Abstract
The Hap4 protein of the budding yeast Saccharomyces cerevisiae activates the transcription of genes that are required for growth on nonfermentable carbon sources. Previous reports suggested the presence of a transcriptional activation domain within the carboxyl-terminal half of Hap4 that can function in the absence of Gcn5, a transcriptional coactivator protein and histone acetyltransferase. The boundaries of this activation domain were further defined to a region encompassing amino acids 359 to 476. Within this region, several clusters of hydrophobic amino acids are critical for transcriptional activity. This activity does not require GCN5 or two other components of the SAGA coactivator complex, SPT3 and SPT8, but it does require SPT7 and SPT20. Contrary to previous reports, a Hap4 fragment comprising amino acids 1 to 330 can support the growth of yeast on lactate medium, and when tethered to lexA, can activate a reporter gene with upstream lexA binding sites, demonstrating the presence of a second transcriptional activation domain. In contrast to the C-terminal activation domain, the transcriptional activity of this N-terminal region depends on GCN5. We conclude that the yeast Hap4 protein has at least two transcriptional activation domains with strikingly different levels of dependence on specific transcriptional coactivator proteins.
Eukaryotic transcriptional activator proteins are typically bipartite in nature, with separable domains for DNA binding and transactivation (49, 73). Transcriptional activation domains (TADs) have historically been classified according to their amino acid composition, with various groups of TADs being rich in acidic or basic residues, glutamine, proline, serine and threonine, or isoleucine (1, 17, 35). However, mutational analyses of a number of TADs have demonstrated that specific patterns of bulky hydrophobic and aromatic amino acids are often more critical for the functioning of the TAD than are the particularly abundant residues (references 72 and 73 and references cited therein). Moreover, activation domains can often be further divided into smaller regions that are each capable of activating transcription independently. For example, the potent activation domain of the herpesvirus VP16 protein can be divided into two regions that can function independently, with each relying on a central cluster of hydrophobic residues (12, 58, 72, 75). The yeast protein Gcn4 relies on multiple overlapping regions containing seven different clusters of hydrophobic residues (13, 33). Collectively, these results and others exploring other activator proteins suggest that activation domains can be composite in nature, containing multiple hydrophobic motifs.
One reason that transcriptional activators possess multiple activation regions could be that different segments of the activation domain interact with different proteins that are necessary for assembly or stimulation of the transcription complex at a gene promoter. Such an ability to interact with numerous targets may be advantageous by allowing a given transcriptional activator to stimulate sets of promoters with intrinsically different rate-limiting steps. For instance, VP16 is reported to interact in vitro with the basal transcription factors TBP (32, 70), TFIIB (43, 44, 60, 61), TFIIA (40), TFIIH (77), and TAF9 (23, 39). Moreover, transcriptional activators can often associate with coactivator protein complexes with enzymatic activities that modulate the chromatin structure. Some of these coactivator complexes can covalently modify histones (and perhaps other proteins) by acetylation, phosphorylation, methylation, ADP-ribosylation, or ubiquitination (for reviews, see references 5, 34, and 69). Other coactivator protein complexes, including the SWI/SNF complex, use energy derived from ATP hydrolysis to alter the positioning of nucleosomes along DNA (for reviews, see references 3, 50, 53, and 71).
In the budding yeast Saccharomyces cerevisiae, a protein complex comprising the Hap2, Hap3, Hap4, and Hap5 proteins positively regulates many of the genes involved in the tricarboxylic acid cycle and oxidative phosphorylation (20, 28, 55, 56; reviewed in references 8, 19, and 41). Hap4 is responsible for the transcriptional activation capability of the Hap2/3/4/5 complex (20) and is not required for the DNA-binding activity (47, 51, 78). Hap2, Hap3, and Hap5 are constitutively expressed, whereas Hap4 expression is strongly induced when any nonfermentable sugar is the sole carbon source (20, 28, 47, 55). Therefore, the activity of this complex transcriptional activator is regulated at least in part by the production of the activation subunit.
The yeast transcriptional coactivator complex known as SAGA comprises the histone acetyltransferase Gcn5, accessory factors Ada2 and Ada3, Tra1, several Spt proteins, and several TBP-associated factors, or TAFs (24, 25, 68). Mutations in the GCN5, ADA1, ADA2, ADA3, and SPT20/ADA5 genes were recovered from a yeast genetic screen for suppressors of the toxic effects of a fusion protein comprising the Gal4 DNA-binding domain and the VP16 TAD (6, 46, 54). Null mutants for these genes fail to support transcriptional activation by VP16 or Gcn4. However, the function of the Hap4 TAD seemed relatively unaffected in such mutants, whether tested with endogenous Hap4-responsive promoters or with a Gal4-Hap4 fusion protein (6, 31). These genetic observations led to the hypothesis that the Hap4 TAD functions by a different mechanism of transcriptional activation that has yet to be fully defined. Paradoxically, however, the TADs of Hap4, VP16, and Gcn5 can each interact directly with Tra1, the largest subunit of the SAGA complex (9, 52). This biochemical evidence for a common mechanism seems discordant with the genetic evidence favoring distinct mechanisms for these various TADs.
As an initial step toward resolving this paradox by defining more explicitly the mechanism of activation by Hap4, we pursued a more complete characterization of the Hap4 TAD. In this pursuit, we discovered the presence of an additional TAD in the N-terminal half of Hap4. Analyses of point mutations constructed in each domain indicated that the activity of each domain was dependent on specific bulky hydrophobic amino acids, consistent with the theme established in studies of other TADs. Intriguingly, the activity of the N-terminal TAD was dependent on Gcn5 activity, whereas the C-terminal TAD was relatively Gcn5 independent. However, the activity of the C-terminal TAD did depend on other components of the SAGA complex, specifically Spt7 and Spt20, but not Spt3 and Spt8. Thus, these results present evidence for multiple distinct regions in Hap4 that activate transcription by apparently distinct mechanisms.
MATERIALS AND METHODS
Yeast strains and media.
Rich and synthetic omission media supplemented with 2% glucose or 2% lactate were prepared by standard methods (66).
The genotypes of yeast strains used in or relevant to this work, insofar as they are known, are listed in Table 1. Strains BWG1-7a and its hap4 derivative SLF401 were used by members of the Guarente laboratory for the discovery and characterization of HAP4 (20, 27). Strain PSY316 was used previously for the analysis of genes encoding the transcriptional coactivators ADA2, ADA3, and GCN5 (9). A hap4 trp1 derivative of PSY316, designated JSY4, was constructed by homologous recombination, using plasmid pKShap4delta (a gift from David McNabb) to disrupt HAP4 and pJL228 (a gift from C. Peterson) to disrupt TRP1. Strain JSY02, with multiple lexA operator sites upstream of both URA3 and a lacZ reporter gene, was obtained by tetrad dissection of a cross between strain RS1254 (a gift from Erik Andrulis and Rolf Sternglanz) and BCY05 (a gift from Barak Cohen and Roger Brent). Strain JSY03 resulted from the introduction of the trp1Δ99 allele into the genome of JSY02 by use of pJL228. The GCN5 gene in JSY03 was disrupted by homologous recombination with a plasmid obtained from Shelley Berger, generating strain JSY05. A hap4 derivative of JSY05, designated JSY45, was obtained by homologous recombination using plasmid pKShap4delta.
