Abstract
We found that the ingestion of Cryptococcus neoformans by Drosophila melanogaster resulted in the death of the fly but that the ingestion of Saccharomyces cerevisiae or the nonpathogenic Cryptococcus kuetzingii or Cryptococcus laurentii did not. The C. neoformans protein kinase A and RAS signal transduction pathways, previously shown to be involved in virulence in mammals, also played a role in killing Drosophila. Mutation of the Toll immune response pathway, the predominant antifungal pathway of the fly, did not play a role in Drosophila defense following ingestion of the yeast. However, the Toll pathway was necessary for the clearance of C. neoformans introduced directly into the hemolymph of D. melanogaster and for the survival of systemically infected flies.
Fungal infections are a common cause of morbidity and mortality among immunocompromised patients, including human immunodeficiency virus-infected individuals (42). Cryptococcal infections are a particularly common and often fatal complication of immune suppression (21, 24, 42, 52). In immunocompromised patients, cryptococcal infection is caused by Cryptococcus neoformans var. neoformans, while the other variety, C. neoformans var. gattii, is usually associated with infections in immunocompetent individuals in tropical and subtropical areas. However, a significant epidemic caused by C. neoformans var. gattii on Vancouver Island has challenged this understanding, and some believe that this outbreak may represent the emergence of a new disease (50; http://ftp.cdc.gov/pub/infectious diseases/iceid/2002/pdf/starr.pdf).
Studies of the pathogenic mechanisms of C. neoformans have been enhanced by the development of transformation protocols, homologous recombination for genetic manipulations, and the establishment of several host models (7, 17, 28, 38, 53). The most important C. neoformans virulence factors identified to date include the polysaccharide capsule (7, 46) and melanin (5, 14, 22, 44, 45). Signal transduction cascades leading to the production of these C. neoformans virulence factors have also been elucidated (1, 15, 40). These and other studies conducted during the past decade have established Cryptococcus as an important human pathogen and a model yeast for the study of fungal pathogenesis (41).
Recently, the development of nonmammalian host models for Cryptococcus infection has emerged as a promising tool to facilitate the study of C. neoformans pathogenesis. Steenbergen et al. reported the use of the free-living amoeba Acanthamoeba castellanii as a model for the study of C. neoformans survival strategies following ingestion by macrophages (48, 49). These investigators found that C. neoformans was phagocytosed by A. castellanii and that, once intracellular, C. neoformans replicated, eventually killing the amoeba. The process was remarkably similar to the events following the phagocytosis of C. neoformans by mammalian macrophages (37). Recently, it was reported that C. neoformans is ingested by and kills the nematode Caenorhabditis elegans. It was shown that the C. neoformans polysaccharide capsule, as well as several C. neoformans genes previously shown to be involved in mammalian virulence, also play a role in the killing of C. elegans (40).
During the past decade, the well-studied fruit fly Drosophila melanogaster has been extensively used to study the host innate response to microbial pathogens, leading to the discovery of a high degree of conservation in the innate immune signaling pathways between mammals and insects (27). In D. melanogaster, activation of the immune response is regulated by at least two pathways: the Toll pathway and the Imd pathway, two parallel signaling cascades which both contribute to the Drosophila response against microbes (30, 33, 54). Following fungal infection, the Toll receptor on the surface of fat body cells is activated by a cleaved form of a cytokine-like protein, Spatzle (Spz), which is present in the Drosophila hemolymph. The physical interaction between Spz and Toll initiates an intracellular cascade that triggers signal transduction through the threonine-serine kinase Pelle (26, 57). This signal leads to the phosphorylation and degradation of Cactus, the release and subsequent nuclear translocation of the Rel family transcription factors Dorsal and Dif, and the synthesis of antifungal and antibacterial peptides (57). Similarly, the Imd pathway leads to the activation of the Rel family transcription factor Relish (29) and the synthesis of antibacterial peptides. The Drosophila Toll and Imd signaling pathways exhibit striking similarities to the Toll-like receptor and tumor necrosis factor alpha pathways, respectively, which regulate NF-κB activity in vertebrates, suggesting common evolutionary roots (27).
Because recent reports suggest that the virulence factors of C. neoformans involved in mammalian pathogenesis may have evolved as a consequence of the interaction of yeast with environmental predators such as amoebae and nematodes (6) and because D. melanogaster has been a valuable model for the study of host-pathogen interactions (3, 9, 12, 31, 33), we studied the interaction between C. neoformans and D. melanogaster. Here, we show that C. neoformans is a potent pathogen of Drosophila when it is ingested but not when it is injected. By analyzing the role of the Toll and Imd innate immune signaling pathways, we show that the Toll pathway does not appear to play any role in conferring resistance to ingested C. neoformans on D. melanogaster but is necessary for protection against systemic C. neoformans infection of the fly following injection.
MATERIALS AND METHODS
Strains and media.
