Significance
Protein O-mannosylation is believed to be an abundant modification of proteins, but only very few glycoproteins with O-mannose have been identified to date. Here, we present a unique strategy for proteome-wide discovery of O-mannosylated glycoproteins, and using this strategy we find that the important cadherin and plexin families of cell membrane receptors are O-mannosylated. The presented strategy invites the opportunity for wider exploration of the O-mannose glycoproteome and studies of the functions of O-mannose glycans.
Keywords: POMGnT1, O-glycosylation, Orbitrap, mass spectrometry, glycoproteomics
Abstract
The metazoan O-mannose (O-Man) glycoproteome is largely unknown. It has been shown that up to 30% of brain O-glycans are of the O-Man type, but essentially only alpha-dystroglycan (α-DG) of the dystrophin–glycoprotein complex is well characterized as an O-Man glycoprotein. Defects in O-Man glycosylation underlie congenital muscular dystrophies and considerable efforts have been devoted to explore this O-glycoproteome without much success. Here, we used our SimpleCell strategy using nuclease-mediated gene editing of a human cell line (MDA-MB-231) to reduce the structural heterogeneity of O-Man glycans and to probe the O-Man glycoproteome. In this breast cancer cell line we found that O-Man glycosylation is primarily found on cadherins and plexins on β-strands in extracellular cadherin and Ig-like, plexin and transcription factor domains. The positions and evolutionary conservation of O-Man glycans in cadherins suggest that they play important functional roles for this large group of cell adhesion glycoproteins, which can now be addressed. The developed O-Man SimpleCell strategy is applicable to most types of cell lines and enables proteome-wide discovery of O-Man protein glycosylation.
Protein O-glycosylation of the O-mannose (O-Man) type found on Ser and Thr residues was initially thought to be restricted to yeast and fungi, and only within the last two decades has it been shown that this glycosylation occurs in metazoans (1, 2). O-Man glycosylation of the basement membrane glycoprotein α-dystroglycan (α-DG) is essential for assembly and function of the dystrophin–glycoprotein complex that links the cytoskeleton with the extracellular matrix, and deficiencies in all of the enzymes involved in the O-Man glycosylation underlie congenital muscular dystrophies (dystroglycanopathies) (3–5).
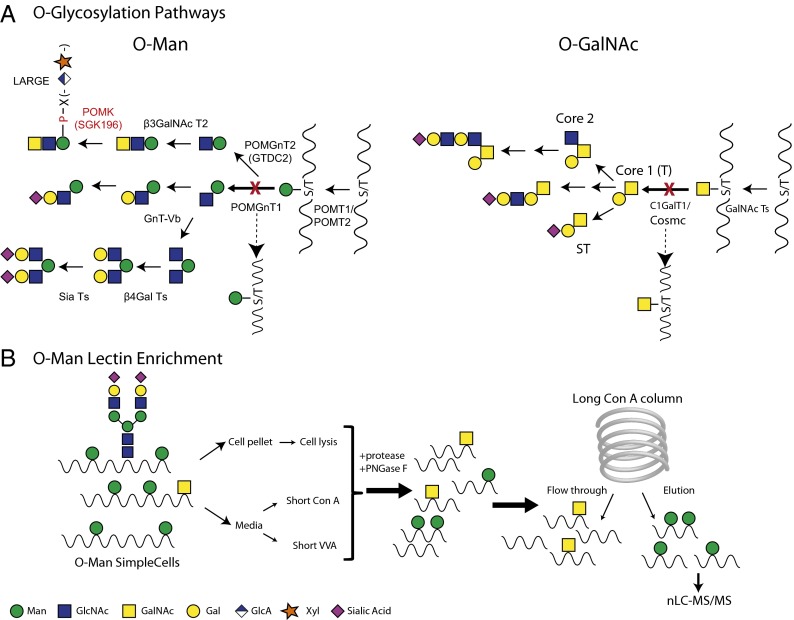
In yeast, O-mannosylation is found on a wide variety of proteins, although this O-glycoproteome is still rather unexplored. In mammals, early studies identified O-Man oligosaccharides released from isolated rat brain proteoglycans by mild alkaline borohydride treatment (6, 7), and subsequent studies clarified these structures as shown in Fig. 1A (8). However, for a long time, α-DG was the only well-characterized protein known to be O-mannosylated in mammals despite evidence that O-Man glycans constitute a major part of the total O-glycans in the brain (8, 9). A few other proteins have been demonstrated or suggested to contain O-Man glycans, including recombinantly expressed IgG2 (10), RPTPβ/ζ (11), CD24 (12), neurofascin 186 (13), as well as lecticans (14), and gel-based analysis has suggested that O-Man glycoproteins are of high molecular weight (9). More recently, it was demonstrated that the brains of α-DG−/− brain-specific knockout mice had unchanged levels of O-Man glycans (15). There are multiple types of metazoan protein O-glycosylation, most of which have been demonstrated to play roles in protein structure and function. However, our knowledge of O-glycoproteomes and especially sites of attachment of O-glycans is still very limited for several types of O-glycosylation including O-Man. Technical constraints primarily related to lack of enrichment strategies and predictive consensus sequence motifs, in addition to heterogeneity in structures of O-glycans, have long hampered identification of O-glycan attachment sites (9, 16).
