Significance
Glycogen is a branched glucose polymer found in most animals, fungi, bacteria, and archaea as an osmotically neutral means of energy storage. Glycogen also contains minor amounts of phosphate which can be removed by a dual specificity phosphatase, laforin. Accumulation of phosphate results in highly insoluble glycogen deposits and underlies Lafora disease, a devastating form of myoclonus epilepsy. In this paper, we present structural and kinetic data that support a plausible mechanism by which phosphate is directly incorporated into glycogen by glycogen synthase.
Keywords: crystallography, glycosyltransfer
Abstract
Glycogen is a glucose polymer that contains minor amounts of covalently attached phosphate. Hyperphosphorylation is deleterious to glycogen structure and can lead to Lafora disease. Recently, it was demonstrated that glycogen synthase catalyzes glucose–phosphate transfer in addition to its characteristic glucose transfer reaction. Glucose-1,2-cyclic-phosphate (GCP) was proposed to be formed from UDP-Glc breakdown and subsequently transferred, thus providing a source of phosphate found in glycogen. To gain further insight into the molecular basis for glucose–phosphate transfer, two structures of yeast glycogen synthase were determined; a 3.0-Å resolution structure of the complex with UMP/GCP and a 2.8-Å resolution structure of the complex with UDP/glucose. Structural superposition of the complexes revealed that the bound ligands and most active site residues are positioned similarly, consistent with the use of a common transfer mechanism for both reactions. The N-terminal domain of the UDP⋅glucose complex was found to be 13.3° more closed compared with a UDP complex. However, the UMP⋅GCP complex was 4.8° less closed than the glucose complex, which may explain the low efficiency of GCP transfer. Modeling of either α- or β-glucose or a mixture of both anomers can account for the observed electron density of the UDP⋅glucose complex. NMR studies of UDP-Glc hydrolysis by yeast glycogen synthase were used to verify the stereochemistry of the product, and they also showed synchronous GCP accumulation. The similarities in the active sites of glycogen synthase and glycogen phosphorylase support the idea of a common catalytic mechanism in GT-B enzymes independent of the specific reaction catalyzed.
Branched glucose polymers, glycogen and starch, are used by nearly all living organisms as an osmotically neutral means of energy storage. These polymers are constructed through the formation of α-1,4-glycosidic bonds and branching points, which use α-1,6-glycosidic linkages (1, 2). Glycogen is found in most animal, fungi, bacteria, and archaea, whereas photosynthetic eukaryotes or their nonphotosynthetic derivatives (such as apicomplexa parasites) use starch (2). Although primarily a glucose reservoir, glycogen also contains minor amounts of both glucosamine and phosphate (3). In animals and yeast, glycogen biosynthesis requires the action of three enzymes: glycogenin, glycogen synthase, and the branching enzyme. Most fungal and animal glycogen synthases use uridine diphosphoglucose (UDP-Glc) as the glucose donor, whereas bacterial, some parasitic, and plant glycogen/starch synthases use adenosine diphosphoglucose (ADP-Glc). Glycogen breakdown is catalyzed by glycogen phosphorylase which uses the cofactor pyridoxal 5′-phosphate (PLP) in combination with inorganic phosphate to phosphorolytically cleave glycogen and generate glucose-1-phosphate.
Glycogen synthase (GS) is classified as a glycosyltransferase (GT), a large superfamily of enzymes that transfer a sugar residue from an activated sugar donor to an acceptor molecule (4). To date, GTs have been grouped into more than 90 families (5) (www.cazy.org) based on sequence similarity. The nucleotide sugar-dependent GTs have been shown to adopt either GT-A or GT-B folds which consist of two associated domains, one of which contains a dinucleotide fold responsible for donor nucleotide recognition (4). The GT-A fold consists of two tightly associated domains, resulting in some describing it as a single-domain fold, whereas the GT-B fold is composed of two structurally distinct dinucleotide folds characteristically separated by a deep interdomain cleft (6). GS enzymes have been classified as GT-B enzymes that are further subdivided into two families, GT3 and GT5 (7). The bacterial, archaeal, and plant glycogen/starch synthase enzymes are grouped into the GT5 family, whereas the mammalian, yeast, and fungal enzymes are grouped into the GT3 family. Although it catalyzes the breakdown of glycogen, glycogen phosphorylase is structurally classified as a GT-B enzyme and is placed in the GT35 family. Functionally, the GTs are classified as either retaining or inverting, in reference to the stereochemistry at the anomeric carbon of the substrates and products; both GS and phosphorylase are retaining-type GTs. Although the catalytic mechanism of inverting GTs is generally well understood, the mechanism used by the retaining GTs remains elusive.