The C. neoformans strains used in these experiments are listed in Table 1. Cryptococcus laurentii strain ATCC 76483 and Cryptococcus kuetzingii strain ATCC 42276 were obtained from the American Type Culture Collection (ATCC). The sources of the other strains are indicated in Table 1. Yeast cultures were maintained on yeast peptone dextrose (YPD; Difco) agar.
TABLE 1.
Yeast strains used in this study and their characteristicsa
| Strain name (reference) | Relevant characteristic(s) or phenotype(s) | LT50 (h) | P value | LB50 (CFU) |
|---|---|---|---|---|
| C. neoformans | ||||
| H99 ATCC 208821 (25) | Serotype A; clinical isolate; genome sequence being determined | 40 | 5 × 104 | |
| H99 pka1 (15) | PKA1 encodes the major cyclic AMP-dependent protein kinase catalytic subunit; mutant attenuated in mammalians | 90 | <0.001 | 5 × 104 |
| H99 pka1 + PKA1 (15) | Complementation of the pka1 mutant with wild-type PKA1 restored virulence in mammals | 38 | 3 × 104 | |
| H99 ras1 (1, 56) | ras1 mutant is avirulent in animal models of cryptococcal meningitis | 60 | <0.001 | 4 × 104 |
| H99 ras1 + RAS1 (1) | Complementation of the ras1 mutant with wild-type RAS1 restored virulence in rabbits | 35 | 2 × 104 | |
| H99 cap59 (43) | A capsular mutant; CAP59 is essential for capsule formation | 48 | 0.01 | 5 × 103 |
| H99 pkr1 (15) | PKR1 encodes the PKA regulatory subunit; in mice, a pkr1 mutant overproduces capsule and is hypervirulent | 32 | 0.01 | 5 × 103 |
| ATCC 62068 (39) | Serotype A; clinical isolate | 50 | ND | |
| ATCC 34877 (47) | Serotype B/C | 40 | ND | |
| ATCC 36556 (32) | Serotype D; clinical isolate | 44 | ND | |
| ACT::GFP (11) | Serotype A; strain H99 containing the inducible GFP gene fused to the actin promoter | 40 | ND | |
| C. laurentii | ||||
| ATCC 18803 (19) | Environmental isolate | >168 | <0.001 | ND |
| C. kuetzingii | ||||
| ATCC 42276 | Clinical isolate | >168 | <0.001 | ND |
| S. cerevisiae | ||||
| YJM145 (23) | Clinically derived strain | >168 | <0.001 | ND |
| YJM237 (23) | Strain isogenic to sequenced strain S288c | >168 | <0.001 | ND |
Time (LT50) and yeast burden per fly (LB50) when 50% of the flies died are shown for each individual strain. Note that although the LT50 values differ between strains, the LB50 values are very similar for most strains. The survival kinetics P value for each strain (LT50) compared to the H99 strain is also displayed where significant.
The Oregon-R (OR) strain of D. melanogaster was used as the wild type. Stocks and crosses were maintained on a standard cornmeal medium. The imd1 and imd1; spzrm7 lines used in this study have been described (36). The spzrm7 (34) and spz197 (Bloomington stock center) alleles were used to obtain spz mutant adults. All experiments were performed at 25°C.
Infection protocols.
We initially assessed different media for the growth of C. neoformans and other yeasts prior to ingestion by the flies. We found that diluted YPD (1/3 YDP) agar medium (17 g of YPD/liter instead of the generally used concentration of 50 g/liter) enhanced the virulence of C. neoformans compared to regular YPD agar (50 g/liter) or brain heart infusion (BHI) medium (Difco); therefore, for all assays involving infection by ingestion, yeasts were grown on 1/3 YPD agar medium and grown overnight to form a lawn covering the medium surface. For the ingestion assay, 50 μl of an overnight liquid YPD medium culture of C. neoformans, C. kuetzingii, C. laurentii, or Saccharomyces cerevisiae was spread in a fly vial containing 2 ml of 1/3 YPD agar medium. For each experimental condition tested, approximately 80 male flies were divided in three vials and transferred to new vials every 24 h. Results for female flies were qualitatively similar to those for males, although some quantitative differences between genders were evident. Each experiment was performed in duplicate or triplicate; similar results were always obtained, and results of representative experiments are shown in the figures.
Injection assay.
The dorsal part of the fly thorax was pricked with a sharpened needle (100-μm diameter) dipped into pelleted yeast cultures that had been grown overnight at 30°C. Approximately 400 yeast cells were introduced into each fly, as judged by counting CFU immediately after injection (see below). For each experimental condition, 50 to 70 male adults aged 2 to 4 days were inoculated. Similar results were obtained with female flies, although minor quantitative differences between genders were evident. Flies that died within 3 h after injection (less than 5% of the total) were not considered in the analysis.
