Abstract
Purpose
Despite the availability of several active combination regimens for advanced colorectal cancer (CRC), the 5-year survival rate remains poor at less than 10%,supporting the development of novel therapeutic approaches. In this study, we focused on the preclinical assessment of a rationally based combination against KRAS-mutated CRC by testing the combination of the MEK inhibitor, selumetinib, and vorinostat, a histone deacetylase (HDAC) inhibitor.
Experimental Design
Transcriptional profiling and gene set enrichment analysis (baseline and post-treatment) of CRC cell lines provided the rationale for the combination. The activity of selumetinib and vorinostat against the KRAS-mutant SW620 and SW480 CRC cell lines was studied in vitro and in vivo. The effects of this combination on tumor phenotype were assessed using monolayer and 3-dimensional cultures, flow cytometry, apoptosis, and cell migration. In vivo, tumor growth inhibition, 18F-fluoro-deoxy-glucose positron emission tomography (FDG-PET), and proton nuclear magnetic resonance were carried out to evaluate the growth inhibitory and metabolic responses, respectively, in CRC xenografts.
Results
In vitro, treatment with selumetinib and vorinostat resulted in a synergistic inhibition of proliferation and spheroid formation in both CRC cell lines. This inhibition was associated with an increase in apoptosis, cell-cycle arrest in G1, and reduced cellular migration and VEGF-A secretion. In vivo, the combination resulted in additive tumor growth inhibition. The metabolic response to selumetinib and vorinostat consisted of significant inhibition of membrane phospholipids; no significant changes in glucose uptake or metabolism were observed in any of the treatment groups.
Conclusion
These data indicate that the rationally based combination of the mitogen-activated protein kinase/extracellular signal-regulated kinase inhibitor, selumetinib, with the HDAC inhibitor vorinostat results in synergistic antiproliferative activity against KRAS-mutant CRC cell lines in vitro. In vivo, the combination showed additive effects that were associated with metabolic changes in phospholipid turnover, but not on FDG-PET, indicating that the former is a more sensitive endpoint of the combination effects.
Introduction
Selumetinib (AZD6244; ARRY-142886) is a small molecule, orally available, noncompetitive inhibitor of mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase 1/2 (MEK1/2; ref. 1). The RAS/RAF/MEK/ERK kinase cascade occupies a central role in mediating signal transduction from extracellular growth factors, cytokines, and proto-oncogenes. Alterations in the control of RAS/MAPK pathway, resulting in its constitutive activation, have a well-established role in oncogenesis and tumor growth, mostly related to uncontrolled cell proliferation and suppression of apoptosis (2). The hyperactivation of this cascade is mediated through various mechanisms, including tyrosine kinase receptor overexpression or mutations, or to KRAS- or BRAF-activating mutations. KRAS mutations are common in many different tumors, particularly in pancreatic (90%) and colon cancers (50%; refs. 3, 4). Although BRAF mutations are more common in melanoma (63%), papillary thyroid cancer (45%), and low-grade ovarian cancers (50%), the BRAF V600E mutation has also been found in colorectal cancers (CRC) that also exhibit defective DNA mismatch repair (5, 6). The RAS/MAPK cascade is one of the major downstream signaling networks linking the epidermal growth factor (EGF) receptor (EGFR), insulin-like growth factor-I (IGF-I) receptor (IGF-1R), and VEGF receptor-2 (VEGFR-2) pathways to nuclear proteins. The safety profile and tolerability of selumetinib has been evaluated in a 2-part, multicenter, ascending dose, phase I clinical study (7). This trial showed the tolerability of selumetinib, with the most common treatment-related toxicities being rash, diarrhea, nausea, and fatigue. Several phase II trials are ongoing to evaluate the activity of selumetinib, as single agent or in combination with chemotherapy in non-small-cell lung carcinoma (NSCLC), melanoma, and CRC (8–10).
Histone deacetylation by histone deacetylases (HDAC) is a posttranslational modification of lysine residues in nucleosomal histone proteins that affects chromatin structure and, thereby, gene regulation (11). Recently, HDAC activity has been shown to be upregulated in cancer cells, and it has been theorized that this results in repression of tumor suppressor gene products such as p53, making HDACs an attractive drug target (12, 13). In cell culture models, HDAC inhibitors (HDACi) have been shown to decrease proliferation and induce apoptosis or autophagy-related death of several cell lines (14–16). Because of their relative specificity toward cancer cells, HDACi represent a new class of cancer treatment agents that are generally well tolerated. One such compound, vorinostat [suberoylanilide hydroxamic acid (SAHA)] has shown encouraging activity in early studies against several cancers, including B-cell lymphoma (17), colon cancer (18, 19), NSCLC (20), and head and neck cancer and is currently approved for the treatment of cutaneous T-cell lymphoma (21). Several phase II studies have been conducted for breast (22), colon, and lung (23, 24), head and neck (25), and ovarian cancer patients (26); however, no consistent antitumor activity of vorinostat as single agent has been observed (11). The prevailing view is that further investigations to evaluate the safety and activity of vorinostat as a combination partner are needed to better evaluate the potential of this agent in cancer treatment (27). Several reports in the literature have shown synergistic interaction between MEK and HDAC inhibitors. Yu and colleagues (28, 29) showed that combining HDAC with MEK inhibitors resulted in increased apoptosis, through the induction of oxidative damage and ROS generation (well-known response markers for HDAC), as well as enhanced lethality in leukemia cells expressing the ABL/BCR mutation and resistance to imatinib.
On the basis of these data, and our own transcriptional profiling and gene set enrichment analysis, we hypothesized that targeting the MEK pathway would inhibit signal transduction pathways involved in CRC tumor cell proliferation, survival, and angiogenesis, and that this could be potentiated by combination with vorinostat (30). Furthermore, we focused these preclinical studies on CRC tumors expressing KRAS mutations, as they are associated with resistance to other signal transduction inhibitors and represent a subtype of CRC that is in dire need of new therapeutic strategies. Finally, we aimed to establish sensitive in vivo endpoints for MEK and HDAC inhibitors in a murine CRC xenograft model.
Materials and Methods
Drugs
Selumetinib (AZD6244) and vorinostat (SAHA) were generously provided by AstraZeneza Pharmaceuticals LP and Merk Sharp and Dohme Corporation, respectively, and National Cancer Institute, NIH. Selumetinib (AZD6244) was prepared as a 10 mmol/L stock solution in dimethyl sulfoxide (DMSO)for the in vitro experiments. For the in vivo studies, selumetinib was prepared in a solution of 0.5% methyl cellulose/0.1% Tween 80 in water. Vorinostat (SAHA) was prepared as a 10 mmol/L stock solution in DMSO for the in vitro studies and as a solution of 45% PEG400 in water for the in vivo studies.