Glycogen is known to contain minor amounts of covalently attached phosphate, and excessive accumulation of covalent phosphate is thought to underlie Lafora disease, a devastating form of myoclonus epilepsy that is ultimately fatal (8–10). Where the phosphate is located on the glucose residues with glycogen is an active area of investigation, as is the mechanism by which it is introduced. Tagliabracci et al. recently showed that GS not only catalyzed the transfer of glucose but also incorporated the β-phosphate of UDP-Glc at a frequency of 1 phosphate per ∼10,000 glucoses, with release of UMP rather than UDP (11). This study also revealed that the phosphate in glycogen was present as C2- and C3-phosphomonoesters (11). It was proposed that the presence of phosphomonoesters in glycogen was due to the ability of GS to use either glucose-1,2-cyclic phosphate (GCP) or glucose-1,3-cyclic phosphate, which can be formed from the breakdown of UDP-Glc (12, 13), as donors in the active site (11). By this mechanism, GS could use the same catalytic mechanism regardless of whether UDP-Glc or the cyclic phosphates were present in the active site. No physiological role has yet been associated with phosphate in glycogen, although the presence of excessive phosphate is known to be detrimental to glycogen structure (14). In contrast, another report has refuted the ability of glycogen synthase to catalyze this incorporation and identified, in addition to the aforementioned 2′ and 3′ phosphate, glucose residues modified at the 6′ position with covalent phosphate (15). The authors suggested that the radioactive phosphate we observed in glycogen elongated by glycogen synthase was, in fact, due to the retention of nonspecifically bound β-32P-UDP in the glycogen (15). However, laforin phosphatase was able to remove the phosphate from this labeled glycogen (11), and there is no evidence to suggest that Laforin can catalyze the removal of phosphate groups from UDP. Our laboratory continues to seek a mechanistic explanation for the ability of glycogen synthase to not only catalyze the incorporation of glucose into glycogen but also elucidate the role of glycogen synthase in covalent phosphate incorporation. The mechanistic enigma surrounding glycosyl transfer and the widespread significance of the reactions catalyzed by GTs make investigation of their catalytic mechanism important because it is central both to a fundamental understanding of their chemistry and toward therapeutic applications in diseases such as Lafora disease.
In this paper, we report the crystal structure of the allosterically activated form of yeast glycogen synthase (Gsy2p) in complexes with either UDP⋅Glc or UMP⋅GCP. In each structure, one of the four subunits of Gsy2p has captured a catalytically relevant form of the nucleotide and donor sugar molecule, whereas the other subunits retain only the nucleotide. NMR studies of UDP-Glc hydrolysis by Gsy2p were used to verify the stereochemistry of the reaction products and unexpectedly revealed the production of GCP. The structural similarities observed in the Gsy2p complexes with either Glc or GCP to the Escherichia coli glycogen synthase and the maltodextrin phosphorylase structure bound to 1-deoxyglucose and maltodextran are consistent with the use of a common mechanism of glucosyl transfer in GT-B enzymes.
Results
We solved the structure of yeast Gsy2p in its activated state with bound G-6-P for a series of complexes with (i) UDP and UDP⋅Glc, (ii) UMP, and (iii) UMP⋅GCP to resolutions between 2.7 and 3.0 Å (Table 1). The overall architecture of the G-6-P activated state of Gsy2p has been described for the R589A/R592A mutant (16). In brief, Gsy2p is a tetrameric protein, and each subunit consists of two domains, an N-terminal dinucleotide-fold domain composed of residues 2–278 and 599–639 and a C-terminal dinucleotide-fold domain comprising residues 279–598. As previously described, residues C-terminal from position 639 are disordered and were not included in the models.
Table 1.
Data collection and refinement statistics
| Enzyme |
|||
| Descriptor | R589/92A | R589/92A | E169Q |
| Ligands | UMP | UMP⋅GCP | UDP⋅Glc |
| Resolution, Å | 50–2.66 | 50–2.95 | 50–2.75 |
| Rmerge, % | 0.081 (0.66) | 0.075 (0.71) | 0.11 (0.50) |
| I/σI | 19.5 (2.1) | 21.6 (2.3) | 13.2 (3.1) |
| Completeness, % | 95 (88) | 99.8 (100) | 99.5 (100) |
| Redundancy | 5.7 (4.4) | 5.6 (5.3) | 6.2 (6.3) |
| Total reflections | 111,217 | 85,877 | 102,312 |
| Rwork/Rfree, % | 0.21/0.26 | 0.23/0.28 | 0.20/0.24 |
| rmsd ideal bonds, Å | 0.008 | 0.007 | 0.008 |
| rmsd bond angles, ° | 1.28 | 1.21 | 1.26 |
| PDB code | 4KQ1 | 4KQ2 | 4KQM |
Space group I222 cell dimensions: a = 192–4, b = 202–4 c = 205–6 Å; α = 90.0°, β = 90.0°, and γ = 90.0°.
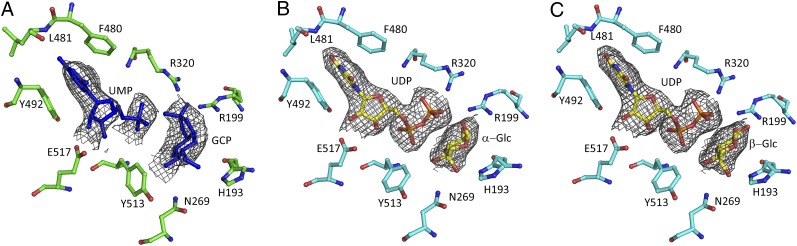
The complex with UMP only and the UMP⋅GCP complex showed clear electron density for the UMP ligand in all subunits, whereas only the crystals soaked with GCP exhibited additional electron density close to the UMP in one subunit (Fig. 1A). Due to the limited resolution and the possibility of water-mediated ring opening of GCP, we attempted to model glucose-1-phosphate (G-1-P) and glucose-2-phosphate (G-2-P) into the observed electron density. The G-1-P ligand could only be modeled and refined after moving R320 out of its electron density to accommodate the phosphate group (Figs. S1 and S2). Although the original density has difficulty accommodating the G-2-P model and clearly shows the phosphate group out of the original electron density (Fig. S1), refinement in REFMAC5 appears to account for the electron density around the G-2-P and R320 (Fig. S2). However, analysis of the refined model with G-2-P shows that the refinement has distorted the chair conformation of the sugar ring. The refined GCP model is the only one that can satisfactorily explain the observed density before and after refinement (Figs. S1 and S2) and maintain good stereochemistry. The C1 atom of the GCP is positioned in an orientation that makes it available for transfer to an activated acceptor. Binding of GCP and the allosteric activator of Gsy2p, G-6-P, was probed by using differential scanning fluorimetry (Fig. S3). The binding of both ligands was characterized by an increase in Tm (Fig. S3A and Table S1), which is consistent with ligand-induced protein stabilization.