The STATA 6 statistical software package (Stata, College Station, Tex.) was used to plot killing curves by the Kaplan-Meier method and to estimate statistical differences in fly survival. The time required for 50% of the flies to die (LT50) was calculated with Prism, version 2.00, software (GraphPad) by using the equation Y = Bottom + (Top − Bottom)/[1 + 10(log ET50−X) −Hill slope], where X is the logarithm of the number of hours, Y is the average number of dead flies, ET50 is the 50% effective time, Top and Bottom are the highest and lowest plateaus, respectively, and Hill slope is the degree of curve inclination.
Yeast CFU were enumerated as previously described (8), by plating serial dilutions of homogenates of five adults on YPD plates containing ampicillin, streptomycin, and kanamycin to prevent bacterial contamination. Prior to homogenization, flies ingesting cryptococci were washed by pipetting in water to remove surface yeast. For the enumeration of yeast in injected flies, the inoculum site was dissected away prior to homogenization and culturing to avoid counting organisms embedded in the wound clot; this process involved the removal of ca. one-fourth of the dorsal thorax with a pair of microsurgical scissors. Plates were incubated at 30°C. Each experiment was performed in duplicate, and the standard deviation was calculated.
Extraction of hemolymph for culture.
To assess the presence or absence of yeast in the hemolymph, a 1-μl sample was extracted as follows. First, a hole was made in the dorsal thoracic cuticle with a sharpened metal needle (diameter, 100 mm). Then, the tip of a Pasteur pipette, manipulated in a flame to decrease its diameter, was inserted through the cuticle hole in a left-to-right direction and dorsal enough to avoid the fly midgut. Negative pressure was applied, and the extracted hemolymph was added to 100 μl of phosphate-buffered saline and spread onto plates.
RESULTS
Killing of D. melanogaster through feeding on C. neoformans.
To investigate the interplay between C. neoformans and D. melanogaster, we fed flies with wild-type C. neoformans (H99) grown on various media differing in richness (1/3 YPD, YPD, and BHI media, described in Materials and Methods). Interestingly, we observed Drosophila killing only when C. neoformans was grown on 1/3 YPD medium prior to feeding (Fig. 1A). This result is in agreement with observations that Cryptococcus virulence is enhanced by growth under nutrient-limiting conditions (1, 2).
FIG. 1.
Killing of D. melanogaster following exposure to C. neoformans. (A) Survival of wild-type D. melanogaster (OR) exposed to lawns of C. neoformans (H99) grown on different agar media (1/3 YPD, YPD, and BHI). (B) Survival of wild-type D. melanogaster (OR) exposed to lawns of C. neoformans serotypes A (ATCC 62068), B/C (ATCC 34877), and D (ATCC 36556), C. laurentii (ATCC 18803), C. kuetzingii (ATCC 42276), and S. cerevisiae (YJM 145 or YJM 237) grown on 1/3 YPD agar medium. P values are less than 0.001 for all C. neoformans strains compared to C. laurentii, C. kuetzingii, or either S. cerevisiae strain.
To assess whether different human pathogenic strains of C. neoformans are virulent following ingestion by flies, we used different serotypes of C. neoformans. All serotypes tested killed 100% of the flies within 2 to 3 days (Fig. 1B). In contrast, two nonpathogenic cryptococci, C. laurentii and C. kuetzingii, killed only approximately 25% of the flies over 7 days (Fig. 1B). Two S. cerevisiae strains (YJM145 and YJM237) had no effect on the life spans of the flies (Fig. 1B), confirming that this yeast, which is often used for D. melanogaster maintenance in the laboratory, is nonpathogenic. Importantly, when fed heat-killed cells of C. neoformans (H99), more than 90% of the flies survived over an 8-day period (data not shown), indicating that only live C. neoformans organisms are virulent for D. melanogaster.
Genes associated with pathogenesis in mammals cause enhanced killing of Drosophila by C. neoformans.
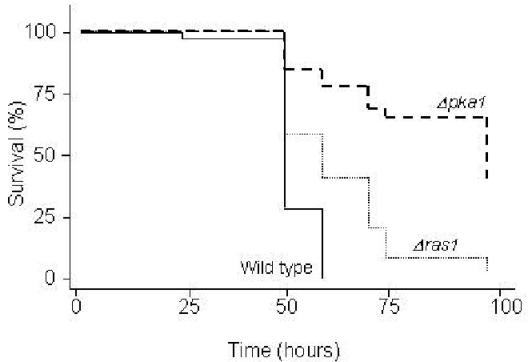
Capsule and melanin production in C. neoformans are regulated by a cyclic AMP-dependent protein kinase A (PKA) signaling pathway. PKA is composed of catalytic and regulatory subunits encoded by the PKA1 and PKR1 genes, respectively (15). Strains in which the PKA1 gene is disrupted display attenuated virulence in murine and Caenorhabditis elegans cryptococcosis infection models, whereas strains with disruptions in the PKR1 gene overproduce capsule and are hypervirulent (15, 16). As shown in Fig. 2, similar to the results obtained for mice and nematodes, the pka1 mutant was significantly less virulent than the isogenic wild type (P < 0.001) or the reconstituted strain, which contains the PKA1 gene integrated into the pka1 mutant genome (2, 15, 16), whereas the pkr1 mutant was hypervirulent (flies fed on pkr1 died 8 h faster than flies fed on the parental strain, H99) (Table 1). In addition to the PKA signaling pathway, another regulatory pathway associated with C. neoformans infection in mammalian hosts involves a RAS1-specific signaling cascade (1). In the D. melanogaster feeding assay, the virulence of a C. neoformans ras1 mutant was also attenuated compared to those of the wild type and a ras1+RAS1 reconstituted strain (Fig. 2 and Table 1). These results suggest that, at least in the case of the PKA and RAS signaling pathways, there is a close correlation between cryptococcal factors required for virulence in three disparate hosts: Caenorhabditis elegans, Drosophila, and mammals (mice and rabbits). The induction of the PKA/PKR and RAS pathways and the formation of a capsule in C. neoformans have previously been associated with growth in minimal media (1, 2). Similarly, in our experiments, C. neoformans strain H99 did not kill flies when grown on a rich medium such as undiluted YPD or BHI agar (Fig. 1A) but did when grown on more minimal media (1/3 YPD).