TABLE 1.
Genotypes of yeast strains used in or relevant to this work
| Strain | Genotype | Reference or source |
|---|---|---|
| BWG1-7a | MATaade1-100 leu2-3,112 his4-519 ura3-52 | 27 |
| SLF401 | BWG1-7a hap4::LEU2 | 20 |
| PSY316 | MATα ade2-101 his3-Δ200 leu2-3,112 lys2 ura3-53 | 6 |
| JSY4 | PSY316 hap4Δ trp1Δ99 | This work |
| BCY05 | MATα ura3 trp1 his3 leu2::(lexAop)6-LEU2 lys2::(lexAop)8-URA3 (derived from EGY48) | B. Cohen and R. Brent, personal communication |
| RS1254 | MATahis3Δ200 trp1-901 leu2-3,112 ade2 lys2-801am URA3::(lexAop)8-lacZ LYS2::(lexAop)4-HIS3 | E. Andrulis and R. Sternglanz personal communication |
| JSY02 | MATα ade2 his3 leu2-3,112 trp URA3::lexA8op-lacZ lys2::lexA8op-URA3 | This work |
| JSY03 | JSY02 trp1Δ99 | This work |
| JSY05 | JSY03 gcn5Δ | This work |
| JSY45 | JSY03 gcn5Δhap4Δ | This work |
| L40 | MATahis3Δ200 trp1-901 leu2-3,112 ade2 lys2-801am URA3::(lexA)8-lacZ LYS2::(lexA)4-HIS3 | 29 |
| FW294 | MATahis4-917δ lys2-173R2 trp1Δ63 leu2Δ1 ura3-52 spt3Δ202 | 21 |
| FW463 | MATahis4-917δ lys2-173R2 trp1Δ63 leu2Δ1 ura3-52 spt8Δ302::LEU2 | 16 |
| FY1093 | MATahis4-917δ lys2-173R2 trp1Δ63 leu2Δ1 ura3-52 ade8 spt7Δ402::LEU2 | 63 |
| FY1106 | MATα his4-917δ lys2-173R2 trp1Δ63 leu2Δ1 ura3-52 spt20Δ100::URA3 | 63 |
Yeast expression plasmids for full-length and truncated Hap4 proteins.
The full-length Hap4 open reading frame (codons 1 to 554) with flanking promoter and terminator sequences from ADC1 was cloned into pRS414 as a BamHI restriction fragment from pDB20-Hap4 (a gift from Leonard Guarente). The truncated Hap4 gene was generated by PCR amplification followed by initial cloning into pDB20 and thence into pRS414 on a BamHI restriction fragment with the ADC1 promoter and terminator.
Yeast expression plasmids for LexA-Hap4 fusion proteins.
DNA fragments encoding various fragments of Hap4 were amplified by PCR and ligated into the NotI restriction site adjacent to the lexA open reading frame in pDB20L (6). Subsequently, DNA fragments encoding the various LexA-Hap4 fusion proteins flanked by the ADH1 promoter and terminator were subcloned into pRS414, a low-copy-number plasmid carrying the TRP1 selectable marker (67), as XmaI-SalI, SacI-SalI, or BamHI fragments. PCR-amplified sequences and cloning junctions were verified by DNA sequencing.
β-Galactosidase assays.
For most experiments, plasmids expressing LexA-Hap4 fusions were transformed into yeast strains JSY03 or L40, both of which have an integrated lacZ gene with eight lexA binding sites upstream. For one experiment, these LexA-Hap4 expression plasmids were cotransformed with high-copy-number plasmids bearing the (lexAop)8-lacZ reporter gene into yeast strains bearing disruptions of individual SPT genes. Cultures were grown in selective medium (8 ml) to an optical density at 595 nm (OD595) of 0.5. β-Galactosidase assays were performed as described previously (26, 48). Three independent assays were performed for each mutant. For the assessment of expression levels of LexA-Hap4 fusion proteins, cell lysates from log-phase cultures were assayed by immunoblotting with a mixture of three mouse monoclonal antibodies raised against LexA (2 μg of YN-lexA-2-12/ml, 4.3 μg of YN-lexA-6-10/ml, and 2 μg of YN-lexA-16-7/ml; Y. Nedialkov and S. J. Triezenberg, unpublished data). Blots were developed by use of enhanced chemiluminescence systems (ECL [Amersham] or Renaissance [DuPont NEN Life Science Products] system).
Construction of substitution mutants.
Clustered point mutations within HAP4 activation domains were constructed by use of a QuikChange mutagenesis kit (Stratagene, La Jolla, Calif.). The mutations were introduced directly into low-copy-number TRP1-marked plasmids expressing particular LexA-Hap4 fusion proteins (described above). The presence of the intended base pair changes and the absence of spurious secondary mutations were confirmed by DNA sequence analysis.
Growth curves.
For comparisons of the ability of Hap4 amino acids (aa) 1 to 330 to support growth on a nonfermentable carbon source with that of full-length Hap4 (aa 1 to 554), yeast strain JSY4 (hap4Δ) or JSY45 (hap4Δ gcn5Δ) was transformed with a plasmid encoding either the wild-type or truncated Hap4 protein. Transformants were initially grown to saturation (OD595 > 1.5) in selective medium supplemented with glucose. A 100-ml culture of selective medium with 2% lactate was inoculated with 0.2 ml of the saturated culture. Growth was monitored by measuring the OD595.
RESULTS
Characterization of an N-terminal activation domain in Hap4.
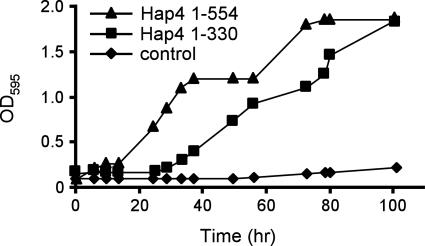
A previous analysis of deletion mutants of HAP4 identified the region comprising aa 330 to 554 as harboring the TAD of Hap4 (20). In that work, carried out using a hap4 derivative of the yeast strain BWG1-7a, the amino-terminal region of Hap4 (aa 1 to 330) was unable to support growth on a nonfermentable carbon source (20). We performed a comparable experiment in a hap4 derivative of strain PSY316 (6). Figure 1 shows that a plasmid expressing the truncated form of Hap4 (aa 1 to 330) was indeed able to support growth on a nonfermentable carbon source significantly better than did an empty vector plasmid, albeit more slowly than did a plasmid expressing the full-length Hap4 protein. Given the assumption that transcriptional activation by Hap4 is important for growth under these conditions, these results imply the existence of an activation domain within Hap4 residues 1 to 330.
