 cells. The decrease of large-budded cells in mcd1-1 ipl1–321 cells in the later time points was also suppressed by dam1–3D (Fig. 5A). We quantified Pds1 protein levels in these cells, confirming the suppression of premature anaphase entry (Fig. S6A). Therefore, dam1–3D mutant blocks the premature anaphase entry in ipl1 mutant cells in the absence of tension.
Fig. 5.
Ipl1 kinase and PP1 phosphatase control SAC silencing through Dam1. (A) dam1–3D mutation blocks premature anaphase entry in mcd1-1 ipl1–321 mutant cells. G1-arrested PDS1–18myc cells with indicated genotypes were released into 37 °C YPD. α-factor was added back after budding. Cells were collected every 15 min for the budding index and the determination of Pds1 protein levels. Pgk1 protein levels are used as a loading control. (B) ipl1–321 dam1–3D cells show compromised Mad1 dephosphorylation. G1-arrested MAD1–3HA cells with indicated genotypes were released into 30 °C YPD medium, and α-factor was added back after budding. Cells were collected at the indicated time points for the determination of budding index and Mad1 protein modification. (C) dam1–3D cells exhibit defective Mad1 dephosphorylation when PP1 is overexpressed. G1-arrested MAD1–3HA and dam1–3D MAD1–3HA cells containing a vector (V) or a PGALGLC7 (GLC7) plasmid were released into galactose medium and incubated at 30 °C. α-factor was added back after budding. Cells were collected over time to determine the budding index and Mad1 protein modification.
We further analyzed the SAC activation and silencing kinetics in synchronized WT, ipl1–321, and ipl1–321 dam1–3D cells by examining the phosphorylation of Mad1. Synchronized ipl1–321 cells showed compromised Mad1 phosphorylation when incubated at 25 °C compared with WT cells, presumably due to the partial loss of Ipl1 kinase activity. However, dam1–3D and ipl1–321 dam1–3D mutant cells exhibited persistent Mad1 phosphorylation as well as increased proportion of large-budded cells (Fig. 5B). Therefore, the phosphorylation of Dam1 can delay anaphase onset in the absence of Ipl1 activity, which supports the notion that Dam1 acts downstream of Ipl1 to regulate SAC silencing.
Recent studies indicate the role of PP1 in SAC silencing (14, 15, 24, 25). Overexpression of glycogen 7 (Glc7), the catalytic subunit of PP1, induces checkpoint silencing in the presence of improper kinetochore attachments (15). To test if PP1 silences the SAC by dephosphorylating Dam1, we analyzed Mad1 phosphorylation kinetics in WT and dam1–3D mutant cells overexpressing GLC7. We found that GLC7 overexpression obviously reduced Mad1 phosphorylation, supporting the positive role of PP1 in SAC silencing. In dam1–3D mutant cells, however, persistent Mad1 dephosphorylation was observed even when Glc7 is overproduced (Fig. 5C), indicating that PP1 likely silences the SAC by dephosphorylating Dam1.
As dam1–3A and sgo1Δ mutants show similar checkpoint defects, we also tested if Sgo1 acts up- or downstream of Dam1. The cell cycle progression in dam1–3D and dam1–3D sgo1Δ mutants were compared by examining the Pds1 levels. Strikingly, the anaphase entry delay in dam1–3D mutant was completely suppressed by sgo1Δ (Fig. S6B). Thus, Ipl1 kinase and phosphatase PP1 act upstream of Dam1, but Sgo1 functions downstream of Dam1.
Discussion
Mistakes in chromosome–microtubule attachment are monitored by the SAC that delays anaphase onset to facilitate error correction, but it remains unclear how the SAC is silenced once all chromosomes are attached correctly. Our data reveal the SSN in budding yeast that ensures correct timing for anaphase onset. Before bipolar attachment that induces tension generation, one branch of the SSN prevents SAC silencing through Ipl1-dependent phosphorylation of a kinetochore protein Dam1. After tension generation, the activation of another SSN branch induces PP1-dependent Dam1 dephosphorylation and the subsequent SAC silencing. Therefore, the Ipl1 kinase, phosphatase PP1, their substrate Dam1, and Sgo1 comprise the SSN that couples the SAC silencing process to chromosome bipolar attachment.
It is well established that cells monitor detached kinetochores by activating the SAC, but the checkpoint response to tension-defective chromosome attachments is much less understood. Previous data suggest that Ipl1 kinase promotes the conversion of tension-defective kinetochores to unattached ones, which in turn activates the SAC and facilitates error correction (10). Our results demonstrate that Ipl1 prevents anaphase entry by phosphorylating a kinetochore protein Dam1, but this function is separable from Ipl1-dependent kinetochore attachment destabilization. In support of this notion, we found that a SAC mutant mad1Δ can suppress the anaphase entry delay in dam1–3D cells without causing a high frequency of chromosome loss. Therefore, in addition to the destabilization of kinetochore attachment, Dam1 phosphorylation also plays a key role in preventing SAC silencing.