Culture of cell lines and assessment of cytotoxicity to selumetinib, vorinostat, or the combination
The following human colon cancer cell lines were obtained from American Type Culture Collection: HCT116 (KRAS mut), HCT15 (KRAS mut), HCT8 (KRAS mut), HT29 (KRAS wt), SW480 (KRAS mut), SW620 (KRAS mut), SW1417 (KRAS wt), LoVo (KRAS mut), LS513 (KRAS mut), LS180 (KRAS mut), LS174T (KRAS mut), LS1034 (KRAS mut), Colo205 (KRAS wt), and the RKO (KRAS wt). Cells were grown in RPMI medium supplemented with 10% FBS, 1% nonessential amino acids, 1% penicillin/streptomycin, and maintained at 37°C in an incubator under an atmosphere containing 5% CO2. The cells were routinely screened for the presence of Mycoplasma (MycoAlert; Cambrex Bio Science). Cytotoxic effects were determined using the sulforhodamine B (SRB) method as previously described (19). Briefly, cells in logarithmic growth phase were transferred to 96-well flat bottom plates with lids. One hundred microliter of cell suspensions containing viable HCT116 (1,500 cells per well), HCT15 (2,000 cells per well), HCT8 (1,500 cells per well), GEO (2,000 cells per well), SW837 (1,500 per well), HT29 (3,000 cells per well), SW480 (2,500 cells per well), SW620 (3,000 cells per well), SW1417 (4,000 cells per well), LoVo (5,000 cells per well), LS513 (5,000 cells per well), GEO (2,000 cells per well), SW837 (1,500 per well), LS180 (3,000 cells per well), LS174T (3,500 cells per well), LS1034 (6,000 cells per well), Colo205 (8,000 cells per well), and RKO (1,500 cells per well) cells were plated into each well and incubated overnight before exposure to selumetinib or vorinostat. Initially, all of the cell lines were exposed for 24, 48, and 72 hours to increasing concentrations of selumetinib (0–5 µmol/L) and vorinostat (0–10 µmol/L). Post drug treatment media was removed and cells were fixed with cold 10% trichloroacetic acid for 30 minutes at 4°C. Cells were then washed with water and stained with 0.4% SRB (Fisher Scientific) for 30 minutes at room temperature, washed again with 1% acetic acid, followed by stain solubilization with 10 mmol/L TRIS at room temperature on shaker for 15 minutes. The plates were then read on a plate reader (Biotek Synergy 2) set at an absorbance wavelength of 565 nm, and cell proliferation curves were derived from the raw absorbance (OD) data. The cells were classified as sensitive to the drug if their IC50 was lower than 1 µmol/L or resistant if their IC50 was higher than 1 µmol/L as previously described (31).
Transcriptional profiling of the CRC cell lines
The baseline gene expression of all of the human CRC cell lines was assessed using the Affymetrix U133 Plus 2.0 microarrays. In addition, posttreatment profiling was carried out on HCT8 (selumetinib-R/vorinostat-S), HT29 (selumetinib-S/vorinostat-R), SW480 (R/R), and SW620 (S/S) cells, treated with vorinostat and selumetinib as single agents and in combination. The cells were plated in T75 flasks for 24 hours and then treated with vorinostat or selumetinib for 72 hours. All cell lines were treated at doses equivalent to the IC50 of the most sensitive cell line. After treatment, RNA was extracted with a RNeasy Plus Mini Kit (Qiagen Inc.) and analyzed by the Gene Array Core at the University of Colorado Cancer Center. Absolute intensity signals from the microarray gene expression profiles were extracted using Affymetrix Power Tools, and probe sets representing the same gene were collapsed based on maximum values. Next, the gene expression levels were converted to a rank-based matrix and standardized (mean = 0, SD = 1) for each microarray. Using this preprocessing method, the same cell lines from different data sets were clustered based on their gene expression profiles. Data analyses were carried out on this rank-based matrix.
Gene set enrichment analysis
Gene set analysis was carried out using the GSEA software Version 2.0.1 obtained from the Broad Institute (http://www.broad.mit.edu/gsea). Gene set permutations were carried out 1,000 times for each analysis. We used the nominal P value and Normalized Enrichment Score (NES) to sort the pathways enriched in each phenotype. We used the 313 pathways defined by BioCarta database as the gene set in this study (www.biocarta.com). One hundred and eighty-eight gene sets passed the gene set size filter criteria (min = 10, max = 500).
In vitro evaluation of the combination effects of selumetinib and vorinostat
Synergy was determined for the KRAS-mut cell lines SW837, LS1034, HCT116, GEO, SW480, and SW620. These cell lines were selected because all of them contain KRAS mutations but exhibit divergent sensitivity to the single agents. To evaluate synergy, cells were exposed for 72 hours to vorinostat at 0.375, 0.75, and 1.5 µmol/L and selumetinib at 0.125, 0.250, 0.5, and 1 µmol/L (SW480), or 0.06, 0.125, 0.250, and 0.5 µmol/L (SW620), in all possible combinations. The results of the combined treatment were analyzed according to the isobolographic method of Chou and Talalay (32), using the Calcusyn software program (Biosoft). The resulting combination index (CI) was used as a quantitative measure of the degree of interaction between the 2 drugs. A CI equal to 1 denotes additivity, CI greater than 1 antagonism, and CI values less than 1 indicate synergism.
The effect of selumetinib and vorinostat was further evaluated in the SW480 cell line which is KRAS mut, resistant to selumetinib as well as vorinostat (R/R), and in the SW620 cell line, which is KRAS mut but sensitive to both drugs (S/S) using a 3-dimensional (3D) culture system. Briefly, 100 µL of Matrigel (BD Biosciences) was plated in 8-well chamber slides (BD Biosciences) to form a base layer. Cells were then seeded at the density of 2,500 cells per well in 2% Matrigel-supplemented media to form the upper layer. After an initial incubation to allow for tumor spheroid formation, vehicle, or drugs were added to media and changed every 3 days for a total of 15 days. The SW480 (R/R) were exposed to vorinostat (0.750 µmol/L) or selumetinib (0.125 µmol/L) or the combination, whereas SW620 (S/S) cells were treated with selumetinib (0.06 µmol/L), vorinostat (0.750 µmol/L), or the combination. After 15 days of treatment, the plates were photographed at 20 × magnification on an inverted microscope. Spheroids with a diameter higher than 1 µm were counted. All of the in vitro experiments were conducted in triplicate.
Flow cytometric analysis of cell-cycle distribution
For cell-cycle analysis, cells were seeded in 6-well plates (2.5 × 105 cells per well) for 24 hours. The cells were then treated with vorinostat at 0.750 µmol/L, or selumetinib at doses of 0.125 µmol/L for SW480 (R/R) and 0.06 µmol/L for SW620 (S/S), or with the respective combination for 24 hours. Culture media was removed and the cells were washed in PBS, harvested, centrifuged for 5 minutes at 1,500 rpm, and resuspended in Krishan’s stain. After incubation at 4°C for 24 hours, the samples were analyzed by flow cytometry at the University of Colorado Cancer Center Flow Cytometry Core Facility. Experiments were conducted in triplicate.
Apoptosis assay
SW480 and SW620 cells were seeded in 96-well, white-walled plates and allowed to plate for 24 hours. SW480 (R/R) cells were treated with vorinostat (0.750 µmol/L) or selumetinib (0.125 µmol/L) or the combination for 24, 48, and 72 hours, after which apoptosis was determined by the measurement of caspase 3/7 activity using a luminometric Caspase-Glo 3/7 assay (Promega) according to the manufacturer’s protocol and read using a 96-well plate reader. Cellular apoptosis was expressed as the fold-increase over untreated control cells. Similarly, SW620 (S/S) cells were treated with selumetinib (0.06 µmol/L) or vorinostat (0.750 µmol/L) or the combination for 24, 48, and 72 hours and apoptosis was measured as described earlier. Experiments were conducted in triplicate.