FIG. 1.
Truncated Hap4 can support yeast growth in lactate medium. A yeast strain (JSY4) bearing a chromosomal disruption of HAP4 was transformed with TRP1-marked ARS/CEN plasmids encoding either full-length Hap4(1-554), truncated Hap4(1-330), or no Hap4 protein. Densities of the yeast cultures (measured as OD595) were monitored after a saturated culture was diluted into synthetic medium lacking tryptophan and containing 2% lactate as the sole carbon source.
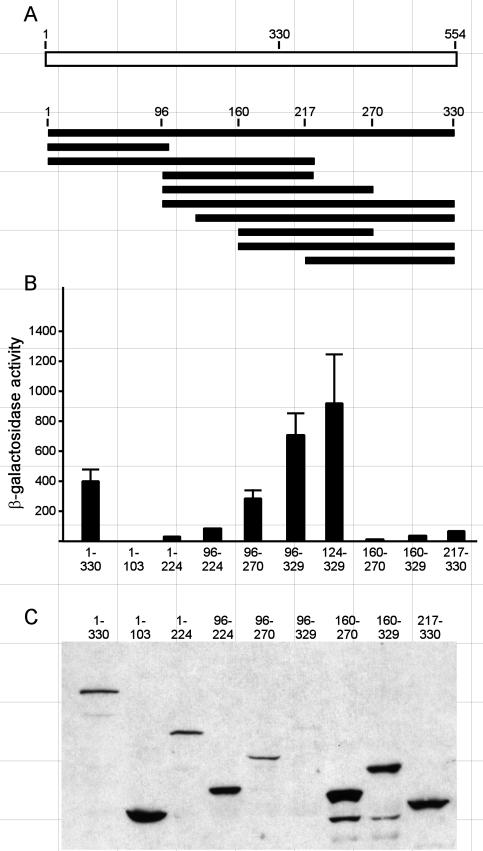
To more precisely define the boundaries of the putative activation domain resident within the aa 1 to 330 region of Hap4, we constructed a series of plasmids to express various portions of Hap4 fused to the DNA-binding domain of the Escherichia coli LexA protein. The LexA-Hap4 fusion proteins were expressed in yeast from a constitutive promoter on a low-copy-number plasmid. The ability of these fusion proteins to activate transcription was assessed by using a chromosomally integrated β-galactosidase (lacZ) reporter gene expressed from a basal promoter with eight lexA binding sites positioned upstream of the transcriptional start site. The β-galactosidase enzyme activity was assayed in crude lysates of yeast cells grown in selective medium to mid-log phase. The results shown in Fig. 2 indicate that the LexA-Hap4(1-330) fusion protein effectively activated expression of the lacZ reporter gene, strengthening the conclusion that this region of HAP4 encompasses a TAD. Deletions of the N terminus to residue 96 or 124 resulted in modest increases in reporter gene activity, but further deletion to aa 160 or 217 resulted in an almost complete loss of activation potential. Thus, the N-terminal boundary of the activation domain present within the region of aa 1 to 330 falls between aa 124 and 160. Carboxyl-terminal deletions to residue 224 or 103 completely destroyed the transcriptional activity of this region. A Hap4 fragment comprising residues 96 to 270 retained much of the ability to activate transcription, and this was diminished markedly by an amino-terminal deletion to aa 160 or a carboxyl-terminal deletion to aa 224. Thus, the region comprising aa 96 to 270 represents the smallest functional fragment that we tested that retained the transcriptional function. Immunoblot analysis using antibodies specific for the LexA DNA-binding domain revealed that the poorly activating deletion mutant proteins were expressed at levels equal to or higher than the levels of the more effective proteins, indicating that the loss of activity could not be attributed to a lack of expression of the corresponding protein. In fact, the protein levels tended to negatively correlate with transcriptional activity for this set of Hap4 deletion mutants.
FIG. 2.
Transcriptional activation by fragments of the Hap4 N-terminal domain (aa 1 to 330). (A) Schematic diagram of the Hap4 regions that were fused to LexA and expressed from the ADH1 promoter on TRP1-marked ARS/CEN plasmids. The host cells (JSY03) contained an integrated lacZ reporter gene with eight upstream lexA binding sites. (B) β-Galactosidase activity was measured in extracts of three parallel cultures grown to mid-log phase and is reported as nanomoles of o-nitrophenol per minute per milligram of total protein (means ± standard deviations). β-Galactosidase activities in extracts of cells expressing only LexA were negligible. (C) Immunoblot of lysates from cells expressing various LexA-Hap4 fusion proteins, detected with a cocktail of monoclonal antibodies specific for the LexA DNA-binding domain.
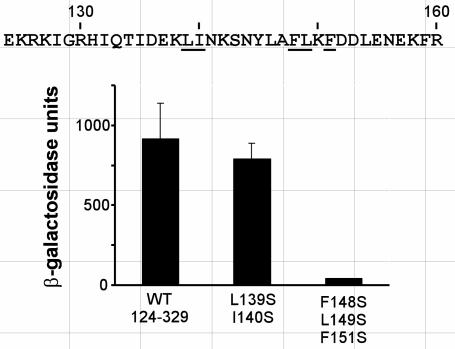
Mutational analyses of the “acidic” TADs of VP16 (12, 58, 72), p53 (30), and Gcn4 (13, 33) have indicated that the activities of these domains are dependent on specific patterns of hydrophobic amino acids. We hypothesized that the transcriptional activation function of the aa 124 to 329 region of Hap4 was also dependent on hydrophobic amino acids. Given that the N-terminal boundary of this region was mapped to the interval between residues 124 and 160, we focused a search for clusters of important hydrophobic amino acids to that interval. For each of two such clusters, point mutations were introduced to replace amino acids having bulky hydrophobic side chains with serine, which has a small hydrophilic side chain, in the context of the LexA-Hap4(124-329) fusion protein. The abilities of these mutant fusion proteins to activate the lacZ reporter gene are shown in Fig. 3. The combined Ser substitutions at Leu139 and Ile140 had little effect on the activity of the aa 124 to 329 region of Hap4, whereas Ser substitutions for Phe148, Leu149, and Phe151 resulted in a dramatic loss of activity. We concluded that the cluster of hydrophobic amino acids near position 150 is important for the transcriptional function of the N-terminal TAD.
FIG. 3.
