Abstract
Genome projects involving Leishmania and other trypanosomatids have revealed that most genes in these organisms are organized into large clusters of genes on the same DNA strand. We have previously shown that transcription of the entire Leishmania major Friedlin (LmjF) chromosome 1 (chr1) initiates bidirectionally between two divergent gene clusters. Here, we analyze transcription of LmjF chr3, which contains two convergent clusters of 67 and 30 genes, separated by a tRNA gene, with a single divergent protein-coding gene located close to the “left” telomere. Nuclear run-on analyses indicate that specific transcription of chr3 initiates bidirectionally between the single subtelomeric gene and the adjacent 67-gene cluster, close to the “right” telomere upstream of the 30-gene cluster, and upstream of the tRNA gene. Transcription on both strands terminates within the tRNA-gene region. Transient-transfection studies support the role of the tRNA-gene region as a transcription terminator for RNA polymerase II (Pol II) and Pol III, and also for Pol I.
Leishmania is a protozoan parasite (order Kinetoplastida) which alternates life-forms between an intracellular amastigote stage residing in vertebrate macrophages and an extracellular promastigote stage living in the digestive tract of sandflies. The numerous human-infective Leishmania species cause a spectrum of disease ranging from asymptomatic to lethal, resulting in widespread human suffering and death, as well as considerable economic loss (35).
Leishmania, as well as other members of the Trypanosomatidae family, possesses unusual mechanisms of gene expression, such as polycistronic transcription (13, 19) and RNA editing of the mitochondrial transcripts (37). In these organisms, the mature nuclear mRNAs are generated from primary transcripts by trans-splicing, a process that adds a capped 39-nucleotide (nt) miniexon or splice leader (SL) to the 5′ termini of the mRNAs (27). The steady-state levels of most of the mature mRNAs appear to be regulated posttranscriptionally by mechanisms that involve their 3′ untranslated region sequences (23). Promoters for RNA polymerase I (Pol I) have been extensively characterized in trypanosomatids (31, 44, 46), as have some Pol III promoters (3, 26). However, little is known about the sequences that drive the expression of protein-coding genes by Pol II.
The Leishmania haploid genome content is ∼34 Mb, consisting of 36 chromosomes which range in size from 0.3 to 2.5 Mb (40). The Leishmania Genome Network was established with the support of the World Health Organization to map and sequence the genome of Leishmania major Friedlin (LmjF), the reference strain of the project. The sequence of chromosome 1 (chr1), the smallest in the parasite, revealed the presence of 79 putative genes, the first 29 of which are in a cluster on the “bottom” DNA strand, while the remaining 50 are in a cluster on the “top” strand (21). Importantly, nuclear run-on analysis of chr1 showed that specific transcription, leading to the production of stable transcripts, initiates within the strand-switch region between these two clusters and proceeds bidirectionally toward the telomeres (19). Stable-transfection studies support the presence of a bidirectional promoter in this region of chr1. It also appears that nonspecific transcription takes place over the entire chr1, but at a level ∼10-fold lower than the specific transcription initiating in the strand-switch area (19). Here we report the transcriptional analysis by nuclear run-on of chr3. This chromosome contains 97 putative protein-coding genes organized into two long convergent clusters, which are separated by a tRNALys gene (42). In addition, a single divergent gene is located at the “left” end of the chromosome. Our data show that Pol II transcription on chr3 initiates bidirectionally between the single subtelomeric gene and the adjacent 67-gene cluster and near the “right” telomere upstream of the 30-gene cluster. The tRNALys gene is transcribed by Pol III. Transcription on both strands terminates in the tRNA-gene region.
MATERIALS AND METHODS
Culture and transfection of Leishmania.
Promastigotes from L. major MHOM/IL/81/Friedlin (LSB-132.1) (LmjF) were grown in supplemented RPMI 1640 medium at 25°C and harvested in the mid-log phase. Electroporation and luciferase assays were performed as previously described (45).
