Significance
Chemotactic cells integrate physical and chemical environmental inputs to determine a direction of migration. Here we report a physical input, the influence of hydraulic pressure, on cell migration. By using bifurcated microfluidic channels with different path lengths to the outlet, we show that confined cells are able to identify the shorter path in the absence of any chemical cues. This decision is made via the hydraulic resistance of the channel. The distinct channel resistances generate a physical asymmetry that determines the directional choice. Remarkably, this physical input can override a locally generated chemotactic signal. Our results define a physical input into cellular directional choice that may be important in navigating interstitial tissues in vivo.
Keywords: cell migration, microfluidics, physical forces, cell confinement, photocaged chemoattractant
Abstract
Cells integrate multiple measurement modalities to navigate their environment. Soluble and substrate-bound chemical gradients and physical cues have all been shown to influence cell orientation and migration. Here we investigate the role of asymmetric hydraulic pressure in directional sensing. Cells confined in microchannels identified and chose a path of lower hydraulic resistance in the absence of chemical cues. In a bifurcating channel with asymmetric hydraulic resistances, this choice was preceded by the elaboration of two leading edges with a faster extension rate along the lower resistance channel. Retraction of the “losing” edge appeared to precipitate a final choice of direction. The pressure differences altering leading edge protrusion rates were small, suggesting weak force generation by leading edges. The response to the physical asymmetry was able to override a dynamically generated chemical cue. Motile cells may use this bias as a result of hydraulic resistance, or “barotaxis,” in concert with chemotaxis to navigate complex environments.
Cell migration and polarization are influenced by a complex interplay of chemical and physical cues that act locally. Chemotactic cells, such as neutrophils and Dictyostelium, integrate chemical cues over the cell surface to move up an attractant gradient (1–5). Stiffness gradients, sensed by adhesion receptors, can also orient migration (durotaxis) (6–9). Cells must balance these and other environmental inputs to determine a direction of polarization and migration (10). Given the complex environment in tissues, it is likely that there are still unknown inputs to directional decision-making.
Microfluidics provides a programmable environment for single cells in which chemical and physical cues can be precisely controlled in space and time, and polarization and migration quantified (11). In particular, cellular confinement has been instrumental in observing many novel cellular behaviors such as integrin-independent motility (12) and EGF gradient sensing (13). Additionally, cellular confinement into narrow microchannels (<10 µm) is thought to mimic the tissue environment better than stiff 2D substrates covered with low-viscosity medium.
Here we use confinement to investigate the role of cell-generated hydraulic pressure in directional decisions. We show that, under strong confinement, migrating neutrophil-like cells push the column of water ahead of the cell, generating a hydraulic pressure. Cells selectively move toward a path of lower hydraulic pressure when presented with multiple paths. The morphological and signaling dynamics of this decision-making process revealed potential mechanisms that diverge from the classical PI3K-mediated chemotactic pathway. Remarkably, this physical input can outcompete conventional chemotactic signals, suggesting an important role in directing migration through tissues.
Results
To challenge single cells with complex hydraulic environments, we used microfluidic devices that harbor bifurcating, fibronectin-coated channels, derived from designs described by Ambravaneswaran et al. (14). These channels are 6 µm or smaller in width and 3 µm in height, forcing differentiated HL-60 cells [a standard neutrophil model (15)] to occupy the entire cross-section. This generates a tight seal with respect to fluid flow across the cell length. Cells enter the channels from a lower cell reservoir by random migration or by gentle pressure imposed during loading.
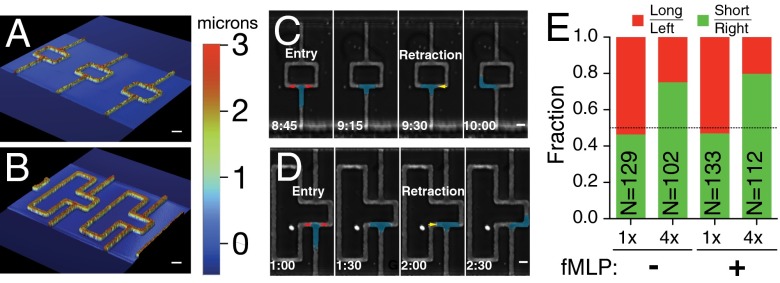
To test whether physical properties of the environment can influence cellular directional decision-making, we manufactured bifurcating microfluidic channels with a symmetric bifurcation (symmetric lengths; Fig. 1A) or an asymmetric bifurcation (4× length asymmetry; Fig. 1B). We observed that, when cells reached the bifurcation, they exhibited two different responses. In the first, the cell extended a pseudopod along each direction of the bifurcation and subsequently retracted the pseudopod from one side while continuing to migrate along the other direction (Fig. 1 C and D and Movies S1 and S2). In the second, the cells, upon reaching the bifurcation, immediately began to migrate along one direction, without extending a pseudopod along the other direction (Fig. S1). Cells that extended two pseudopods showed symmetric decision-making (0.5:0.5, expressed as a fraction of total cells) in the symmetric channels but displayed a strong bias in the asymmetric bifurcations (4×) to the shorter side (0.75:0.25; P < 0.0001, Fisher exact test; Fig. 1E). This behavior was independent of the presence of a chemokine gradient across the channel generated by the introduction of 100 nM N-formyl-methionyl-leucyl-phenylalanine (fMLP) in the upper reservoir (no significant difference by Fisher exact test; Fig. 1E). Interestingly, cells that did not extend two pseudopods, the number of which varied greatly across the different geometries, exhibited no significant directional bias in any of the geometries (Fig. S1), suggesting that the extension of two pseudopods, and competition between them, is essential for the bifurcation geometry to bias directional cellular migration.