Immunoblotting of downstream effector proteins
SW480 and SW620 cells were seeded into 6-well plates (2.5 ×105 cells per well) for 24 hours before treatment and exposed to each drug alone or in combination for an additional 24 hours. After treatment, cells were recovered in radioimmunoprecipitation assay buffer containing protease inhibitors, EDTA, NaF, and sodium orthovana-date. The total protein in samples was determined using the BioRad DC Protein Assay (BioRad). Fifty micrograms of total protein was loaded onto a 4% to 20% gradient gel, electrophoresed, and then transferred to nitrocellulose using the I-Blot (Invitrogen). Membranes were blocked for 1 hour in blocking buffer (0.1% casein solution in 0.2 PBS), before overnight incubation at 4°C with one of the following primary antibodies: phospho-ERK1/2, cleaved PARP, acetylated histone H3, or β-actin (Cell Signaling Technology). Following primary antibody incubation, membranes were then washed 4 times (10 minutes each) in TBS-Tween (0.1%) and then incubated with the appropriate secondary anti-rabbit or anti-mouse IgG1 horseradish peroxidase–linked antibody at 1:15,000 (Jackson ImmunoResearch) for 1 hour at room temperature. After 4 additional washes, blots were developed using the Odyssey Infrared Imaging System (LI-COR Biosciences). Immunoblot experiments were done in triplicate for each antibody.
Effects of selumetinib and vorinostat on ligand-induced cell migration
The effects of selumetinib and vorinostat on SW620 and SW480 cell migration were measured using the modified Boyden chamber assay. Assays were carried out using uncoated 8.0 µm transwell inserts placed in 24-well plates (Becton Dickinson). Cells were resuspended in 250 µL of standard growth media (RPMI supplemented with 10% FBS, 1% nonessential amino acids, 1% penicillin/streptomycin) and seeded at a density of 10 × 104 cells per well onto the inserts (upper chamber of the well), and the lower chambers of each well were filled with 600 µL of standard growth media. After 24 hours, the media in the upper chambers was replaced with media containing 0.1% FBS, in presence or absence of selumetinib or vorinostat or the combinations (selumetinib at the doses of 0.125 µmol/L and vorinostat at the dose of 0.750 µmol/L). The media in the bottom of the 24-well plate was replaced with fresh media containing 10% FBS with or without the drugs (selumetinib 0.125 µmol/L or vorinostat, at the dose of 0.750 µmol/L). After 48 hours, the filters were fixed with 4% formaldehyde for 15 minutes, washed 3 times with D-PBS, and migrating cells were stained with 4’,6-diamidino-2-phenylindole (DAPI) for 30 minutes, washed with D-PBS 3 × 15 minutes, and photographed at 20 × magnification on an inverted microscope. Five fields per well were counted manually and averaged. Selumetinib, vorinostat, and the combination effects on EGF- or IGF-I–induced migration was explored used SW480 (R/R) cells because the SW620 cells were not sufficiently migratory. Briefly, 10 × 104 cells per well were resuspended in 250 µL of standard growth media and plated in the upper chamber. The lower chamber of the wells was filled with 600 µL of standard growth media. After 24 hours, the media in the upper and lower chambers of the plate was replaced with 0.1% FBS media and incubated overnight in the presence or absence of treatment. Cell migration was then stimulated by replacing media in the lower chamber with media containing either 10% FBS or EGF (100 ng/mL), IGF (200 ng/mL) in absence or presence of treatment (selumetinib at the dose of 0.125 µmol/L and vorinostat at the dose of 0.750 µmol/L). Cell migration was evaluated after 48 hours from growth factor stimulation, as described earlier. After 48 hours, the filters were fixed with 4% formaldehyde for 15 minutes and washed 3 times with D-PBS, and migrating cells were stained with DAPI for 30 minutes, washed with D-PBS 3 × 15 minutes, and photographed at 20 × magnification on an inverted microscope. Five fields per well were counted manually and averaged.
Assessment of the selumetinib and vorinostat combination in vivo
Female athymic nude mice, 4 to 6 weeks old, were purchased from Harlan Laboratories (Harlan Laboratories online). Animals were housed in polycarbonate cages and maintained on a 12-hour light/dark cycle in the University of Colorado Center for Comparative Medicine, a facility accredited by the American Association for Accreditation of Laboratory Animal Care. Animals were housed 3 to 5 per cage and food and water were provided ad libitum. All studies were conducted in accordance with the NIH Guidelines for the Care and Use of Laboratory Animals. SW620 (S/S) and SW480 (R/R) cell lines were harvested in exponential growth phase and resuspended in a 1:1 mixture of media:Matrigel. One million SW620 and 5 million of SW480 per injection were injected subcutaneously in both flanks of each mouse. When tumors reached an average of 150 to 300 mm3 of volume, mice were randomized into 4 groups (n = 8 tumor per group). Animals received either vehicle, 25 mg/kg selumetinib, 75 mg/kg vorinostat, or a combination of 25 mg/kg selumetinib and 75 mg/kg vorinostat. Selumetinib was administered by oral gavage twice a day, 7 days per week. Vorinostat was administered by intraperitoneal injection daily, 5 days per week. For combination treatment, animals received selumetinib followed immediately by vorinostat injection. Mice were monitored daily for signs of toxicity and were weighed twice weekly. Tumor size was evaluated twice per week by caliper measurements using the following formula: tumor volume = [length × width2]/0.52. Tumor volume and body weight data were collected using the Study Director Software package (Studylog Systems). Animals were treated for 28 days and then euthanized by isoflurane anesthesia. These studies were carried out under an approved University of Colorado Animal Care (IACUC) protocol.
Tumor metabolic response of SW480 xenografts by 18FDG-PET and 1H-NMR
18F-fluoro-2-deoxy-D-glucose positron emission tomography (FDG-PET) was carried out on SW620 (S/S) xenografts to measure the effect of selumetinib, vorinostat, or the combination on tumor metabolic activity. When tumors reached an average of 150 to 200 mm3, mice were randomized into 4 groups (n = 8 tumors per group). Treatment groups were the same as the first in vivo study and animals were treated for a total of 21 days as described in the previous section. FDG-PET was carried out the day before the start of treatment (baseline; BL) and on days 4 (D4) and 21 (D21) of treatment. Animals were fasted for 4 to 8 hours before FDG injection and glucose blood levels were monitored. Approximately 250 µCi of FDG, obtained through the University of Colorado Hospital (PetNet Solutions), was administered by tail vein injection (intravenously) to conscious animals. Animals were maintained in cages on a heated water pad for 1 hour to allow for FDG uptake in tumors. Under isoflurane anesthesia (2.5%), animals were placed on a warm pad (m2m Imaging) and a 10-minute emission scan was acquired with a Siemens Inveon micro-PET scanner (Siemens Medical). Image analysis was conducted with Asi-ProVM (Concorde Microsystems) software. Regions of interest were drawn with the trace command around the tumors on axial slices, and the total activity of all tumor slices was summed. Total activity was divided by the time-corrected dose delivered (time-corrected dose = dose injected × exp(−0.006317 × t), where t is the time between the injection and scan time and is presented as the percentage of the baseline scan of respective tumor.
By the end of the study (day 22), CRC xenografts were collected, snap frozen in liquid nitrogen, and extracted for an expanded metabolic profile by proton nuclear magnetic resonance (1H-NMR). The exact extraction and NMR acquisition protocols were extensively reported previously by our group (33, 34). All quantitative data sets (presented as µmol of metabolite per gram tissue) were included in custom-build metabolic fluxes analyzers and data interfaces and presented as metabolic heatmaps (35).
Statistical analysis
The statistical analyses of the in vitro and in vivo data were carried out using Prism version 4.02 program (GraphPad Software, Inc.). A 1-way ANOVA test was carried out to evaluate statistically significant changes between the study groups. P < 0.05 was considered statistically significant. Bar represents the mean of 3 independent experiments carried out in triplicate.