Fig. 1.
All of the electron density maps were calculated from the initial protein models before the addition of ligands and are displayed upon the respective final refined structures. (A) Original σA-weighted 2Fo – Fc electron density maps contoured at 1σ around the UMP and GCP. (B and C) Original σA-weighted 2Fo – Fc electron density contoured at 1σ for the hydrolyzed UDP-Glc within the active site of Gsy2p E169Q. The figures were generated by PyMOL.
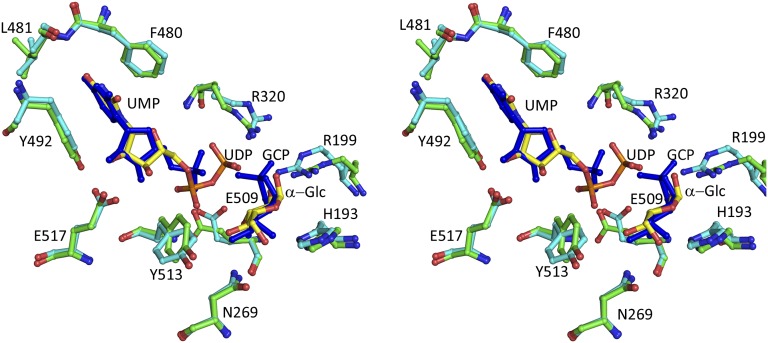
The Gsy2p-E169Q⋅G-6-P⋅UDP⋅Glc complex was obtained by cocrystallization in the presence of UDP-Glc and G-6-P. The Gsy2p-E169Q mutant retains less than 1% catalytic activity relative to the wild-type enzyme and was used for cocrystallization experiments with UDP-Glc in hopes of limiting turnover. As with the GCP structure, only one of the subunits retained both UDP and the glucosyl moiety; the remaining subunits retained only UDP. The lack of an available acceptor molecule and the time frame for crystallization likely led to the nonproductive hydrolysis of UDP-Glc to UDP and Glc. Modeling of either α- and β-Glc or a mixture of both C1 anomers can account for the observed electron density illustrated in Fig. 1 B and C. The distance between the closest β-phosphate oxygen and the C1 of either the α- or β-Glc is longer (3.8 or 3.3 Å, respectively) than would be expected if the covalent bond was still intact. The GCP and Glc moieties are bound within the same relative subunit (subunit C), and their respective C1 atoms occupy almost identical positions within their respective active sites (Fig. 2). Additionally, most of the active site residues are similarly positioned around these two glucosyl moieties.
Fig. 2.
Stereo representation of selected active site residues in the glycogen synthase active sites. Superposition of Gsy2pE169Q (cyan) in complex with UDP and glucose (yellow) with Gsy2pR589/92A (green) in complex with UMP and GCP using green for the protein residues and blue for the ligands.
Structural alignment using the program DynDom (8) of the UDP⋅Glc bound subunit with a subunit containing only UDP revealed a rotation of 13.3° (Fig. S4). The rmsd between the N-terminal domain α-carbons (4–205, 210–274, and 597–637) and the C-terminal domain α-carbons (206–209 and 275–596) are 0.52 and 0.41 Å, suggestive of a global rigid body domain movement. The active site domain closure creates an additional interaction across the interdomain cleft between the 3′OH of the ribosyl moiety of UDP with the side chain of S26 that is absent in the more open domain conformations found in other subunits. Comparison of the UDP molecules in the presence and absence of glucose shows that there is a difference in the orientation of the α- and β-phosphate groups (Figs. S4 and S5). In contrast to the fully closed active site structure observed in the UDP⋅Glc complex, the N-terminal domain position in the UMP⋅GCP bound subunit is intermediate in positioning, with 4.8° less closure (Fig. S6).
Glucose Binding Pocket.
The interactions between Gsy2p and the bound glucosyl and GCP molecules are similar (Fig. 2 and Fig. S5). Their 6′OH groups interact with the universally conserved side chains of H193 and N269. The 4′OH group is within hydrogen bonding distance to the peptide nitrogen of G512 and to one of the β-phosphate oxygens. However, the interactions at the 3′OH and 2′OH groups differ due to the presence of the cyclic phosphate on GCP. The 3′OH groups in the glucose moiety and GCP interact with the peptide nitrogen of W511, but the additional interaction with the side chain of E509 observed in the Glc structure is lacking in the GCP structure (Fig. 2 and Fig. S5). The latter motion is likely induced by the presence of the charged α-phosphate group of UMP. In the Glc structure, the 2′OH interacts with both the side chain of R199 and the β-phosphate of UDP. In the GCP structure, the 2′OH is now bonded to the phosphate, and the side chain of R199 interacts with the phosphoryl group.