Fig. 1.
Confined cells can identify shorter paths independent of chemical cues. (A and B) Images from an optical profilometer showing 3D geometry of bifurcating microchannel features on a silicon master for 1× (A) and 4× (B) length ratios. (Scale bar, 10 µm.) The color bar gives the height of the features, as measured by the profilometer. (C and D) Time course montage of a cell migrating through a bifurcation in 1× (C) and 4× (D) length ratio geometries. Cells are false-colored for visibility. Frames are labeled to emphasize the processes of entering the bifurcation (red arrows) and retracting one of the two leading edges (yellow arrow). (Scale bar, 10 µm.) (E) Directional decision statistics for cells in 1× and 4× length ratio geometries and in the absence or presence of chemokine (fMLP). Green denotes cells that migrated toward the shorter length (or right) side, and red denotes cells that migrated toward the longer length (or left) side. N indicates number of cells measured.
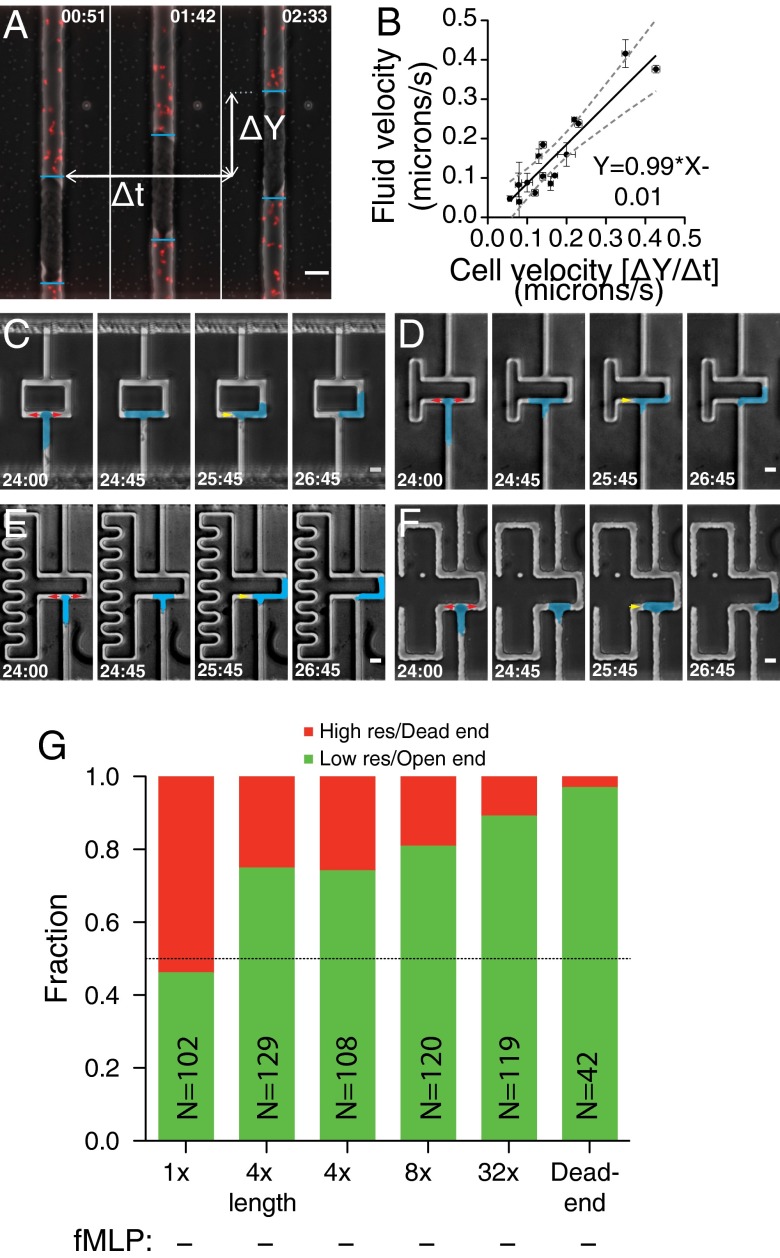
Given the tight seal formed by the cells in the microchannel (16), we speculated that the cells push water as they migrate in the channels. As a result, they would be able to detect the length difference in the asymmetric bifurcation through the different hydraulic loads presented to each extending pseudopod. To test if cells push water as they migrate in the microchannels, we loaded cells into straight microchannels with small fluorescent polystyrene beads in the medium (500 nm). We used a spatiotemporal image correlation spectroscopy (STICS) method (17) to measure the positional fluctuations of the beads (Fig. 2A and Fig. S2). From these measurements, we were able to extract the flow velocity of the beads, and therefore the bulk fluid. Fluid velocities in the channel measured this way were identical, within our measurement error, to the velocity of the cells themselves (Fig. 2B), indicating that these cells do indeed push water as they move.
Fig. 2.