Fig. 1.
Mining the O-Man glycoproteome by ZFN gene targeting of POMGNT1. (A) Knockout of POMGNT1 and COSMC abrogates elongation of O-Man (Left) and O-GalNAc (Right) type glycosylation, respectively, resulting in truncated homogenous O-glycan structures limited to O-Man and O-GalNAc (in some cells NeuAcα2–3GalNAc). Note that POMGNT2-mediated elongation of O-Man will not be affected by this gene targeting strategy. (B) Double knockout of POMGNT1 and COSMC in MDA-MB-231 cells allows for enrichment of O-Man glycoproteins from the cell culture supernatant using either a short Con A mannose-binding lectin column (captures both O-Man and N-glycoproteins) or for O-GalNAc glycoprotein enrichment on a short VVA αGalNAc-binding lectin column. Enriched eluates from these columns or total cell lysates can then be treated with proteases and PNGase F to remove N-glycans before glycopeptides are isolated by LWAC with a long Con A column and sequenced by nanoflow liquid chromatography-mass spectrometry (nLC-MS/MS).
O-mannosylation of proteins is initiated in endoplasmic reticulum (ER) by transfer of mannose from dolichol monophosphate-activated mannose to serine and threonine by homologous protein O-mannosyltransferases (PMTs). Yeast has at least six PMTs grouped into three subfamilies, PMT1, PMT2, and PMT4, while metazoans only have two PMT orthologs, POMT1 and POMT2, grouped in subfamilies PMT4 and PMT2, respectively (17). The specific contribution of each PMT isoform to O-mannosylation is predicted to be different but detailed information on substrate specificities of individual isoenzymes is largely missing (1). Studies have shown, however, that members of the PMT4 subfamily may act selectively on membrane bound protein substrates (18). In human O-Man glycans are generally elongated sequentially with N-acetylglucosamine (GlcNAc) and galactose (Gal) and capped by sialic acid (NeuAc), glucuronic acid (GlcA), or fucose (Fig. 1A). The first step in this elongation is the addition of GlcNAc to the O-Man glycans, catalyzed by a single enzyme, protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 (POMGnT1) (4). An alternative elongation pathway, which has so far only been identified at one specific O-Man site in α-DG, involves elongation of O-Man by the UDP-GlcNAc: Manα1-O-Ser/Thr β1,4GlcNAc-transferase (POMGnT2/GTDC2) (19). This pathway involves further elongation by β3GalNAc-T2 with subsequent phosphorylation of the mannose residue by POMK (SGK196) which is extended with GlcA and xylose by like-acetylglucosaminyltransferase to form the structure required for DG function (19, 20).
We previously developed a strategy to analyze GalNAc-type O-glycoproteins on a proteome-wide level using zinc finger nuclease (ZFN) gene engineered human cell lines with simplified O-glycan structures, the so-called “SimpleCell” (16). “Bottom-up” higher-energy collision-induced dissociation–electron-transfer dissociation (HCD/ETD)-based mass spectrometric analysis has allowed us to provide a first draft of the human O-GalNAc glycoproteome where we expanded the knowledge of O-glycoproteins and O-glycosites many fold (21). Here, we designed a similar strategy to simplify O-Man glycans in a human breast cancer cell line, which allowed us to probe the O-Man glycoproteome. The strategy led to identification of over 50 unique O-Man glycoproteins with more than 230 glycosites, and the surprising finding that the large cadherin family of membrane receptors is the major carrier of O-Man glycans.
Results and Discussion
Development of O-Man SimpleCells for O-Glycoproteomics.
We used the human MDA-MB-231 breast cancer cell line because we previously identified O-GalNAc glycosites in the mucin-linker domain of α-DG in MDA-MB-231 O-GalNAc SimpleCells (21). α-DG contains both O-Man and O-GalNAc glycans in this region (22–24), and some sites are potentially occupied by either type of glycosylation (25). This dual-occupancy was also proposed in neurofascin 186 (13). We reasoned that we could identify O-Man sites and the interplay between the two types of O-glycosylation on α-DG and other proteins in MDA-MB-231 with both types of glycosylation simplified, making both O-Man and O-GalNAc glycopeptides easily identifiable. O-Man was simplified by ZFN targeting of the POMGNT1 gene, which encodes for POMGnT1 controlling the first step in elongation of O-Man glycans (Fig. 1A) (4). Knockout of POMGnT1 is expected to truncate most O-Man glycosylation, although the function of POMGnT2 recently shown to extend O-Man by β1,4GlcNAc on α-DG could potentially compensate (19) (Fig. 1A). So far this pathway has only been associated with a few O-Man glycosites (Thr317, Thr319, and Thr379) in α-DG (20, 26), and the POMGnT2 enzyme has only been tested with a peptide derived from the site at Thr317. Moreover, we did not identify glycosites in these regions in the present study despite finding many of the expected O-Man glycosites in the N-terminal region of the mucin domain of α-DG (Fig. 2). Nevertheless, mice deficient in POMGnT1 have previously been shown to produce truncated O-glycans, suggesting that the majority of O-Man glycosylation is elongated by POMGnT1 (15). O-GalNAc was simplified by targeting the private chaperone, COSMC, for the core UDP-Gal: GalNAcα1-O-Ser/Thr β1,3galactosyltransferase (C1GalT1) controlling the first step in O-GalNAc elongation (16). We previously developed a workflow for isolation and characterization of GalNAc O-glycopeptides from O-GalNAc SimpleCells (16, 27), which we modified for Man O-glycopeptides using Concanavalin A (Con A) lectin chromatography (Fig. 1B). Total cell lysates as well as secretomes were analyzed after enrichment of the growth medium on a short Con A column. Because Con A binds mannose and also will enrich for N-glycopeptides the total protease digests were treated with PNGase F to remove N-glycans before isolation of O-Man glycopeptides on a long Con A column and subsequent separation and analysis by nanoflow liquid chromatography-mass spectrometry (nLC-MS/MS). Using this strategy O-Man glycopeptides from α-DG and a total of 51 unique O-Man glycoproteins were identified, comprising a total of 235 O-Man glycosites (Table 1 and Dataset S1). We did not identify any of the individual proteins more recently suggested to carry O-Man glycans (10–14), which could be due to a number of reasons including experimental limitations as well as lack of expression of these proteins in MDA-MB-231 cells. Both site and peptide data for the identified O-Man glycosites have been made available on the GlycoDomainViewer (21). The GlycoDomainViewer illustrates identified glycosites in the context of the entire protein amino acid sequence and assigned domain structures. The Viewer shows the O-Man glycosites as well as other glycosylation sites identified in previous studies. The GlycoDomainViewer can be accessed online (http://glycodomain.glycomics.ku.dk/doi/10.1073/pnas.1313446110/).