Specific hydrophobic amino acids are essential for transcriptional activation by the Hap4 aa 124 to 329 fragment. The amino acid sequence of Hap4(124-160) is shown in single-letter code; underlined positions show sites of serine substitutions. LexA-Hap4(124-329) fusion proteins, with or without the indicated substitutions, were expressed in yeast strain L40 and assayed for activation of the integrated lacZ reporter gene as described for Fig. 2. The histograms indicate means (with standard deviations) of β-galactosidase activity in yeast cell extracts from three parallel cultures.
Characterization of the Hap4 C-terminal activation domain.
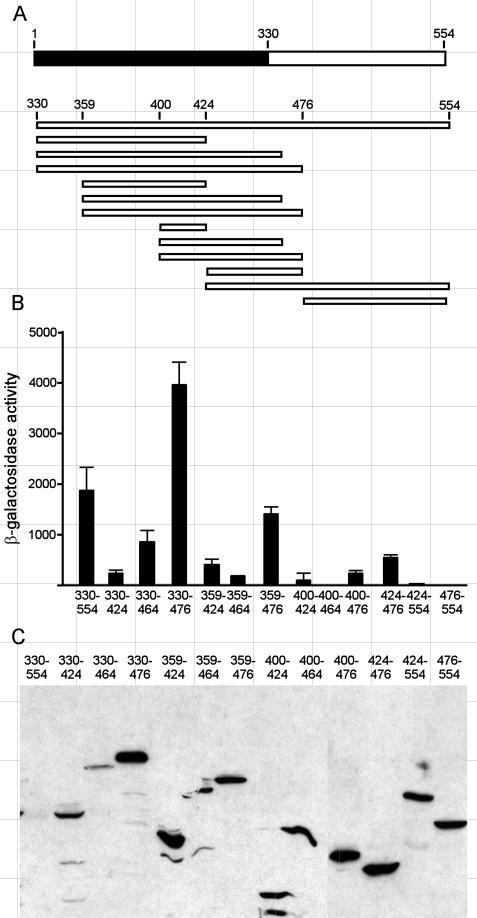
Previous work indicated the presence of a TAD within the aa 330 to 554 region of Hap4 (20). To more narrowly define the boundaries of this domain, we expressed various portions of this region as LexA fusion proteins and assayed them in yeast for the ability to activate expression of the lacZ reporter gene with upstream lexA binding sites. As shown in Fig. 4, deletion of the C-terminal residues to position 476 resulted in an approximate doubling of the β-galactosidase activity, suggesting that an inhibitory activity within the C-terminal 80 aa was lost with the deletion. Further deletions to positions 464 or 424 resulted in a considerable loss of reporter gene expression, suggesting that a C-terminal boundary of sequences important for strong transcriptional activity resides between aa 476 and 464. Hap4 fragments with N termini at residue 359 or 400 and various C termini were also tested. In each case, the activity of the fragment extending to residue 476 was much stronger than the corresponding fragment extending to residue 464 or 424, reinforcing the conclusion that a C-terminal boundary lies between aa 476 and 464. Moreover, fragments with N termini at position 359 had stronger activities than corresponding fragments with N termini at residue 400, indicating that an N-terminal boundary for high-level transcriptional activity resides between positions 359 and 400. Several nonoverlapping fragments displayed discernible activities, including a region from aa 359 to 424 and another comprising residues 424 to 476, suggesting that discrete segments in this region are sufficient for activating transcription. The C-terminal 80 aa had no intrinsic activity, but they did depress the activities of adjacent activating fragments (compare the region of aa 424 to 476 to aa 424 to 554), supporting the earlier suggestion that an inhibitory activity resides in the C-terminal 80 aa. Immunoblot assays (Fig. 4C) indicated that all of the LexA fusion proteins bearing portions of the C-terminal region of Hap4 were present at detectable and roughly comparable levels, except the largest fragment (aa 330 to 554), which was barely detectable.
FIG. 4.
Transcriptional activation by fragments of the Hap4 C-terminal domain (aa 330 to 554). (A) Schematic diagram depicting Hap4 fragments that were fused to LexA and expressed from the ADH1 promoter on TRP1-marked ARS/CEN plasmids in yeast strain JSY03. (B) β-Galactosidase activities were measured in extracts of three parallel log-phase cultures as described for Fig. 2; means and standard deviations are reported. (C) Immunoblot of lysates from cells expressing various LexA-Hap4 fusion proteins, detected with monoclonal antibodies specific for the LexA DNA-binding domain.
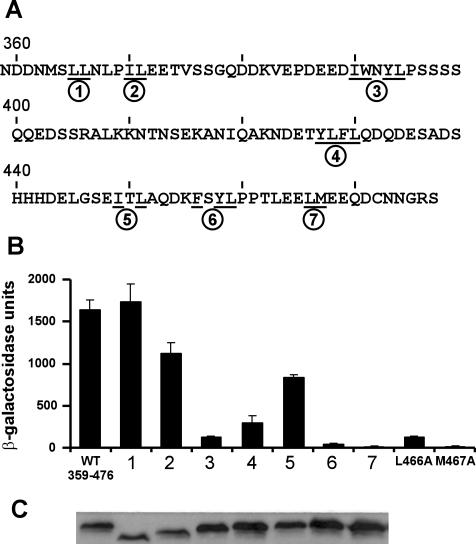
Given that the Hap4 segment from residues 359 to 476 was the smallest fragment with transcriptional activity approximating that of the entire Hap4 C terminus, we next tested whether the activity of this region depended on specific clusters of bulky hydrophobic amino acids. Within the aa 359 to 476 region of Hap4, we identified seven different clusters of hydrophobic amino acids: L365/L366, I370/L371, I390/W391/Y393/L394, Y427/L428/F429/L430, I450/L452, F456/Y458/L459, and L466/M467, designated clusters 1 to 7 in Fig. 5A. We hypothesized that some of these clusters would be essential for this region of Hap4 to activate transcription. Therefore, mutants were generated [in the context of the LexA-Hap4(359-476) construct] in which each hydrophobic amino acid within any given cluster was replaced with serine. The effects of these mutations on the ability of cells to activate the LexA-driven lacZ reporter gene were tested as described above. The results shown in Fig. 5B reveal that the mutations in cluster 4 resulted in a 5-fold reduction in activity and the mutations in clusters 3, 6, and 7 resulted in a >10-fold reduction in activation potential. Interestingly, cluster 3 lies within the region (aa 359 to 400) encompassing the N-terminal boundary of transcriptional activity, as defined with deletion mutants, and cluster 7 falls within the region (aa 464 to 476) encompassing the C-terminal boundary (Fig. 2). In contrast, multiple serine mutations in three of the hydrophobic clusters (mutants 1, 2, and 5) had little or no effect on transcriptional activation by Hap4(359-476). An immunoblot of cell extracts expressing the various substitution mutant proteins indicated that all of these proteins, including those showing diminished expression of the reporter gene, were expressed at comparable levels (Fig. 5C).