Cells push water and can identify the path of least resistance. (A) Time course montage of a cell in a microchannel with fluorescent beads. ΔY shows the distance traveled by the cell, and Δt shows time taken to travel ΔY. Image was acquired with a 40× oil 1.0 NA immersion objective. Correlation data were acquired with a 10× air 0.25 NA objective. (Scale bar, 10 µm.) (B) Plot and fit of fluid velocity calculated by STICS (y axis) compared with measured cell velocity (x axis). Gray dashed lines show 95% CI of fit, shown by black line. A nonlinear regression gave a slope of 0.99 ± 0.26 and an intercept of 0.01 ± 0.05. Fit equation is shown. (C–F) Time course montages of cell migrating in channels with resistance ratios of 4× (C), 8× (D), 32× (E), and dead end (F). Cells are false-colored for visibility. (Scale bar, 10 µm.) Red arrows indicate the process of entering the microchannel, while the yellow arrow indicates the process of retracting the nonpersistent leading edge. (G) Directional decision statistics of cells in all geometries in the absence of chemokine.
To further test if cells responded to hydraulic resistance, and not simply channel length, we used the equation given by Fuerstman et al. (18) to fabricate devices of three different hydraulic resistance ratios. For the first (Fig. 2C and Movie S3), the lengths of the two bifurcations were equal, but the hydraulic resistance of the left side was fourfold higher, accomplished by reducing the width of the downstream channel far from the bifurcation. Approximately 75% of the cells migrated along the lower-resistance side (Fig. 2G), similar to the statistics from the 4× length ratio bifurcations. For the next two geometries, a combination of increased channel length and decreased channel width was used to achieve resistance ratios of eightfold and 32-fold (Fig. 2 D and E and Movies S4 and S5). Cells in these geometries exhibited an even stronger bias, with 80% and 90% of the cells migrating toward the lower hydraulic resistance in the 8× and 32× geometries, respectively (Fig. 2G). Finally, we fabricated microchannel devices in which one side of the bifurcation ends in a dead end (Fig. 2F and Movie S6). When loaded into these channels, we observed that cells still extended two pseudopods, but the pseudopod that extended along the dead-end direction did so for only a short distance, resulting in an asymmetrical extension biased toward the open-ended side of the bifurcation. Correspondingly, nearly all cells (41 of 42 total, ∼98%) migrated away from the dead end (Fig. 2G).
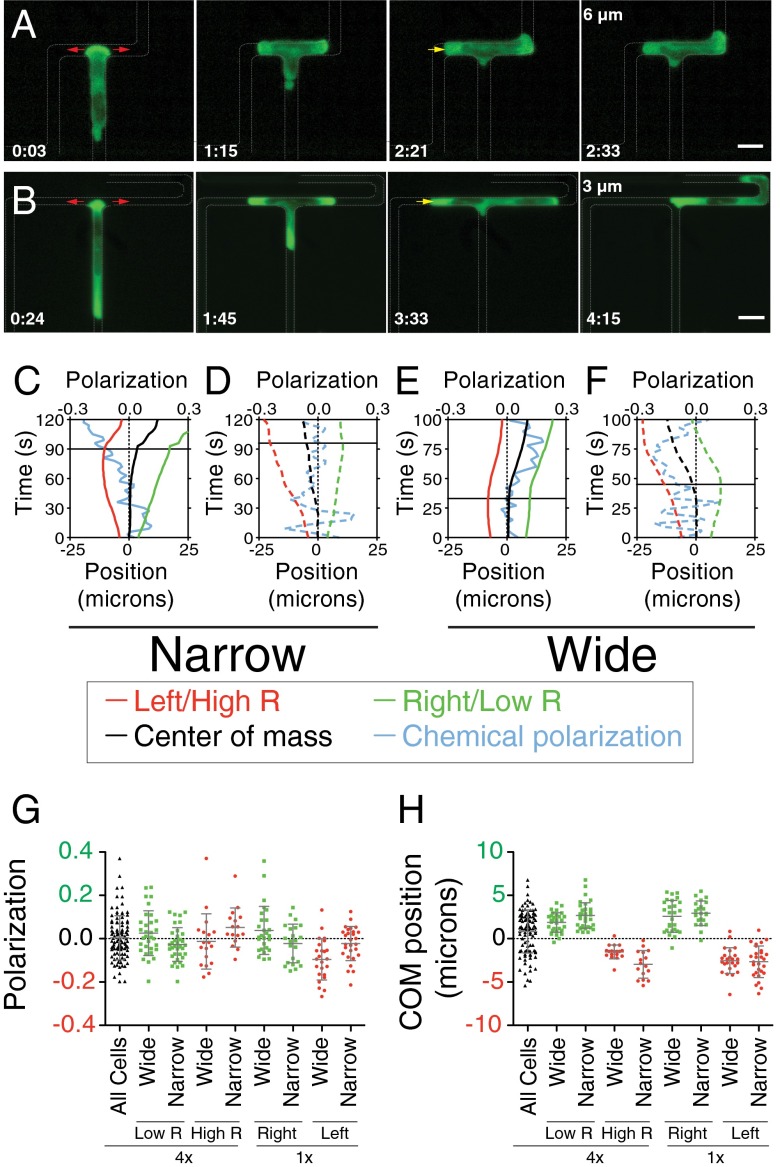
To investigate a potential molecular basis of the directional decision-making, we imaged HL-60 cells stably expressing 3xPH-Akt-GFP (PH-Akt), a marker of phosphoinositol 3-phospholipids and an indirect readout of PI3K activity. This marker is strongly enriched at the leading edge during chemotaxis in neutrophils and the slime mold Dictyostelium discoideum (19, 20). Bifurcating pseudopods exhibited PH-Akt–positive leading edges in microchannels of width 6 µm (wide) and 3 µm (narrow) (Fig. 3 A and B and Movies S7 and S8). In many cells, the leading edge that eventually retracted was enriched for PH-Akt right until the time of retraction, with no obvious decrease in brightness. We quantified the maximum intensity of leading-edge PH-Akt in a 3-μm window at each leading edge. When observed in space–time plots, the normalized chemical polarization showed no obvious bias over the decision-making time (Fig. 3 C–F, blue line). Moreover, we observed no correlation between the initial or final asymmetry in leading-edge chemical polarization and the ultimate direction chosen (Fig. S3B and Fig. 3G, respectively). Thus, the competition between the extended pseudopods did not appear to be mediated by differences in strength of canonical leading edge signaling.