Results
Gene array and pathway analyses
To identify genes and pathways that are correlated with single-agent vorinostat sensitivity, we compared the baseline gene expression across the CRC cell line panel using GSEA. HDAC pathway was among the top enriched pathways in the sensitive lines as compared with the resistant lines. We next sought to identify which genes and pathways were enriched in the resistant cell lines after vorinostat treatment. Comparing the baseline and posttreatment expression profiles of the 2 resistant cell lines (HT29 and SW480) revealed that MAPK signaling pathway is among the top enriched pathways after vorinostat treatment (Fig. 1A). The core genes of the MAPK signaling pathway are illustrated in Fig. 1B and C. This suggested that treating these vorinostat-resistant cell lines with selumetinib may synergize the combination drug effects. Indeed, when we compared the core genes in the MAPK signaling pathway of the 2 vorinostat-resistant cell lines after selumetinib and the combination, the gene expression patterns were altered (Fig. 1B).
Figure 1.
Pathway analysis of CRC cell lines at baseline and after treatment with vorinostat, selumetinib, or the combination. A, top 5 pathways enriched in the vorinostat-resistant cell lines (HT29 and SW480) after vorinostat treatment. B, heatmap of the core genes in the HDAC and MAP kinase signaling pathway at baseline, after treatment with vorinostat, selumetinib, and the combination. Pathway diagram derived from BioCarta where the core genes are circled. Red and green in the heatmap represent gene overexpression and underexpression, respectively. C, pathway diagram derived from BioCarta where the core genes are colored in red.
Effects of the selumetinib and vorinostat combination on proliferation of KRAS-mutant CRC cell lines in vitro
To seek for a new active treatment for KRAS-mut CRC patient, we studied the combination of selumetinib and vorinostat on GEO, HCT116, SW837, SW620, and LS1034. In an initial study, conducted on a panel of CRC cell lines to evaluate the activity of selumetinib and vorinostat as single agent, we observed that the SW837 and the SW480 were resistant to both selumetinib and vorinostat (R/R), the GEO were resistant to selumetinib and sensitive/intermediate for vorinostat (R/I-R), the LS1034 were sensitive to selumetinib and resistant to vorinostat (S/R), and the SW620 were sensitive to both drugs (S/S) as shown in Supplementary Fig. S1. Then, cells were treated concurrently for 72 hours at clinically relevant doses, according to phase I trial results (7, 36, 37). Our results, as calculated by the method of Chou and Talalay (32), showed that selumetinib and vorinostat treatment resulted in clear synergy for the all cell lines analyzed (Supplementary Fig. S2), with a strongest effect observed for the SW620 (S/S) cell line (Fig. 2A and B) with CI values between 0.354 and 0.736, and for the SW480 (R/R) cell line (Fig. 2C and D) with CI values of 0.07 to 0.777. On the basis of these results, we chose to use for the subsequent experiments, the SW620 and the SW480, treated at the following doses of selumetinib and vorinostat for subsequent experiments: SW620 (S/S): 0.06 µmol/L selumetinib and 0.750 µmol/L vorinostat; SW480 (R/R): 0.125 µmol/L selumetinib and 0.750 µmol/L vorinostat.
Figure 2.
Selumetinib in combination with vorinostat shows synergistic inhibition against CRC cell lines. SW620 S/S (A, B) and SW480 R/R (C, D) were plated in 96-well plates for 24 hours and treated for 72 hours with varying doses of selumetinib and vorinostat. Proliferation was assessed by the SRB assay. Raw proliferation data were expressed as percent of viable cells and CI values were analyzed according to the Chou and Talalay method for drug interactions using the Calcusyn software package.
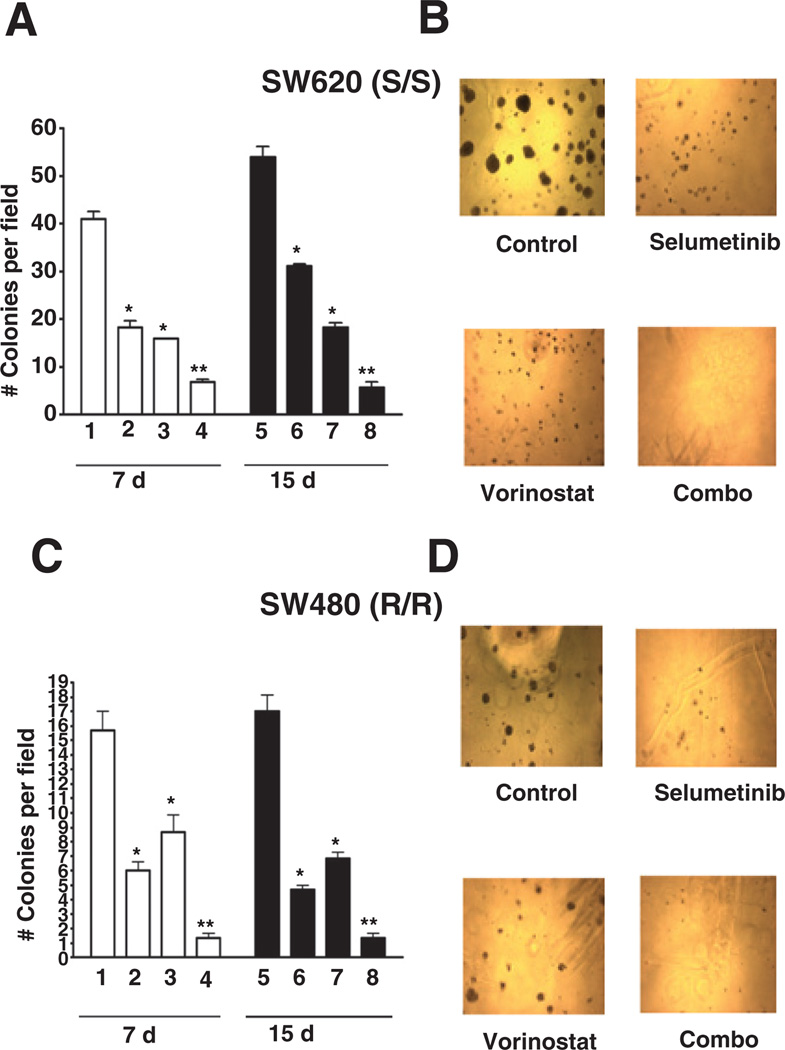
To assess the efficacy of selumetinib, vorinostat, and the combination in a more physiologic model, we next carried out a 3D culture analysis with tumor spheroid formation in Matrigel as the endpoint. As depicted in Fig. 3A–D, the SW620 S/S and SW480 R/R cells generated spheroids when grown in Matrigel, which were inhibited by both single agents and, more strikingly, by the combination. Interestingly, the SW480 R/R cells displayed a sensitive phenotype to selumetinib and vorinostat in 3D culture, but a resistant phenotype in monolayer growth, suggesting that these agents require more intact tumor architecture to exert their full effects.
Figure 3.
Effects of selumetinib and vorinostat on CRC spheroid formation in 3D culture. CRC cell lines were plated in 8-well chamber slides on Matrigel pads at a density of 2,500 cell per well resuspended in 2% Matrigel-supplemented RPMI plus 10% FBS. SW620 (S/S) and SW480 R/R cells were treated with indicated doses of selumetinib, vorinostat, or the combination for 15 days. SW620 (A, B) and SW480 (C, D) spheroids were photographed and counted after 7 and 15 days of treatment. Spheroids with a diameter greater than 1 µm were counted. Data represent the mean of 3 independent experiments. *P < 0.05 selumetinib, vorinostat, or combination treatment versus control; **P <0.05 combination versus control, and single agents. Bars represent the mean of 3 independent experiments carried out in triplicate.