FIG. 2.
C. neoformans virulence factors for mammalian infection also enhance the killing of D. melanogaster. Shown is the survival of D. melanogaster (OR) flies exposed to wild-type (H99) or mutant C. neoformans. Mutations refer to disruptions in the genes encoding the PKA- or the Ras1-controlled signal transduction cascades. P values are less than 0.001 for each of the mutants compared to the parental strain, H99.
In mammalian hosts, the C. neoformans polysaccharide capsule, which distinguishes C. neoformans from many other pathogenic fungi, protects against phagocytosis and killing by immune effector cells and also blocks the presentation of antigen to T cells and the production of cytokines (4, 13, 18). To determine the role of the capsule in Drosophila killing, we tested cap59, an acapsular derivative of H99 (43). Interestingly, although differences between cap59 and H99 are evident (flies fed on cap59 died 8 h later than flies fed on the parental strain H99 [Table 1]), the cap59 mutation did not cause a major diminution in virulence, indicating that the capsule is not a major virulence factor in the Drosophila-C. neoformans interaction.
Cryptococcal accumulation in the fly following ingestion.
We determined the number of CFU of C. neoformans that had accumulated in the fly following ingestion as described in Materials and Methods. Cryptococcal cells accumulated in wild-type flies, reaching titers of 1.4 × 103, 3.3 × 103, 1.1 × 104, and 5 × 104 after 6, 12, 24, and 48 h of exposure, respectively. We selected time points only up to 48 h because more than 50% of wild-type flies were already dead by that time (Fig. 2). Independent experiments showed that 50% mortality correlated with a fungal burden of 2 × 104 to 5 × 104 per fly. Similar results were obtained with the pka1 and ras1 mutants (Table 1). In contrast, 50% mortality for the cap59 and pkr1 mutants corresponded to a fungal burden of approximately 5 × 103 cells per fly (Table 1).
Role of starvation in killing of Drosophila by C. neoformans.
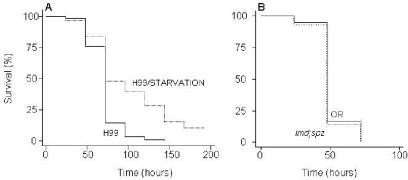
To rule out the possibility that flies feeding on C. neoformans were dying simply due to starvation, because this yeast is a nonnutritional source of food, we compared the survival of flies continuously exposed to C. neoformans with the survival of flies exposed for 18 h per day and starved for the remaining 6 h. Reduction of the exposure of flies to C. neoformans from 24 to 18 h per day yielded significantly longer survival times (Fig. 3A), demonstrating that starvation per se is not the sole mechanism by which Cryptococcus kills the fly.
FIG. 3.
Role of starvation and D. melanogaster immune system in survival following intestinal exposure to C. neoformans. (A) Killing of wild-type (OR) flies feeding continuously on C. neoformans (H99) or feeding for 18 h per day and then starved for the remaining 6 h. (B) Killing rates for wild-type (OR) and mutant (imd; spz) flies following feeding on C. neoformans (H99) are identical. P values are less than 0.001 for the comparison of the curves in panel A.
Role of Drosophila innate immune signaling pathways in defense against ingested C. neoformans.
To evaluate whether previously described innate immune signaling pathways of D. melanogaster are involved in an effective response to the ingestion of C. neoformans, we tested various fly strains mutated in the Imd and/or Toll pathways, the two most prominent pathways dictating immune responses, for survival following the ingestion of C. neoformans. Surprisingly, strains with a mutation in either the Imd or the Toll pathway were not more susceptible to killing than wild-type flies (data not shown). Among the strains tested, imd; spz double mutants were defective in both antifungal and antibacterial responses. Still, these flies survived as well as the wild type (Fig. 3B). This finding shows that either the fly immune system does not provide resistance to C. neoformans following ingestion or C. neoformans effectively blocks the immune response.