FIG. 5.
Specific hydrophobic amino acids are essential for transcriptional activation by the Hap4 aa 359 to 476 fragment. (A) Amino acid sequence of Hap4(359-476) shown in single-letter code. Underlined positions indicate sites of serine substitutions; circled numbers identify the seven multiple-substitution mutants. (B) LexA-Hap4(359-476) fusion proteins, with or without the indicated substitutions, were expressed in L40 and assayed for activation of the integrated lacZ reporter gene as described for Fig. 2. The histograms indicate means (with standard deviations) of β-galactosidase activity in yeast cell extracts from three parallel cultures. (C) Immunoblot of cell lysates expressing LexA-Hap4 fusion proteins (wild-type or substitution mutants 1 through 7) probed with a panel of monoclonal antibodies recognizing the LexA DNA-binding domain. Mutant proteins 1 and 2 consistently showed slightly retarded mobility by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
We were particularly interested in mutant 7, since this mutant Hap4 fragment had the lowest transcriptional activity yet bore substitutions of only two amino acids (L466S/M467S). Moreover, these hydrophobic amino acids are flanked by acidic amino acids, as is typically found in TADs. Therefore, individual mutations of L466 and M467 to alanine were constructed and tested as LexA-Hap4(359-476) proteins. The L466A mutation caused a 10-fold reduction in transcriptional activity, and the M467A mutation completely abolished this activity (Fig. 5). Thus, M467 is an essential residue for the transcriptional activity of this segment of Hap4. An immunoblot analysis revealed comparable expression levels for the wild-type and mutant fusion proteins (data not shown), and thus the differences in transactivation potential cannot be attributed to differences in the expression or stability of the mutant proteins.
Given that residues 148 to 151 seem to be key elements of the N-terminal activation domain and that residues 466 to 467 seem to be key elements of the C-terminal activation domain when tested as LexA fusion proteins, we tested whether these mutations were sufficient to disrupt transcriptional activation in the native Hap4 protein. In a hap4 deletion strain, we ectopically expressed Hap4 variants bearing the clustered Ser substitutions affecting either residues 148 to 151, residues 466 to 467, or both sets of changes. Yeast cells expressing these variants showed no defect in growth on lactate plates and no significant change in the activation of a Hap4-responsive lacZ reporter gene (data not shown). We concluded that although the residues identified in our mutational analysis may be important for the function of the minimal activation regions, the Hap4 protein in its entirety has sufficient redundancy to retain its transcriptional function even when those sites are altered.
The Hap4 region from aa 124 to 329 is a GCN5-dependent activation domain.
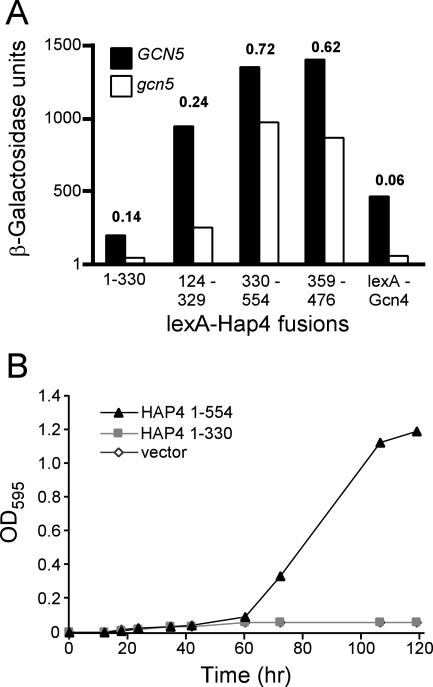
The ability of Hap4 to activate transcription is less dependent on Ada2, Ada3, and Gcn5 than are other transcriptional activators, such as Gcn4 and VP16 (6, 22, 31, 54). In an effort to assign this GCN5 independence to one or the other of the two Hap4 activation domains defined above, we tested the ability of various LexA-Hap4 fusion proteins to function as transcriptional activators in the gcn5Δ strain JSY05. As shown in Fig. 6A, the activity of the N-terminal activation domain (aa 1 to 330 or 124 to 329) was markedly reduced in the gcn5 null mutant. In contrast, the C-terminal activation domain (aa 330 to 554 or 359 to 476) was only modestly affected by the absence of Gcn5. As an additional assay of the GCN5 dependence of the N-terminal domain of Hap4, we tested whether the region from aa 1 to 330 of Hap4 would be able to support growth on a nonfermentable carbon source in the absence of Gcn5. Plasmids expressing either full-length Hap4 or the N-terminal domain only (aa 1 to 330) were transformed into the gcn5Δ hap4Δ double deletion strain JSY45. Transformants were grown in selective glucose medium and then diluted into selective lactate medium, and the density of the culture was monitored for several days. As shown in Fig. 6B, Hap4(1-330) was unable to support growth in this medium whereas Hap4(1-554) was able to support growth. Thus, it appears that the N-terminal region of Hap4 harbors a GCN5-dependent activation domain whereas the C-terminal region harbors a GCN5-independent activation domain.
FIG. 6.
Transcriptional activities of various Hap4 fragments are differentially affected by gcn5 deletion. (A) The indicated Hap4 fragments fused to LexA were assayed for the ability to activate expression of a (lexAop)8-lacZ reporter gene in yeast strains JSY03 (GCN5) and JSY05 (Δgcn5). Bars indicate mean β-galactosidase activities in yeast cell extracts from three parallel cultures. The calculated relative values of activity in JSY05 compared to that in JSY03 are indicated for each Hap4 fragment. (B) Hap4(1-330) fails to support growth in lactate medium in gcn5 yeast. Growth curves were monitored as described for Fig. 1, using yeast strain JSY45 (hap4 gcn5). The relative activities of various Hap4 fragments in JSY05 compared to JSY03 are indicated above each pair of histograms.
The transcriptional activity of Hap4(330-554) requires SPT7 and SPT20 but not SPT3 and SPT8.