Fig. 3.
Asymmetries in leading edge extension predict cellular directional decision. (A and B) Time course montage of a cell expressing PH-Akt migrating in 4× length ratio bifurcation geometry with widths of 6 µm (A) and 3 µm (B). (Scale bar, 10 µm.) The red arrows denote the process of entering the bifurcation, and the yellow arrow denotes the retraction of the nonpersistent leading edge. (C–F) Trajectories of the position of the low-resistance leading edge (green), high-resistance leading edge (red), center of mass (black), and PH-Akt polarization (cyan) vs. time. Plots are grouped according to whether the cell migrated in narrow channels (C and D) or wide channels (E and F). Dashed trajectories represent a cell choosing the high-resistance channel; solid trajectories represent a cell choosing the low-resistance channel. Horizontal lines show the retraction time for each cell. (G and H) Column scatter dot plots of the position of the PH-Akt polarization (G) and center of mass (COM) (H) at retraction time for 4× and 1× geometries. All plots show distribution for cells in narrow and wide geometries for the 4× resistance ratio in black. The distribution for cells that migrate toward the low-resistance/right side is shown in green, grouped by resistance ratio. The distribution for cells that migrated toward the high-resistance/left side is shown in red, grouped by resistance ratio.
We then turned to careful geometric analysis of the competing pseudopods. In asymmetric channels, pseudopods became asymmetrical in length over time, with the low-resistance branch longer, indicating that extension rates were faster on the low-resistance side (Fig. 3 A and B). When plotted in space–time plots, we observed that, in individual cells, the center of mass position, with respect to the center of the bifurcation, rapidly biased to the final directional choice (Fig. 3 C–F, black line). This occurred even in the low percentage of cells that chose the high-resistance channel. Thus, a physical asymmetry resulting from differential extension rates predicted the directional choice.
By normalizing initial pseudopod velocities, we found that a bias toward the low-resistance side was observed within the first five frames (15 s) after the cell entered the bifurcation (Fig. S3A). Comparing the initial extension bias to the final direction chosen for each individual cell, we found that the initial extension bias correlated well with the final decision of the cell (Fig. S3A). Notably, the cells in the wide channel showed a stronger bias in the correlation of initial extension velocity with final decision compared with the cells in the narrow channel. The bias in the center of mass, in the frame directly before retraction, exhibited an even stronger correlation with the direction chosen by the cell (Fig. 3H). In this case, the cells in narrow channels exhibited a modestly stronger correlation.
To verify that the observed correlation between extension asymmetry and direction chosen was not a result of the physical asymmetry of the bifurcating channels, we imaged HL-60 cells in wide and narrow symmetrical bifurcating channels. As shown in Fig. 3H, cells in symmetrical bifurcations exhibited a very similar correlation, with only a small number of cells migrating in a direction opposite that of the center of mass, and, in those cases, the center of mass was quite close to the bifurcation. Similarly, cells in symmetrical bifurcations exhibited no obvious correlation between polarization of PIP3 and the direction chosen (Fig. 3G).
Observations of the retraction timing indicated that long cells, in wide or narrow channels, were delayed in making the directional choice in comparison with shorter cells (Fig. S4A). Measurements of pseudopod length (Fig. S4B) and uropod velocity (Fig. S4C) indicated that long cells moved at similar speeds compared with short cells, but extended more pseudopod into the bifurcation before retracting one leading edge, and thus are delayed in making the directional choice. Together, these observations indicated that there may be a characteristic distance between the rear of the cell and the bifurcation to determine decision timing. Analysis of the fraction of cell length that remained in the entry channels (cell rear) showed a trend consistent with this hypothesis (SI Text and Fig. S4D).
Having observed no apparent correlation between PIP3 chemotactic signaling and directional choice in asymmetric channels, we investigated competition between physical and chemical inputs. A “caged,” or photoactivatable, version of fMLP (cfMLP) was synthesized (Fig. S5A) by appending a nitrobenzyl moiety to the formyl group. This derivative appeared to lack any chemotactic activity. When exposed to UV light, the bond to the nitrobenzyl is cleaved, liberating active fMLP. We tested cfMLP by uncaging it in a dead-end channel that cells rarely enter, and observed the effect on cells expressing PH-Akt. Measurements of fluorescence intensity made at each leading edge immediately before as well as 18 s following uncaging showed a dramatic increase in chemical polarization toward the dead end (Fig. S5B), indicating that photoliberated fMLP activates canonical signaling pathways.