Nucleotide Binding Pocket.
The interactions between Gsy2p and the UDP/UMP molecules are similar to those reported previously (16), although the higher resolution of the activated state structures reported here permits better visualization of those interactions. In all our nucleotide bound structures, the uridine ring is sandwiched between the aromatic side chains of F480 and Y492. The O4 group of the uridine moiety is within hydrogen-bonding distance to the peptide nitrogen of L481, and the 2′- and 3′OH groups of the ribosyl moiety are within hydrogen-bonding distance to the side chain E517. Although the conformations for the uridine and ribose rings are identical in all of the structures, we observe two different conformations of the diphosphates, depending on whether Glc is also bound within the active site. The conformations of the α- and β-phosphates are essentially switched. For convenience we will refer to UDP conformation with bound Glc as UDPa (pretransfer) and the other conformation as UDPb (posttransfer), illustrated in Fig. S5 B–D.
In UDPa, the diphosphate group of UDP lies parallel to the plane of the Glc, and the 1′OH of α-Glc is within hydrogen bonding distance to one of the UDP β-phosphate oxygens. However, when modeled in the β-configuration, the 1′OH is incapable of making this interaction. The α-phosphate group is within hydrogen-bonding distance to the peptide nitrogens of Y513 and T514 and the side chain hydroxyl of T514. The β-phosphate group interacts with the side chains of R320 and K326 and the peptide nitrogen of G23, located across the interdomain cleft. In contrast, it is the α-phosphate of the UDPb conformation that interacts with the side chain of K326 and the peptide nitrogen and side chain of R320. The bridging oxygen of the diphosphate is also within hydrogen-bonding distance to R320. The β-phosphate in the UDPb conformation interacts with the peptide nitrogens from Y513 and T514 and the hydroxyl group of T514.
The uridine O2 in UMP appears to make an additional interaction with R20 which is in the N terminus in a region that was generally found to be highly disordered; this interaction is only observed in the subunit where GCP is bound. The phosphate of UMP in the presence of GCP appears to interact with the helical dipole provided by helix α19. However, the phosphate group in the active site with UMP only bound appears to interact with the main chain amide nitrogen of R320, the R320 side chain, and the T514 hydroxyl group. The side chain of K326 is also within hydrogen-bonding distance to the phosphate of UMP.
1H-NMR Monitoring of Stereochemical Course of UDP-Glc Hydrolysis by Gsy2p.
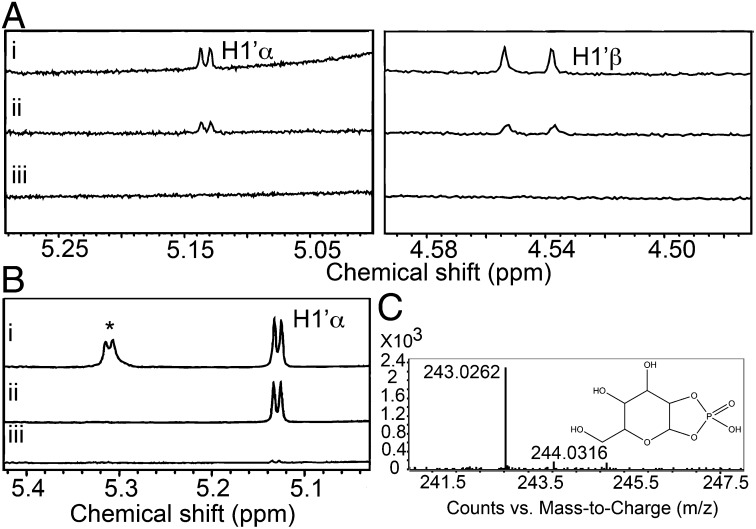
Our crystal structures with bound Glc are consistent with the formation of α- and/or β-Glc from hydrolysis of UDP-Glc. However, given the time frame of crystallographic experiments, it is not clear if the anomeric mixture was produced catalytically or through mutarotation of one anomer over time. To assess whether the nonproductive hydrolytic reaction results in anomerically selective products, NMR analysis was carried out. The production of Glc using the wild-type Gsy2p enzyme was evident within 3 h of incubation, and both α- and β-Glc appeared simultaneously, consistent with a nonstereoselective hydrolytic activity (Fig. 3A). An unexpected resonance was also observed to accumulate during the same experiment. The spectrum following an overnight incubation shows the presence of a doublet between 5.2 and 5.4 ppm (Fig. 3B). This doublet was also present in the 1D 1H NMR of the chemically synthesized GCP used for the crystallographic experiments. Mass spectroscopy was used to determine the identity of the small molecule(s) present in the NMR reaction mix. A small molecule with an m/z of 243.02 (Fig. 3C) was detected, consistent with the presence of GCP. A time course analysis revealed that detectable amounts of GCP were formed within 3 h of incubating the enzyme with substrate (Fig. S7).
Fig. 3.
