Significance
Fluorescence microscopy is an enormously powerful tool for investigating structural organization and dynamical processes on the cellular level because of its noninvasiveness, sensitivity, and specificity. However, classical fluorescence microscopy is limited in resolution by the diffraction of light. In recent years, structural illumination microscopy has succeeded in doubling this resolution without requiring any special sample preparation or labeling dyes. However, it is technically very challenging and complex. Here, we present an alternative that achieves the same resolution enhancement by using a standard confocal spinning-disk microscope with minimal modifications. This method, in principle, allows one to double the resolution of any existing confocal microscope.
Abstract
We demonstrate how a conventional confocal spinning-disk (CSD) microscope can be converted into a doubly resolving image scanning microscopy (ISM) system without changing any part of its optical or mechanical elements. Making use of the intrinsic properties of a CSD microscope, we illuminate stroboscopically, generating an array of excitation foci that are moved across the sample by varying the phase between stroboscopic excitation and rotation of the spinning disk. ISM then generates an image with nearly doubled resolution. Using conventional fluorophores, we have imaged single nuclear pore complexes in the nuclear membrane and aggregates of GFP-conjugated Tau protein in three dimensions. Multicolor ISM was shown on cytoskeletal-associated structural proteins and on 3D four-color images including MitoTracker and Hoechst staining. The simple adaptation of conventional CSD equipment allows superresolution investigations of a broad variety of cell biological questions.
Fluorescence microscopy is an extremely powerful research tool in the life sciences. It combines highest sensitivity with molecular specificity and exceptional image contrast. However, as with all light-based microscopy techniques, its resolution is limited by the diffraction of light to a typical lateral resolution of ∼200 nm and an axial resolution of ∼500 nm (for 500-nm wavelength light). Only recently, this diffraction limit was broken by using the quantum, or nonlinear, character of fluorescence excitation and emission. The first of these superresolution methods was stimulated emission depletion (STED) microscopy (1). Later, methods based on single-molecule localization, such as photoactivated localization microscopy (PALM) (2) and stochastic optical reconstruction microscopy (STORM) (3), joined the field. These methods “break” the diffraction limit because they all use principles that operate beyond the diffraction of light.
Although still bound to light diffraction, increased spatial resolution can be achieved in a class of advanced resolution methods that exploit a clever combination of excitation and detection modalities (4–7). Although these methods do not reach the resolution achievable with STED, PALM, STORM, and related techniques, they do not require any specialized labels or high excitation intensities, and they may be applied to any fluorescent sample at any excitation/emission wavelength. The most prominent example of this class is structured illumination microscopy (SIM) (5), in which one scans a sample with a structured illumination pattern while taking images with a wide-field imaging system. Meanwhile, several commercial instruments for SIM have become available. The disadvantages of SIM are its technical complexity, reflected in the rather large cost of the commercially available systems, and its sensitivity to optical imperfections and aberrations, which are unavoidable in biological samples.
In a theoretical study in 1988, Sheppard (8) pointed out that it is possible to double the resolution of a scanning confocal microscope in a manner closely related to SIM. In SIM, one starts with a conventional wide-field imaging microscope, and by implementing a scanning structured illumination, one subsequently obtains, after appropriate deconvolution of the recorded images, an image with increased resolution. In image-scanning microscopy (ISM), as proposed by Sheppard, one starts with a conventional confocal microscope that uses a diffraction-limited laser focus for scanning a sample but replaces the point detector typically used for recording the excited fluorescence signal with an imaging detector. Also here, an image with enhanced resolution is obtained after applying an appropriate algorithm to the recorded images.
We experimentally realized this idea first in 2010 (4), indeed demonstrating a substantial increase in resolution. The major drawback of this implementation was the slowness of the imaging. At each scan position of the laser focus, an image of the excited region had to be recorded, limiting the scan speed by the frame rate of the imaging camera used. For the small scan area of 2 µm × 2 µm shown with the original ISM setup, data acquisition took 25 s. In 2012, York et al. (6) demonstrated that this limitation may be overcome by using a multifocal excitation scheme. They generated an array of multiple excitation foci by implementing a digital micro-mirror device (DMD) into the excitation path of a wide-field microscope. Using this system, ISM images can be obtained with excellent speed, in two excitation/emission wavelengths (two-color imaging) and in three dimensions. However, this approach requires the incorporation of a DMD with all the necessity of perfect optical alignment.
Here, we demonstrate that existing imaging detector-based confocal systems can be converted easily into a doubly resolving ISM system. This mainly includes two kinds of microscopes that are widely available in research laboratories: confocal spinning-disk (CSD) microscopes and rapid laser scanning confocal microscopes with an imaging camera as the detector. We present the results obtained with a CSD system.
Results
CSD-Based Implementation of ISM.