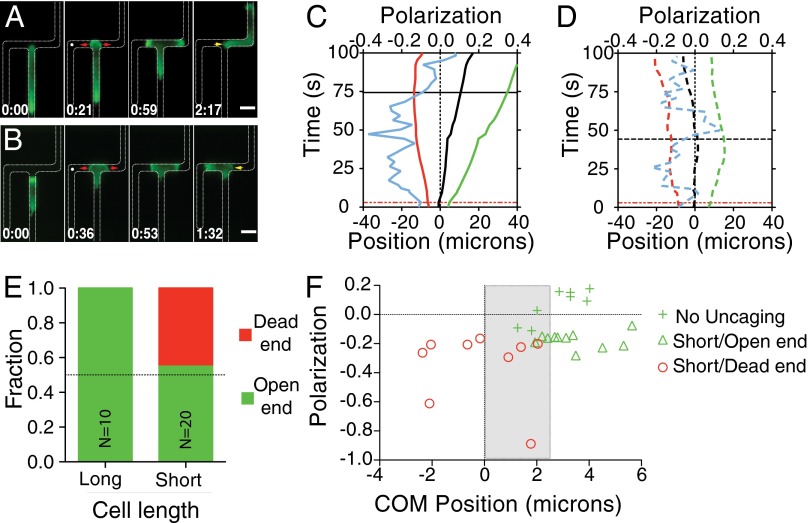
To probe the competition between chemical and hydraulic stimuli, cfMLP was uncaged in the dead-end side of a dead-end bifurcation soon after cells started to extend two pseudopods into the channels. We found that many cells (70%) migrated down the open channel despite exhibiting strong PIP3 polarization toward the dead-end side (Fig. 4A and Movie S9). Thus, the chemical signal polarized the signaling machinery toward the high chemoattractant side, but the response to hydraulic resistance overrode this signal to direct cell migration. Dynamics of the response of the cells that entered the open-end channel were similar to those in the absence of uncaging, with the notable exceptions of chemical polarization toward the dead end after uncaging (Fig. 4 C and D) and further extension of the pseudopod down the dead end following uncaging (Fig. S5D). A subset of cells (30%) did enter the dead-end channel when cfMLP was uncaged (Fig. 4B and Movie S10). These cells retracted the leading edge that had extended toward the open end and migrated a short distance into the dead end, at which point they ceased to continue migrating, but did not retreat from the dead end. This effect was dependent on the presence of cfMLP and laser activation, as neither one alone caused cells to enter the dead-end channels (Fig. S5C). When the directional choice statistics were analyzed, we found that the cells that entered the dead-end channel were all short (<40 µm; Fig. 4E). Interestingly, the cells that entered the dead-end channel exhibited center-of-mass biases both into and away from the final directional choice, implicating a competition between chemical and physical inputs at small physical asymmetries (Fig. 4F).
Fig. 4.
Physical asymmetry can override chemical activation. (A and B) Time course montages of cells in the presence of caged fMLP exposed to laser excitation. Location of uncaging (white dot) and uncaging time are shown in the second image. Cells migrated toward the open end (A) or toward the dead end (B), albeit with different statistics. (Scale bar, 10 µm.) The red arrows denote the process of entering the bifurcation, and the yellow arrow denotes the retraction of the nonpersistent leading edge. (C and D) Leading edge trajectories as shown previously for cells exposed to uncaging of caged fMLP and migrating away from the dead end (C) or toward the dead end (D). Note the increase in polarization magnitude after cfMLP uncaging. Red line represents the uncaging time, while the black line represents the retraction time. (E) Directional decision statistics for cells that were exposed to laser excitation in the presence of caged fMLP. Cells were grouped according to whether they were longer (“long”) or shorter (“short”) than 40 µm. (F) Scatter plot of the maximum polarization (negative is toward the dead end) following uncaging and the center of mass (COM) bias directly before retraction. Shown are all short cells exposed to uncaging and cells in dead-end bifurcations not exposed to uncaging. Gray box marks the region in which cells with a physical bias toward the open end migrate toward the dead end.
Discussion
Here we have investigated the role of hydraulic pressure generated by cell migration as a physical input to cellular directionality. The influence of this input was revealed by observing confined, chemotactic cells presented with asymmetric hydraulic environments and demonstrating that they were able to determine the path of lesser hydraulic resistance, a phenomenon consistent with the term “barotaxis.” Notably, this choice did not require chemical cues, unlike previous work using similar bifurcated channels (21). The decision-making dynamics involved two phases: (i) generation of an initial physical asymmetry manifest as differential extension of two leading edges and (ii) retraction of the leading edge closest to the bifurcation. Together, these activities resulted in biased migration toward the lower-resistance channel. Timing of decision-making showed a correlation between the time to make a decision and cell length (SI Text). By using a caged form of a chemoattractant, we demonstrated that hydraulic and chemical inputs compete, with hydraulic overriding chemical under some circumstances. This competition occurred downstream of polarized synthesis of PIP3, a canonical chemotactic polarization marker.
From our measurements of the leading edge extension and the fluid flow in a microchannel, we propose that, as the leading edge extends, it experiences an external load, namely the column of fluid ahead of it. By using the equation provided by Fuerstman et al. (18) [channel dimensions 6 µm wide and 3 µm high, viscosity of water 1 mPa⋅s, channel length 100 µm, and cell (fluid) velocity of 0.3 μm/s], we found that the net force to move this column of water was on the order of 1 pN. Usami et al., observing confined neutrophils in micropipettes (22), and others in unconfined settings (23–25), have shown that forces on the order of tens of nanonewtons are required to stall cell motion.
To understand this potential paradox of how the small forces associated with water column movement might bias migration, we note that the forces exerted by the neutrophil are balanced by the sum of the forces that result from adhesion, hydrodynamic friction in the thin layer, and the external pressure. Although the leading edge is relatively labile and free of forces, the rest of the cell is not. Indeed, forces are likely nonuniform along the neutrophil, which is free at either end; in fact, scaling considerations suggest that there is a characteristic dynamic length scale associated with the balance between contraction and friction that screens the front end of the cell from the back in the straight channel. Thus, for a sufficiently long cell in a straight channel, the front and back could become mechanically decoupled. When the cell has bifurcated into the T-junction, depending on the lengths of the three segments, one can envisage different sensitivities to the pressure differences along each segment. The longer the cell, the more sensitive it would be to small pressure differences.