(A) 1H NMR spectrum of UDP-Glc and wild-type Gsy2p following (A, i) overnight incubation, (A, ii) 3 h incubation, and (A, iii) no incubation. Left is from 5.0 to 5.3 ppm highlighting H1′α protons downfield of the water signal, and Right highlights H1′β protons (4.48–4.59 ppm) upfield of the water signal. (B) 1H NMR spectrum highlighting region near the H1′α proton (5.0–5.4 ppm) of UDP-Glc, wild-type Gsy2p, and G6P following (B, i) an overnight incubation, (B, ii) no incubation, and (B, iii) overnight incubation of UDP-Glc and Gsy2p shown for comparison. Asterisk highlights peak for H1′ of GCP. The concentration of GCP was approximated by measuring the relative area under the peak at 5.2–5.4 and comparing this to the area under the curve for the H1′ of UDP-glucose of known concentration from the beginning of the reaction. (C) Mass spectrum of unknown peak in NMR spectrum demonstrating a mass peak at 243.03.
Discussion
Lafora disease is an autosomal recessive juvenile-onset myoclonus epilepsy, characterized by abnormal glycogen deposits known as Lafora bodies, which contain an insoluble and poorly branched glycogen-like polysaccharide, termed polyglucosan. It is estimated that ∼50% of patients with Lafora disease have mutations in the gene that codes for laforin (9, 10, 17), a phosphatase that dephosphorylates glycogen (18). Analysis of the glycogen from Lafora bodies revealed the presence of abnormally high levels of phosphate (14, 15, 18, 19), raising the question of how phosphate is incorporated into glycogen. Here we present results that provide strong evidence for a plausible mechanism by which glycogen synthase can catalyze the transfer of 2-phosphoglucose into glycogen using a similar reaction scheme for both glucosyl and phosphoglucosyl transfer, albeit with considerably different efficiencies. Whether a similar direct mechanism for introduction of 3ʹ or 6ʹ phosphate exists is currently unknown, but the tendency of phosphomonoesters to migrate to neighboring hydroxyl groups is well established (20–22) and may provide an explanation for the occurrence of the other phosphomonoesters as possibly derived from 2ʹ-phosphomonoesters.
Comparison of the structures of GCP and Glc bound to the active site of Gsy2p reveals that the binding is nearly identical even though the GCP bound structure was obtained by soaking, whereas the Glc bound structure was obtained from hydrolysis of UDP-Glc during cocrystallization. The occupation of nearly identical positions in the active site by these two compounds supports the proposal that phosphate incorporation is due to the formation of cyclic phosphates within the active site and that GS uses a similar mechanism during Glc or G-2-P transfer. The anomeric carbons of both GCP and Glc are in a location that is accessible for transfer to an acceptor. At a resolution of 2.7 Å it was difficult to distinguish between α- and β-Glc as the products of UDP-Glc because both anomers appeared to fit the electron density equally well. Given that Glc undergoes mutarotation at a rate of ∼0.015 min−1 (23), we chose to use proton NMR to more definitively determine the stereochemistry of the products of UDP-Glc hydrolysis. Collecting the NMR data over a time course revealed the appearance of peaks consistent with the formation of both α- and β-Glc synchronously, suggesting that either anomer is released in the absence of an acceptor. It seems likely that there are fewer steric constraints on a bound water in the active site such that either the α- or β-face of Glc is accessible. Overnight incubation of G-6-P–activated Gsy2p with UDP-Glc revealed the accumulation of GCP, previously proposed to be an intermediate in the reaction in the incorporation of phosphate into glycogen. The identity of this peak in the NMR spectrum was verified by LC-MS.
We believe that several factors contribute to the generation of GCP and also to limiting the incorporation of GCP into glycogen. We propose that the conformation in which UDP-Glc is held within the active site promotes the formation of GCP because the 2′OH is positioned in close proximity to the β-phophorus atom (Fig. S5 B and C). Attack by the 2′OH on the β-phosphate yields GCP and UMP. However, the formation of this intermediate is incompatible with a fully closed active site structure. Compared with the UDP⋅Glc structure, the UMP⋅GCP bound structure shows two distinct structural adaptations to the presence of GCP. First, the N-terminal domain is 4.8° less closed than the Glc structure. This domain positioning may be due to the inability of the peptide nitrogen of G23 to interact with the β-phosphate after the formation of GCP. Without this interaction it is possible that the interaction between S26 and the 3′OH of the uridine ribosyl group is insufficient to hold the domain closed. Second, the presence of UMP and GCP also results in the repositioning of E509, which is likely repelled by the more highly charged α-phosphate group of UMP. A more open domain conformation and the loss of the interaction between the 3′OH of GCP and E509 could affect GCP retention within the active site and lower its efficiency of transfer to the nonreducing end of a glycogen chain.
Based on the peak heights for GCP in the NMR spectrum, we would conservatively estimate that ∼700 µM GCP was generated over a 12-h time frame by 15 µM Gsy2p, giving a rate of 0.065 min−1. The concentration of GCP was approximated by measuring the relative area under the peak at 5.2–5.4 and comparing this to the area under the curve for the H1′ of UDP-glucose of known concentration from the beginning of the reaction. Because the glucosyl transfer rate for Gsy2p to glycogen is ∼1,600 min−1, there is generally insufficient time between productive catalytic cycles to generate enough GCP to detect with the methods currently used. The ratio of GCP production to that of glucosyl transfer is ∼1:25,000, and the calculated rate of phosphate incorporation into glycogen by glycogen synthase was about 1:10,000 cycles. Given the limitations of both experimental approaches, the fact that they give ratios in the same range is suggestive that this process is likely the one operating in cells. In addition, GCP was only detectable when the enzyme was present as the G-6-P activated state. An NMR spectrum of a control overnight incubation between Gsy2p and G-6-P alone did not show the presence of GCP. Our attempts to detect GCP production in the absence of G-6-P or in the presence of acceptor substrates were unsuccessful. However, even though GCP did not accumulate in the absence of acceptors or activators, we did observe the production of free glucose. This suggests that in the absence of an acceptor, detectable GCP production occurs only in the activated state, whereas the hydrolysis of UDP-Glc can occur in either activity state. Our attempts to characterize GCP binding in the presence or absence of UMP/G6P revealed that the presence of these ligands did not change the observed EC50 of ∼10 mM (Table S1), consistent with the binding of GCP to a distinct site that does not overlap with either UMP or G6P.