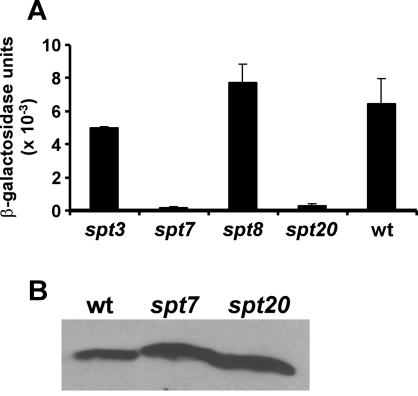
Gcn5 is a component of at least three multiprotein complexes in yeast, designated ADA, SAGA, and SLIK (15, 24, 25, 57). Several sets of proteins are present in the SAGA complex but not in the ADA complex, including four Spt proteins (Spt3, -7, -8, and -20) and some TAFs (TAFs 5, 6, 9, 10, and 12). These proteins apparently have functions in addition to their roles in the SAGA complex, but the precise nature of those functions remains to be defined. We tested the ability of LexA-Hap4(330-554) to activate transcription in yeast strains that lacked SPT3, SPT7, SPT8, or SPT20 (Table 1). For these experiments, the LexA-driven lacZ reporter gene was introduced in a high-copy-number plasmid, resulting in more β-galactosidase activity than in previous experiments. Deletion of SPT3 or SPT8 had little or no effect on the transcriptional activity of LexA-Hap4(330-554) (Fig. 7A). In contrast, deletion of SPT7 or SPT20 completely abolished that activity. Immunoblots showed that the levels of LexA-Hap4 protein were not significantly diminished in the spt7 or spt20 strains (Fig. 7B). Therefore, the SPT7 and SPT20 gene products represent potential mechanistic targets of Hap4 aa 330 to 554.
FIG. 7.
Disruptions of some SPT genes abolish transcriptional activation by the C-terminal domain of Hap4. A low-copy-number plasmid expressing the LexA-Hap4(330-554) fusion protein was transformed together with a high-copy-number plasmid carrying a (lexAop)8-lacZ reporter gene into yeast strains bearing disruptions of either SPT3, SPT7, SPT8, or SPT20. (A) Bars indicate mean β-galactosidase activities (with standard deviations) in yeast cell extracts from three parallel cultures. (B) Immunoblot of cell lysates from wild-type, spt7, and spt20 strains probed with monoclonal antibodies recognizing the LexA DNA-binding domain.
DISCUSSION
The molecular genetic studies presented in this report help define two TADs within the yeast Hap4 protein that can function independently and suggest that these two domains may function by distinct mechanisms. One activation domain had been previously mapped to the C-terminal half of Hap4 (residues 330 to 554) (20). We have more narrowly defined the boundaries of this domain to the region within residues 359 to 476, and in that region we identified several clusters of hydrophobic amino acids critical to this function. We also identified a previously unknown transcriptional activation function in the N-terminal half of Hap4 (aa 1 to 330). A variant of Hap4 was able to support yeast growth in medium containing a nonfermentable carbon source, contrary to a previous report (20). Within this region, too, we identified a cluster of hydrophobic amino acids critical to the function of this activation domain.
The two activation domains that we have identified appear to function through different mechanisms and may interact with different target proteins. For the Gcn5-dependent N-terminal TAD of Hap4, those targets are likely to be within the Gcn5-containing coactivator complexes SAGA or Ada (24, 25, 68). Although early studies implicated the Ada2 protein as a potential interaction target for activation domains (2), more recent reports suggested that the very large Tra1 protein makes direct contact with the activation domains of Gcn4 and VP16 as well as the C-terminal domain of Hap4 (9, 52). The N-terminal domain of Hap4 has not been tested for direct interactions with Tra1 or Ada2.
The case for a role of the SAGA complex in activation by the C-terminal activation domain of Hap4 remains somewhat ambiguous. This domain of Hap4 has been shown to directly interact with Tra1, as noted above, but Tra1 is also present in the NuA4 protein complex that includes the histone acetyltransferase (HAT) enzyme Esa1. Thus, one explanation could be that Hap4 has separate domains for recruiting two different HAT complexes. Paradoxically, the activity of the C-terminal Hap4 domain requires several proteins that are known to be associated with the SAGA complex (Spt7 and Spt20), but not others (Gcn5, Spt3, and Spt8). Distinctive functions of Spt3 and Spt8, but not Spt7 and Spt20, on the function of TATA-binding protein have been previously noted (4, 16). Moreover, spt7 and spt20 mutant alleles show synthetic lethality with mutant alleles affecting the SWI/SNF complex or the Srb/mediator complex (62). These results could suggest that the Spt7 and Spt20 proteins (but not the Spt3 and Spt8 proteins) are components of an additional, as yet unidentified complex on which Hap4C activity depends. These Spt proteins are not known to be present in the NuA4 complex implicated by the Tra1 interaction. The notion that the proteins found in the SAGA complex do not act monolithically is further supported by evidence that the Gcr1 and Gcn4 activators require Spt3 and Spt20 but not Gcn5 to activate a reporter gene (14). Moreover, repression of the Arg1 promoter requires some SAGA components (Gcn5, Ada2, Spt7, and Spt8), but not others (notably Spt3) (59). Another possibility is suggested by the observation that deletion of Spt7 or Spt20 destabilizes the SAGA complex (68). Thus, Hap4C activity may require an intact SAGA complex but not its only known catalytic component, Gcn5. These hypotheses can be addressed genetically in future work by seeking suppressors of the substitution mutants of Hap4C or biochemically by using affinity tagging strategies to isolate and identify proteins that physically interact with the Hap4C domain.
For deletion variants of the N-terminal activation domain of Hap4, we noted a negative correlation between transcriptional activity and protein abundance. This suggests that this region of Hap4 is both a TAD and a signal for protein degradation. A similar association has been established for the activation domains of VP16 and c-myc (65). Moreover, for certain activators, specific E3 ubiquitin ligases have been identified as important coactivator proteins for particular activators (38, 64, 74), leading to the hypothesis that ubiquitination, and perhaps protein degradation, is inherently coupled to mechanisms of transcriptional activation (11, 45). Curiously, this correlation was not observed for deletion variants of the C-terminal activation domain, suggesting that it may function by a ubiquitin-independent mechanism.
Homologs of HAP4 have now been identified in the genomes of a number of yeast species, including Saccharomyces sensu stricto species that are closely related to S. cerevisiae (including S. bayanus, S. mikatae, and S. paradoxus) (10, 37), two Saccharomyces species that are more distantly related (S. castellii and S. kluyveri) (10), and one species from a different genus, Kluyveromyces lactis (7). As expected, the predicted amino acid sequences for Hap4 homologs from the sensu stricto species are very similar to each other. An alignment that includes the more distant homologs revealed three well-conserved regions in these proteins (Fig. 8). One conserved motif, corresponding to residues 64 to 79 of S. cerevisiae Hap4, includes amino acids that are essential for the interaction of Hap4 with the DNA-binding complex of Hap2, Hap3, and Hap5 (7). A second conserved motif corresponds to the N-terminal activation domain identified by our mutational analysis (residues 140 to 155 in S. cerevisiae). Although no clear similarity was evident for the S. castellii or K. lactis homologs, the Hap4 protein from S. kluyveri contained the core motif AFLKFEE that was identified as a key element by our point mutation analysis (Fig. 3). The third element corresponds to residues 452 to 472 of S. cerevisiae Hap4, with a motif of PTLEELMEEQ that is strongly conserved even in the distantly related homologs. Our deletion mutations mapped a boundary of the C-terminal activation domain to this region (Fig. 4). Moreover, double or single amino acid substitutions at positions 458, 459, 466, and 467 dramatically reduced the activity of this domain (Fig. 5). This conservation may point further to the importance of this region in the transcriptional activities of Hap4.