Fig. 2.
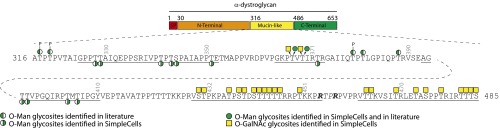
Schematic representation of the identified O-Man and O-GalNAc glycosylation sites in the mucin-like domain of human α-dystroglycan. Glycopeptides covered by the nLC-MS/MS analysis are underlined.
Table 1.
Identified O-Man glycoproteins and glycosites
| Superfamily | No. of proteins | No. of glycosites |
| Cadherins | 37 | 133 |
| Classical | 7 | 37 |
| Solitary | 1 | 2 |
| Desmocollin | 1 | 1 |
| Desmoglein | 1 | 3 |
| Flamingo | 2 | 5 |
| CR-1b FAT4 | 1 | 16 |
| CR-1b Daschous | 1 | 1 |
| Clustered protocadherin | 13 | 38 |
| Nonclustered | 7 | 20 |
| Nonclustered Delta-1 | 2 | 10 |
| Nonclustered Delta-2 | 4 | 8 |
| CR-2 | 1 | 3 |
| CR-3 FAT-like | 1 | 6 |
| Cadherin fragment | 1 | 1 |
| Plexins | 6 | 8 |
| KIAA1549 | 1 | 64 |
| Others | 8 | 30 |
| α-DG | 1 | 13 |
| Total | 52 | 235 |
Identification of O-Man Glycosites on α-DG.
For α-DG we obtained the greatest coverage of human O-glycosites so far, and we could confirm that O-Man and O-GalNAc glycosites are distributed in N- and C-terminal regions, respectively, of the mucin domain (Fig. 2) (25). Four O-Man glycopeptides from α-DG were identified allowing assignment of 11 additional unique human O-Man glycosites in the N-terminal region of the mucin domain (Fig. S1). While only a few of these sites have been identified in man, the majority of the sites have previously been identified in mice and rabbits (20, 25, 28–30). No glycopeptides were identified in the region previously shown to carry the phosphoryl-Man glycan (Fig. 2), which was expected if simple O-Man glycans are not available on digested peptide fragments for capture by Con A. These data also support the notion that the phosphoryl modification of O-Man at Thr317, Thr319, and Thr379 is specific for these residues. We previously identified a number of O-GalNAc glycosites using the SimpleCell strategy (Fig. 2) (21), and in the present study no glycopeptides with mixtures of O-Man and O-GalNAc were identified, but one glycopeptide (Pro361-Arg373) with three O-Man glycosites (Thr367, Thr369, and Thr372) in the middle were previously found with two O-GalNAc O-glycosites (Thr367 and Thr369). This confirms that some glycosites serve as acceptors for both O-Man and O-GalNAc glycosylation.
Cadherins Are Major O-Man Glycoproteins.
The major surprise was identification of the large cadherin family of cell membrane receptors as the major carrier of O-Man glycans (Table 1 and Dataset S1). We identified 37 distinct members of the cadherin superfamily with O-Man glycosites, and further sequence analysis suggests distinct evolutionary conservation in distribution of glycosites, in particular in the extracellular cadherin (EC) domains EC2-5 of classical type 1 and 2 cadherins (Fig. 3). No O-Man glycosites were found in the EC1 domains of classical cadherins, but we previously identified an O-GalNAc glycosite in EC1 of E- and T-cadherins that appears conserved in several cadherins (Fig. 3). Some of the O-Man sites identified in the cadherin EC2-5 domains were also found in the crystal structure of secreted E- and N-cadherins expressed in human HEK293 cells, although these were not concluded to represent O-Man (31). Although less conserved, the protocadherins exhibit similar distribution and conservation of glycosites. In protocadherins, the majority of sites were found in the largest subgroup of the diverse cadherin superfamily, the clustered protocadherins. The clustered protocadherins are predominantly expressed in the brain (32), and glycosites were mainly found in EC2-3 and to some degree in EC5-6 but not EC4. The function of O-Man glycans in cadherins is clearly unknown at present, but the conserved distribution and orientation of glycans on the surface of the membrane proximal EC domains offer a plausible scenario for positive or negative guidance of trans- and cis-interactions required for the assembly of the network of cadherin ectodomains that is believed to be the basis for the extracellular architecture of adherens junctions (31, 33).