How this small pressure difference might be detected is not known, but a potential candidate might be membrane tension sensors that respond to the stretch induced in the membrane by the pressure difference across it. Future work that uses a range of bifurcation geometries and pressure differences should help us clarify the mechanism, sensitivity, and robustness of the cellular response to asymmetrical hydraulic resistance.
By using the dead-end channel geometry and the caged fMLP compound, we were able to activate the chemotactic machinery in opposition to the external physical input. The chemotactic signal prevailed only at small physical asymmetries seen in short cells, whereas long cells (>40 µm) were refractory to concomitant chemical polarization. This is consistent with previous work showing that neutrophils can enter high-resistance channels under strong chemotactic activation (21). How the activation of the chemotactic pathway was overridden by the input of the hydraulic resistance is unknown. One potential mechanism is that the chemotactic signaling network is able to enhance the activity of the physical motility. Consistent with this idea, it was observed that uncaging increased pseudopod entry into the dead-end channels. However, in a small subset of cells that entered the dead-end side, the center of mass was biased toward the open channel, indicating that the chemical polarization induced by chemoattractant uncaging can act to override a small physical bias. It is appealing to think that, in complex environments, such as the interstitial tissue, cells use chemotactic and barotactic cues to find their target and reduce the likelihood of local entrapment. Further studies of the cellular response to physical inputs more broadly and their integration with chemotactic pathways may provide some clues of how cells balance these many environmental inputs.
Materials and Methods
Cell Culture and Stable Line Production.
HL-60 cultures were maintained in RPMI 1640 media (no. 11875119; Invitrogen) supplemented with 20% (vol/vol) FBS (no. 26104; Invitrogen), 1% Penstrep (30-002-CI; Cellgro), and 25 mM sterile filtered HEPES (BP310; Fisher Scientific), and were passaged every 2 to 3 d. Cells were differentiated in complete media supplemented with DMSO to a final concentration of 1.3% (vol/vol). Cells which were differentiated for at least 5 d and not more than 7 d were used for experiments.
Cell lines stably expressing PH-Akt were generated via retroviral infection, as described previously (10). Briefly, Plat-GP cells (Cell Biolabs) were used to produce retrovirus by cotransfection with a pBABE packaging vector (pBABEblast, pJag98) (26) containing the cDNA for PH-Akt (cDNA obtained from Lew Cantley, Harvard Medical School, Boston, MA) and VSV-G, using Fugene 6 (Roche). The viral supernatant was collected after 48 to 72 h and filtered by using a 0.45-μm syringe filter. Polybrene was added to the supernatant at a final concentration of 16 µg/mL. Cells were suspended in one well of a six-well plate at a concentration of 106/mL in a volume of 2 mL. To this suspension was added 5 mL of viral supernatant, and the cells were spun at 1,000 × g for 90 min in a swinging bucket rotor. Cells were then incubated for 5 h at 37 C. Following this incubation, cells were pelleted and resuspended into 2 mL fresh media. The next day, 2 mL of fresh media was added to the cells. The following day, cells were placed under selection in 1 µg/mL Blasticidin. When a stable pool of Blasticidin-resistant cells had been obtained, the cells were then sorted for GFP expression (Systems Biology Flow Cytometry Facility, Harvard Medical School).
Microfluidic Device Fabrication.
Microfluidic devices were generated as described previously (14). High-resolution film (Fineline Imaging) or chrome masks (Advanced Reproductions or Front-Range Photomasks) were used to generate photoresist masters. A negative photo resist, SU-8 (Microchem), was spun onto silicon wafers at a rate according to the feature height desired. The photoresist was then exposed to UV light by using a mask aligner (Neutronix Quintel). Unexposed SU-8 was developed away. (Poly) dimethyl-siloxane (PDMS) was mixed at a ratio 10:1 (elastomer:curing agent) and then spun (for membranes) or poured onto the wafers, and cured overnight at 70 C. The top layer of the devices was then cut out, access holes were punched for pneumatic valve control by using a 0.75-mm-diameter punch, and the devices were then bonded to the spun membrane by using oxygen plasma. These devices were again cut out and additional fluid access holes were punched, and the devices were then bonded to 1 × 3 in glass slides (Fisher Scientific) or 45 × 50 mm no. 1.5 coverslips (Fisher Scientific).
Microfluidic Device Operation.
Tygon tubing (part no. 2007900S-54HL; Greene Rubber Company) and 30-gauge blunt syringe needles (BN2505; Brico Medical Supplies) were used to connect syringes to the microfluidic devices. The outer diameter of the Tygon tubing (0.03 in) is slightly larger than the diameter of the hole punched, such that, when the tubing was inserted into the hole, a seal was created. Each of the pneumatic valve control chambers was connected to a 1-mL syringe. To close the valves, the syringe was compressed from a volume of 0.2 mL to ∼0.05 mL. To open the valves, the syringes were pulled open from a volume of 0.2 mL to the maximum volume of the syringe.
Fibronectin was adsorbed onto the channel walls by filling the device with a solution containing 50 µg/mL fibronectin (F0895; Sigma-Aldrich) in 50 mM Tris⋅HCl. Any air bubbles present were removed by pressurizing the device. The solution was incubated in the device for 1 h and then rinsed out with HBSS supplemented with 25 mM HEPES and 0.2% human serum albumin. The same buffer was used during all phases of the device operation.