FIG. 8.
Localized sequence similarities among Hap4 homologs in yeasts. Selected portions of amino acid sequences of Hap4 homologs from S. cerevisiae (Scer), S. bayanus (Sbay), S. mikatae (Smik), S. paradoxus (Spar), S. castellii (Scas), S. kluyveri (Sklu), and K. lactis (Klac) were aligned based in part on an alignment displayed at the SGD website (http://db.yeastgenome.org/cgi-bin/FUNGI/nph-showAlign?locus = YKL109W). Numbers refer to amino acid residues; no number is assigned for the homolog from S. kluyveri because this open reading frame has not been rigorously defined. (A) Similarity in a domain of Hap4 that interacts with the Hap2, Hap3, and Hap5 DNA-binding protein complex. (B) Similarity in the region defined in this report as the N-terminal activation domain. Bold underlined letters represent residues which, when mutated, affected the activity of LexA-Hap4(124-329). (C) Similarity in the region defined in this report as the C-terminal activation domain. Bold underlined letters represent residues which, when mutated, affected the activity of LexA-Hap4(359-476).
Regulators such as Hap4 with multiple output mechanisms may be particularly capable of regulating the expression of diverse types of responsive promoters that are challenged by different levels of inhibition by chromatin. The use of such parallel strategies need not be limited to transcriptional activators such as Hap4 and VP16, but may also be appropriate for repressors. For example, the yeast Ume6 protein silences the early meiosis-specific genes by recruiting two different corepressors, the Sin3p-Rpd3p complex and the Isw2p complex (18, 36, 42). Genomic analysis of gene expression and of the physical association of regulators with promoters will likely be useful for testing this hypothesis (18, 76).
Acknowledgments
We thank Fred Winston, Shelley Berger, Barak Cohen, Roger Brent, Erik Andrulis, Rolf Sternglanz, Leonard Guarente, Craig Peterson, and David McNabb for yeast strains and plasmids. David McNabb, Dennis Thiele, Kevin Morano, Suzanne Kleff, and Peter Horn contributed helpful advice and fruitful discussions. David McNabb, Min-Hao Kuo, and Jeffrey Regier critically read drafts of the manuscript.
This work was supported by Public Health Service grant AI27323 (to S.J.T.) and by funds from Michigan State University.
REFERENCES
- 1.Attardi, L. D., and R. Tjian. 1993. Drosophila tissue-specific transcription factor NTF-1 contains a novel isoleucine-rich activation motif. Genes Dev. 7:1341-1353. [DOI] [PubMed] [Google Scholar]
- 2.Barlev, N. A., R. Candau, L. Wang, P. Darpino, N. Silverman, and S. L. Berger. 1995. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270:19337-19344. [DOI] [PubMed] [Google Scholar]
- 3.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 4.Belotserkovskaya, R., D. E. Sterner, M. Deng, M. H. Sayre, P. M. Lieberman, and S. L. Berger. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20:634-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 6.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2—a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 7.Bourgarel, D., C. C. Nguyen, and M. Bolotin-Fukuhara. 1999. HAP4, the glucose-repressed regulated subunit of the HAP transcriptional complex involved in the fermentation-respiration shift, has a functional homologue in the respiratory yeast Kluyveromyces lactis. Mol. Microbiol. 31:1205-1215. [DOI] [PubMed] [Google Scholar]
- 8.Brakhage, A. A., A. Andrianopoulos, M. Kato, S. Steidl, M. A. Davis, N. Tsukagoshi, and M. J. Hynes. 1999. HAP-like CCAAT-binding complexes in filamentous fungi: implications for biotechnology. Fungal Genet. Biol. 27:243-252. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 10.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 11.Conaway, R. C., C. S. Brower, and J. W. Conaway. 2002. Emerging roles of ubiquitin in transcription regulation. Science 296:1254-1258. [DOI] [PubMed] [Google Scholar]
- 12.Cress, W. D., and S. J. Triezenberg. 1991. Critical structural elements of the VP16 transcriptional activation domain. Science 251:87-90. [DOI] [PubMed] [Google Scholar]
- 13.Drysdale, C. M., E. Duenas, B. M. Jackson, U. Reusser, G. H. Braus, and A. G. Hinnebusch. 1995. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol. Cell. Biol. 15:1220-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudley, A. M., L. J. Gansheroff, and F. Winston. 1999. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912delta promoter in Saccharomyces cerevisiae. Genetics 151:1365-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberharter, A., D. E. Sterner, D. Schieltz, A. Hassan, J. R. Yates III, S. L. Berger, and J. L. Workman. 1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenmann, D. M., C. Chapon, S. M. Roberts, C. Dollard, and F. Winston. 1994. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics 137:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estruch, J. J., L. Crossland, and S. A. Goff. 1994. Plant activating sequences: positively charged peptides are functional as transcriptional activation domains. Nucleic Acids Res. 22:3983-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fazzio, T. G., C. Kooperberg, J. P. Goldmark, C. Neal, R. Basom, J. Delrow, and T. Tsukiyama. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21:6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsburg, S. L., and L. Guarente. 1989. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 5:153-180. [DOI] [PubMed] [Google Scholar]
- 20.Forsburg, S. L., and L. Guarente. 1989. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 3:1166-1178. [DOI] [PubMed] [Google Scholar]
- 21.Gansheroff, L. J., C. Dollard, P. Tan, and F. Winston. 1995. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics 139:523-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgakopoulos, T., and G. Thireos. 1992. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 11:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodrich, J. A., T. Hoey, C. J. Thut, A. Admon, and R. Tjian. 1993. Drosophila TAF(II)40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75:519-530. [DOI] [PubMed] [Google Scholar]
- 24.Grant, P. A., and S. L. Berger. 1999. Histone acetyltransferase complexes. Semin. Cell. Dev. Biol. 10:169-177. [DOI] [PubMed] [Google Scholar]
- 25.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 26.Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101:181-191. [DOI] [PubMed] [Google Scholar]
- 27.Guarente, L., and T. Mason. 1983. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32:1279-1286. [DOI] [PubMed] [Google Scholar]
- 28.Hahn, S., J. Pinkham, R. Wei, R. Miller, and L. Guarente. 1988. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol. Cell. Biol. 8:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horikoshi, N., A. Usheva, J. Chen, A. J. Levine, R. Weinmann, and T. Shenk. 1995. Two domains of p53 interact with the TATA-binding protein, and the adenovirus 13S E1A protein disrupts the association, relieving p53-mediated transcriptional repression. Mol. Cell. Biol. 15:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horiuchi, J., N. Silverman, G. A. Marcus, and L. Guarente. 1995. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol. Cell. Biol. 15:1203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingles, C. J., M. Shales, W. D. Cress, S. J. Triezenberg, and J. Greenblatt. 1991. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature 351:588-590. [DOI] [PubMed] [Google Scholar]