Fig. 3.

Cadherins are the major class of O-mannosylated proteins. (A) Schematic drawing of extracellular domains of classical type 1 and type 2 cadherins. White circles illustrate potential O-Man glycosites predicted based on sequence conservation of Ser/Thr residues in alignments. Green-white circles show glycosites that were ambiguously identified, i.e., a glycopeptide was identified, but localization of the glycan was not identifiable by ETD. The O-GalNAc site identified by the O-GalNAc SimpleCell strategy is shown as a yellow square (21). Predicted O-GalNAc sites based on sequence identity are depicted as white squares. The O-Man sites are located either on or at the border of β-strand B and G. (B) Evolutionary conservation of predicted O-Man glycosites in type 1 and 2 cadherins. Representative analysis of CADH1 and CAD11 are shown with prediction of highly conserved and distinct patterns of distribution of glycosites from human to zebra fish in type 1 and type 2 cadherins. O-Man glycosites identified are depicted as green circles. These also include glycosites found in the crystal structures of mouse CADH1 and CADH2 (31).
Plexins Are O-Man Glycoproteins.
Another large family of cell membrane receptors identified was the plexins, for which we found conserved O-Man glycans on Ig-like, plexin and transcription factor (IPT) domains (Fig. S2). Plexins are large transmembrane glycoproteins that function as the receptors for semaphorins serving as axon guidance cues for neural development, but they also have nonneural roles (34). The extracellular domains of plexins have one sema domain, two or three Met-related sequences, and several glycine–proline-rich Ig-domains that are shared by plexins and transcription factors, and all these domains are shared with the Met family tyrosine kinases. The O-Man sites were identified in three out of four IPT domains of the plexins, and the sites are largely conserved in all four plexin subgroups. We also found O-Man sites in the IPT domains of two members of the Scatter Factor Receptor family, HGFR and MST1R.
A Unique O-Man Mucin-Like Membrane Glycoprotein.
The study also identified an uncharacterized large membrane protein (KIAA1549) with a high number of O-Man glycosites (Dataset S1). The function of this protein is unknown and it has only been implicated in a tandem duplication event at 7q34 that leads to a fusion between KIAA1549 and BRAF expression of a fusion protein with constitutive kinase activity in a majority of pilocytic astrocytomas (35). This protein is predicted to have a 936-aa N-terminal extracellular domain, and we identified 64 O-Man glycosites on 24 glycopeptides covering 268 amino acids evenly spread over the 936 extracellular domain. This exceptional pattern of O-Man glycosylation is illustrated using the online resource GlycoDomainViewer in Fig. S3. While this protein has not been identified in our O-GalNAc SimpleCells as a GalNAc O-glycoprotein, the O-GalNAc prediction algorithm NetOGlyc4.0 (21) in fact predicts the entire extracellular domain to be evenly O-glycosylated including all of the glycosites identified in this study with O-Man. Similarly, NetOGlyc4.0 predicts the whole mucin-like region of α-DG to be glycosylated—also predicting identified O-Man sites as O-GalNAc glycosylation sites, highlighting that we currently cannot predictively distinguish O-Man and O-GalNAc glycosylation sites using this predictor. Given that the extracellular domain of KIAA1549 displays an evenly distributed high density of Ser and Thr residues and no other assigned or apparent structural features, it is likely that the entire ectodomain carries O-Man glycans in the regions not identified in the present study. Thus, KIAA1549 may be representative of a mucin-like molecule with dense O-Man glycosylation similar to traditional mucins with heavy O-GalNAc glycosylation.
Finally, we identified one O-Man glycosite in one of the disulphide-isomerases, PDIA3 (Dataset S1), which we have previously also identified with O-GalNAc (21). All of the protein disulphide-isomerases (PDIs) have C-terminal KDEL-like ER retrieval signals and hence will be exposed to both the ER and Golgi, which offers the possibility of acquiring either O-Man or O-GalNAc glycans. O-Man glycosylation is initiated co- and posttranslationally in ER and can interfere with N-glycosylation and protein folding (36, 37), while O-GalNAc glycosylation is initiated in cis-Golgi (38). However, the initiating polypeptide GalNAc-transferases can function in ER (38) and they can relocate to ER by activation of Src kinases, opening up the possibility for direct competition (39, 40). This may represent another example of common substrate recognition between two forms of O-glycosylation similar to α-DG as discussed above.