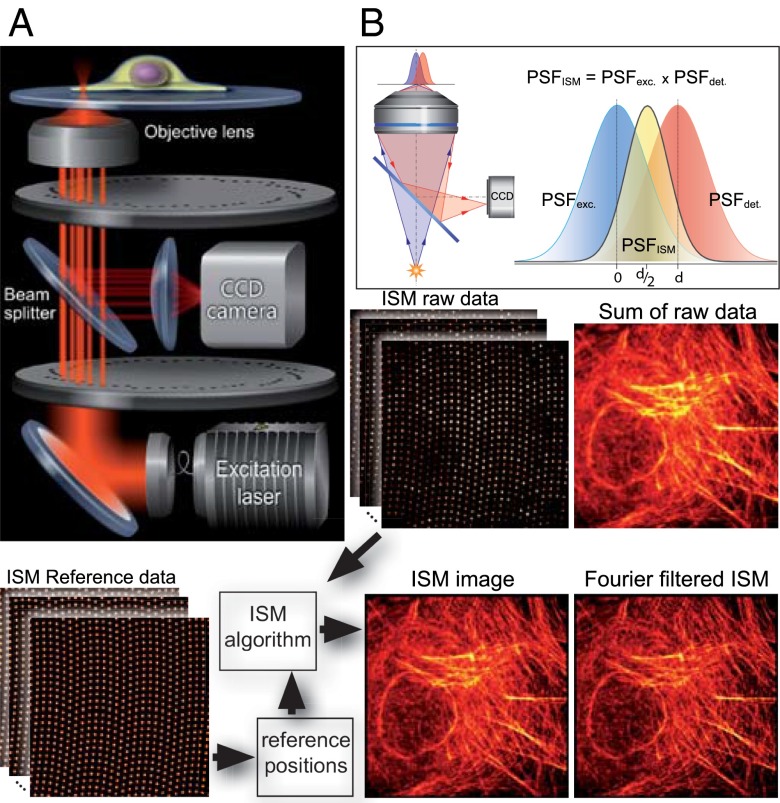
In ISM, the sample is excited by a tightly focused laser spot and the fluorescence light emerging from the focus is imaged by means of a camera. If the pixels of the camera are small enough, they may be considered “infinitely small pinholes” for the purpose of enhancing the lateral resolution. The beauty of CSD-based ISM (CSD-ISM) as a 3D superresolution technique lies in the simplicity of the optical setup. The principle behind the use of CSD microscopy for ISM is based on the use of a camera and the inherent presence of many excitation foci that rapidly scan the sample. Here we show how the CSD can be upgraded easily to be used for ISM. Although it has been known for a while that a very small pinhole enhances the resolution in confocal microscopy (9), this approach is not practical, because almost all the fluorescent light is rejected by the pinhole. In the case of a camera, however, in which there are many pixels, we do not lose light because every (detectable) photon from the sample will be recorded on the camera chip. In this fashion, every pixel records a “perfect” confocal image. To obtain a final image that contains all the light from every pixel, the confocal images gathered by every pixel must be combined. For this, we need to consider the offset from the optical axis of each pixel and its effect on the image that this pixel records. If a pixel is shifted by a distance d from the optical axis, the image this pixel records will be shifted by d/2 with respect to an image recorded on the optical axis (Theory of Image-Scanning Microscopy). Fig. 1B explains graphically the origin of this factor 1/2. Thus, the fluorescence intensity information from each pixel must be shifted half its distance toward the optical axis, and summed to obtain the final image. Alternatively, and equivalently, the unmodified raw data can be registered in an image in which the scan coordinates are multiplied by two. This approach naturally leads to twice as many pixels in each dimension. For detailed information about the basics of ISM and the image calculation, please see Theory of Image Scanning Microscopy and Fourier Reweighting and Figs. S1 and S2.
Fig. 1.
Basics of CSD-ISM. (A) Schematic drawing of the optical setup. The excitation light (400 nm, 488 nm, 561 nm, or 647 nm) is coupled into the spin-disk confocal unit via an optical fiber. A Nipkow disk with microlenses focuses the light onto pinholes of an opposing disk. The microscope objective focuses the light emerging from the pinholes to diffraction-limited spots in the sample. The fluorescence emission from these spots is collected by the same objective and passes through the pinholes onto a CCD camera via a dichroic beam splitter. (B) A pixel on the CCD camera, if it is small enough, detects light from a diffraction-limited region of the sample. Pixels offset from the optical axis will detect an area of the sample that does not coincide with the excitation; thus, the product of these two point-spread functions, which is proportional to the amount of light detected, will be shifted. This way, each pixel on the CCD records a confocal image that is shifted by half its distance from the optical axis. The bottom of the figure illustrates the work flow in CSD-ISM. Reference data from a homogenously fluorescing sample is recorded to determine the center position of each pinhole. These reference positions are fed into the ISM algorithm that shifts the raw data from every pinhole, as indicated in B. Finally, Fourier filtering is performed to gain the full resolution enhancement of ISM.
To be able to apply the ISM method to data gathered with a CSD, one has to make sure the camera captures the light from individual pinholes without being smeared out while the disk spins. In our setup, this is achieved by means of stroboscopic illumination. The illuminating laser is active for only a few microseconds at a time; in this time, the disk has not rotated noticeably. The lasers and the camera are synchronized with the motion of the spinning disk. The synchronization, which is realized using programmable electronics [field-programmable gate array (FPGA)], allows an image to be recorded at a highly reproducible position of the disk during its motion. The whole image is “scanned” by introducing a delay to the laser trigger, which moves the illuminated spots through the sample. The positions of the spots are prerecorded for each time point by imaging a homogenously fluorescing sample. Details on the synchronization and calibration of the system may be found in Methods and in ISM Algorithm and Image Calculation Using Matlab.
The raw data collected by the CSD in ISM mode consists of a set of about 250 images. The sum of these images is equivalent to a regular CSD image. One CSD-ISM image can be acquired within 0.3–1 s, depending on the speed of the spin disk (1,500–5,000 rpm). For weakly fluorescing samples or to accommodate for a slower camera readout for larger fields of view, the camera can be read out only once while multiple laser triggers with the same delay are produced during multiple spin disk cycles. In this fashion, the signal-to-noise ratio is enhanced (ISM Algorithm and Image Calculation Using Matlab; Fig. S3). When presenting the experimental data below, we give the degree of averaging by noting the exposure time per frame, which is the laser pulse length times the number of cycles over which the image is accumulated, and the number of frames from which one ISM image is calculated. For a better comparison of all data, the total acquisition time of one ISM image also is given.
We applied the ISM algorithm to each focal spot in each image, as indicated in Fig. 1. After applying the ISM algorithm to the raw data, the resolution can be enhanced further by Fourier reweighting, as described in ref. 4 (compare Figs. 1 and 2C, and Fourier Reweighting).