Molecular cloning into M13 and preparation of single-stranded DNA.
DNA fragments from chr3 were amplified by PCR and cloned into M13mp18 and mp19 replicative-form DNA. Insert I was amplified with oligonucleotides 501-5′ and 501-3′ (Table 1), and fragment II was amplified with 502-5′ and 502-3′. LmjF3.0010 (gene 1) was amplified with oligonucleotides LRRP1-1 and LRRP1-2, and fragment III was amplified with 1000-5′ and 1000-3′. LmjF3.0020 (gene 2) was amplified with oligonucleotides 1001-5′ and 1001-3′, and fragment IV was amplified with oligonucleotides 1002-5′ and 1002-3′. LmjF3.0030 (gene 3) was amplified with D3PGDH-5′ and D3PGDH-3′, and LmjF3.0040 (gene 4) was amplified with 2AEPAT-5′ and 2AEPAT-3′. LmjF3.0050 (gene 5) was amplified with oligonucleotides L952.4-5′ and L952.4-3′, and LmjF3.0190 (gene 19) was amplified with U2AF23-5′ and U2AF23-3′. LmjF3.0270 (gene 27) was amplified with oligonucleotides L6202.3-5′ and L6202.3-3′, and LmjF3.0670 (gene 67) was amplified with L6910.7-5′ and L6910.7-3′. Fragment V was amplified with primers 2-int.L6910.7/L6290.1-5′ and 2-int.L6910.7/L6290.1-3′. LmjF3.0680 (gene 68) was amplified with oligonucleotides L6290.1-5′ and L6290.1-5′, and fragment VI was amplified with int.L6290.1/tRNA-5′ and int.L6290.1/tRNA-3′. LmjF3.tRNALys.01 (tRNALys gene) was amplified with oligonucleotides tRNA-Lys-5′ and tRNA-Lys-3′, and LmjF3.0690 (gene 69) was amplified with HEL2-5′ and HEL2-3′. LmjF3.0700 (gene 70) was amplified with oligonucleotides L505.2-5′ and L505.2-3′, and LmjF3.0880 (gene 88) was amplified with L7234.2-5′ and L7234.2-3′. LmjF3.0950 (gene 95) was amplified with oligonucleotides MCO1-5′ and MCO1-3′, and LmjF3.0980 (gene 98) was amplified with EIF-2a-5′ and EIF-2a-3′. Fragment VII was amplified with oligonucleotides 2001-5′ and 2001-3′, and fragment VIII was amplified with 2002-5′ and 2002-3′. Fragment IX was amplified with oligonucleotides 2003-5′ and 2003-3′. LmjF chr1 genes 27 to 32 and fragments AC and DE and fragments R-1 to R-4 from the rRNA locus of LmjF were previously described (19). The tRNA fragment from chr23 (306 bp), which contains two tRNA genes (tRNAMet and tRNALeu), was amplified by PCR with primers tRNA cluster-5′ and tRNA cluster-3′. After transfection of DH5αF′ competent cells (Invitrogen), single-stranded DNA was purified from colorless plaques with the QIAprep Spin M13 columns (QIAGEN) as specified by the supplier. The orientation of each insert was confirmed by sequencing using the M13 −21 primer.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| 501-5′ | ATGTAAGCTTAGGACGCTCTCGATCCAGTAAG |
| 501-3′ | ATGTGAGCTCCCAGTTGTCTCTTCCTAACGGC |
| 502-5′ | ATGTAAGCTTAAGAGAACGGAAACCGATGACG |
| 502-3′ | ATGTGAGCTCTTGCTTTCTCTCGATCACCACG |
| LRRP1-1 | ATTCGAGAGAGGATGCGACTGGCAC |
| LRRP1-2 | TCTGGAGGTGCTGGACATTGGAGGA |
| 1000-5′ | ATGTAAGCTTACCGCGAATGATAGAGAGGAAC |
| 1000-3′ | ATGTGAGCTCCTTGCGACTGTTCCACAGAGAC |
| 1001-5′ | ATGTAAGCTTTGCGATTTGTATCGGAACGAGG |
| 1001-3′ | ATGTGAGCTCCGCTATGTTCTTTGGCACGTGG |
| 1002-5′ | ATGTAAGCTTCGTATTTCTCCGTTCGGGTATC |
| 1002-3′ | ATGTGAGCTCAAGCGAGAGCTGATGCAAAGTG |
| D3PGDH-5′ | ATGTAAGCTTTGGGATCGGCTGTTTCTGCATC |
| D3PGDH-3′ | ATGTGAGCTCTGCAGCTCCTTGTTCGACCCAG |
| 2AEPAT-5′ | ATGTAAGCTTATCAGTGCCTTTGGCGGTATCC |
| 2AEPAT-3′ | ATGTGAGCTCGTCAATGCCCATGGATTTGAGC |
| L952.4-5′ | ATGTAAGCTTGAGGCCAACCGTCGTGTATGAC |
| L952.4-3′ | ATGTGAGCTCAAGATCAGCAATGCGGACGAAC |
| U2AF23-5′ | ATGTAAGCTTCATTGACTGTGCGACACTTCAC |
| U2AF23-3′ | ATGTGAGCTCACTGCTCCTTCTCCAGCTTCTC |
| L6202.3-5′ | ATGTAAGCTTGCGCCCTTCAAGTATTATACGC |
| L6202.3-3′ | ATGTGAGCTCCTCCACAGAGAGCAGACAAGGC |
| L6910.7-5′ | ATGTAAGCTTACTCCACTACAATGCACTGTGC |
| L6910.7-3′ | ATGTGAGCTCTAGAACGGTGATAATTCAACGG |
| 2-int.L6910.7/L6290.1-5′ | ATGTAAGCTTTGTTGTTGGTTTCGCTTTGCTC |
| 2-int.L6910.7/L6290.1-3′ | ATGTGAGCTCGATCGGCATTTGTGTAGGCATG |
| L6290.1-5′ | ATGTAAGCTTTAACCGCGCCTGAGGTGTACTC |
| L6290.1-5′ | ATGTGAGCTCTTCCATTCTTCAGCGCCGTAAC |
| int.L6290.1/tRNA-5′ | ATGTAAGCTTGGTGTGGACACGCTGACGAAAG |
| int.L6290.1/tRNA-3′ | ATGTGAGCTCCGGCGTAATGTCAACGAAGACC |
| tRNA-Lys-5′ | ATGTAAGCTTTGCCTACGGCTTTGTCCAGGAG |
| tRNA-Lys-3′ | ATGTGAGCTCTTCAACACCCTCCCACCCACAG |
| HEL2-5′ | ATGTAAGCTTGTGGCGGAAGAAGACGATGTGG |
| HEL2-3′ | ATGTGAGCTCCTTCGACGCCACCGTTGAGATC |
| L505.2-5′ | ATGTAAGCTTTGTCTGACGGCACCGTCGATTG |
| L505.2-3′ | ATGTGAGCTCGGGCGCATCACTGGATGATTGG |
| L7234.2-5′ | ATGTAAGCTTTCCATCGACGACGAGCTTGTAC |