Given the large number of O-Man glycosites now available, we sought to identify potential acceptor sequence motifs for O-mannosylation. Among all of the sites identified in cadherins and plexins, there did not appear to be discernible common sequence features suggestive of a glycosylation motif (Fig. S4). Instead, we found that all but one of the O-Man glycosites identified in proteins for which structures are deposited in structural databases are located on β-strands (Figs. S5–S7). O-Man glycosites in α-DG and KIAA1549 are located in disordered regions and looking at these separately did also not produce any clear sequence motifs. Interestingly, the NetOGlyc4.0 predictor showed overlap with O-Man glycosites only in these proteins and not in cadherins and plexins, which is in agreement with the finding that O-GalNAc glycosites are mainly predicted to be located in disordered regions (21). Human only has two O-mannosyltransferases, POMT1 and POMT2, and in yeast an ortholog of POMT1 in the PMT4 subfamily was demonstrated to selectively function with membrane proteins (18). It is thus tempting to speculate that POMT1 and POMT2 have different substrate specificities and that POMT1 primarily functions with membrane proteins with a preference for β-strands, while POMT2 primarily recognize linear sequence motifs in disordered regions similar to polypeptide GalNAc-transferases. Deficiencies in both genes underlie similar congenital muscular dystrophy phenotypes, but this may be associated with a need for POMT2 to function in a heteromeric complex (17, 41).
O-Man glycans are clearly essential for ligand binding to α-DG, as demonstrated by the severe dystroglycanopathies caused by hypoglycosylation (3). Congenital deficiencies in all of the genes involved in O-Man glycosylation produce muscular dystrophy phenotypes (42), but it is premature to identify phenotypic characteristics ascribable to other O-Man glycoproteins such as cadherins and plexins. However, it is clear that both families of proteins serve a range of essential functions that could be part of the phenotypes observed. O-Man glycosylation in yeast has multiple functions including those ascribed to O-GalNAc glycosylation in metazoans, i.e., roles for conformation, stability, and secretion of proteins (1). In yeast Pmt1p/Pmt2p, directed O-Man glycosylation has been demonstrated to play a role for ER protein quality control (43), and more recently the yeast Pmt1/2 mannosyltransferase complex was demonstrated to serve as a termination sensor of futile protein folding in ER (37).
In conclusion, the proteome-wide analysis of the O-Man glycoproteome presented here provides an entirely unique view of distinct classes of O-Man glycoproteins such as the cadherins and plexins, and the study provides insight into the structural feature (β-strands) governing this type of glycosylation. We applied the O-Man SimpleCell engineering strategy to a breast cancer cell line that we had already engineered for simplification of O-GalNAc glycosylation to be able to compare O-Man and O-GalNAc glycosites on α-DG. The O-Man glycoproteome is therefore limited to proteins expressed and O-mannosylated in this cell line. Given that O-Man glycans appear to be particularly abundant in brain, further exploration of the O-Man glycoproteome is clearly warranted, and the presented SimpleCell engineering strategy for exploration of the O-Man glycoprotome can now be widely applied to other cell lines and transgenic animals.
Materials and Methods
ZFN Gene Targeting.
ZFN targeting constructs for COSMC and POMGNT1 were custom produced (Sigma-Aldrich) with the following binding and (cutting) sites: COSMC 5′-CCCAACCAGGTAGT(AGAAGGCT)GTTGTTCAGATATGGCTGTT-3′; and POMGNT1 5′-AGCCAAGGCTCT(GCTGA)GGAGCCTGGGCAGCCAGG-3′. The MDA-MB-231 COSMC knockout O-GalNAc SimpleCell was generated as previously described (21). The MDA-MB-231 COSMC/POMGNT1 double knockout was generated by three sequential transfections with POMGNT1 ZFN pairs in the MDA-MB-231 COSMC knockout cell line. Transfected pools were single cell cloned by limiting dilution and clones identified by size difference in PCR product amplified around the ZFN cut site. Mutations were identified by sequencing of TOPO-cloned PCR fragments using the following primer pairs: 5′-AGGGAGGGATGATTTGGAAG-3′ and 5′-TTGTCAGAACCATTTGGAGGT-3′for COSMC; and 5′-TAGTTCGTGCTCTGTGAGGC-3′ and 5′-CACTGCCACTGGCTCCTATT-3′ for POMGNT1 (Fig. S8).
Lectin Weak Affinity Chromatography Isolation of O-Man Glycopeptides.
The lectin weak affinity chromatography (LWAC) protocol for isolation of O-Man glycopeptides was modified from our previously described method for isolation of O-GalNAc glycopeptides (16). In brief, a total of 140 mL conditioned media (secretome) or 0.5 mL packed cells (total cell lysate) was harvested from the MDA-MB-231 COSMC/POMGNT1 double knockout cell line grown in DMEM supplemented with 10% (vol/vol) FBS and 1% glutamine and processed as detailed hereafter.
Secretome.