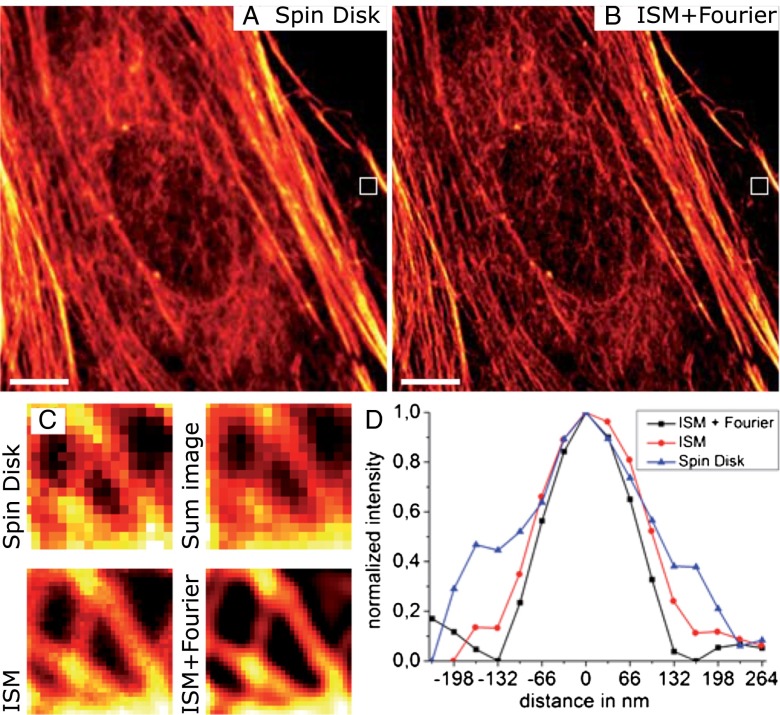
Fig. 2.
Doubling the resolution for actin imaging in mammalian cells with CSD-ISM. Rat embryonal fibroblasts (REF52) were labeled for actin with Alexa 488–phalloidin. The exposure time for the conventional spin-disk image (A) was 10 ms. The CSD-ISM image (B) was calculated from 250 single images with an exposure time of 138 µs per frame (∼20 s acquisition time). (C) Comparison of a small area (white frame) where individual actin filaments can be distinguished. The size of the magnified region is 1 μm. As expected, the conventional spin-disk image and the sum over all 250 single shots of the CSD-ISM dataset are of comparable quality, whereas the CSD-ISM image and the CSD-ISM image after Fourier reweighting show more detail. (D) The graphs show a section through the actin filament in the center of the zoom-in. Gaussian fits to these sections exhibit FWHMs of 201 nm, 162 nm, and 130 nm for the spin-disk image, the CSD-ISM image, and the Fourier reweighted CSD-ISM image, respectively. The scale bars in A and B are 5 μm.
From the point of view of the optical setup, no modifications must be made for using a CSD for ISM. However, because the size of a pixel of the camera is a determining factor for the achievable resolution enhancement in ISM, the choice of camera and the magnification must be considered. In our case, we used a 100× objective and a camera adapter with 1.2× magnification. For our 16-μm pixel EM CCD, this translates to about a 133-nm effective pixel size. To obtain optimal ISM resolution (9), we added an additional 2× magnification to achieve a 67-nm pixel size. We also used a scientific complementary metal–oxide–semiconductor (sCMOS) camera with 6.5-μm pixels, which translates to about a 54-nm effective pixel size without additional magnification.
Resolution in CSD-ISM.
The resolution in ISM was estimated theoretically to be twice that of conventional wide-field microscopy (4). Experimentally, resolution enhancement factors of 1.63 (4) and 2 (6) were determined by measuring the apparent width of fluorescent beads (4, 6) and microtubules (6). To evaluate the resolution enhancement of our CSD-based ISM, we used single fluorescent molecules (Atto 655) and actin filaments labeled with Alexa 488 in a cell. Fig. 2A shows a regular CSD image of the actin network. The corresponding CSD-ISM image (Fig. 2B) clearly reveals a finer structure. The apparent width of the filaments was evaluated by zooming in on a region where individual actin filaments can be distinguished. Whereas the regular CSD image and the sum image of the ISM raw data are qualitatively similar, the CSD-ISM image and the Fourier reweighted CSD-ISM image clearly show more distinct structures (Fig. 2C). A section through the central filament in the indicated magnified region was used to estimate the enhanced resolution of CSD-ISM (Fig. 2D). For the CSD image, the FWHM, determined by a Gaussian fit, is 201 ± 20 nm (SE). From the CSD-ISM image, we measure an FWHM of 162 ± 10 nm, and 130 ± 10 nm for the Fourier reweighted CSD-ISM image, which translates to a resolution enhancement factor of 1.55. The theoretical values for the FWHMs of the point-spread functions of confocal and Fourier reweighted ISM (Fourier Reweighting) are 201 nm and 129 nm, respectively. These values were calculated assuming excitation at 488 nm and detection at 525 nm.
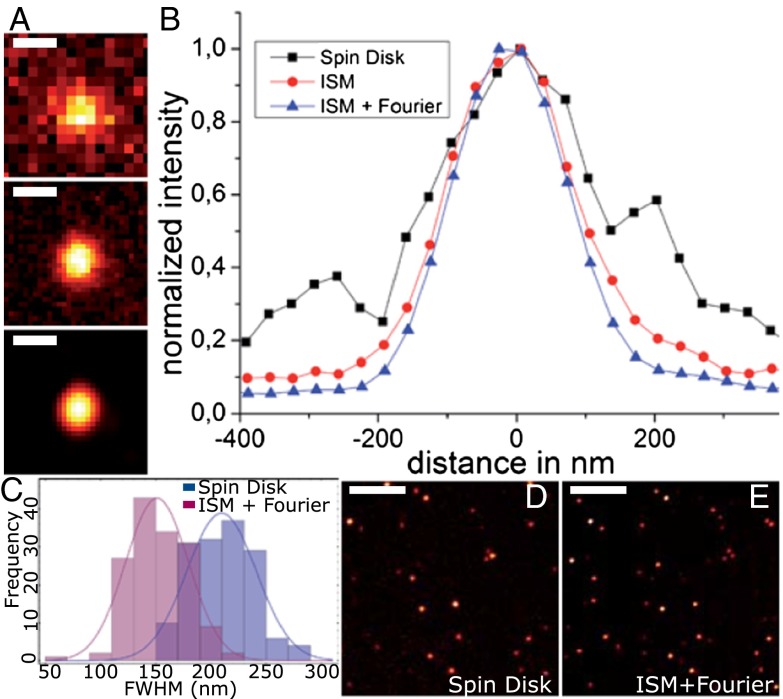
For a more rigorous evaluation of the resolution enhancement, we chose single molecules because they may be considered ideal point sources of fluorescence light. Because the brightness of the single molecules is too weak for high-speed imaging, averaging over longer exposure times was necessary. We imaged individual molecules of Atto 655, a fluorescence label shown to be very stable and to exhibit little blinking on glass surfaces under dry conditions (10). Two-dimensional Gaussians were fit to the single-molecule emission spots in the CSD image and in the CSD-ISM image for multiple scans. An example of the evolution from the CSD image, over the CSD-ISM image, to the final Fourier reweighted CSD-ISM image of a single Atto 655 molecule is given in Fig. 3A. The distribution of resolution values was determined from 86 molecules (histogram in Fig. 3B). For the CSD image, the mean of the distribution of FWHMs is 265 ± 27 nm (SD), and 197 ± 17 nm (SD) for the final CSD-ISM image. This corresponds to a resolution enhancement factor of 1.35. These values compare with theoretical values of 246 nm for confocal and 172 nm for Fourier reweighted ISM. The calculations were performed assuming excitation at 647 nm and detection at 690 nm.