| L7234.2-3′ | ATGTGAGCTCGTGGACGCCCTTCTCACCTATG |
| MCO1-5′ | ATGTAAGCTTAGCGACATGTTTATGTAGCACG |
| MCO1-3′ | ATGTGAGCTCTGTCTTGACGCTAGTGAACGAC |
| EIF-2a-5′ | TCCTTCAGCTCTGTATTAGTCCG |
| EIF-2a-3′ | CAATAACGAGCGTCCAGAGTGGA |
| 2001-5′ | ATGTGAATTCTTATTGCTACCCTTCGTCTCAC |
| 2001-3′ | ATGTGCATGCGCCAATTTCAACTGTTCAAGTC |
| 2002-5′ | ATGTGAATTCACCGTGGCACGAACAATAAACG |
| 2002-3′ | ATGTGCATGCGACAAGCTTCCGCTCAGCAGAC |
| 2003-5′ | ATGTGAATTCGTCGTCGCAGTTTGTTCTCCTG |
| 2003-3′ | ATGTGCATGCCGCACTCCTATGGGTTAACGTG |
| tRNA cluster-5′ | GAAGGACAGACAGTTGGGACCA |
| tRNA cluster-3′ | TACCAGGTTCAGGTGGGAGGAA |
| LRRP1-RT′ | CAGATGTAGTGGTCGCAGTGATTG |
| LRRP1-nested | GCACGTGTATCGGTCGTCAAGATT |
| LRRP1-nested 2 | TCTGGTTGTGCTTGCAATCGTC |
| LRRP1-nested 3 | TTGTCACAGTCCTGTGTGCGAGGT |
| L952.3-nested 1 | AAGCGATTGAAACGGATCAAA |
| L952.3-nested 2 | AGGACAGAAGGATACACCAGATGC |
| L952.3-nested 3 | AACAGTGCCTGCGCACCACATCT |
| L952.3-nested 4 | GGATAGTAGTGGCACGACCTTT |
| Miniexon | AACGCTATATAAGTATCAGTT |
| D3PGDH-nested 3 | CTGTCAGGTGACAGAGATGATG |
| D3PGDH-nested 4 | GCGACACCTTTCGTAAGTACTT |
| D3PGDH-nested 5 | ACTTGAGCCAACAATCACCTCC |
| EIF-2a-RT | TGCGGCCTACCTTGATTAGCTTTC |
| EIF-2a-nested | TCACCTCCGTGTACGGAATAATGC |
| EIF-2a-21 | CAGAGCAGAAGCGACACACCATT |
| Nested(dT) | CCTCTGAAGGTTCACGGATCCACATCTAGA(T)18VN |
| B1 | CCTCTGAAGGTTCACGGAT |
| B2 | CACGGATCCACATCTAGAT |
| L6290.1-PA-1 | GTTACGGCGCTGAAGAATGGAA |
| L6290.1-PA-2 | CAGTGCCTGATGAACATCGATG |
| HEL2-PA-1 | CGGCTGCTGTCGAACATATCA |
| HEL2-PA-2 | GGAGGAGGACCTCAAATTCTAC |
| tRNA-Bam-5′ | ATGTGGATCCTGCCTACGGCTTTGTCCAGGAG |
| tRNA-Bam-3′ | ATGTGGATCCTTCAACACCCTCCCACCCACAG |
| 72-Bam-5′ | ATGTGGATCCCAACGTCGTCACAACGTACAGC |
| 72-Bam-3′ | ATGTGGATCCTGGTGAATGTAGCAGCGACACG |
| tRNA-dTTTT-5′ | CGATCCCCACGGAGTGCGCCCCCCTTGGCTGCGCCAAGCC |
| tRNA-dTTTT-3′ | GGCTTGGCGCAGCCAAGGGGGGCGCACTCCGTGGGGATCG |
| chr3-tss2-5′ | ACACAAGCACGGGAGCTGCGGTGA |
| tRNA-5′ end | TTGCGAAGCATTCCTAGCTCAGT |
Nuclear run-on assays.
These experiments were performed with mid-log LmjF promastigotes, as described elsewhere (19). In the assays carried out in the presence of UV light, promastigotes (in a total volume of 15 ml) were irradiated in petri dishes, with agitation, in a Stratalinker UV cross-linker (Stratagene). After irradiation, cells were incubated for at least 1.5 h at 28°C to allow the clearing of RNA polymerases engaged prior to irradiation. Elongation of nascent RNA in the presence of transcription inhibitors was performed by preincubating the nuclei with α-amanitin (Roche Molecular Biochemicals) or tagetitoxin (Tagetin; Epicentre Biotechnologies) for 15 min on ice in lysis buffer. Nuclei were next pelleted and resuspended in elongation buffer in the presence of the drugs. Labeled nascent RNA was hybridized to Hybond filters (Amersham) containing dots of 1 μg of single-stranded M13 DNA. Hybridization was performed for 48 h at 50°C in a solution containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.2% sodium dodecyl sulfate, 4× Denhardt's reagent, and salmon sperm DNA (100 μg/ml). Posthybridization washes were carried out in 0.1× SSC and 0.1% sodium dodecyl sulfate at 65°C.
5′ RACE analysis.
5′ rapid amplification of cDNA ends (5′ RACE) experiments were performed with 5 μg of total RNA from LmjF with a kit from Life Technologies, Inc. For LmjF3.0010, the first-strand cDNA was synthesized with primer 1000-5′, and the PCR amplifications were carried out with nested primers LRRP1-nested 2 and LRRP1-nested 3. For LmjF3.0020, the first-strand cDNA was synthesized with primer L952.3-nested 1 and the PCR amplifications were done with L952.3-nested 2 and L952.3-nested 3. For LmjF3.0990, the cDNA was produced with primer 2001-5′ and the PCR amplifications were performed with primers EIF-2a-21 and 2002-5′. The nested PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced. In a control experiment designed to detect transcripts further upstream of the most 5′ transcription start sites (TSS) for LmjF3.0010, the first-strand cDNA was synthesized with primer LRRP1-nested 3, and the PCR amplification was carried out with primer chr3-tss2-5′. In the control for LmjF3.0020, the first-strand cDNA was generated with primer L952.3-nested 4, and the PCR amplification was done with primer 1000-3′. In the control for LmjF3.0980, the first-strand cDNA was synthesized with primer 2002-5′, and the PCR was performed with primer 2003-5′.
RT-PCR assays.