Conditioned media obtained from 4× T175 flasks (4× 35 mL) cultured for 72–96 h, harvested by centrifugation (2,500 × g, 10 min) were dialyzed (Mw cutoff, 3,500 Da) twice against 5 L 20 mM Tris⋅HCl, pH 7.4, centrifuged (2,500 × g, 10 min), and diluted in 140 mL 2× Con A binding buffer A (40 mM Tris⋅HCl, pH 7.4, 300 mM NaCl, 2 mM CaCl2/MgCl2/MnCl2/ZnCl2, 1 M urea) or 2× Vicia villosa agglutinin (VVA) buffer (40 mM Tris⋅HCl, pH 7.4, 300 mM NaCl, 2 mM CaCl2/MgCl2/MnCl2/ZnCl2, 2 M urea) before subjected to either Con A or VVA lectin chromatography for enrichment of O-mannosylated and N-glycosylated glycoproteins or O-GalNAc glycoproteins, respectively. Con A and VVA agarose (Vector Laboratories) 0.5–0.7 mL in 2 mL syringes were equilibrated in Con A buffer A or VVA buffer A, respectively. The sample was loaded twice followed by 10–20 column volumes (CVs) wash in Con A buffer A or VVA buffer A, 2–4 CVs in 50 mM ammonium bicarbonate, and enriched glycoproteins were eluted by heating (2× 90 °C 10 min, and 2× wash of the beads) in the presence of 0.05% RapiGest (Waters) in 50 mM ammonium bicarbonate. The eluates were further processed as for cell lysates for digestion and N-glycan deglycosylation described below.
Total cell lysates.
Packed cells from 2× T175 flasks at confluency were lysed in 0.1% RapiGest in 50 mM ammonium bicarbonate with a sonic probe and the solution cleared by centrifugation (1,000 × g for 10 min). The cleared lysate and secretome samples were heated for 10 min at 80 °C, followed by reduction (5 mM DTT, 60 °C, 0.5 h) and alkylation (10 mM iodoacetamide, room temperature, 0.5 h), and digestion with trypsin (25 μg, Roche), Chymotrypsin (25 μg, Roche) or GluC (20 μg, Promega) (37 °C on). Proteases were heat inactivated (95 °C, 20 min) before N-glycanase treatment with PNGase F (8 U, Roche) (37 °C on), and then incubated 4 h more with additional PNGase F (3 U). N-deglycosylated digests were treated with TFA (8 μL, 37 °C, 20 min), cleared by centrifugation, purified on C18 Sep-Pak (Waters), concentrated by Speedvac, and resuspended in Con A buffer A to 1 mL before loading onto a preequilibrated (Con A buffer A 20 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 1 mM CaCl2/MgCl2/MnCl2/ZnCl2, 0.5 M urea) 2.8-m long Con A lectin agarose column for isolation of O-man glycopeptides. The column was washed with 10 CVs Con A buffer A (100 μL⋅min−1) before elution with Con A buffer B (20 mM Tris⋅HCl, pH 7.4, 300 mM NaCl, 2 mM CaCl2/MgCl2/MnCl2/ZnCl2, 0.5 M methyl-α-d-glucopyranoside/methyl-α-d-mannopyranoside) 5 CVs, 50 μL⋅min−1, 1-mL fractions); glycopeptide-containing fractions were purified by Stage Tips (Thermo Scientific) for analysis.
nLC-MS/MS Analysis.
Mass spectrometric analyses were performed essentially as previously described (27). Samples were analyzed either on a setup composed of an EASY-nLC II (Thermo Fisher Scientific) interfaced via a nanoSpray Flex ion source to an LTQ-Orbitrap XL hybrid spectrometer (Thermo Fisher Scientific) or an EASY-nLC 1000 (Thermo Fisher Scientific) interfaced via a nanoSpray Flex ion source to an LTQ-Orbitrap Velos Pro hybrid spectrometer (Thermo Fisher Scientific). The instruments were equipped with capabilities for both HCD- and ETD-MS2 fragmentation modes.
The conditions of LC analysis were essentially as previously described (27). The EASY-nLC II was equipped with a single analytical column set up using PicoFrit Emitters (New Objectives, 75-µm inner diameter) packed in-house with Reprosil-Pure-AQ C18 phase (Dr. Maisch, 3-µm particle size). The EASY-nLC 1000 was also operated in a single analytical column set up (28-cm length, 75-μm inner diameter, and 1.9-μm particle size). Briefly, a precursor MS1 scan (m/z 350–1,700 for LTQ-Orbitrap XL or 350–1,500 for LTQ-Orbitrap Velos Pro) of intact peptides was acquired in the Orbitrap at a nominal resolution setting of 30,000, followed by Orbitrap HCD-MS2 and ETD-MS2 (m/z of 100–2,000) of the three (LTQ-Orbitrap XL) or five (LTQ-Orbitrap Velos Pro) most abundant multiply charged precursors in the MS1 spectrum; a minimum MS1 signal threshold of 5,000 ions (LTQ-Orbitrap XL) or 20,000 (LTQ-Orbitrap Velos Pro) was used for triggering data-dependent fragmentation events; MS2 spectra were acquired at a resolution of 7,500 (LTQ-Orbitrap XL) or 15,000 (LTQ-Orbitrap Velos Pro).
Data Analysis.
Data processing was carried out using Proteome Discoverer 1.4 software (Thermo Fisher Scientific) as previously described (27) with minor changes in preprocessing and processing procedures. The monosaccharide subtraction routine for correctly interpreting HCD spectra was used as previously described (27), i.e., the exact masses of 1, 2, 3, and 4 hexose (Hex) units were subtracted from the corresponding precursor ion mass, generating four distinct files. The .raw files and subtracted .mgf files were searched against the human-specific UniProt database downloaded on Feb. 13, 2013, using semispecific trypsin, chymotrypsin, or GluC proteolytic cleavage. Carbamidomethyl was set as fixed modification for cysteine residues; methionine oxidation and asparagine deamidation were set as variable modifications; Hex was allowed as an additional variable modification when considering ETD-MS2 spectra. Fragmentation spectra of candidate-matched glycopeptides associated with each protein were inspected to verify accuracy of sequence and site assignments.