Fig. 3.
Quantification of resolution enhancement using single Atto 655 molecules on glass. (A) Comparison among conventional spin disk, CSD-ISM, and CSD-ISM with Fourier reweighting of a single Atto 655 molecule. The spin-disk image (D) was taken at an exposure time of 1 s. The CSD-ISM image (E) was calculated from 1,000 single images, each with an exposure time of 138 µs (∼80 s acquisition time). (B) Plot of a section through the center of the molecule for the different images in A. The FWHMs (Gauss fit) of the sections are 248 ± 12 nm, 186 ± 3 nm, and 171 ± 2 nm (SE), respectively. The histogram of FWHMs of single-molecule fluorescence (C) is determined by a 2D Gauss fit. The histogram contains both the x and y values of the FWHM of 86 single-molecule spots. The scale bar is 300 nm in A and 4 μm in D and E.
The difference in the resolutions determined with the two test samples stems from their different excitation and emission wavelengths as the resolution of confocal microscopy, and thus ISM still depends on the wavelength of the light used.
In our present implementation of ISM, we use a simple but well-defined Fourier-reweighting procedure that enhances higher vs. lower frequencies in Fourier space for approaching the ideal optical transfer function of a microscope system with doubled resolution (4). More sophisticated algorithms, such as the popular Richardson–Lucy deconvolution (11–13), may give better results by better handling the inherent noisiness of real data but are not as straightforward and transparent as the Fourier-reweighting procedure used here.
ISM does not significantly enhance the axial resolution of a confocal microscope with a well-sized pinhole (Theory of Image-Scanning Microscopy), which also is what we observe in our data recorded with the CSD microscope.
Imaging of the Nuclear Pore Complex on the Nuclear Membrane.
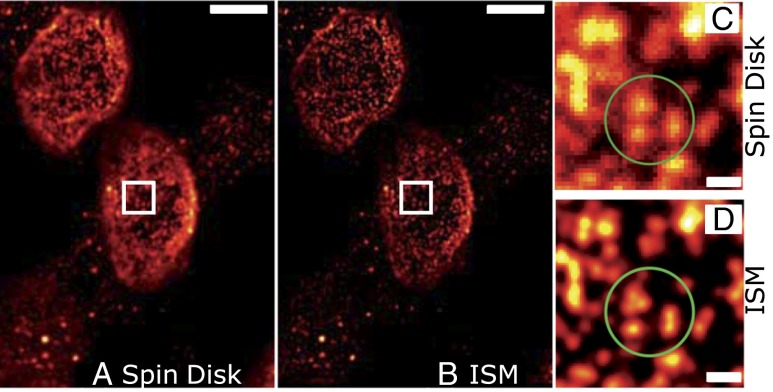
The nuclear pore complex (NPC) is an aqueous channel in the nuclear envelope that has a central role in the selective nuclear transport between cyto- and nucleoplasm. Accumulating evidence shows that the NPC has various regulatory functions, e.g., during development and aging (14–17). The structure of the metazoan NPC has been studied intensively by (cryo)electron microscopy, which revealed the hourglass-like shape whose diameter ranges from ∼95 nm at the widest point to ∼50 nm at the narrowest point, at the center of the NPC (18, 19). The eightfold arrangement of the NPC, which consists of multimeric subunits of more than 30 nucleoporins, was demonstrated recently by superresolution imaging by direct STORM (20). With a size close to the resolution of ISM at 488-nm excitation, single NPCs can be distinguished more clearly and accurately than from spin-disk images (Fig. 4). The NPC number and its distribution on the nuclear envelope change during the cell cycle (21); therefore, precise quantification is crucial. Whereas the measured NPC density from spin-disk images is only about 5.34 ± 0.28 NPC/µm2, quantifications from CSD-ISM images yielded in an ∼1.3-fold higher NPC density (7.01 ± 0.75 NPC/µm2), which agrees very well with previously reported NPC densities in HeLa cells (21), confirming a relevant information gain with the superior CSD-ISM method.
Fig. 4.
Improved identification of single NPCs with CSD-ISM. Image of NPCs immunolabeled by mAb414 and Alexa 488. (A) Conventional spinning-disk image with an exposure time of 50 ms. (B) Final CSD-ISM image, calculated from 500 single images with an exposure of 138 µs per image (∼40 s acquisition time). (C and D) Direct comparison of a smaller area between conventional spin disk (C) and CSD-ISM (D). The superior performance of CSD-ISM is immediately clear. Single NPCs, which were obscured in the confocal image, can be distinguished clearly (green circled area). The scale bars are 5 μm in the overview and 500 nm in the indicated magnified region.
Multicolor Imaging of the Cellular Cytoskeleton.