The location of SL and polyadenylation sites for selected genes was determined by reverse transcription (RT)-PCR. For LmjF3.0010, the cDNA was prepared with oligonucleotide LRRP1-RT, and the PCR was carried out with primers LRRP1-nested and miniexon. For LmjF3.0020, the cDNA was synthesized with primer D3PGDH-nested 3, and the PCRs were carried out with primers D3PGDH-nested 4 and D3PGDH-nested 5, together with miniexon. The cDNA for LmjF3.0980 was prepared with primer EIF-2a-RT and the PCR with primers EIF-2a-nested and miniexon. To map the polyadenylation sites for genes 68 and 69, the cDNA was prepared with oligonucleotide Nested(dT). For LmjF3.0680, the first PCR was carried out with primers L6290.1-PA-1 and B1. The second PCR was done with primers L6290.1-PA-2 and B2. For LmjF3.0690, the first PCR was performed with primers HEL2-PA-1 and B1, and the second PCR was performed with primers HEL2-PA-2 and B2. The final PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced. Transcription termination sites were mapped by poly(A) tailing of total RNA, which was performed by mixing 2 μg of total RNA, 1 μl of 25 mM ATP, 2 μl of 5× Poly (A) Polymerase reaction buffer (U.S. Biochemicals), and 1,200 U of Yeast Poly (A) Polymerase (U.S. Biochemicals) in a final volume of 20 μl. The mixture was incubated for 20 min at 37°C, and the reaction was terminated by heating at 65°C for 10 min. The cDNA was prepared with oligonucleotide Nested(dT). For the tRNA gene (LmjF3.tRNALys.01), the first PCR was done with primers tRNA-5′end and B1, and the second PCR was done with primers tRNA-5′end and B2. For Pol II transcripts that originate upstream of the tRNA gene, the first PCR was performed with primer tRNA-Lys-5′ and B1, and the second PCR was performed with primers tRNA-5′end and B2.
Plasmid constructs.
To generate the constructs employed in the transient- transfection experiments, a 339-bp fragment containing LmjF3.tRNALys.01 was amplified by PCR with primers tRNA-Bam-5′ and tRNA-Bam-3′. The PCR product was then digested with BamHI and cloned into BamHI-digested pNBUC (44) and pLMRIB (18). Clones containing the tRNA-gene fragment in the sense orientation (pNT-5′ and pLMT-5′) and in the antisense orientation (pNT-3′ and pLMT-3′) were identified by sequencing. To generate pLM72-5′ and pLM72-3′, a 435-bp fragment from Chr1_0650 from LmjF chr1 was amplified by PCR with oligonucleotides 72-Bam-5′ and 72-Bam-3′. The BamHI-digested PCR product was cloned into pLMRIB cut with BamHI. The QuikChange XL Site-Directed Mutagenesis kit (Stratagene) was used to delete the four Ts located downstream of the tRNA gene in pNT-5′, pNT-3′, pLMT-5′, and pLMT-3′, generating plasmids pNT-5′-ΔT, pNT-3′-ΔT, pLMT-5′-ΔT, and pLMT-3′-ΔT, respectively. Primers tRNA-dTTTT-5′ and tRNA-dTTTT-3′ were used in these experiments. The absence of the Ts was confirmed by sequencing.
RESULTS
Nuclear run-on analysis of LmjF chr3.
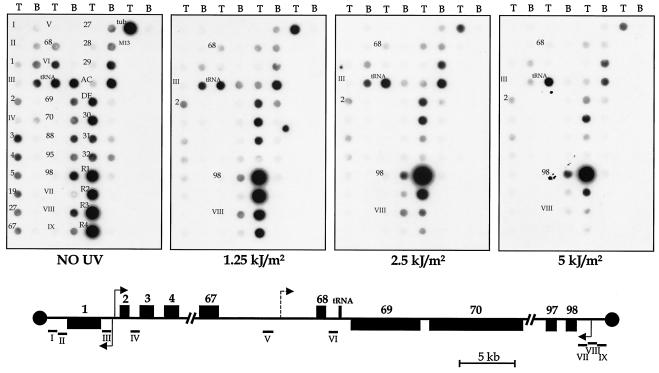
The organization of the genes on chr3 suggests that transcription may initiate in three different areas of the chromosome: between LmjF3.0010 and LmjF3.0020 (genes 1 and 2), upstream of LmjF3.0980 (gene 98), and in the LmjF3.tRNALys.01 region (tRNA gene) (Fig. 1). Therefore, the nuclear run-on analysis used several single-stranded M13 DNA probes in and around these regions, as well as a smaller number of probes from other regions (genes 19, 27, 88, and 95) of chr3. In the absence of UV irradiation (Fig. 1), the hybridization signal was, on average, eightfold more intense on the coding strand (bottom strand for genes 1 and 69 to 98, top strand for genes 2 to 68) than the noncoding strand for most of the fragments. The single exception is the tRNA-gene (LmjF3.tRNALys.01) fragment which showed strong signal on both strands (Fig. 1). The difference in signal intensity between coding and noncoding strands for chr3 is similar to that (11-fold) found for LmjF chr1 (19), as confirmed by the chr1 controls (genes 27 to 32) shown in Fig. 1. The signal obtained with some intergenic fragments (I, II, IV, V, and VII) was weaker than that obtained with most of the fragments that contain coding region, although other intergenic fragments (III, VI, VIII, and IX) showed signal as strong or stronger than some of the coding-region fragments. Thus, the hybridization signal likely reflects the rates of both transcription and RNA abundance, due to the rapid RNA processing in Leishmania, as well as other factors, such as the size and G+C content of the fragment, and secondary structure within the RNA.
FIG. 1.