Effects of selumetinib and vorinostat on cell-cycle distribution, apoptosis, and downstream signaling pathways
To further elucidate the mechanism(s) responsible for the observed synergistic effects on CRC cell proliferation, we investigated the impact of selumetinib in combination with vorinostat on cell-cycle distribution, apoptosis, and intracellular signaling pathways. By flow cytometry, cell-cycle analysis following treatment of SW620 S/S cells with single-agent selumetinib or vorinostat resulted in a significant increase in the percentage of cells in G1, which was further increased in the combination and associated with a reduction of cells in S phase (Fig. 4A). By contrast, in the SW480, R/R cells, G1 arrest was induced by treatment with selumetinib which was maintained in the combination (Fig. 4B). Next, we evaluated the ability of selumetinib, vorinostat, or the combination to induce apoptosis in SW620 and SW480 cells after 24 hours of exposure to these agents using a caspase 3/7 activity assay. Although apoptosis was modestly increased by the single agents, the combination showed a significant induction of apoptosis in both cell lines, as depicted in Fig. 4C and D.
Figure 4.
Selumetinib and vorinostat induce G1 cell-cycle arrest and apoptosis in CRC cell lines. SW620 S/S (A, C) and SW480 R/R (B, D) CRC cell lines were treated with selected doses of selumetinib and vorinostat for24 hours. For cell-cycle analysis, cells were stained with Krishanaposs stain followed by flow cytometric analysis. Apoptosis was analyzed by fluorometric measurement of caspase-3 and caspase-7 activity. *P < 0.05 selumetinib, vorinostat, or combination treatment versus control; **P <0.05 combination versus control and single agents. Bars represent the mean of 3 independent experiments carried out in triplicate.
Analysis of downstream signaling pathways by immunoblotting showed that in both cell lines selumetinib reduced ERK phosphorylation and vorinostat induced histone H3 acetylation, as expected (Supplementary Fig. S3). Interestingly, H3 acetylation was markedly increased in the combination treatment (Supplementary Fig. S3). Increased PARP cleavage was observed in both cell lines after treatment with vorinostat or selumetinib, however, levels of cleaved PARP were markedly increased in SW620 S/S line after the combination treatment and, to a lesser extent, in the resistant cell line, SW480 (R/R), consistent with an increase in apoptosis as observed using the caspase 3/7 activity assay (Supplementary Fig. S3; Fig. 4C and D).
Effects of selumetinib, vorinostat, and the combination on ligand-induced migration of SW480 cells
As cancer cell migration is mediated by growth factor pathways such as the RAS/RAF/MEK/ERK pathway (38–44), we evaluated the effects of selumetinib and vorinostat alone and in combination on cell migration using a modified Boyden chamber assay. As depicted in Supplementary Fig. S4, the combination of selumetinib and vorinostat significantly reduced both FBS- and IGF-stimulated migration compared with EGF, which exhibited little induction of migration, regardless of treatment.
Antitumor activity of selumetinib in combination with vorinostat in SW620 S/S and SW480 R/R xenograft models
To further evaluate the efficacy of selumetinib in combination with vorinostat as a potential treatment option for CRC, we tested the tumor growth inhibitory potential of these drugs in a murine xenograft model. Tumor growth curves for these studies are depicted in Supplementary Fig. S5 for the SW480 R/R xenograft and in Fig. 5A for the SW620 S/S xenograft. In the SW620 S/S model, vorinostat alone showed little antitumor growth effects, whereas selumetinib and the combination were similar, showing a trend toward significant tumor growth inhibition compared with control (*, P < 0.05). Treatment of the SW480 R/R xenograft resulted in more modest growth inhibitory effects, which also appeared to be dominated by the single-agent effects of selumetinib.
Figure 5.
Antiproliferative and metabolic responses of the SW620 S/S cell line xenograft in vivo. A, curves represent tumor growth rate, expressed as “percent of day one,” when treated with either vehicle, selumetinib (25 mg/kg), vorinostat (75 mg/kg), or the combination. B, metabolic heatmaps based on quantitative FDG-PET and 1H-NMR spectroscopic data sets in SW620 xenografts on day 22. The data represents mean ± SEM of 8 biopsies per group. The metabolites, their ratios, and metabolic fluxes were grouped on the basis of their biochemical relevance. For the control group, all intracellular metabolite levels are presented as µmol per cell wet weights; metabolite ratios and glucose uptake are unitless. Metabolic pathways, which were undisturbed by treatment, are presented as yellow blocks. A decrease in a metabolic endpoint is indicated by red, whereas an increase is depicted in green. Statistical significance for metabolite changes are based on multivariate analysis of metabolic fluxes with *P < 0.02. The interactive metabolic profile array database was custom based (35). C, representative PET images and their quantification (D, presented as the percent of normalized glucose uptake to the baseline) acquired on days 4 and 21 posttreatment. Decreases of 18FDG uptake in tumors can be measured as early as 4 days posttreatment, with no significant differences between either treatment group compared with control but with a trend of selumetinib compared with vorinostat measured at day 21.
Assessment of SW620 S/S xenograft tumor metabolic response by 1H-NMR and 18FDG-PET
An expanded metabolic profile, calculated from 1H-NMR extracts of treated and untreated tumors, showed that no changes in lactate, the end product of glycolysis, were observed in the treatment groups (Fig. 5B). In fact, no significant metabolic changes were detected for the HDAC inhibitor, vorinostat. The most striking changes observed with MEK inhibition by selumetinib were related to highly decreased levels of membrane phospholipids (such as phosphatidylcholine, its precursors phosphocholine, and phosphatidylinositol). Accumulation of nucleosides and adenosines were also seen in the selumetinib-treated group. The combination group mostly reflected the metabolic effects of single-agent selumetinib, with broader decreases in membrane phospholipids. To assess the effects of selumetinib in combination with vorinostat on glucose uptake, 18FDG-PET was carried out on mice bearing SW620 tumors at baseline, before starting treatment, after 4 days of treatment to evaluate early drug effects and after 21 days of treatment with either vehicle, selumetinib, vorinostat, or the combination. No significant decreases in glucose uptake were observed in either study group when compared with untreated controls after 3 days of treatment (data not shown), whereas a trend of decrease in glucose uptake was observed on day 21 in the selumetinib and combo group compared with vorinostat (Fig. 5C and D; *, P < 0.05).
Discussion
Recent efforts in cancer therapeutics have focused on the development of targeted therapies directed toward specific molecules related to cancer cell biology to achieve a more specific and effective treatment, and at the same time, avoid some of the systemic toxicities associated with chemotherapy (45, 46).To optimize this therapeutic approach, patient selection for clinical trials has become of critical importance (47–50). For example, recent studies have shown a correlation between CRC tumors with activating KRAS mutations and resistance to EGFR-targeted therapies. Therefore, there is an emerging need for alternative therapeutic approaches for this patient population (5, 50, 51). In this study, we utilized in vitro and in vivo models of CRC to evaluate the efficacy of selumetinib in combination with vorinostat as a potential therapeutic strategy for the treatment of CRC patients expressing mutant KRAS. Our data show that this combination exerts potent antitumor effects on in vitro models of CRC, including synergistic inhibition of cell proliferation, G1 cell-cycle arrest, induction of apoptosis, and spheroid formation in 3D culture. Interestingly, this combination inhibited CRC cell migration, which suggests that this treatment not only inhibits tumor growth but may play a role in the metastatic process as well. In vivo studies using KRAS-mutant CRC xenograft models in nude mice confirmed the antitumor effects of the combination of selumetinib and vorinostat, although synergy was not observed, likely because of the single-agent effects of selumetinib.