Previous data show that the dephosphorylation of Dam1 depends on tension at sister kinetochores (26). Here we present evidence showing that the dephosphorylation of Dam1 is necessary for SAC silencing, supporting a model that tension at kinetochores silences the SAC by inducing Dam1 dephosphorylation. Therefore, kinetochore attachment and tension may regulate anaphase onset in different ways. It is likely that unattached kinetochores activate the SAC robustly to delay anaphase entry, but tension-defective attachments maintain metaphase arrest by preventing SAC silencing. In support of this conclusion, we found that Sgo1 protein and Ipl1-dependent phosphorylation of Dam1 are required to retain the phosphorylation status of SAC proteins when tension is absent. It is possible that Dam1 and Sgo1 regulate the activity of a phosphatase or kinase at the kinetochore to control the timing of SAC silencing, although further work is needed to elucidate the detailed mechanism.
In summary, our research work reveals the SSN in budding yeast, which includes Ipl1 kinase, phosphatase PP1, kinetochore protein Dam1, and a centromere-associated Sgo1. Our genetic analysis suggests that Ipl1 and PP1 act upstream of Dam1, but Sgo1 functions downstream of Dam1. Our data support the model that biorientation, once achieved, enables PP1 to dephosphorylate Dam1, which extinguishes the SAC signal individually on each sister kinetochore. Given the fact that all of the SSN components are conserved in eukaryotes, human cells likely have a similar network to coordinate cell cycle progression and chromosome attachments. Deregulation of this mechanism will lead to premature anaphase onset, thereby resulting in chromosome missegregation and aneuploidy, a cause for cancer development.
Materials and Methods
Yeast Strains, Growth, and Media.
The relevant genotypes and sources of the yeast strains used in this study are listed in Table S1. All of the strains listed are isogenic to Y300, a W303 derivative. The BUB1-13myc strains and some deletion mutants were constructed by using a PCR-based protocol. The yeast cell growth, synchronization, and CIK1-CC overexpression were described previously (9).
Western Blot Analysis.
We collect 1.5 mL of yeast cells by centrifugation and prepare yeast protein samples. Protein samples were resolved by 8% (vol/vol) SDS/PAGE. Proteins were detected with ECL (Perkin-Elmer LAS, Inc.) after probing with anti-myc and anti- HA primary antibodies (Covance Research Products, Inc.) and HRP-conjugated secondary antibody (Jackson ImmunoResearch, Inc.). Antiphosphoglycerate kinase 1 (Pgk1) antibody (Molecular Probes) was used to determine the protein levels of Pgk1, which are used as a loading control.
Cytological Techniques.
Collected cells were fixed with 3.7% formaldehyde for 20 min at room temperature. The cells were washed once with 1xPBS (pH 7.2) and then resuspended in PBS buffer to examine fluorescence signals with a microscope (Carl Zeiss MicroImaging, Inc.). Live-cell microscopy was carried out with the Andor Revolution SD imaging system. Cells synchronized in G1 were spotted onto an agarose pad filled with synthetic complete medium. All live-cell images were acquired at 25 °C with a 100× objective lens. A z-stack with 20 planes separated by 0.4 μm was acquired every 5 min and converted to maximum projection using Andor IQ2 software.
Supplementary Material
Acknowledgments
We thank Drs. Georjana Barnes, Frank Uhlmann, and Sue Biggins for providing yeast strains. We are grateful to Kelly McKnight and Drs. Daniel Kaplan and Tim Megraw, who read through the manuscript. We also thank the yeast community at Florida State University (FSU) for reagents and suggestions. This work was supported by the R15GM097326-01 and RO1GM102115-01A1 from National Institutes of Health (to Y.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307595111/-/DCSupplemental.
References
- 1.Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66(3):507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 2.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66(3):519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 3.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273(5277):953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 4.Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol. 2004;164(4):535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274(5285):242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10(24):3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 7.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15(23):3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307(5706):130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- 9.Jin F, Liu H, Li P, Yu HG, Wang Y. Loss of function of the Cik1/Kar3 motor complex results in chromosomes with syntelic attachment that are sensed by the tension checkpoint. PLoS Genet. 2012;8(2):e1002492. doi: 10.1371/journal.pgen.1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8(1):78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108(3):317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 12.Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci. 2011;124(Pt 22):3905–3916. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howell BJ, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155(7):1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanoosthuyse V, Hardwick KG. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol. 2009;19(14):1176–1181. doi: 10.1016/j.cub.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19(14):1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick KG, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131(3):709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirchenko L, Uhlmann F. Sli15(INCENP) dephosphorylation prevents mitotic checkpoint reengagement due to loss of tension at anaphase onset. Curr Biol. 2010;20(15):1396–1401. doi: 10.1016/j.cub.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Jin F, Liang F, Tian X, Wang Y. The Cik1/Kar3 motor complex is required for the proper kinetochore-microtubule interaction after stressful DNA replication. Genetics. 2011;187(2):397–407. doi: 10.1534/genetics.110.125468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111(2):163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 20.Farr KA, Hoyt MA. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol Cell Biol. 1998;18(5):2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol. 2000;10(11):675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- 22.Hieter P, Mann C, Snyder M, Davis RW. Mitotic stability of yeast chromosomes: A colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 23.Liang F, Wang Y. DNA damage checkpoints inhibit mitotic exit by two different mechanisms. Mol Cell Biol. 2007;27(14):5067–5078. doi: 10.1128/MCB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meadows JC, et al. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev Cell. 2011;20(6):739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol. 2011;21(11):942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keating P, Rachidi N, Tanaka TU, Stark MJ. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. J Cell Sci. 2009;122(Pt 23):4375–4382. doi: 10.1242/jcs.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.