Multicolor fluorescence imaging allows one to distinguish proteins and structures of interest and to study their colocalization and interactions. CSD-ISM is readily extendible to the imaging of multiple spectrally separable fluorescent labels. If no pulsed lasers are available in the existing CSD setup, the stroboscopic illumination of the sample can be achieved straightforwardly using an acousto-optical tunable filter (AOTF). In our setup, we used pulsed diode lasers at 400 nm, 488 nm, and 647 nm, and a 561-nm continuous wave (cw) laser with AOTF modulation. The different emission wavelengths may be imaged in sequential order, or beam splitters may be used to image different colors onto separate areas on the camera chip or onto separate cameras. Another way to realize simultaneous multicolor imaging is to modulate the lasers in quick succession so that their corresponding emission is separated properly on the camera (Synchronization and Data Acquisition).
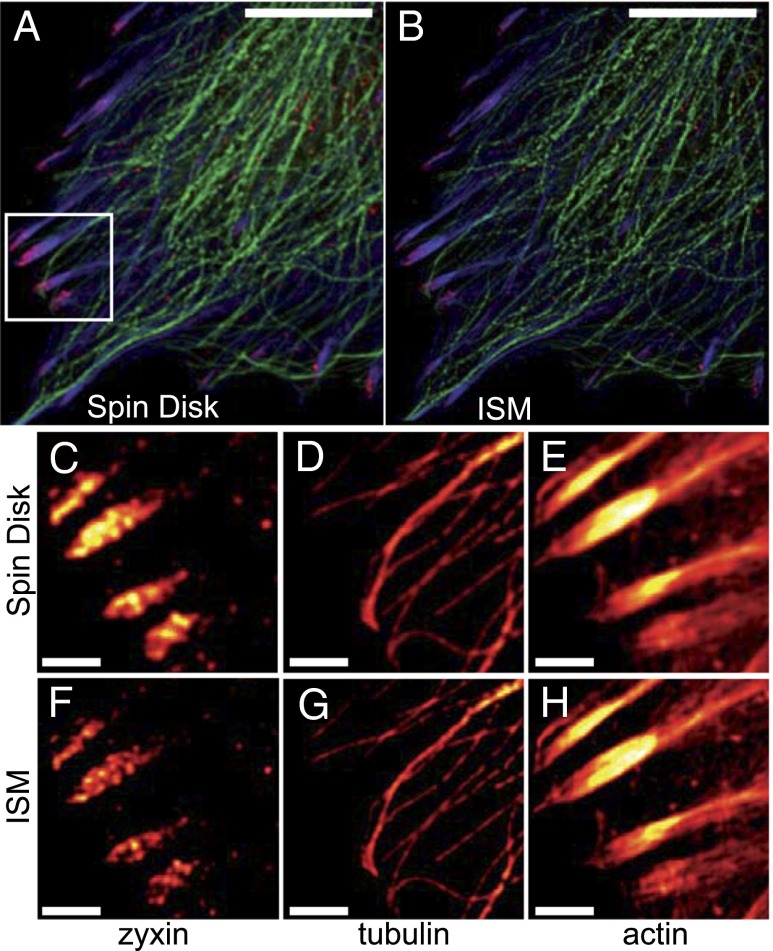
To show the multicolor imaging potential of CSD-ISM, we labeled different cytoskeletal structures in rat embryonal fibroblasts (REF52). Actin was labeled with phalloidin–Alexa 488 tubulin by immunolabeling with TRITC, and the focal adhesion protein zyxin was labeled with Alexa 647. Comparison with the respective confocal images shows the increase in contrast and the resolution of substructures, especially in the macromolecular focal adhesion complexes to which the actin cables dock. At the higher resolution, the actin stress fiber in the focal adhesion ending appears to split to match rows of punctate zyxin structures (Fig. 5).
Fig. 5.
ISM allows higher resolution in multicolor imaging. Multicolor ISM of fixed REF52 cells showing Alexa 488-labeled actin (blue) and TRITC-labeled tubulin (green) cytoskeletal networks together with the Alexa 647-labeled focal adhesion protein zyxin (red). A confocal image is shown in A. The corresponding CSD-ISM image from 378 single images is shown in B. For CSD-ISM images of Alexa 488 and TRITC, each single image was exposed for 66 µs (∼15 s acquisition time), and for Alexa 647 an exposure of 210 µs (∼45 s acquisition time) was used. Magnifications of the indicated regions of interest are shown in C–H. The scale bars are 10 μm for the overview and 2 μm for the regions of interest.
We corrected chromatic aberration using the ImageJ plugin bUnwarpJ (22). Images of whole cells of the same sample, acquired using an sCMOS camera, may be found in CSD-ISM with an sCMOS Camera (also see Fig. S4).
Fast 3D Imaging of Tau Protein.