Each inlet was connected to a syringe, with the plunger removed. One syringe contained a solution of HBSS, whereas the other contained a solution of HBSS with fMLP. Fluorescein was added to the syringe containing fMLP to visualize the interface of the two inlet streams for balancing the stream pressures. For experiments in which no chemokine was introduced, the inlet tubing was clamped off to eliminate any flow through the device. To reduce the introduction of air bubbles while loading cells, a drop of HBSS was placed onto the cell inlet hole before removing the priming tubing.
To load cells into the device, 2 to 3 mL of differentiated cells were pelleted, and 30 μL of the cell pellet was taken and placed into a blunt needle with a short section of tubing. The cells were then pushed through the tubing by applying pressure to the top of the needle with a finger, until a small drop appeared at the end of the tubing to provide a liquid–liquid interface while inserting the tubing to prevent the introduction of air bubbles. The cells were then pushed into the device by applying pressure with a finger and the on-chip valves were manipulated to ensure that the cells were loaded on only one side of the microchannels, and were forced into the entrance of the microchannel.
STICS Image Acquisition and Analysis.
Devices were prepared and cells were loaded as described earlier, with the addition of 500-nm-diameter red fluorescent polystyrene beads (F8812; Invitrogen). Images were acquired with a 10× Ph1 ADL objective at a frame rate of one frame every 1.1 s in RFP and bright-field channels. Each microchannel was then extracted from the image and averaged across the width of the microchannel, to generate a 1D array at each time point. The resulting 2D array (one spatial and one temporal) was analyzed, and a spatiotemporal correlation function was calculated by using a modified version of the STICS algorithm (26). A Gaussian curve was then fit to the spatial correlation function for each time-lag. The center of each Gaussian fit was extracted, and a line was fit to the center position vs. time lag data to determine the fluid velocity. The range of time lags for fitting the line was selected so that the diffusion constant output by the algorithm was no larger than 1.1 μm2/s (near the expected diffusion constant of the beads). To estimate the measurement error in the fluid velocity, the inverse of the variance for each Gaussian fit was used in a weighted nonlinear least-squares fit of the Gaussian position vs. time lag. The velocity of the cell was determined by measuring the distance the front of the cell had traveled over the time period that was analyzed and dividing by the duration of the time period. Only cells that persistently migrated were analyzed. All analysis was performed using Matlab (MathWorks). Images were analyzed by using only a portion of the channel in which the cell is not present (Fig. 2A), as well as using the entire region for certain channels (Fig. S2F). Both methods gave similar results.
Polystyrene Bead Functionalization.
The polystyrene beads were covalently modified with human serum albumin (HSA) by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)-mediated crosslinking. Briefly, 500 μL of the beads were incubated for 15 min with 500 μL of HSA in MES buffer at 5mg/mL. Next, 200 μL of EDC in water at 10 mg/mL was added to the solution, covered with aluminum foil, and incubated for 2 h at room temperature. Following this incubation, 300 μL of glycine in MES buffer at 500 mM was added to the solution and incubated for 30 min at room temperature to quench the reaction. The suspension was pelleted and washed with water three times, and finally resuspended in 500 μL of HBSS supplemented with 25 mM HEPES and 0.2% HSA.
Caged fMLP Assay.
For the cfMLP (synthesized by New England Peptide) experiments, cells were loaded into the device as described earlier, with the exception that, before loading the cells, the device was flushed with an HBSS solution containing 100 nM cfMLP. In addition, cfMLP was added to the needle with the cells to a final concentration of 100 nM. To uncage the cfMLP, a two-photon IR laser (Mai Tai; Spectra Physics) was used at a wavelength of 780 nm and a power of 100 mW. The laser excitation was performed for a duration of 3 s at various times after the cell had blocked off the bifurcation to ensure stimulation of only one side of the cell. All uncaging experiments were performed on a Nikon Ti (Nikon) microscope with a 60× Ph3 Plan Apo (Nikon) objective. The microscope was controlled with the Elements software package (Nikon).
Image Acquisition and Analysis.
Cells were imaged on a TE2000 microscope (Nikon) with a 20× Ph1 ADL (Nikon) or 60× Ph3 Plan Apo (Nikon) objective. The microscope was controlled with the Volocity software package (Perkin-Elmer). For images taken with the 20× objective, 10 x-y positions (XY Controlled Stage; LUDL BioPrecision) were taken at a frame rate of one frame every 15 s, acquiring only bright-field images. For images taken with the 60× objective, images were acquired at a frame rate of one frame every 3 s in the GFP and bright-field channels.
For each cell, the central 30% of the channel was extracted, and the image was averaged over the width of the channel for each time point, generating a 1D intensity profile. The edge of the cell was then determined, at each time point, by applying a nonlinear filter (27) and thresholding based on maximum and minimum intensities of this intensity profile. All images were analyzed with in-house Matlab (MathWorks) algorithms.
Fluorescence measurements of Ph-Akt were taken by measuring the integrated intensity of the first 3 µm of the extended pseudopod and then normalized by subtracting the value for the high-resistance side from the low-resistance side and then dividing by the sum of the values. The result is a value that can vary from −1, polarized to the high-resistance side, to +1, polarized to the low-resistance side. Pseudopod velocities were also normalized by using the same approach.