Effect of UV irradiation on chr3 transcription. Nuclear run-on assays were performed with nuclei isolated from 2 × 108 promastigotes which were irradiated with three different intensities of UV light (1.25, 2.5, and 5 kJ/m2, as indicated below each panel). Run-on RNA was radiolabeled by 6-min incubation, extracted, and hybridized to dot blots of single-stranded M13 DNAs (1 μg) that contain inserts which are complementary to the top (T) or bottom (B) strand of several regions of chr3. A control experiment with nonirradiated cells is shown in the left panel. Control DNAs include genes 27 to 32 and fragments AC and DE from chr1, fragments R-1 to R-4 from the rRNA locus on chr27, and α-tubulin. A map of chr3 is shown below the figure. Arrows indicate the regions of Pol II transcription initiation. The dashed arrow denotes the putative initiation region for gene 68.
Transcription initiation.
At the left end of chr3 (genes 1 to 5), transcription on the bottom strand appears to initiate within or near fragment III, since signal on this strand is limited to I, II, 1, and III (Fig. 1). Similarly, transcription on the top strand seems to initiate near gene 2. At the right end, signal is seen on the bottom strand up to and including fragment IX, suggesting that transcription initiates within this fragment, close to the telomeric repeats.
In order to confirm these results, nuclear run-on experiments were performed with cells previously irradiated with UV light. RNA elongation is arrested with UV irradiation by the production of pyrimidine dimers in the DNA and inactivation of transcription of a given sequence is a function of the dose of UV and the distance to the transcription initiation site (33). In trypanosomatids, irradiation with low doses of UV also inhibits RNA processing, resulting in the accumulation of transcripts from sequences situated adjacent to a promoter (6). We have previously used UV irradiation and nuclear run-on analysis to map the transcription initiation sites of LmjF chr1 (19). The results obtained with chr1 and rRNA-gene promoter (18) controls (Fig. 1) confirm that UV irradiation had the expected effect, i.e., the hybridization signal was enhanced or not significantly reduced with UV irradiation for fragments closest to the initiation region (AC and DE for chr1 and R1 for rRNA), while the signal was progressively reduced with high UV doses (Fig. 1) for fragments further from the TSSs.
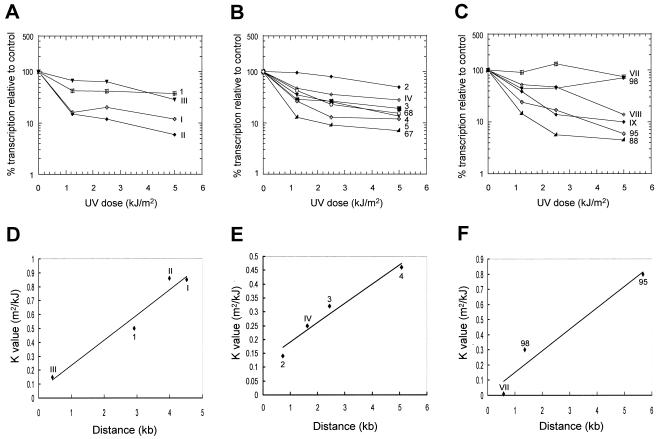
At the left end of chr3, the hybridization signal for fragments within or close to the strand-switch region (1, III, and 2) remained high even at the maximum dose, even increasing slightly with low doses of UV irradiation (Fig. 1). Conversely, the signal of the genes and fragments that are more distant from the strand-switch area was progressively reduced (Fig. 1). This can be seen more clearly when the relative signal intensity is plotted against UV dose (Fig. 2A and B). This supports the argument for the presence of transcription initiation on the top strand (for genes 2 to 68) just upstream of gene 2 and on the bottom strand (for gene 1) within or just upstream of fragment III. Indeed, a linear relationship was observed between the log of the relative signal intensity divided by UV dose and distance from the putative TSSs (Fig. 2D and E). Therefore, it is likely that a bidirectional transcription initiation region (or two closely juxtaposed regions) is present between genes 1 and 2, similar to the situation between genes 29 and 30 (LmjF1.0310/XPP and LmjF1.0320/PAXP) on LmjF chr1 (19).
FIG. 2.
Relative rates of transcription as a function of UV intensity. (A to C) The results shown in Fig. 1 and another similar experiment (not shown) were quantified, and the transcription signal for each clone, relative to the nonirradiated control, was plotted against UV dose. The average slope (K) of the curves from each fragment was calculated using the equation K = (ln R0/R)/d, where R0 is nascent RNA synthesis in nuclei made from nonirradiated cells and R is nascent RNA synthesis at UV dose d (13). (D to F) K is plotted versus the distance from the putative promoter to the midpoint of each fragment. (A and D) Gene 1 and fragments I, II, and III; (B and E) genes 2 to 5, 67, and 68 and fragment IV; (C and F) genes 98, 95, and 88 and fragments VII to IX.
In contrast to genes 3 to 67, the hybridization signal for gene 68 was not reduced with UV exposure as much as it would be expected if its transcription started upstream of gene 2 (Fig. 1 and 2B). While transcription of gene 67 was reduced by 87% with 1.25 kJ of UV/m2, transcription of gene 68 was reduced by only 57% (Fig. 2B), and the pattern of signal reduction with increasing UV is more similar to that seen with genes only 3 to 5 kb from a transcription initiation site (e.g., genes 3 and 4) than that for gene 65 (Fig. 2B). Moreover, fragment V, which is located between genes 67 and 68, did not show any appreciable signal on either strand (Fig. 1). To determine if the lack of signal observed with fragment V is an artifact, a riboprobe corresponding to this fragment was generated in vitro and hybridized to one of the filters containing chr3 single-stranded DNA. Hybridization signal was detected in the corresponding dot (data not shown), indicating that the RNA is capable of hybridizing to that region. Thus, the lack of signal in the nuclear run-on experiments most likely indicates that fragment V is not transcribed and that there might be a transcription start region upstream of gene 68. It is notable that the intergenic region between genes 67 and 68 is considerably larger (10 kb) than any others seen on chr3 or chr1. However, it is also possible that the relative resistance to UV irradiation of the hybridization signal for gene 68 may be due to cross-hybridization with transcripts from another region of the LmjF genome that is located close to a TSS.