Yeast cells are undetectable in fly hemolymph following ingestion of C. neoformans.
The fact that the Drosophila immune system did not affect killing in the C. neoformans ingestion assay suggests that killing may not be a consequence of the systemic growth of C. neoformans in the Drosophila hemolymph. To test this hypothesis, we extracted samples of hemolymph (∼1 μl each) at 24 and 48 h following the initiation of feeding. Analyzing six flies at each time point, we detected virtually no yeast (an average of 0.8 CFU per fly) in the hemolymph. As a control, in parallel experiments (see below) in which ca. 400 CFU of H99 was injected directly into the hemolymph, ca. 20 yeast cells per fly could be recovered 3 h later.
As an alternative approach to detecting the systemic spread of C. neoformans following its ingestion by D. melanogaster flies, we fed flies a strain of C. neoformans that constitutively expresses green fluorescent protein (GFP) (Table 1) (11). Following the ingestion of this strain, flies were examined by fluorescence microscopy at 24 and 48 h. While we detected fluorescence in the fly gastrointestinal tract (this fluorescence could be due to yeast cells and/or the autofluorescence of consumed yeast media), no light emission was detected in any other part of the fly (data not shown). These experiments, in addition to the enumeration of CFU (detailed above) and the finding that wild-type flies are able to clear significant numbers of systemically introduced cryptococci (detailed below), suggest that there is either no systemic spread or very limited systemic spread of yeast cells following the ingestion of Cryptococcus by D. melanogaster flies.
Clearance of systemic infection by C. neoformans depends on the innate immune response of the fly.
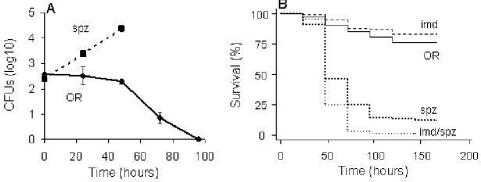
The immune system of Drosophila has proved to be extremely efficient in clearing systemic infections of various fungi (35). To investigate the effectiveness of the innate immune signaling pathways against C. neoformans, we tested whether Drosophila can clear C. neoformans (H99) cells directly injected into the fly hemolymph and survive systemic infection. Following the injection of ca. 400 CFU of C. neoformans into the hemolymph, wild-type flies eliminated yeast cells in 3 to 4 days (Fig. 4A) and most of these flies survived the infection (Fig. 4B); this result indicates that the wild-type fly is capable of clearing a systemic infection. As was the case for wild-type flies, no killing was observed when imd mutant flies were injected with C. neoformans (Fig. 4B) and the imd mutant flies rapidly cleared the yeast (data not shown). In contrast to wild-type and imd mutant flies, however, spz mutant flies and spz; imd double mutants were highly susceptible to C. neoformans killing following injection (Fig. 4B) and C. neoformans multiplied in these mutants following injection (Fig. 4A and data not shown). Taken together, these findings provide evidence that the well-established antifungal pathway of the fly plays a pivotal role following systemic infection with C. neoformans.
FIG. 4.
The D. melanogaster immune response is necessary for clearing systemic C. neoformans infection. (A) Accumulation of C. neoformans within mutant (spz) or wild-type (OR) flies following injection with ∼400 CFU. CFU in surviving flies were enumerated. (B) Survival of wild-type (OR) and mutant (spz, imd, or imd; spz) flies following injection with ∼400 CFU of C. neoformans.
DISCUSSION
D. melanogaster typically feeds on yeast as a natural source of nutrients (51). In this paper, we report the novel observation that strains of the pathogenic yeast C. neoformans kill D. melanogaster, in contrast to the nonpathogenic yeasts C. laurentii, C. kuetzingii, and S. cerevisiae. We excluded the possibility that the killing of flies is simply a consequence of starvation, because flies that were exposed to C. neoformans for a portion of the day and starved for the rest of the day lived significantly longer than flies exposed to C. neoformans continuously. Also, when fed heat-killed cells of C. neoformans (H99), more than 90% of the flies survived over an 8-day period, indicating that C. neoformans can provide adequate nutrients. Hence, it is likely that live C. neoformans organisms cause an infection-like process leading to mortality.
Following its ingestion by D. melanogaster, C. neoformans was restricted to the digestive tract and did not gain access to the hemolymph. Different observations support this notion: (i) we detected few if any yeast cells in extracted hemolymph samples, and (ii) we failed to detect fluorescent cryptococci outside the fly gut. Nevertheless, wild-type flies are able to clear significant numbers of systemically introduced cryptococci; thus, we cannot exclude the possibility that a number of yeast cells adhere to or penetrate into the fly during feeding.