Note Added in Proof.
The authors were made aware of an upcoming paper with the related finding of O-mannose residues identified on rabbit cadherins (44).
Supplementary Material
Acknowledgments
We thank Lawrence Shapiro and members of his laboratory for discussions and critical review of the manuscript. We also thank Frederic Bard for sharing cell lines. This work was supported by A. P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til Almene Formaal, Kirsten og Freddy Johansen Fonden, The Carlsberg Foundation, The Novo Nordisk Foundation, The Danish Research Councils, the University of Copenhagen Program of Excellence, and the Danish National Research Foundation (DNRF107).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 20858.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313446110/-/DCSupplemental.
References
- 1.Gentzsch M, Tanner W. Protein-O-glycosylation in yeast: Protein-specific mannosyltransferases. Glycobiology. 1997;7(4):481–486. doi: 10.1093/glycob/7.4.481. [DOI] [PubMed] [Google Scholar]
- 2.Endo T. Structure, function and pathology of O-mannosyl glycans. Glycoconj J. 2004;21(1-2):3–7. doi: 10.1023/B:GLYC.0000043740.26062.2c. [DOI] [PubMed] [Google Scholar]
- 3.Barresi R, Campbell KP. Dystroglycan: From biosynthesis to pathogenesis of human disease. J Cell Sci. 2006;119(Pt 2):199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida A, et al. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev Cell. 2001;1(5):717–724. doi: 10.1016/s1534-5807(01)00070-3. [DOI] [PubMed] [Google Scholar]
- 5.Jae LT, et al. Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science. 2013;340(6131):479–483. doi: 10.1126/science.1233675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krusius T, Finne J, Margolis RK, Margolis RU. Identification of an O-glycosidic mannose-linked sialylated tetrasaccharide and keratan sulfate oligosaccharides in the chondroitin sulfate proteoglycan of brain. J Biol Chem. 1986;261(18):8237–8242. [PubMed] [Google Scholar]
- 7.Finne J, Krusius T, Margolis RK, Margolis RU. Novel mannitol-containing oligosaccharides obtained by mild alkaline borohydride treatment of a chondroitin sulfate proteoglycan from brain. J Biol Chem. 1979;254(20):10295–10300. [PubMed] [Google Scholar]
- 8. Chai W, et al. (1999) High prevalence of 2-mono- and 2,6-di-substituted manol-terminating sequences among O-glycans released from brain glycopeptides by reductive alkaline hydrolysis. Eur J Biochem/FEBS 263(3):879–888. [DOI] [PubMed]
- 9.Breloy I, Pacharra S, Aust C, Hanisch FG. A sensitive gel-based global O-glycomics approach reveals high levels of mannosyl glycans in the high mass region of the mouse brain proteome. Biol Chem. 2012;393(8):709–717. doi: 10.1515/hsz-2012-0214. [DOI] [PubMed] [Google Scholar]
- 10.Martinez T, Pace D, Brady L, Gerhart M, Balland A. Characterization of a novel modification on IgG2 light chain. Evidence for the presence of O-linked mannosylation. J Chromatogr A. 2007;1156(1-2):183–187. doi: 10.1016/j.chroma.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 11.Dwyer CA, Baker E, Hu H, Matthews RT. RPTPζ/phosphacan is abnormally glycosylated in a model of muscle-eye-brain disease lacking functional POMGnT1. Neuroscience. 2012;220:47–61. doi: 10.1016/j.neuroscience.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bleckmann C, et al. O-glycosylation pattern of CD24 from mouse brain. Biol Chem. 2009;390(7):627–645. doi: 10.1515/BC.2009.044. [DOI] [PubMed] [Google Scholar]
- 13.Pacharra S, Hanisch FG, Breloy I. Neurofascin 186 is O-mannosylated within and outside of the mucin domain. J Proteome Res. 2012;11(8):3955–3964. doi: 10.1021/pr200996y. [DOI] [PubMed] [Google Scholar]
- 14.Pacharra S, et al. The Lecticans of Mammalian Brain Perineural Net Are O-Mannosylated. J Proteome Res. 2013;12:1764–1771. doi: 10.1021/pr3011028. [DOI] [PubMed] [Google Scholar]
- 15.Stalnaker SH, et al. Glycomic analyses of mouse models of congenital muscular dystrophy. J Biol Chem. 2011;286(24):21180–21190. doi: 10.1074/jbc.M110.203281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steentoft C, et al. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8(11):977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 17.Lommel M, Strahl S. Protein O-mannosylation: Conserved from bacteria to humans. Glycobiology. 2009;19(8):816–828. doi: 10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- 18.Hutzler J, Schmid M, Bernard T, Henrissat B, Strahl S. Membrane association is a determinant for substrate recognition by PMT4 protein O-mannosyltransferases. Proc Natl Acad Sci USA. 2007;104(19):7827–7832. doi: 10.1073/pnas.0700374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida-Moriguchi TWT, et al. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science. 2013;341(6148):896–899. doi: 10.1126/science.1239951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida-Moriguchi T, et al. O-mannosyl phosphorylation of alpha-dystroglycan is required for laminin binding. Science. 2010;327(5961):88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steentoft C, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiba A, et al. Structures of sialylated O-linked oligosaccharides of bovine peripheral nerve alpha-dystroglycan. The role of a novel O-mannosyl-type oligosaccharide in the binding of alpha-dystroglycan with laminin. J Biol Chem. 1997;272(4):2156–2162. doi: 10.1074/jbc.272.4.2156. [DOI] [PubMed] [Google Scholar]