Supplementary Material
Acknowledgments
We thank members of the laboratory of J.V.S., especially Yinghua Guan and Mikhail Sergeev; Guillaume Charras (University College London), Tony Yu-Chen Tsai (Stanford University), and Gaudenz Danuser (Harvard Medical School) for helpful discussions and constructive comments; Lew Cantley (Harvard Medical School), Guillaume Charras (University College London), Paul Wiseman (McGill University), and Orion Wiener (University of California, San Francisco) for generously sharing reagents and software. All microfabrication was carried out in the National Institutes of Health-funded facility (Grant P41 EB002503) at the Charlestown Navy Yard at Massachusetts General Hospital. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (C.-H.C.). Work in the laboratory of J.V.S. is supported by a grant from the Beckman Laser Institute Foundation and funds from Brigham and Women’s Hospital.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317441110/-/DCSupplemental.
References
- 1.Swaney KF, Huang C-H, Devreotes PN. Eukaryotic chemotaxis: A network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Insall RH. Understanding eukaryotic chemotaxis: A pseudopod-centred view. Nat Rev Mol Cell Biol. 2010;11(6):453–458. doi: 10.1038/nrm2905. [DOI] [PubMed] [Google Scholar]
- 3.Parent CA. Making all the right moves: Chemotaxis in neutrophils and Dictyostelium. Curr Opin Cell Biol. 2004;16(1):4–13. doi: 10.1016/j.ceb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Fuller D, et al. External and internal constraints on eukaryotic chemotaxis. Proc Natl Acad Sci USA. 2010;107(21):9656–9659. doi: 10.1073/pnas.0911178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kay RR, Langridge P, Traynor D, Hoeller O. Changing directions in the study of chemotaxis. Nat Rev Mol Cell Biol. 2008;9(6):455–463. doi: 10.1038/nrm2419. [DOI] [PubMed] [Google Scholar]
- 6.Bruinsma R. Theory of force regulation by nascent adhesion sites. Biophys J. 2005;89(1):87–94. doi: 10.1529/biophysj.104.048280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CS. Mechanotransduction - a field pulling together? J Cell Sci. 2008;121(Pt 20):3285–3292. doi: 10.1242/jcs.023507. [DOI] [PubMed] [Google Scholar]
- 8.Nicolas A, Geiger B, Safran SA. Cell mechanosensitivity controls the anisotropy of focal adhesions. Proc Natl Acad Sci USA. 2004;101(34):12520–12525. doi: 10.1073/pnas.0403539101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harland B, Walcott S, Sun SX. Adhesion dynamics and durotaxis in migrating cells. Phys Biol. 2011;8(1):015011. doi: 10.1088/1478-3975/8/1/015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah JV, et al. Dynamics of centromere and kinetochore proteins; Implications for checkpoint signaling and silencing. Curr Biol. 2004;14(11):942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 11.Lautenschläger F, Piel M. Microfabricated devices for cell biology: All for one and one for all. Curr Opin Cell Biol. 2013;25(1):116–124. doi: 10.1016/j.ceb.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Lämmermann T, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453(7191):51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 13.Scherber C, et al. Epithelial cell guidance by self-generated EGF gradients. Integr Biol (Camb) 2012;4(3):259–269. doi: 10.1039/c2ib00106c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambravaneswaran V, Wong IY, Aranyosi AJ, Toner M, Irimia D. Directional decisions during neutrophil chemotaxis inside bifurcating channels. Integr Biol (Camb) 2010;2(11-12):639–647. doi: 10.1039/c0ib00011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breitman TR, Selonick SE, Collins SJ. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc Natl Acad Sci USA. 1980;77(5):2936–2940. doi: 10.1073/pnas.77.5.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irimia D, Charras G, Agrawal N, Mitchison T, Toner M. Polar stimulation and constrained cell migration in microfluidic channels. Lab Chip. 2007;7(12):1783–1790. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hebert B, Costantino S, Wiseman PW. Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys J. 2005;88(5):3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuerstman MJ, et al. The pressure drop along rectangular microchannels containing bubbles. Lab Chip. 2007;7(11):1479–1489. doi: 10.1039/b706549c. [DOI] [PubMed] [Google Scholar]
- 19.Wang F, et al. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat Cell Biol. 2002;4(7):513–518. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Charest PG, Firtel RA. Spatiotemporal regulation of Ras activity provides directional sensing. Curr Biol. 2008;18(20):1587–1593. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler KL, et al. Burn injury reduces neutrophil directional migration speed in microfluidic devices. PLoS ONE. 2010;5(7):e11921. doi: 10.1371/journal.pone.0011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usami S, Wung SL, Skierczynski BA, Skalak R, Chien S. Locomotion forces generated by a polymorphonuclear leukocyte. Biophys J. 1992;63(6):1663–1666. doi: 10.1016/S0006-3495(92)81745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann J, et al. Actin filament elasticity and retrograde flow shape the force-velocity relation of motile cells. Biophys J. 2012;102(2):287–295. doi: 10.1016/j.bpj.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobelli J, et al. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA-regulated adhesions. Nat Immunol. 2010;11(10):953–961. doi: 10.1038/ni.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renkawitz J, et al. Adaptive force transmission in amoeboid cell migration. Nat Cell Biol. 2009;11(12):1438–1443. doi: 10.1038/ncb1992. [DOI] [PubMed] [Google Scholar]
- 26.Morgenstern JP, Land H. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18(12):3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung SH, Kennedy RA. Forward-backward non-linear filtering technique for extracting small biological signals from noise. J Neurosci Methods. 1991;40(1):71–86. doi: 10.1016/0165-0270(91)90118-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.