Interestingly, the virulence of C. neoformans for flies was reduced when the yeast was grown on rich media, suggesting that factors inducible in minimal media affect virulence. Indeed, genes associated with the PKA and RAS signal transduction pathways that are known to be induced in minimal media and that were previously shown to be involved in the mammalian virulence of C. neoformans were also shown to play a significant role in the killing of Drosophila by C. neoformans. Despite the fact that the PKA and RAS signaling pathways are involved in capsule production, however, the virulence of the acapsular cap59 mutant was not dramatically reduced in Drosophila, a result that was similar to our findings for Caenorhabditis elegans (40). In addition to capsule formation, the PKA pathway is known to affect melanin production and the alpha mating factor, and RAS1 affects at least the last process as well (1, 16). Because mutations in these pathways have a more severe effect on virulence in Drosophila than the cap59 mutation does, we conclude that PKA- and Ras1-regulated factors unrelated to C. neoformans capsule formation could be primarily responsible for fly killing.
In contrast to the observation that the virulence of the C. neoformans acapsular mutant is not profoundly affected, the observation that the C. neoformans pkr1 mutant, which overproduces capsule, is hypervirulent and lethal in lower doses (Table 1) suggests that the capsule may play a role in the killing of Drosophila. On the other hand, C. laurentii also forms a capsule, and this organism is rarely identified in clinical specimens and is not pathogenic to nematodes or flies. Thus, it is possible that factors unrelated to the capsule, which may also be altered in the pkr1 mutant, are responsible for the hypervirulence of this strain.
Our data suggest that Cryptococcus-mediated killing of Drosophila following ingestion involves a variety of factors relevant to mammalian pathogenesis and is therefore a promising model with which to study fungal pathogenesis. This notion is strengthened by the recent appreciation that C. neoformans pathogenesis may be a consequence of adaptations that have evolved during the interaction of C. neoformans with environmental predators such as free-living nematodes and amoebae and that these adaptations may have selected for virulence factors that also play key roles in human disease (6). Thus, either Drosophila could be a natural predator for C. neoformans, since it is known to feed on decayed fruits in the wild (54), or it may resemble a natural predator, as has been proposed for Caenorhabditis elegans (40).
In Drosophila, fungi and bacteria activate the Toll receptor pathway, which in turn leads to the production of antimicrobial peptides. Gram-negative bacteria primarily activate a parallel pathway (the Imd pathway), which also leads to the induction of antibacterial peptides. Additional evidence indicates that cross talk occurs between the two pathways (10) and that there may be activation of distinct sets of effector molecules in response to different categories of pathogens. In our experiments, the antifungal immune response mediated by the Toll pathway in flies was important in clearing systemic infection by C. neoformans, but neither the Toll nor the Imd pathway protected against killing after the ingestion of C. neoformans. Our results are in agreement with previous work reporting that the expression of the antifungal effector Drosomycin in the fly digestive system is not dependent on Toll signaling (20) and that the Toll-mediated host responses, which are highly expressed in the fat body following systemic infection (34), are apparently absent from the fly intestine (20, 55).
In conclusion, C. neoformans pathogenesis may be a consequence of adaptations that have evolved during the interaction of C. neoformans with environmental predators; our data suggest that the ingestion of C. neoformans by D. melanogaster may help to model certain aspects of mammalian pathogenesis of this yeast. Our data also suggest that there are significant differences in the systemic and intestinal responses of D. melanogaster to pathogens. Comparative studies of Drosophila responses to systemic versus intestinal exposure to C. neoformans may lead to a deeper understanding of host-fungus interactions and innate immunity.
Acknowledgments
We thank Andy Alspaugh, Gary Cox, Jenny Lodge, and John McCusker for generous gifts of strains.
Financial support was provided by the Howard Hughes Medical Institute; by the Pfizer fellowship in medical mycology of the Infectious Disease Society of America; by the Massachusetts General Hospital Center for the Study of Inflammatory Bowel Disease; by the Harvard Medical School Center for AIDS Research; by the New Scholar Award in the Global Infectious Diseases of the Ellison Medical Foundation; by the Charles E. Culpeper Biomedical Pilot Initiative to E.M; by a grant from Aventis SA to F.M.A., L.G.R., and S.B.C.; and by NIH grant GM48707, awarded to F.M.A. The authors have no commercial or other association that might pose a conflict of interest.
REFERENCES
- 1.Alspaugh, J. A., L. M. Cavallo, J. R. Perfect, and J. Heitman. 2000. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36:352-365. [DOI] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basset, A., R. S. Khush, A. Braun, L. Gardan, F. Boccard, J. A. Hoffmann, and B. Lemaitre. 2000. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. USA 97:3376-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, D.C.