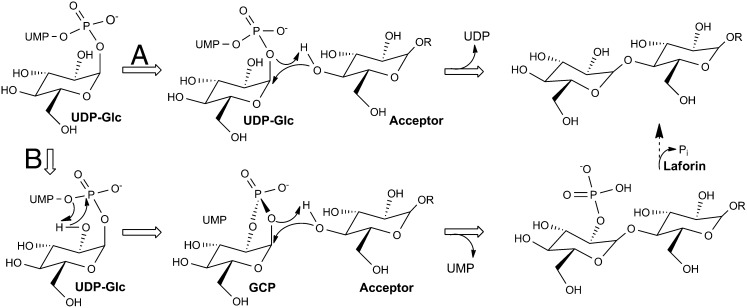
The chemical mechanism underlying retaining glycosyl transferases generates considerable debate, and although these structures do not necessarily settle the debate, they do provide compelling evidence that the incorporation of glucose and glucose-2′-phosphate into glycogen may have similar mechanistic underpinnings. Our UDP⋅Glc structure is very similar to that obtained for the E. coli glycogen synthase structure prepared by incubation with ADP-Glc. Consequently, the mechanism used by all glycogen synthases will undoubtedly use identical chemistries. Most retaining glycosyl transferases lack an identifiable nucleophile within their respective active sites, suggesting that they use an SN1/SNi mechanism for glucosyl transfer (Fig. 4). The active sites of glycogen synthases not only lack an identifiable amino acid side chain as a nucleophile, they also lack an easily identifiable amino acid serving as a general base. Structural alignment of the closed ADP⋅Glc structure of E. coli glycogen synthase active site with the Gsy2p structures (Fig. S8) reported here leads to the conclusion that the diphosphate of the UDP leaving group likely deprotonates and activates the incoming acceptor 4′OH group. In the case of GCP, the cyclic phosphate may perform this same function, albeit with lowered efficiency. Because glycogen synthase is a non–metal-dependent glycosyl-transferase, stabilization of the UDP leaving group is accomplished through hydrogen-bonding interactions, which include the side chains of R199, R320, and K326 and the helical dipole and peptide nitrogens contributed by residues 513–521. In contrast, stabilization of the 2′-phosphate leaving group in GCP appears to be accomplished by R199 and R320. Last, the peptide carbonyl oxygen of the residue equivalent to H193 in Gsy2p has been proposed to stabilize the charge distribution of the oxo-carbenium ion intermediate during glucosyl transfer (24, 25). Our structures are consistent with that proposal.
Fig. 4.
Proposal for the catalytic mechanism of glycogen synthase. The acceptor is deprotonated by the phosphate oxygens, and attack proceeds at the same face from which the phosphate leaves. The reaction that occurs within the glycogen synthase active site is illustrated using the block arrows, whereas dephosphosphorylation of glycogen by Laforin is shown by the dashed arrow. The standard glucosyl transfer reaction follows route A, whereas the formation of GCP and its subsequent transfer follows route B.
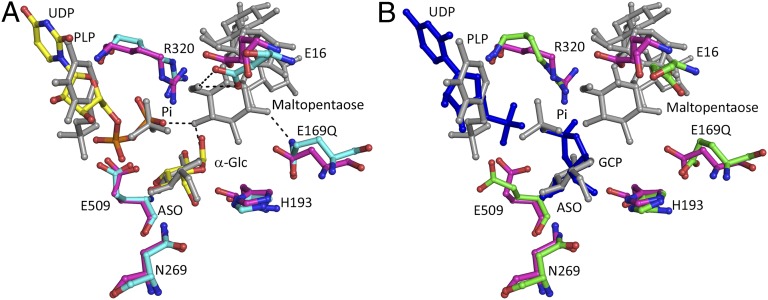
In addition to the remarkable similarity between the active sites of the yeast and E. coli GS complexes with nucleotide and Glc, striking similarities with the active site of E. coli maltodextran phosphorylase (MalP) (26) were also observed. When our Gsy2p complexes are aligned to the MalP⋅PLP⋅Pi⋅1-deoxy-Glc⋅maltopentaose complex (PDB 2asv) (Fig. 5), the juxtaposition of the PLP coenzyme in MalP with either UDP or UMP in Gsy2p and the similarity in positioning of the hydrolytic phosphate with either the β-phosphate of UDP or with the cyclic phosphate of GCP are compelling. These similarities underscore their functional relationship and suggest a plausible evolutionary path between the glycogen synthetic and degradative enzymes. The strong conservation of the functional residues in immediate contact with these substrates is clearly seen in this alignment, which stands in stark contrast to the overall sequence identity between these two enzymes (<10% pairwise identity). Thus, even though these enzymes have evolved to optimally use different substrates for their opposing action on the glycogen polymer, the amino acid contributions to the catalytic events are remarkably well conserved. In this context, it is worth noting that in vitro at supraphysiologic concentrations of G-1-P, phosphorylase can catalyze the transfer of Glc to the nonreducing end of a glycogen chain and that this reverse reaction serves as the standard assay for glycogen phosphorylase. It is also notable that rabbit glycogen phosphorylase was also crystallized with GCP bound in the active site (27) where it acts as a transitition-state mimic and inhibitor rather than a substrate of phosphorylase (28).