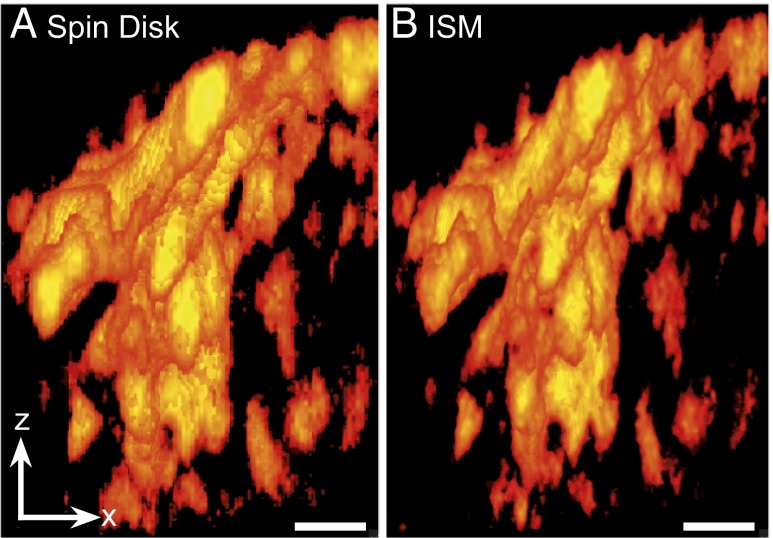
Tau is a neuronal microtubule-binding protein whose aggregation in Alzheimer’s disease is thought to contribute to cell toxicity and neuronal loss in the affected brain. The size and distribution of these aggregates might hold important clues to understanding the still poorly understood mechanism of toxicity. Three-dimensional CSD-ISM operates at the length scale at which aggregates may be studied for fragmentation or fusion, and for potential clearance routes using the autophagosomal or exosomal compartment. Using a genetic expression model based on full-length (441-aa) Tau that was structurally optimized for β-sheet formation by three amino acid modifications in the microtubule binding repeat (3PO-Tau), we imaged the formation of morphological aggregates in three dimensions. This mutant Tau rapidly forms aggregates with associated toxicity and pathological hallmarks of Alzheimer’s disease (23, 24). To this end, GFP-fused 3PO-Tau was expressed in N1E115 neuroblastoma cells, and the GFP label was imaged using CSD-ISM. Volume rendering of aggregated Tau in a selection of the cell shows the increased level of detail that may be observed (Fig. 6) with CSD-ISM. Each of the 85 CSD-ISM images in the stack was acquired within 1 s, demonstrating that superresolution images can be obtained on a sufficiently short time scale for biological imaging. A maximum-intensity projection movie of this data after 3D deconvolution may be found as Movie S1. For details, see Fast 3D Imaging of Tau Protein.
Fig. 6.
ISM gives enhanced resolution in three dimensions. Three-dimensional representation of confocal (A) and CSD-ISM (B) z-stack data of GFP-fused 3PO-Tau aggregates in an N1E115 neuroblastoma cell. The increased level of detail in the ISM data allows for better characterization of different types and progression of aggregates. The data were acquired using the 16-pulse laser sequence (Synchronization and Data Acquisition). The images were calculated from 125 single images with an exposure time of 6 µs per image (∼0.8 s acquisition time). The scale bar (1 μm) is the same for x- and z-direction. A movie of these data may be found as Movie S1.
Four-Color 3D Imaging.
Three-dimensional and multicolor imaging capabilities can be combined in CSD-ISM. Four-color 3D stacks of HeLa cells were acquired using the commonly used organelle stains Hoechst 33258 and MitoTracker M-7510, together with Alexa 488-labeled α-tubulin and Alexa 633-labeled Nup358, a protein of the NPC (25, 26). Images and 3D-projection movies may be found in Four-Color 3D Imaging, Movie S2, and Fig. S5. The ability to use commonly used fluorophores from selective organic stains to typical immunostaining labels and fluorescent proteins makes CSD-ISM a useful tool in cell biology. The morphology of a cell and its organelles can be evaluated in greater detail compared with the currently used routine fluorescence microscopy technique with highest resolution: confocal microscopy. The examples have shown that new biological information lies in the resolution window just beyond confocal microscopy.
Discussion
Like existing SIM techniques, CSD-ISM can deliver superior image quality in resolution and contrast compared with confocal imaging. We see its significant advantage over other techniques in its straightforward implementation, potentially opening up superresolution imaging capabilities to all camera-based confocal microscopy users. An existing confocal spinning-disk setup can be upgraded easily for ISM by modulating the laser light and synchronizing the spinning disk with the illumination and imaging camera. Modulation of any laser can be achieved using an AOTF, and here we have shown the performance of such a device for CSD-ISM. The synchronization with lasers and camera of the Yokogawa CSU-X1, which is used in most commercially available spinning-disk systems, is achieved simply by using an FPGA and the readily accessible trigger signals the CSU-X1 generates. The optical resolutions we could achieve using this system compare remarkably well with theoretical predictions. The lateral resolution and optical sectioning ability of the CSD-ISM approach the theoretical limit for any far-field microscope using linear optics, such as SIM, multifocal SIM, and related techniques.
Besides single-molecule measurements for evaluation of the improved resolution, we have demonstrated the capabilities of our method by imaging various biological systems. CSD-ISM does not put any restrictions on the used fluorophores, allowing its use in any experiment that is also accessible to normal wide-field or confocal fluorescence imaging. The fluorophores we used span the range from commonly used specific stains for organelles and subcellular structures, such as Hoechst for the nucleus, MitoTracker for mitochondria, and phalloidin for the actin cytoskeleton, to conventional fluorescent labels such as Alexa dyes, rhodamine, and fluorescent proteins (GFP).
We anticipate that the exceptional ease of use of CSD-ISM, its robustness, and its versatility, will make it a commonly used tool in the life sciences.
Methods
Microscopy System.
Data acquisition was performed on an Olympus IX-71 microscope with a Yokogawa CSU-X1 (M1N-E) mounted to the left side port. The objective used was an Olympus 100× UAPON, N.A. 1.49. The dichroic mirror in the spinning-disk unit was a Di01-T405/488/568/647 from Semrock Inc. As emission filters, either a quad-band filter (BrightLine HC 446/523/600/677; Semrock) or individual bandpass filters (593-40, 525-45, 685-70, 447-60 BrightLine HC; Semrock) were used. After the spinning-disk unit, a telescope system for doubling the magnification (MAP1075150-A achromatic matched pair doublet; Thorlabs) was introduced to decrease the effective pixel size of the EM CCD camera [iXon ×3–897D (Andor Technology) with 512 × 512 pixels and 16-μm pixel size). The magnification of the objective, 1.2× magnification of the spinning-disk camera adapter, and 2× magnification before the camera resulted in an effective pixel size of 67 nm. Alternatively, an sCMOS camera [Neo (Andor Technology), 2,560 × 2,160 pixels, 6.5-μm pixel size] was mounted after the spin-disk unit.
Laser light (400, 488, 561, or 647 nm) was coupled into the CSU-X1 with an optical fiber. For illumination, the following laser sources were used: for 400 nm, a Cube 405–100C, 60 mW (Coherent); for 488 nm, a PhoxX 488–60, 60 mW; and for 647 nm, a PhoxX 647–147, 140 mW (Omicron Laserage). For illumination at 561 nm, a Cobolt Jive, 50 mW (Cobolt), modulated by an AOTFnC-400.650-TN (AA Optoelectronic) was used.