- 5.Casadevall, A., A. L. Rosas, and J. D. Nosanchuk. 2000. Melanin and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 3:354-358. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall, A., J. N. Steenbergen, and J. D. Nosanchuk. 2003. ‘Ready made' virulence and ‘dual use' virulence factors in pathogenic environmental fungi—the Cryptococcus neoformans paradigm. Curr. Opin. Microbiol. 6:332-337. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Currie, B. P., L. F. Freundlich, and A. Casadevall. 1994. Restriction fragment length polymorphism analysis of Cryptococcus neoformans isolates from environmental (pigeon excreta) and clinical sources in New York City. J. Clin. Microbiol. 32:1188-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Gregorio, E., P. T. Spellman, P. Tzou, G. M. Rubin, and B. Lemaitre. 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21:2568-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Poeta, M., D. L. Toffaletti, T. H. Rude, S. D. Sparks, J. Heitman, and J. R. Perfect. 1999. Cryptococcus neoformans differential gene expression detected in vitro and in vivo with green fluorescent protein. Infect. Immun. 67:1812-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dionne, M. S., N. Ghori, and D. S. Schneider. 2003. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect. Immun. 71:3540-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doering, T. L. 2000. How does Cryptococcus get its coat? Trends Microbiol. 8:547-553. [DOI] [PubMed] [Google Scholar]
- 14.Doering, T. L., J. D. Nosanchuk, W. K. Roberts, and A. Casadevall. 1999. Melanin as a potential cryptococcal defence against microbicidal proteins. Med. Mycol. 37:175-181. [PubMed] [Google Scholar]
- 15.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Souza, C. A., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349-364. [DOI] [PubMed] [Google Scholar]
- 17.Edman, J. C., and K. J. Kwon-Chung. 1990. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol. Cell. Biol. 10:4538-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldmesser, M., J. Rivera, Y. Kress, T. R. Kozel, and A. Casadevall. 2000. Antibody interactions with the capsule of Cryptococcus neoformans. Infect. Immun. 68:3642-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50:1351-1371. [DOI] [PubMed] [Google Scholar]
- 20.Ferrandon, D., A. C. Jung, M. Criqui, B. Lemaitre, S. Uttenweiler-Joseph, L. Michaut, J. Reichhart, and J. A. Hoffmann. 1998. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 17:1217-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.French, N., K. Gray, C. Watera, J. Nakiyingi, E. Lugada, M. Moore, D. Lalloo, J. A. Whitworth, and C. F. Gilks. 2002. Cryptococcal infection in a cohort of HIV-1-infected Ugandan adults. AIDS 16:1031-1038. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Rivera, J., and A. Casadevall. 2001. Melanization of Cryptococcus neoformans reduces its susceptibility to the antimicrobial effects of silver nitrate. Med. Mycol. 39:353-357. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein, A. L., and J. H. McCusker. 2001. Development of Saccharomyces cerevisiae as a model pathogen. A system for the genetic identification of gene products required for survival in the mammalian host environment. Genetics 159:499-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gumbo, T., G. Kadzirange, J. Mielke, I. T. Gangaidzo, and J. G. Hakim. 2002. Cryptococcus neoformans meningoencephalitis in African children with acquired immunodeficiency syndrome. Pediatr. Infect. Dis. J. 21:54-56. [DOI] [PubMed] [Google Scholar]
- 25.Heitman, J., A. Casadevall, J. K. Lodge, and J. R. Perfect. 1999. The Cryptococcus neoformans genome sequencing project. Mycopathologia 148:1-7. [DOI] [PubMed] [Google Scholar]
- 26.Hetru, C., L. Troxler, and J. A. Hoffmann. 2003. Drosophila melanogaster antimicrobial defense. J. Infect. Dis. 187(Suppl. 2):S327-S334. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann, J. A., and J. M. Reichhart. 2002. Drosophila innate immunity: an evolutionary perspective. Nat. Immunol. 3:121-126. [DOI] [PubMed] [Google Scholar]
- 28.Hua, J., J. D. Meyer, and J. K. Lodge. 2000. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 7:125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hultmark, D. 2003. Drosophila immunity: paths and patterns. Curr. Opin. Immunol. 15:12-19. [DOI] [PubMed] [Google Scholar]
- 30.Khush, R. S., F. Leulier, and B. Lemaitre. 2002. Immunology. Pathogen surveillance—the flies have it. Science 296:273-275. [DOI] [PubMed] [Google Scholar]
- 31.Kirsanova, R. V., M. M. Levitin, L. P. Lekarkina, L. I. Usenko, and V. I. Sharygin. 1975. Drosophila and the entomopathogenic fungus Beauveria bassiana as a model for the study of host and parasite interrelationships. Zh. Obshch. Biol. 36:251-258. (In Russian.) [PubMed] [Google Scholar]