- 33.Jackson, B. M., C. M. Drysdale, K. Natarajan, and A. G. Hinnebusch. 1996. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol. Cell. Biol. 16:5557-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, P. F., E. Sterneck, and S. C. Williams. 1993. Activation domains of transcriptional regulatory proteins. J. Nutr. Biochem. 4:386-398. [Google Scholar]
- 36.Kassir, Y., N. Adir, E. Boger-Nadjar, N. G. Raviv, I. Rubin-Bejerano, S. Sagee, and G. Shenhar. 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224:111-171. [DOI] [PubMed] [Google Scholar]
- 37.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 38.Kim, S. Y., A. Herbst, K. A. Tworkowski, S. E. Salghetti, and W. P. Tansey. 2003. Skp2 regulates Myc protein stability and activity. Mol. Cell 11:1177-1188. [DOI] [PubMed] [Google Scholar]
- 39.Klemm, R. D., J. A. Goodrich, S. L. Zhou, and R. Tjian. 1995. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc. Natl. Acad. Sci. USA 92:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi, N., P. J. Horn, S. M. Sullivan, S. J. Triezenberg, T. G. Boyer, and A. J. Berk. 1998. DA-complex assembly activity required for VP16C transcriptional activation. Mol. Cell. Biol. 18:4023-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwast, K. E., P. V. Burke, and R. O. Poyton. 1998. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201:1177-1195. [DOI] [PubMed] [Google Scholar]
- 42.Lamb, T. M., and A. P. Mitchell. 2001. Coupling of Saccharomyces cerevisiae early meiotic gene expression to DNA replication depends upon RPD3 and SIN3. Genetics 157:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, Y. S., and M. R. Green. 1991. Mechanism of action of an acidic transcriptional activator in vitro. Cell 64:971-981. [DOI] [PubMed] [Google Scholar]
- 44.Lin, Y. S., I. Ha, E. Maldonado, D. Reinberg, and M. R. Green. 1991. Binding of general transcription factor TFIIB to an acidic activating region. Nature 353:569-571. [DOI] [PubMed] [Google Scholar]
- 45.Lipford, J. R., and R. J. Deshaies. 2003. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat. Cell Biol. 5:845-850. [DOI] [PubMed] [Google Scholar]
- 46.Marcus, G. A., N. Silverman, S. L. Berger, J. Horiuchi, and L. Guarente. 1994. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 13:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNabb, D. S., Y. Xing, and L. Guarente. 1995. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 9:47-58. [DOI] [PubMed] [Google Scholar]
- 48.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 49.Mitchell, P. J., and R. Tjian. 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245:371-378. [DOI] [PubMed] [Google Scholar]
- 50.Olave, I. A., S. L. Reck-Peterson, and G. R. Crabtree. 2002. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71:755-781. [DOI] [PubMed] [Google Scholar]
- 51.Olesen, J., S. Hahn, and L. Guarente. 1987. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell 51:953-961. [DOI] [PubMed] [Google Scholar]
- 52.Park, J., S. Kunjibettu, S. B. McMahon, and M. D. Cole. 2001. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 15:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 54.Pina, B., S. Berger, G. A. Marcus, N. Silverman, J. Agapite, and L. Guarente. 1993. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol. Cell. Biol. 13:5981-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinkham, J. L., and L. Guarente. 1985. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:3410-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinkham, J. L., J. T. Olesen, and L. P. Guarente. 1987. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol. Cell. Biol. 7:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pray-Grant, M. G., D. Schieltz, S. J. McMahon, J. M. Wood, E. L. Kennedy, R. G. Cook, J. L. Workman, J. R. Yates III, and P. A. Grant. 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22:8774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Regier, J. L., F. Shen, and S. J. Triezenberg. 1993. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 90:883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ricci, A. R., J. Genereaux, and C. J. Brandl. 2002. Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol. Cell. Biol. 22:4033-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts, S. G. E., B. Choy, S. S. Walker, Y. S. Lin, and M. R. Green. 1995. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr. Biol. 5:508-516. [DOI] [PubMed] [Google Scholar]
- 61.Roberts, S. G. E., I. Ha, E. Maldonado, D. Reinberg, and M. R. Green. 1993. Interaction between an acidic activator and transcription factor IIB is required for transcriptional activation. Nature 363:741-744. [DOI] [PubMed] [Google Scholar]
- 62.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roberts, S. M., and F. Winston. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 65.Salghetti, S. E., M. Muratani, H. Wijnen, B. Futcher, and W. P. Tansey. 2000. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. USA 97:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 67.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 70.Stringer, K. F., C. J. Ingles, and J. Greenblatt. 1990. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature 345:783-786. [DOI] [PubMed] [Google Scholar]
- 71.Sudarsanam, P., and F. Winston. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 72.Sullivan, S. M., P. J. Horn, V. A. Olson, A. H. Koop, W. Nu, R. H. Ebright, and S. J. Triezenberg. 1998. Mutational analysis of a transcriptional activation region of the VP16 protein of herpes simplex virus. Nucleic Acids Res. 26:4487-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Triezenberg, S. J. 1995. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5:190-196. [DOI] [PubMed] [Google Scholar]
- 74.von der Lehr, N., S. Johansson, and L. G. Larsson. 2003. Implication of the ubiquitin/proteasome system in Myc-regulated transcription. Cell Cycle 2:403-407. [PubMed] [Google Scholar]
- 75.Walker, S., R. Greaves, and P. O'Hare. 1993. Transcriptional activation by the acidic domain of Vmw65 requires the integrity of the domain and involves additional determinants distinct from those necessary for TFIIB binding. Mol. Cell. Biol. 13:5233-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams, R. M., M. Primig, B. K. Washburn, E. A. Winzeler, M. Bellis, C. Sarrauste de Menthiere, R. W. Davis, and R. E. Esposito. 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA 99:13431-13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao, H., J. D. Friesen, and J. T. Lis. 1995. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol. Cell. Biol. 15:5757-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xing, Y., S. Zhang, J. T. Olesen, A. Rich, and L. Guarente. 1994. Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short alpha-helix in HAP2. Proc. Natl. Acad. Sci. USA 91:3009-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]