For 3D scanning, a PIFOC P725-2DC with E709-CRD controller from Physik Instrumente (PI) GmbH & Co. KG was used.
Stroboscopic Laser Excitation.
Details on the synchronization of the CSD with the laser illumination and the camera using an FPGA may be found in Synchronization and Data Acquisition. Briefly, one ISM image was divided into typically 250 single camera exposures. A trigger signal from the CSU-X1 was used to modulate the laser and to time the camera exposure. After each of the 250 single images, the laser pulses were offset in time to achieve an offset in space as the disk spun, which accomplished scanning of the sample. Four laser pulses per offset were used, which produced four times as many spots on the camera image. These spots did not overlap. The x-distance between spots between shots was about 100 nm. The y-distance between spots (after the next row was at same x-position) was about 66 nm. For most images shown here, each shot was averaged over 23 cycles of the CSD, which corresponds to an acquisition time of 20 s for one CSD-ISM image. For all images recorded, the software waited for one trigger from the spin disk before starting the next cycle.
Image Calculation and Analysis.
The calculation of the CSD-ISM image from the raw data stack, as well as the Fourier reweighting of the resulting image, was performed using Matlab (The MathWorks Inc.). For the Fourier reweighting, optical transfer functions were calculated using the wave optical theory by Wolf (27) and Richards and Wolf (28). The positions of the pinholes of the CSD were determined using rapidSTORM 3.2 software (29) with maximum likelihood estimator (MLE) improvement of the fit. Details of the image calculation are described in ISM Algorithm and Image Calculation Using Matlab. An ImageJ plugin for calculation of the CSD-ISM image from raw data and reference data may be found on our Web site under www.joerg-enderlein.de/ISM along with sample datasets.
Nuclear pore density was measured with the ITCN ImageJ plugin, which finds the peak fluorescence intensity of each pore in a manually selected area on the nuclear envelope of seven samples for each method. For the spinning-disk samples, the parameters were set as follows: width of four pixels, minimum distance of five pixels, and a threshold of 1.0. With a twofold resolution enhancement, the parameters for the ISM samples were adjusted as follows: width of eight pixels, minimum distance of six pixels, and a threshold of 0.5. Errors for peak detection were tested manually (spin disk, maximum 4%; ISM, maximum 9%). In two of seven samples, the NPC density was measured from images in which NPCs were labeled with Alexa 568. An example of the NPC density measurements may be found in Measurement of Nuclear Pore Complex Density (Fig. S6).
Sample Preparation.
Atto 655 single-molecule samples.
For recording images of single fluorescent molecules on a glass surface, cover slides were cleaned by ultrasonication in acetone, ethanol, and ultrapure water. Single molecules of Atto 655 on glass were prepared by drying a very dilute solution onto a cleaned glass cover slide. The concentration of the dye was adjusted to result in a surface density of molecules in which individual molecules could be distinguished clearly. The samples were imaged without any further treatment.
NPC samples.
HeLa s/a cells (14) were maintained in DMEM (GlutaMAX; Invitrogen) supplemented with 10% (vol/vol) FCS (Gibco) under 5% CO2 at 37 °C. For staining, cells were seeded onto 12-mm poly-l-lysine–coated (Sigma) coverslips and incubated until they reached 50% cell density. The sample was washed with PBS, fixed with 3.7% (vol/vol) formaldehyde in PBS, and permeabilized with 0.5% Triton X-100 in PBS for 5 min. After washing, the sample was blocked with 10% (wt/vol) BSA in PBS for 30 min and incubated with mAb414 (Sigma; 1.5 µg/mL) diluted in PBS with 1% (wt/vol) BSA for 2 h. The sample was rinsed with PBS three times and incubated with the secondary antibody (goat anti-mouse Alexa 488, 4 µg/mL; Molecular Probes) diluted in PBS with 1% (wt/vol) BSA for 2 h. After the final washing step, the sample was mounted in Aqua-Poly/Mount (Polyscience) and sealed with nail polish.
Cytoskeletal and focal adhesion samples.
REF52 cells were cultured in DMEM supplemented with GlutaMAX and 10% (vol/vol) FCS in a humidified atmosphere of 5% CO2. For microscopy, cells were plated on fibronectin-coated (10 μg/mL) coverslips and grown to a confluency of 40–50%. Cells were fixed with 4% (vol/vol) formaldehyde, quenched with 100 mM glycine in PBS, and permeabilized with 0.1% Triton X-100 in PBS. Blocking, washing, and antibody incubations were performed using 0.2% gelatin in PBS. Tubulin and zyxin proteins were detected using the monoclonal anti–α-tubulin (Clone DM1A; Sigma) antibody at a dilution of 1:250 and the polyclonal antizyxin antibody (Sigma) at a dilution of 1:400, respectively. TRITC-conjugated goat anti-mouse and Alexa Fluor 647-conjugated goat anti-rabbit secondary antibodies (both minimized for cross-reactivity; Jackson ImmunoResearch Laboratories) were used for visualization. The actin cytoskeleton was stained using phalloidin–Alexa 488 (Invitrogen) in a dilution of 1:200. Cells were mounted in Mowiol. The images of actin filaments shown in Figs. 1 and 2 were prepared in the same fashion.
Tau protein samples.
N1E115 neuroblastoma cells were cultured in DMEM supplemented with 10% (vol/vol) FCS in a humidified atmosphere of 5% CO2. Cells were plated on coverslips, grown to a confluency of 60–70%, and transfected with GFP-3PO-Tau using Lipofectamine LTX (Invitrogen) according to the manufacturer’s instructions. After 24 h of expression, cells were fixed in 4% (vol/vol) formaldehyde, quenched with 100 mM glycine in PBS, and washed with PBS before mounting in Mowiol.
Four-color samples.