In our initial studies, we evaluated selumetinib and vorinostat as single agents against a panel of CRC cell lines that were selected based on differential KRAS, BRAF, and p53 mutational status. Several reports have suggested a relationship between KRAS and BRAF mutation status and response to MEK inhibitors (1). Other investigators show that the blockage of the MAPK pathway with a MEK inhibitor induced cell death only in cells that were KRAS mutant but p53 wild type (52). Although we failed to show any significant correlations between cell line sensitivity to selumetinib or vorinostat and KRAS, BRAF, or p53 status (data not shown), a recent study by Dry and colleagues showed that a gene signature of functional MEK activation was consistently elevated in cell lines sensitive to selumetinib, whereas BRAF/KRAS mutation or ERK activation varied across cell lines, indicating that expression profiling may be a better predictor of drug response than individual biomarkers or gene mutations for this class of agents (53). Consistent with this report, we observed constitutive overexpression of HDAC and MAPK pathways in the cell lines sensitive to vorinostat or selumetinib, respectively. Interestingly, our data showed that cell lines resistant to vorinostat exhibited a constitutive overexpression of the MAPK pathway and that the MAPK pathway was upregulated in response to treatment with vorinostat, providing a functional rationale for exploring the use of vorinostat in combination with selumetinib in our preclinical models of CRC.
To further evaluate the synergistic effects of this combination, we chose to focus on several KRAS-mutant CRC cell lines. Our results showed that the combination of selumetinib and vorinostat resulted in a synergistic antiproliferative effects in all cell lines studied. Mechanistically, our results suggest that this antiprolifterative effect of combination of selumetinib and vorinostat is mediated by an increase in apoptotic cell death, as measured by caspase 3/7 activity, cleaved PARP, and, perhaps, increased acetylated histone H3 levels. This is in accordance with previous reports showing that MEK inhibition was synergistic with HDAC inhibitor through the induction of cell death via apoptosis (29). We also evaluated the effects of the combination on cell-cycle distribution and apoptotic events. It has been previously reported that vorinostat increases the expression of cyclin-independent kinases such as p21CIP/WAF1 in bladder cancer, p27KIP1 in gliomas, and induces G2/M cell-cycle arrest in CRC cell lines, whereas selumetinib induced G1 cell-cycle arrest and induction of apoptosis in a CRC xenograft model (1, 19, 54). In our study, we observed a G1 cell-cycle arrest in the SW620 S/S cell line after each single-agent treatment that was significantly increased after exposure to the combination. In the SW480 R/R cell line, vorinostat showed no effects on cell-cycle distribution, whereas single-agent selumetinib and combination treatment resulted in a G1 arrest, suggesting that the combination effects were mediated by selumetinib.
Because cell proliferation is only one phenotypic endpoint of malignant behavior, we used other in vitro models such as a modified Boyden chamber and 3D culture and found that the combination of these agents resulted in significant decreases in cell motility and spheroid formation, respectively. As MEK is a downstream effector of the EGFR, VEGFR, and IGF-1R signaling pathways, we hypothesized that MEK activation could play a role in ligand– induced cell migration. For example, previous data have shown that treatment of NSCLC with HDAC inhibitors induces reexpression of E-cadherin mRNA, induction of the epithelial phenotype, and decreased migratory activity (55). By contrast, our findings indicated that stimulation of EGFR pathway did not affect the migration of SW480 cells, although both FBS and IGF-1 induced cell migration that was inhibited by both single agents and substantially by the combination. The IGF1R and other components of the IGF system have been shown to be overexpressed in colon cancer and are associated with advanced stage of disease, metastasis, and reduced survival (56). These results suggest that the combination of vorinostat and selumetinib may act downstream of this pathway to block the phenotypic effects of IGF-induced CRC migration. In 3D culture, cancer cells are able to form tumor spheroids that are thought to more closely recapitulate the cancer cell–extracellular matrix interactions that play a fundamental role in tumor growth and metastasis (57). The inhibitory effects of the combination were confirmed in the 3D model in both the sensitive (SW620) and resistant (SW480) cell lines. Of interest was the finding that the SW480 cells appeared to be sensitive to selumetinib in the 3D model, supporting the limitation of 2D assays in assessing more global impacts on cellular phenotype in vitro.
In our in vivo studies of both SW620 (S/S) and SW480 (R/R) cell line xenografts, combination treatment showed significant tumor growth inhibition compared with untreated controls or vorinostat, but not to single-agent selumetinib. As would be expected from the 3D results, the SW480 xenograft was sensitive to selumetinib in vivo, which we hypothesize could be related to MEK-dependent effects on the tumor microenvironment and tumor cell–matrix interactions. Although the single-agent activity of selumetinib at 25 mg/kg obfuscated the detection of synergy in vivo, our assessment of metabolic endpoints led to some interesting findings. Surprisingly, no significant inhibition of glucose uptake (FDG-PET) or metabolism (lactate from 1H-NMR) was observed for either selumetinib or vorinostat, alone or in combination, compared with untreated controls. Although HDAC inhibition did not appear to induce any metabolic changes, the MEK inhibitor selumetinib had a profound inhibitory effect on membrane phospholipid turnover (decrease in total phospholipids as well as phosphatidyl-choline and phosphatidyl-inositol). This, together with an accumulation of adenosine and nucleotides, is reflective of the antiproliferative effects observed in this study for selumetinib and the combination. These data show that effects on glucose metabolism are not observed with all targeted agents and that preclinical metabolic studies should be conducted to more precisely determine the most appropriate readout for functional imaging. For example, these results suggest that in human clinical trials, incorporation of 18F-choline PET, rather than FDG-PET, might be the most relevant imaging endpoint for treatment response to MEK inhibitors.
In summary, we utilized gene array and gene set enrichment analyses to develop a rational basis for testing the combination of selumetinib and vorinostat as a novel therapeutic option for KRAS-mutant CRC. Moreover, the preclinical findings described in this article, utilizing both in vitro and in vivo models of CRC, show that dual inhibition of MAPK signaling and HDAC activity results in inhibition of several biologic processes associated with tumor growth and progression. Together, these data suggest that clinical testing of selumetinib in combination with vorinostat should be explored as a potential therapy for KRAS-mutant CRC.
Supplementary Material
Translational Relevance.
Colorectal cancer (CRC) is the third leading cause of cancer worldwide. Although current therapeutic regimens incorporate active chemotherapy and biologic agents, patients eventually relapse without options for effective salvage therapy. Recent analysis of numerous clinical trials evaluating response rates of advanced CRC to epidermal growth factor receptor inhibitors showed a correlation between activating mutations of the Kristen rat sarcoma viral oncogene homolog (KRAS) gene and resistance to these agents, thus identifying a large subset of patients in urgent need of alternative therapeutic approaches. In this study, we utilized gene set enrichment analyses to support the rational combination of the mitogen-activated protein kinase/extracellular signal-regulated kinase 1/2 inhibitor, selumetinib (AZD6244), and the histone deacetylase inhibitor, vorinostat (SAHA), against preclinical models of KRAS-mutant CRC. Our data show that this combination exhibits potent, synergistic antitumor effects both in vitro and in vivo. These findings strongly support the clinical development of this rational combination in CRC patients with KRAS-mutated tumors.
Acknowledgments
The authors thank Heather Selby and Kelly McPhillips for their excellent technical support.
Grant Support
This work was supported by research grants CA106349 (S.G. Eckhardt), CA079446 (S.G. Eckhardt), CA46934 (NCI Cancer Center Support Grant), and an Astra Zeneca Research Agreement (M.P. Morelli, S.G. Eckhardt).