At the right end of chr3, the hybridization signal for gene 98 and fragment VII was not significantly reduced by UV irradiation (Fig. 1 and 2C). The signal for fragment VIII was reduced only at the highest UV dose. By contrast, signals of the other genes in the same cluster (genes 95 to 69) were strongly reduced, even at the lowest UV dose (Fig. 1 and 2C). Therefore, transcription of that polycistronic unit appears to initiate on the bottom strand within fragment VIII, which is supported by the observed linear relationship between UV dose and distance from the putative TSS (Fig. 2F). The rapid decrease in signal intensity for fragment IX after irradiation with UV suggests that the hybridization signal may be due to nonspecific transcription (that is, transcription that does not originate from a specific initiation site) or cross-hybridization with transcripts from elsewhere in the genome.
The hybridization signal on the coding (top) strand of the tRNA-gene fragment (LmjF3.tRNALys.01) remained high even at 5 kJ of UV/m2, while the signal for the region immediately upstream (fragment VI) was rapidly reduced by UV irradiation (Fig. 1). Thus, it is likely that transcription of the tRNA gene is initiated within the tRNA fragment, quite possibly by a promoter within the tRNA itself. By contrast, hybridization signal on the noncoding (bottom) strand of this region was considerably diminished with UV irradiation (Fig. 1), suggesting that transcription of this strand starts some distance upstream and probably represents a continuation of the polycistronic transcription initiated within fragment VIII.
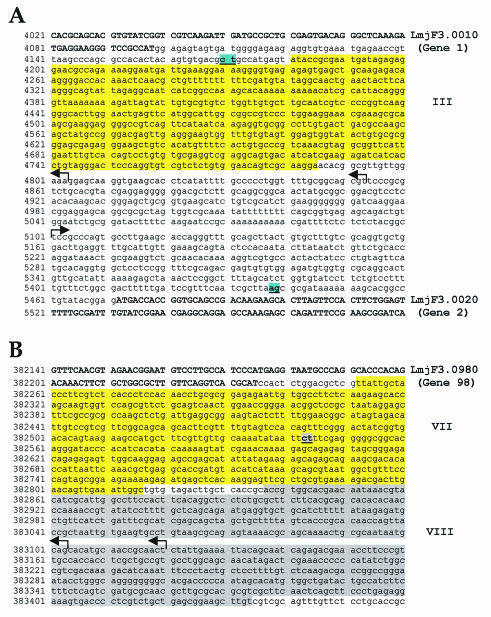
To accurately localize TSSs on chr3, the SL addition sites for genes 1, 2, and 98 were first mapped (by RT-PCR) for each of these genes and found to be 72, 33, and 308 bp upstream of the corresponding start codons, respectively (Fig. 3A and B). Subsequent 5′ RACE analysis using primers upstream of the SL site mapped two putative TSSs for gene 1, 705 and 755 bp upstream of the start codon (Fig. 3A). These TSSs are located immediately upstream of fragment III, in agreement with the nuclear run-on results. A single TSS was localized 371 bp upstream of gene 2 (Fig. 3A), again in agreement with the results from the nuclear run-on. Two putative TSSs were located 868 and 885 bp upstream of gene 98 (Fig. 3B), within fragment VIII in agreement with the nuclear run-on data. Control experiments designed to detect transcripts further upstream of the most 5′ TSS for genes 1, 2, and 98 did not show any amplified bands, confirming that there is very little, if any, transcription in these regions of chr3.
FIG. 3.
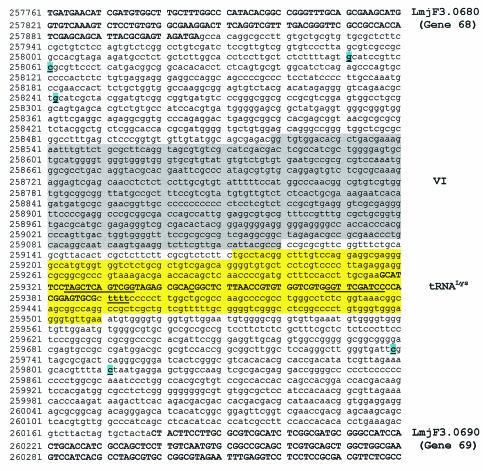
Pol II transcription on chr3 initiates in two different regions. (A) Sequence between genes 1 (LmjF3.0010) and 2 (LmjF3.0020). (B) Sequence upstream of gene 98 (LmjF3.0980). The two putative TSSs for gene 1, the putative TSS for gene 2 and the two putative TSSs for gene 98 are indicated with the arrows. The SL acceptor sites (CT for genes 1 and 98, and AG for gene 2) are underlined and highlighted. Protein-coding sequences are shown in capital letters and boldface type. The sequences of fragments III, VI, I and VIII are boxed.
The 247-bp region which separates the most 5′ TSSs for gene 1 and gene 2 does not show any substantial sequence identity with the region upstream of the TSSs of gene 98, the putative promoter region for LmjF chr1 (19), or the SL promoter region of chr2 (16). As previously seen for chr1, no TATA box or any other typical Pol II promoter elements are apparent, and the GC content (55%) is similar to that of the entire strand-switch region (51% GC), although it is somewhat lower than chr3 (and the genome) as a whole (63%).
Pol II and Pol III transcription.