- 32.Kozel, T. R., and J. Cazin, Jr. 1972. Immune response to Cryptococcus neoformans soluble polysaccharide. I. Serological assay for antigen and antibody. Infect. Immun. 5:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau, G. W., B. C. Goumnerov, C. L. Walendziewicz, J. Hewitson, W. Xiao, S. Mahajan-Miklos, R. G. Tompkins, L. A. Perkins, and L. G. Rahme. 2003. The Drosophila melanogaster Toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa. Infect. Immun. 71:4059-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemaitre, B., E. Nicolas, L. Michaut, J. M. Reichhart, and J. A. Hoffmann. 1996. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86:973-983. [DOI] [PubMed] [Google Scholar]
- 35.Lemaitre, B., J. M. Reichhart, and J. A. Hoffmann. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94:14614-14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leulier, F., A. Rodriguez, R. S. Khush, J. M. Abrams, and B. Lemaitre. 2000. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 1:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levitz, S. M., S. Nong, M. K. Mansour, C. Huang, and C. A. Specht. 2001. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 98:10422-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lodge, J. K., E. Jackson-Machelski, D. L. Toffaletti, J. R. Perfect, and J. I. Gordon. 1994. Targeted gene replacement demonstrates that myristoyl-CoA: protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 91:12008-12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell, T. G., and L. Friedman. 1972. In vitro phagocytosis and intracellular fate of variously encapsulated strains of Cryptococcus neoformans. Infect. Immun. 5:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mylonakis, E., F. M. Ausubel, J. R. Perfect, J. Heitman, and S. B. Calderwood. 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 99:15675-15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mylonakis, E., F. M. Ausubel, R. J. Tang, and S. B. Calderwood. 2003. The art of serendipity: killing of C. elegans by human pathogens as a model of bacterial and fungal pathogenesis. Expert Rev. Anti-infect. Ther. 1:89-95. [DOI] [PubMed] [Google Scholar]
- 42.Mylonakis, E., N. A. Merriman, J. D. Rich, T. P. Flanigan, B. C. Walters, K. T. Tashima, M. D. Mileno, and C. M. van der Horst. 1999. Use of cerebrospinal fluid shunt for the management of elevated intracranial pressure in a patient with active AIDS-related cryptococcal meningitis. Diagn. Microbiol. Infect. Dis. 34:111-114. [DOI] [PubMed] [Google Scholar]
- 43.Nelson, R. T., J. Hua, B. Pryor, and J. K. Lodge. 2001. Identification of virulence mutants of the fungal pathogen Cryptococcus neoformans using signature-tagged mutagenesis. Genetics 157:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nosanchuk, J. D., A. L. Rosas, S. C. Lee, and A. Casadevall. 2000. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 355:2049-2050. [DOI] [PubMed] [Google Scholar]
- 45.Nosanchuk, J. D., P. Valadon, M. Feldmesser, and A. Casadevall. 1999. Melanization of Cryptococcus neoformans in murine infection. Mol. Cell. Biol. 19:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perfect, J. R., B. Wong, Y. C. Chang, K. J. Kwon-Chung, and P. R. Williamson. 1998. Cryptococcus neoformans: virulence and host defences. Med. Mycol. 36(Suppl. 1):79-86. [PubMed] [Google Scholar]
- 47.Schmeding, K. 1981. Sexual compatibility between serotypes of Filobasidiella neoformans (Cryptococcus neoformans). Curr. Microbiol. 5:133-138. [Google Scholar]
- 48.Steenbergen, J. N., and A. Casadevall. 2003. The origin and maintenance of virulence for the human pathogenic fungus Cryptococcus neoformans. Microbes Infect. 5:667-675. [DOI] [PubMed] [Google Scholar]
- 49.Steenbergen, J. N., H. A. Shuman, and A. Casadevall. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 98:15245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephen, C., S. Lester, W. Black, M. Fyfe, and S. Raverty. 2002. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can. Vet. J. 43:792-794. [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan, W., M. Ashburner, and R. S. Hawley. 2000. Drosophila protocols, p. 590. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, N.Y.
- 52.Thomas, C. J., J. Y. Lee, L. A. Conn, M. E. Bradley, R. W. Gillespie, S. R. Dill, R. W. Pinner, and P. G. Pappas. 1998. Surveillance of cryptococcosis in Alabama, 1992-1994. Ann. Epidemiol. 8:212-216. [DOI] [PubMed] [Google Scholar]
- 53.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzou, P., E. De Gregorio, and B. Lemaitre. 2002. How Drosophila combats microbial infection: a model to study innate immunity and host-pathogen interactions. Curr. Opin. Microbiol. 5:102-110. [DOI] [PubMed] [Google Scholar]
- 55.Tzou, P., S. Ohresser, D. Ferrandon, M. Capovilla, J. M. Reichhart, B. Lemaitre, J. A. Hoffmann, and J. L. Imler. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13:737-748. [DOI] [PubMed] [Google Scholar]
- 56.Waugh, M. S., C. B. Nichols, C. M. DeCesare, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191-201. [DOI] [PubMed] [Google Scholar]
- 57.Weber, A. N., S. Tauszig-Delamasure, J. A. Hoffmann, E. Lelievre, H. Gascan, K. P. Ray, M. A. Morse, J. L. Imler, and N. J. Gay. 2003. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat. Immunol. 4:794-800. [DOI] [PubMed] [Google Scholar]