HeLa P4 cells (31) were grown in DMEM (1X; Gibco) supplemented with 10% (vol/vol) calf serum (Gibco), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine (Gibco) under 5% CO2 at 37 °C. For staining, cells were seeded onto poly-l-lysine–coated (Sigma) 12-mm coverslips and incubated until they reached 50% confluency. Cells were incubated with 20 ng/mL MitoTracker (M-7510; Invitrogen) for 15 min, washed with PBS, fixed with 3.7% (vol/vol) formaldehyde in PBS, and permeabilized with 0.5% Triton X-100 in PBS for 5 min. After washing, the samples were blocked with 2% (wt/vol) BSA in PBS for 30 min and incubated with rabbit anti–α-tubulin (111224-AP; Proteintech) and goat anti-Nup358 (25, 26) diluted in PBS with 2% (wt/vol) BSA for 1 h. The samples were rinsed three times with PBS and incubated with secondary antibodies (donkey anti-rabbit Alexa 488 and donkey anti-goat Alexa 633, 4 µg/mL; Molecular Probes) diluted in PBS with 1% (wt/vol) BSA for 1.5 h. DNA was stained with Hoechst 33258 (Molecular Probes). After the final washing step, samples were mounted in mounting medium (Dako).
Supplementary Material
Acknowledgments
We thank Dr. Alexey Chizhik for his kind help with the schematic drawing of the setup and ISM principle. We also thank Katrina Don Paul for technical assistance with sample preparations. This work was supported by the Deutsche Forschungsgemeinschaft Cluster of Excellence “Center for Nanoscale Microscopy and Molecular Physiology of the Brain.” M.C. thanks the Dorothea Schlözer Programme of the Georg-August-University Göttingen for support.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315858110/-/DCSupplemental.
References
- 1.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19(11):780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 2.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 3.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3(10):793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller CB, Enderlein J. Image scanning microscopy. Phys Rev Lett. 2010;104(19):198101. doi: 10.1103/PhysRevLett.104.198101. [DOI] [PubMed] [Google Scholar]
- 5.Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198(Pt 2):82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 6.York AG, et al. Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nat Methods. 2012;9(7):749–754. doi: 10.1038/nmeth.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gustafsson MGL, Agard DA, Sedat JW. I5M: 3D widefield light microscopy with better than 100 nm axial resolution. J Microsc. 1999;195(Pt 1):10–16. doi: 10.1046/j.1365-2818.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- 8.Sheppard CJR. Super-resolution in confocal imaging. Optik (Stuttg) 1988;80:53–54. [Google Scholar]
- 9.Van der Voort HTM, Brakenhoff GJ. 3-D image formation in high-aperture fluorescence confocal microscopy: A numerical analysis. J Microsc. 1990;158:43–54. [Google Scholar]
- 10.Schulz O, et al. Tip induced fluorescence quenching for nanometer optical and topographical resolution. Optical Nanoscopy. 2013;2:1. [Google Scholar]
- 11.Lucy L. An iterative technique for the rectification of observed distributions. Astron J. 1974;79:745–754. [Google Scholar]
- 12.Richardson WH. Bayesian-based iterative method of image restoration. J Opt Soc Am. 1972;62(1):55–59. [Google Scholar]
- 13.Sarder P, Nehorai A. Deconvolution methods for 3-D fluorescence microscopy images. IEEE Signal Process Mag. 2006;23:32–45. [Google Scholar]
- 14.Weidenfeld I, et al. Inducible expression of coding and inhibitory RNAs from retargetable genomic loci. Nucleic Acids Res. 2009;37(7):e50. doi: 10.1093/nar/gkp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Annu Rev Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- 16.Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: An evolving relationship. Cell Mol Life Sci. 2010;67(13):2215–2230. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hetzer MW. The role of the nuclear pore complex in aging of post-mitotic cells. Aging (Albany, NY Online) 2010;2(2):74–75. doi: 10.18632/aging.100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20(6):998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 19.Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449(7162):611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 20.Löschberger A, et al. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J Cell Sci. 2012;125(Pt 3):570–575. doi: 10.1242/jcs.098822. [DOI] [PubMed] [Google Scholar]
- 21.Maeshima K, et al. Cell-cycle-dependent dynamics of nuclear pores: pore-free islands and lamins. J Cell Sci. 2006;119(Pt 21):4442–4451. doi: 10.1242/jcs.03207. [DOI] [PubMed] [Google Scholar]
- 22.Arganda-Carreras I, et al. Consistent and elastic registration of histological sections using vector-spline regularization. In: Beichel R, Sonka M, editors. Computer Vision Approaches to Medical Image Analysis. Berlin: Springer; 2006. pp. 85–95. [Google Scholar]
- 23.Dassie E, et al. Focal expression of adeno-associated viral-mutant tau induces widespread impairment in an APP mouse model. Neurobiol Aging. 2013;34(5):1355–1368. doi: 10.1016/j.neurobiolaging.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Iliev AI, Ganesan S, Bunt G, Wouters FS. Removal of pattern-breaking sequences in microtubule binding repeats produces instantaneous tau aggregation and toxicity. J Biol Chem. 2006;281(48):37195–37204. doi: 10.1074/jbc.M604863200. [DOI] [PubMed] [Google Scholar]
- 25.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108(1):109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 26.Hutten S, Flotho A, Melchior F, Kehlenbach RH. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol Biol Cell. 2008;19(5):2300–2310. doi: 10.1091/mbc.E07-12-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf E. Electromagnetic diffraction in optical systems. I. An integral representation of the image field. Proc R Soc Lond A. 1959;253(1274):349–357. [Google Scholar]
- 28.Richards B, Wolf E. Electromagnetic diffraction in optical systems. II. Structure of the image field in an aplanatic system. Proc R Soc Lond A. 1959;253(1274):358–379. [Google Scholar]
- 29.Wolter S, et al. Real-time computation of subdiffraction-resolution fluorescence images. J Microsc. 2010;237(1):12–22. doi: 10.1111/j.1365-2818.2009.03287.x. [DOI] [PubMed] [Google Scholar]
- 30.Charneau P, et al. HIV-1 reverse transcription. A termination step at the center of the genome. J Mol Biol. 1994;241(5):651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.