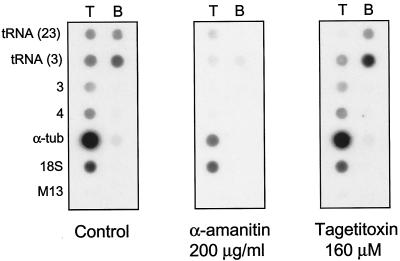
In order to identify which RNA polymerase(s) is involved in transcription of chr3, nuclear run-on experiments were performed in the presence of α-amanitin, which inhibits Pol II, and tagetitoxin, which selectively inhibits Pol III (36). High concentrations of α-amanitin (1 mg/ml) totally abolished the signal on all the chr3 fragments, while the rRNA-gene controls remained unaffected (data not shown), indicating that chr3 is not transcribed by Pol I. This concentration of α-amanitin, however, does not distinguish between Pol II and III transcription, since Pol III is also inhibited by high concentrations of α-amanitin. In experiments carried out with α-amanitin (200 μg/ml), the signal for genes 3 and 4 was totally abolished and that of α-tubulin control was reduced by 92%, while the signal for 18S rRNA was not affected (Fig. 4). In contrast, the signal intensity obtained with the coding (top) strand of the tRNA-gene fragment (LmjF3.tRNALys.01) and a control fragment that contains two tRNA genes (tRNAMet and tRNALeu) from chr23 of LmjF was substantially (85 and 65%, respectively), but not completely, reduced by α-amanitin (200 μg/ml), as would be expected for Pol III transcription. This was confirmed by the use of 160 μM tagetitoxin, which reduced the signal intensity on the tRNA coding strand by 52 and 87%, respectively, for the chr3 and chr23 tRNA genes (Fig. 4). In contrast, the signal for the noncoding (bottom) strand of the chr3 and chr23 tRNA genes was not affected by tagetitoxin but was strongly reduced (88 and 98%, respectively) with α-amanitin (Fig. 4). Transcription of the chr3 protein-coding (genes 3 and 4) and rRNA (18S) genes was not inhibited by tagetitoxin, as expected for Pol II and Pol I, respectively. Therefore, these results suggest that LmjF3.tRNALys.01 is transcribed by Pol III while the remaining chr3 genes are transcribed by Pol II.
FIG. 4.
Identification of the RNA polymerases that transcribe chr3. Nuclear run-on RNA was radiolabeled in the presence of α-amanitin (200 μg/ml) or tagetitoxin (160 μM) and hybridized to filters containing single-stranded DNAs from the tRNALys gene and genes 3 and 4 from chr3. A tRNA fragment from chr23 was used as a control, together with α-tubulin, small-subunit rRNA (18S), and mp18 vector with no insert (M13). The left panel shows a control experiment performed in the absence of any transcription inhibitor. Abbreviations: T, DNA complementary to top strand; B, DNA complementary to bottom strand.
Transcription termination.
The nuclear run-on analysis of chr3 (Fig. 1) suggests that transcription of both the top and bottom strands terminates within the tRNA-gene fragment, since signal intensity was substantially reduced for the top strand fragments to the right of this region and for bottom strand fragments to the left. To determine the transcription termination sites on chr3, RT-PCR was performed with total RNA that was poly(A)-tailed in vitro. Transcription of the tRNA gene was found to end within the run of four Ts located downstream of the gene, as has been reported in other organisms (1). In three of the clones analyzed, the poly(A) tail was added after the third T, and in one clone it was added after the fourth T (Fig. 5). When the first PCR was carried out using a primer 5′ to the tRNA gene (presumably upstream of the Pol III promoter) we found that the 3′ termini were located within the tRNA coding sequence: out of 24 clones analyzed, 16 end at position 259345 (C), 4 end at position 259344 (A), 3 end at position 259346 (G), and 1 ends at position 259341 (C). Thus, it appears that Pol II transcripts that originate upstream of gene 68 (or upstream of gene 2) terminate within the anticodon stem of the tRNA gene.
FIG. 5.
Sequence around the tRNALys gene (LmjF3.tRNALys.01). The tRNALys gene fragment used in the nuclear run-on assays and cloned into pNBUC and pLMRIB is highlighted. Also highlighted is the sequence of fragment VI. Coding sequences are shown in capital letters and boldface type. The putative internal control elements for the tRNA gene, boxes A and B, are underlined, as well as the run of four Ts located downstream of the gene and the C located at position 259345. The three putative polyadenylation sites for gene 68 and the two for gene 69 are indicated.
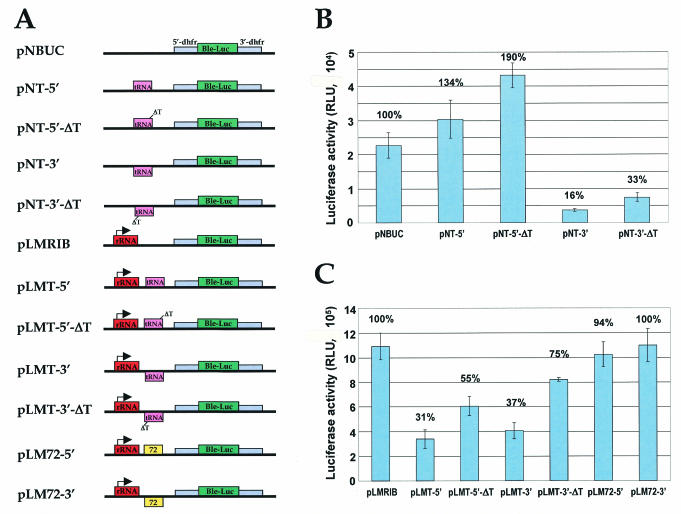
To functionally test the role of the tRNA-gene region in Pol III and Pol II transcription termination, a 339-bp fragment (the same one used for the nuclear run-on experiments) containing LmjF3.tRNALys.01 (Fig. 5) was cloned, in both orientations, upstream of the phleomycin-luciferase (ble-luc) fusion gene in pNBUC, which does not contain any promoter, and between the LmjF rRNA promoter and the ble-luc gene in pLMRIB (Fig. 6A). The resultant constructs, pNT-5′, pNT-3′, pLMT-5′, and pLMT-3′, were transfected into LmjF promastigotes, and transient luciferase activity was measured (Fig. 6B and C). The luciferase signal obtained for the control which contains the rRNA promoter alone (pLMRIB) was considerably (48-fold) higher than that obtained for the control with no promoter (pNBUC), as expected for transcription mediated by Pol I and Pol II, respectively. In the constructs with no promoter, the presence of the tRNA gene in the sense orientation (pNT-5′) produced a 34% increase in luciferase activity, while the antisense orientation (pNT-3′) caused a strong reduction (74%) in luciferase activity (Fig. 5B). In contrast, both constructs containing the tRNA fragment downstream of the rRNA-gene promoter, pLMT-5′ and pLMT-3′, showed a 69 and 63% decrease in luciferase activity, respectively (Fig. 6C). Control experiments with vectors containing a fragment from the Chr1_0650 gene in place of the tRNA fragment downstream of the rRNA-gene promoter (pLM-72-5′ and pLM-72-3′) did not show any significant reduction in luciferase activity (Fig. 