Footnotes
Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Davies BR, Logie A, McKay JS, Martin P, Steele S, Jenkins R, et al. AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1/2 kinases: mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship, and potential for combination in preclinical models. Mol Cancer Ther. 2007;6:2209–2219. doi: 10.1158/1535-7163.MCT-07-0231. [DOI] [PubMed] [Google Scholar]
- 2.Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96:1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balko JM, Black EP. A gene expression predictor of response to EGFR-targeted therapy stratifies progression-free survival to cetuximab in KRAS wild-type metastatic colorectal cancer. BMC Cancer. 2009;9:145. doi: 10.1186/1471-2407-9-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Au HJ, Karapetis CS, O'Callaghan CJ, Tu D, Moore MJ, Zalcberg JR, et al. Health-related quality of life in patients with advanced colorectal cancer treated with cetuximab: overall and KRAS-specific results of the NCIC CTG and AGITG CO.17 Trial. J Clin Oncol. 2009;27:1822–1828. doi: 10.1200/JCO.2008.19.6048. [DOI] [PubMed] [Google Scholar]
- 5.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 6.Friday BB, Yu C, Dy GK, Smith PD, Wang L, Thibodeau SN, et al. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase and Raf proteins. Cancer Res. 2008;68:6145–6153. doi: 10.1158/0008-5472.CAN-08-1430. [DOI] [PubMed] [Google Scholar]
- 7.Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennouna J, Lang I, Valladares-Ayerbes M, Boer K, Adenis A, Escudero P, et al. A phase II, open-label, randomized study to assess the efficacy and safety of AZD6244 (ARRY-142886) versus capecitabine monotherapy in patients with colorectal cancer who have failed one or two prior chemotherapeutic regimens. Invest New Drug. 2011;29:1021–1028. doi: 10.1007/s10637-010-9392-8. [DOI] [PubMed] [Google Scholar]
- 9.Dummer R, Robert C, Chapman PB, Sosman JA, Middleton M, Bastholt L, et al. AZD6244 (ARRY-142886) vs temozolomide (TMZ) in patients (pts) with advanced melanoma: an open-label, randomized, multicenter, phase II study. J Clin Oncol. 2008;26(15 Suppl) Abstract 9033. [Google Scholar]
- 10.Hainsworth JD, Cebotaru CL, Kanarev V, Ciuleanu TE, Damyanov D, Stella P, et al. A phase II, open-label, randomized study to assess the efficacy and safety of AZD6244 (ARRY-142886) versus pemetrexed in patients with non-small cell lung cancer who have failed one or two prior chemotherapeutic regimens. J Thorac Oncol. 2010;5:1630–1636. doi: 10.1097/JTO.0b013e3181e8b3a3. [DOI] [PubMed] [Google Scholar]
- 11.Al-Janadi A, Chandana SR, Conley BA. Histone deacetylation: an attractive target for cancer therapy? Drugs R D. 2008;9:369–383. doi: 10.2165/0126839-200809060-00003. [DOI] [PubMed] [Google Scholar]
- 12.Shankar S, Srivastava RK. Histone deacetylase inhibitors: mechanisms and clinical significance in cancer: HDAC inhibitor-induced apoptosis. Adv Exp Med Biol. 2008;615:261–298. doi: 10.1007/978-1-4020-6554-5_13. [DOI] [PubMed] [Google Scholar]
- 13.Kouraklis G. HDAC inhibitors in leukemia: current status and perspectives. Leuk Res. 2009;33:207–208. doi: 10.1016/j.leukres.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 14.Crisanti MC, Wallace AF, Kapoor V, Vandermeers F, Dowling ML, Pereira LP, et al. The HDAC inhibitor panobinostat (LBH589) inhibits mesothelioma and lung cancer cells in vitro and in vivo with particular efficacy for small cell lung cancer. Mol Cancer Ther. 2009;8:2221–2231. doi: 10.1158/1535-7163.MCT-09-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buglio D, Georgakis GV, Hanabuchi S, Arima K, Khaskhely NM, Liu YJ, et al. Vorinostat inhibits STAT6-mediated TH2 cytokine and TARC production and induces cell death in Hodgkin lymphoma cell lines. Blood. 2008;112:1424–1433. doi: 10.1182/blood-2008-01-133769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zopf S, Neureiter D, Bouralexis S, Abt T, Glaser KB, Okamoto K, et al. Differential response of p53 and p21 on HDAC inhibitor-mediated apoptosis in HCT116 colon cancer cells in vitro and in vivo . Int J Oncol. 2007;31:1391–1402. [PubMed] [Google Scholar]
- 17.Crump M, Coiffier B, Jacobsen ED, Sun L, Ricker JL, Xie H, et al. Phase II trial of oral vorinostat (suberoylanilide hydroxamic acid) in relapsed diffuse large-B-cell lymphoma. Ann Oncol. 2008;19:964–969. doi: 10.1093/annonc/mdn031. [DOI] [PubMed] [Google Scholar]
- 18.Fedier A, Dedes KJ, Imesch P, Von Bueren AO, Fink D. The histone deacetylase inhibitors suberoylanilide hydroxamic (Vorinostat) and valproic acid induce irreversible and MDR1-independent resistance in human colon cancer cells. Int J Oncol. 2007;31:633–641. [PubMed] [Google Scholar]
- 19.Pitts TM, Morrow M, Kaufman SA, Tentler JJ, Eckhardt SG. Vorinostat and bortezomib exert synergistic antiproliferative and proapoptotic effects in colon cancer cell models. Mol Cancer Ther. 2009;8:342–349. doi: 10.1158/1535-7163.MCT-08-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witta SE, Dziadziuszko R, Yoshida K, Hedman K, Varella-Garcia M, Bunn PA, Jr, et al. ErbB-3 expression is associated with E-cadherin and their coexpression restores response to gefitinib in non-small-cell lung cancer (NSCLC) Ann Oncol. 2009;20:689–695. doi: 10.1093/annonc/mdn703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 22.Luu TH, Morgan RJ, Leong L, McNamara M, Portnow J, Frankel P, et al. A phase II trial of vorinostat (suberoylanilide hydroxamic acid) in metastatic breast cancer: a California Cancer Consortium study. Clin Cancer Res. 2008;14:7138–7142. doi: 10.1158/1078-0432.CCR-08-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vansteenkiste J, Van Cutsem E, Dumez H, Chen C, Ricker JL, Randolph SS, et al. Early phase II trial of oral vorinostat in relapsed or refractory breast, colorectal, or non-small cell lung cancer. Invest New Drugs. 2008;26:483–488. doi: 10.1007/s10637-008-9131-6. [DOI] [PubMed] [Google Scholar]
- 24.Traynor AM, Dubey S, Eickhoff JC, Kolesar JM, Schell K, Huie MS, et al. Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J Thorac Oncol. 2009;4:522–526. doi: 10.1097/jto.0b013e3181952478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blumenschein GR, Jr, Kies MS, Papadimitrakopoulou VA, Lu C, Kumar AJ, Ricker JL, et al. Phase II trial of the histone deacetylase inhibitor vorinostat (Zolinza, suberoylanilide hydroxamic acid, SAHA) in patients with recurrent and/or metastatic head and neck cancer. Invest New Drugs. 2008;26:81–87. doi: 10.1007/s10637-007-9075-2. [DOI] [PubMed] [Google Scholar]
- 26.Modesitt SC, Sill M, Hoffman JS, Bender DP. A phase II study of vorinostat in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2008;109:182–186. doi: 10.1016/j.ygyno.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 28.Yu C, Friday BB, Lai JP, McCollum A, Atadja P, Roberts LR, et al. Abrogation of MAPK and Akt signaling by AEE788 synergistically potentiates histone deacetylase inhibitor-induced apoptosis through reactive oxygen species generation. Clin Cancer Res. 2007;13:1140–1148. doi: 10.1158/1078-0432.CCR-06-1751. [DOI] [PubMed] [Google Scholar]