Fig. 5.
Similarities of Gsy2p and maltodextran phosphorylase active sites. (A) Structural comparison of the active site of the Gsy2p E169Q⋅UDP⋅Glc (cyan and yellow) with maltodextran phosphorylase PLP⋅Pi⋅1-anhydroglucose⋅maltopentaose (MalP, PDB 2ASV; magenta and gray). The black dashes represent inferred hydrogen bonds. The residues are labeled according to Gsy2p sequence position. (B) Structural comparison of the Gsy2p E169Q⋅UMP⋅GCP complex (green and blue) and MalP (magenta and gray). Structural alignment of the active sites was performed in PyMOL.
In conclusion, we have solved the structure of yeast glycogen synthase 2 in complexes with both UDP⋅Glc and UMP⋅GCP. These structures provide plausible mechanisms by which glycogen synthase transfers Glc from UDP-Glc to glycogen and, as a rare side reaction, introduces phosphate at the 2′ position of a growing polyglucose chain, thereby providing another step toward explaining the phosphorylation of glycogen.
Methods
Mutagenesis, Expression, and Purification of Yeast Glycogen Synthase.
The Saccharomyces cerevisiae glycogen synthase, Gsy2p, gene in the pET-28A vector (16) was used as the template for the PCR to generate the point mutations E169Q and R589A/R592A. Site-directed mutagenesis was carried out by QuikChange site-directed mutagenesis. The identity and sequence integrity of all clones produced was verified by DNA sequencing. The expression and purification of His-tagged Gsy2p was carried out as described for the wild-type protein (16). Purified protein was flash frozen in liquid nitrogen and stored in 20 mM Tris·HCl (pH 8.0) and 1 mM DTT at –80 °C.
Crystallization and Data Collection.
Crystals of the activated state conformations of Gsy2p; E169Q and R589/92A were prepared, cryoprotected, and frozen as described previously (16). Crystals of Gsy2p-R589A/R592A with UMP and G-6-P were grown in the presence of 2 mM UMP and 25 mM G-6-P. GCP crystals were obtained by adding GCP (∼10 mM) briefly to the UMP cocrystals followed by immediate freezing. Crystals of Gsy2pE169Q were grown in the presence of 2 mM UDP-Glc and 25 mM G-6-P. The data sets were collected at the Advanced Photon Source at beamline 19-ID operated by Structural Biology Center – Collaborative Access Team. The data sets were indexed, integrated, and scaled using the HKL3000 program suite (29). The structures were solved by molecular replacement using MOLREP, part of the Collaborative Computational Project Number 4 (CCP4) program package using the Gsy2pR589/92A2 activated state (Protein Data Bank ID code 3NB0) as the search model. The structures were initially refined with rigid body refinement followed by refinement of Translation/Libration/Screw (TLS) tensors (30, 31) and restrained refinement using REFMAC5 (32) model building was carried out using the program COOT (33).
NMR.
The 1D 1H NMR were acquired using WATERGATE (34) water suppression of samples containing 10% (vol/vol) D2O in a Shigemi NMR tube at room temperature using a Bruker 500 MHz NMR. The 1D 1H NMR were recorded (2 s relaxation delay) with a 6,100-Hz sweep width using 512 scans. The data were processed using Topspin 3.0. Gys2p (15 μM concentration) was incubated with varying G-6-P (15 mM or 500 μM for the time course experiment) and UDP-Glc (30 mM).
Mass Spectrometry.
The mixture of Gys2p (15 μM), UDP-Glc (30 mM), and G-6-P was injected (1 μL volume) into an Agilent 6520 quadrupole time-of-flight mass spectrometer operating in positive ion electrospray ionization-time of flight mode (from 100 to 1,100 m/z). The data were analyzed with formula assignments for low-molecular weight compounds using the Mass Hunter software suite.
Supplementary Material
Acknowledgments
The authors thank the Structural Biology Center Beamline staff, especially Marianne Cuff and Steve Ginell. MS and NMR analyses were acquired at the instrumentation facility of the Department of Chemistry and Chemical Biology at Indiana University-Purdue University Indianapolis with the help of Karl Dria. The authors wish to acknowledge support from National Institutes of Health (NIH) Grant R01-DK079887 (to T.D.H.), NIH Grant R01-NS056454 (to P.J.R.), and NIH Grant R37-DK027221 (to P.J.R.). V.M.C. was supported by NIH Grant T32-DK064466 (to P.J.R.). Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research, under Contract DE-AC02-06CH11357.
Footnotes
Conflict of interest statement: T.D.H. holds significant financial equity in SAJE Pharma, LLC. However, none of the work described in this study is related to, based on, or supported by the company.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the RCSB Protein Data Bank (accession nos. 4KQ1, 4KQ2, and 4KQM).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310106111/-/DCSupplemental.