- 23.Brancaccio A, Schulthess T, Gesemann M, Engel J. Electron microscopic evidence for a mucin-like region in chick muscle alpha-dystroglycan. FEBS Lett. 1995;368(1):139–142. doi: 10.1016/0014-5793(95)00628-m. [DOI] [PubMed] [Google Scholar]
- 24.Endo T. O-mannosyl glycans in mammals. Biochim Biophys Acta. 1999;1473(1):237–246. doi: 10.1016/s0304-4165(99)00182-8. [DOI] [PubMed] [Google Scholar]
- 25.Gomez Toledo A, et al. O-Mannose and O-N-acetyl galactosamine glycosylation of mammalian α-dystroglycan is conserved in a region-specific manner. Glycobiology. 2012;22(11):1413–1423. doi: 10.1093/glycob/cws109. [DOI] [PubMed] [Google Scholar]
- 26.Hara Y, et al. Like-acetylglucosaminyltransferase (LARGE)-dependent modification of dystroglycan at Thr-317/319 is required for laminin binding and arenavirus infection. Proc Natl Acad Sci USA. 2011;108(42):17426–17431. doi: 10.1073/pnas.1114836108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vakhrushev SY, et al. Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol Cell Proteomics. 2013;12(4):932–944. doi: 10.1074/mcp.O112.021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson J, Nilsson J, Larson G, Grahn A. Characterization of site-specific O-glycan structures within the mucin-like domain of alpha-dystroglycan from human skeletal muscle. Glycobiology. 2010;20(9):1160–1169. doi: 10.1093/glycob/cwq082. [DOI] [PubMed] [Google Scholar]
- 29.Harrison R, et al. Glycoproteomic characterization of recombinant mouse α-dystroglycan. Glycobiology. 2012;22(5):662–675. doi: 10.1093/glycob/cws002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stalnaker SH, et al. Site mapping and characterization of O-glycan structures on alpha-dystroglycan isolated from rabbit skeletal muscle. J Biol Chem. 2010;285(32):24882–24891. doi: 10.1074/jbc.M110.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison OJ, et al. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 2011;19(2):244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yagi T. Clustered protocadherin family. Dev Growth Differ. 2008;50(Suppl 1):S131–S140. doi: 10.1111/j.1440-169X.2008.00991.x. [DOI] [PubMed] [Google Scholar]
- 33.Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: How cadherins drive adhesion. Trends Cell Biol. 2012;22(6):299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perälä N, Sariola H, Immonen T. More than nervous: The emerging roles of plexins. Differentiation. 2012;83(1):77–91. doi: 10.1016/j.diff.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Jones DT, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ecker M, et al. O-mannosylation precedes and potentially controls the N-glycosylation of a yeast cell wall glycoprotein. EMBO Rep. 2003;4(6):628–632. doi: 10.1038/sj.embor.embor864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu C, Wang S, Thibault G, Ng DT. Futile protein folding cycles in the ER are terminated by the unfolded protein O-mannosylation pathway. Science. 2013;340(6135):978–981. doi: 10.1126/science.1234055. [DOI] [PubMed] [Google Scholar]
- 38.Röttger S, et al. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J Cell Sci. 1998;111(Pt 1):45–60. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- 39.Gill DJ, Chia J, Senewiratne J, Bard F. Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J Cell Biol. 2010;189(5):843–858. doi: 10.1083/jcb.201003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gill DJ, Clausen H, Bard F. Location, location, location: New insights into O-GalNAc protein glycosylation. Trends Cell Biol. 2011;21(3):149–158. doi: 10.1016/j.tcb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 41.Lommel M, Willer T, Cruces J, Strahl S. POMT1 is essential for protein O-mannosylation in mammals. Methods Enzymol. 2010;479:323–342. doi: 10.1016/S0076-6879(10)79018-2. [DOI] [PubMed] [Google Scholar]
- 42.Godfrey C, Foley AR, Clement E, Muntoni F. Dystroglycanopathies: Coming into focus. Curr Opin Genet Dev. 2011;21(3):278–285. doi: 10.1016/j.gde.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Goder V, Melero A. Protein O-mannosyltransferases participate in ER protein quality control. J Cell Sci. 2011;124(Pt 1):144–153. doi: 10.1242/jcs.072181. [DOI] [PubMed] [Google Scholar]
- 44.Winterhalter PR, Lommel M, Ruppert T, Strahl S. O-glycosylation of the noncanonical T-cadherin from rabbit skeletal muscle by single mannose residues. FEBS Letters. 2013;587(22):3715–3721. doi: 10.1016/j.febslet.2013.09.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.