- 29.Yu C, Dasmahapatra G, Dent P, Grant S. Synergistic interactions between MEK1/2 and histone deacetylase inhibitors in BCR/ABL+ human leukemia cells. Leukemia. 2005;19:1579–1589. doi: 10.1038/sj.leu.2403868. [DOI] [PubMed] [Google Scholar]
- 30.Ciuffreda L, Del Bufalo D, Desideri M, Di Sanza C, Stoppacciaro A, Ricciardi MR, et al. Growth-inhibitory and antiangiogenic activity of the MEK inhibitor PD0325901 in malignant melanoma with or without BRAF mutations. Neoplasia. 2009;11:720–731. doi: 10.1593/neo.09398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tentler JJ, Nallapareddy S, Tan AC, Spreafico A, Pitts TM, Morelli MP, et al. Identification of predictive markers of response to the MEK1/2 inhibitor selumetinib (AZD6244) in KRAS-mutated colorectal cancer. Mol Cancer Ther. 2010;9:3351–3362. doi: 10.1158/1535-7163.MCT-10-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morelli MP, Cascone T, Troiani T, Tuccillo C, Bianco R, Normanno N, et al. Anti-tumor activity of the combination of cetuximab, an anti-EGFR blocking monoclonal antibody and ZD6474, an inhibitor of VEGFR and EGFR tyrosine kinases. J Cell Physiol. 2006;208:344–353. doi: 10.1002/jcp.20666. [DOI] [PubMed] [Google Scholar]
- 33.Kominsky DJ, Klawitter J, Brown JL, Klawitter J, Christians U, Leibfritz D, et al. Abnormalities in glucose uptake and metabolism in imatinib-resistant human BCR-ABL-positive cells. Clin Cancer Res. 2009;15:3442–3450. doi: 10.1158/1078-0432.CCR-08-3291. [DOI] [PubMed] [Google Scholar]
- 34.Serkova NJ, Glunde K. Metabolomics of cancer. Methods Mol Biol. 2009;520:273–295. doi: 10.1007/978-1-60327-811-9_20. [DOI] [PubMed] [Google Scholar]
- 35.Serkova N, Boros LG. Detection of resistance to imatinib by metabolic profiling: clinical and drug development implications. Am J Pharmacogenomics. 2005;5:293–302. doi: 10.2165/00129785-200505050-00002. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Manero G, Yang H, Bueso-Ramos C, Ferrajoli A, Cortes J, Wierda WG, et al. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood. 2008;111:1060–1066. doi: 10.1182/blood-2007-06-098061. [DOI] [PubMed] [Google Scholar]
- 37.Richardson P, Mitsiades C, Colson K, Reilly E, McBride L, Chiao J, et al. Phase I trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) in patients with advanced multiple myeloma. Leuk Lymphoma. 2008;49:502–507. doi: 10.1080/10428190701817258. [DOI] [PubMed] [Google Scholar]
- 38.Chen H, Zhu G, Li Y, Padia RN, Dong Z, Pan ZK, et al. Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 2009;69:9228–9235. doi: 10.1158/0008-5472.CAN-09-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng GZ, Zhang W, Wang LH. Regulation of cancer cell survival, migration, and invasion by Twist: AKT2 comes to interplay. Cancer Res. 2008;68:957–960. doi: 10.1158/0008-5472.CAN-07-5067. [DOI] [PubMed] [Google Scholar]
- 40.Cui X, Kim HJ, Kuiatse I, Kim H, Brown PH, Lee AV. Epidermal growth factor induces insulin receptor substrate-2 in breast cancer cells via c-Jun NH(2)-terminal kinase/activator protein-1 signaling to regulate cell migration. Cancer Res. 2006;66:5304–5313. doi: 10.1158/0008-5472.CAN-05-2858. [DOI] [PubMed] [Google Scholar]
- 41.Price JT, Tiganis T, Agarwal A, Djakiew D, Thompson EW. Epidermal growth factor promotes MDA-MB-231 breast cancer cell migration through a phosphatidylinositol 3′-kinase and phospholipase C-dependent mechanism. Cancer Res. 1999;59:5475–5478. [PubMed] [Google Scholar]
- 42.Scotton CJ, Wilson JL, Milliken D, Stamp G, Balkwill FR. Epithelial cancer cell migration: a role for chemokine receptors? Cancer Res. 2001;61:4961–4965. [PubMed] [Google Scholar]
- 43.Xiong S, Grijalva R, Zhang L, Nguyen NT, Pisters PW, Pollock RE, et al. Up-regulation of vascular endothelial growth factor in breast cancer cells by the heregulin-beta1-activated p38 signaling pathway enhances endothelial cell migration. Cancer Res. 2001;61:1727–1732. [PubMed] [Google Scholar]
- 44.Morelli MP, Brown AM, Pitts TM, Tentler JJ, Ciardiello F, Ryan A, et al. Targeting vascular endothelial growth factor receptor-1 and −3 with cediranib (AZD2171): effects on migration and invasion of gastrointestinal cancer cell lines. Mol Cancer Ther. 2009;8:2546–2558. doi: 10.1158/1535-7163.MCT-09-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein B, Gottfried M. Targeted agents to improve treatment results in colon cancer:bevacizumab and cetuximab. J BUON. 2007;12(Suppl 1):S127–S136. [PubMed] [Google Scholar]
- 46.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 47.Hirsch FR, Varella-Garcia M, Cappuzzo F, McCoy J, Bemis L, Xavier AC, et al. Combination of EGFR gene copy number and protein expression predicts outcome for advanced non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2007;18:752–760. doi: 10.1093/annonc/mdm003. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Zhang H, Yu P, Liu Q, Liu K, Duan H, et al. Effects of matrine against the growth of human lung cancer and hepatoma cells as well as lung cancer cell migration. Cytotechnology. 2009;59:191–200. doi: 10.1007/s10616-009-9211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 50.Cappuzzo F, Varella-Garcia M, Finocchiaro G, Skokan M, Gajapathy S, Carnaghi C, et al. Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients. Br J Cancer. 2008;99:83–89. doi: 10.1038/sj.bjc.6604439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jimeno A, Messersmith WA, Hirsch FR, Franklin WA, Eckhardt SG. KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: practical application of patient selection. J Clin Oncol. 2009;27:1130–1136. doi: 10.1200/JCO.2008.19.8168. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Van Becelaere K, Jiang P, Przybranowski S, Omer C, Sebolt-Leopold J. A role for KRAS in conferring resistance to the MEK inhibitor, CI-1040. Neoplasia. 2005;7:336–347. doi: 10.1593/neo.04532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dry JR, Pavey S, Pratilas CA, Harbron C, Runswick S, Hodgson D, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010;70:2264–2273. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, et al. Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res. 2007;13:1576–1583. doi: 10.1158/1078-0432.CCR-06-1150. [DOI] [PubMed] [Google Scholar]
- 55.Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, et al. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 2006;66:944–950. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 56.Morgillo F, Woo JK, Kim ES, Hong WK, Lee HY. Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer Res. 2006;66:10100–10111. doi: 10.1158/0008-5472.CAN-06-1684. [DOI] [PubMed] [Google Scholar]
- 57.Rieber M, Rieber MS. Signalling responses linked to betulinic acid-induced apoptosis are antagonized by MEK inhibitor U0126 in adherent or 3D spheroid melanoma irrespective of p53 status. Int J Cancer. 2006;118:1135–1143. doi: 10.1002/ijc.21478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.