References
- 1.Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: Some new developments and old themes. Biochem J. 2012;441(3):763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball SG, Morell MK. From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- 3.Fontana JD. The presence of phosphate in glycogen. FEBS Lett. 1980;109(1):85–92. doi: 10.1016/0014-5793(80)81317-2. [DOI] [PubMed] [Google Scholar]
- 4.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 5.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unligil UM, Rini JM. Glycosyltransferase structure and mechanism. Curr Opin Struct Biol. 2000;10(5):510–517. doi: 10.1016/s0959-440x(00)00124-x. [DOI] [PubMed] [Google Scholar]
- 7.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328(2):307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 8.Qi G, Lee R, Hayward S. A comprehensive and non-redundant database of protein domain movements. Bioinformatics. 2005;21(12):2832–2838. doi: 10.1093/bioinformatics/bti420. [DOI] [PubMed] [Google Scholar]
- 9.Delgado-Escueta AV. Advances in Lafora progressive myoclonus epilepsy. Curr Neurol Neurosci Rep. 2007;7(5):428–433. doi: 10.1007/s11910-007-0066-7. [DOI] [PubMed] [Google Scholar]
- 10.Andrade DM, Turnbull J, Minassian BA. Lafora disease, seizures and sugars. Acta Myol. 2007;26(1):83–86. [PMC free article] [PubMed] [Google Scholar]
- 11.Tagliabracci VS, et al. Phosphate incorporation during glycogen synthesis and Lafora disease. Cell Metab. 2011;13(3):274–282. doi: 10.1016/j.cmet.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paladini AC, Leloir LF. Studies on uridine-diphosphate-glucose. Biochem J. 1952;51(3):426–430. doi: 10.1042/bj0510426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nunez HA, Barker R. The metal ion catalyzed decomposition of nucleoside diphosphate sugars. Biochemistry. 1976;15(17):3843–3847. doi: 10.1021/bi00662a030. [DOI] [PubMed] [Google Scholar]
- 14.Tagliabracci VS, et al. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J Biol Chem. 2008;283(49):33816–33825. doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nitschke F, et al. Hyperphosphorylation of glucosyl C6 carbons and altered structure of glycogen in the neurodegenerative epilepsy Lafora disease. Cell Metab. 2013;17(5):756–767. doi: 10.1016/j.cmet.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Baskaran S, Roach PJ, DePaoli-Roach AA, Hurley TD. Structural basis for glucose-6-phosphate activation of glycogen synthase. Proc Natl Acad Sci USA. 2010;107(41):17563–17568. doi: 10.1073/pnas.1006340107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minassian BA, et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet. 1998;20(2):171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 18.Tagliabracci VS, et al. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci USA. 2007;104(49):19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai M, Austin J, Witmer F, Trueb L. Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology. 1970;20(2):160–176. doi: 10.1212/wnl.20.2.160. [DOI] [PubMed] [Google Scholar]
- 20.MacDonald DL. Phosphates and Other Inorganic Esters. New York: Elsevier; 1972. pp. 256–257. [Google Scholar]
- 21.Oivanen M, Schnell R, Pfleiderer W, Lonnberg H. Interconversion and hydrolysis of monomethyl and monoisopropyl esters of adenosine 2′- and 3′-monophosphates: Kinetics and mechanisms. J Org Chem. 1991;56(11):3623–3628. [Google Scholar]
- 22.Tomlinson RV, Ballou CE. Complete characterization of the myo-inositol polyphosphates from beef brain phosphoinositide. J Biol Chem. 1961;236(7):1902–1906. [PubMed] [Google Scholar]
- 23.Livingstone GF, Franks F, Aspinall LJ. The effects of aqueous solvent structure on the mutarotation kinetics of glucose. J Solution Chem. 1977;6(3):203–216. [Google Scholar]
- 24.Sheng F, Jia X, Yep A, Preiss J, Geiger JH. The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase. J Biol Chem. 2009;284(26):17796–17807. doi: 10.1074/jbc.M809804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Díaz A, et al. Lyase activity of glycogen synthase: Is an elimination/addition mechanism a possible reaction pathway for retaining glycosyltransferases? IUBMB Life. 2012;64(7):649–658. doi: 10.1002/iub.1048. [DOI] [PubMed] [Google Scholar]
- 26.Geremia S, Campagnolo M, Schinzel R, Johnson LN. Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase. J Mol Biol. 2002;322(2):413–423. doi: 10.1016/s0022-2836(02)00771-4. [DOI] [PubMed] [Google Scholar]
- 27.Withers SG, Madsen NB, Sprang SR, Fletterick RJ. Catalytic site of glycogen phosphorylase: Structural changes during activation and mechanistic implications. Biochemistry. 1982;21(21):5372–5382. doi: 10.1021/bi00264a039. [DOI] [PubMed] [Google Scholar]
- 28.Hu HY, Gold AM. Inhibition of rabbit muscle glycogen phosphorylase by alpha-D-glucopyranose 1,2-cyclic phosphate. Biochim Biophys Acta. 1978;525(1):55–60. doi: 10.1016/0005-2744(78)90199-7. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 31.Painter J, Merritt EA. A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):465–471. doi: 10.1107/S0907444905001897. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piotto M, Saudek V, Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2(6):661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.