6C). Thus, these data indicate that the region containing LmjF3.tRNALys.01 is capable of termination or attenuation of Pol I transcription in either orientation and of Pol II transcription in at least one orientation. The increase in activity seen in the other (sense) orientation with pNT-5′ presumably reflects the presence of a Pol III promoter within this fragment, as suggested by the nuclear run-on experiments with UV. RNase protection assays performed with total RNA isolated from transiently transfected cultures showed a 52% reduction in the abundance of the luciferase transcripts with pNT-3′ and an increase of 45% with pNT-5′ (compared to the amount of luciferase transcripts with the control pNBUC) (data not shown). This indicates that the observed changes in luciferase activity are a result of an increase or decrease in the abundance of reporter gene transcripts. RT-PCR experiments carried out with poly(A)-tailed RNA isolated from cells transfected with pNT-5′ and pLMT-5′ showed that transcription terminates at similar positions within the tRNA-gene fragment, including the T tract and the C at position 259345, as described above for the parental cell lines, although there was somewhat more heterogeneity in the 3′ ends. Thus, it appears the transcription termination in these constructs accurately reflects the situation within the wild-type locus.
FIG. 6.
The tRNALys-gene region is involved in termination of transcription. (A) Maps of plasmids used in the transfection experiments. Ble-Luc represents the phleomycin-luciferase fusion gene, which is flanked by 5′- and 3′-dihydrofolate reductase intergenic regions from L. major. The tRNA box represents the tRNALys gene from chr3. The arrow on the rRNA box indicates the initiation site of the LmjF rRNA genes. A fragment from open reading frame 72 from chr1 (Chr1_0650) was cloned into pLM72-5′ and -3′. (B and C) Transient-transfection assays. Luciferase activity was tested 24 h after LmjF promastigotes were transfected by electroporation with the constructs indicated. The results shown were obtained from three independent experiments, each performed in triplicate. The numbers above each column represent a relative percentage of activity, with the results obtained with the parental plasmids pNBUC (no promoter) and pLMRIB (with the rRNA promoter) set at 100%. Error bars, standard deviations.
In other systems, deletion of the short T tracts immediately downstream of tRNA genes results in read-through transcription (1). To determine the consequences of the loss of the four Ts located downstream of the LmjF3.tRNALys.01 gene (Fig. 5), they were deleted from pNT-5′, pNT-3′, pLMT-5′ and pLMT-3′, to generate the vectors pNT-5′-ΔT, pNT-3′-ΔT, pLMT-5′-ΔT and pLMT-3′-ΔT, respectively (Fig. 6A). Deletion of the T tract in the promoterless sense orientation construct resulted in a 42% increase in luciferase activity (compare pNT-5′-ΔT and pNT-5 [Fig. 5B]), while deletion of the T tract in the corresponding antisense construct (pNT-3′-ΔT) caused a 106% increase in activity (Fig. 6B). Similar results were obtained with the rRNA promoter constructs (pLMT-5′-ΔT and pLMT-3′-ΔT), where deletion of the T tract resulted in 77 and 103% increases in luciferase activity, respectively (Fig. 6C). Therefore, the deletion of the T tract resulted in an increase in the activity of the reporter gene in all cases, supporting its role in termination of transcription on chr3. However, deletion of the four Ts did not restore luciferase activity to the control levels, suggesting that other sequences such as anticodon stem region also play an important role in transcription termination and/or attenuation.
DISCUSSION
We have used nuclear run-on, RT-PCR, and transient-transfection analyses to characterize transcription initiation and termination of LmjF chr3. Our data suggest that, like chr1 (19), specific Pol II transcription starts upstream of the most-5′ gene of the two long polycistronic clusters. We have also identified a region where Pol III transcription starts for a tRNA gene located at the convergence of these two gene clusters. Termination of Pol II transcription on both DNA strands, as well as Pol III transcription of the tRNA, seems to occur within this region. Thus, we have now characterized the transcriptional organization of two entire chromosomes and have identified sequences involved in both Pol II and Pol III transcription initiation and termination.
Identification and characterization of the Pol II promoters that drive the expression of protein-coding genes in trypanosomatids has proven to be an elusive goal, complicated by factors such as relatively low transcriptional activity and rapid processing of primary transcripts. Putative promoters for actin (2), hsp70 (15), and GARP (11) genes have been reported in Trypanosoma brucei and Trypanosoma congolense, although these conclusions remain controversial (5, 7, 20). Until recently, the only Pol II promoter region that has been extensively characterized is the one driving the expression of the SL RNA (10). These findings, along with the apparent lack of regulation of Pol II transcription and the observation that episomal molecules are transcribed on both strands in Leishmania (41), have led to the hypothesis that Pol II has low specificity in trypanosomatids and that transcription of protein-coding genes can start indiscriminately throughout the genome (14, 20, 39). However, transcriptional analysis of the entire LmjF chr1 (19) showed that the coding strand-specific Pol II transcription that initiated in the strand-switch region between the two long polycistronic gene clusters was at least 10-fold higher than any nonspecific transcription which may have initiated randomly throughout the chromosome. These studies localized several transcription initiation sites within a <100-bp region, and transfection studies support the presence of a bidirectional promoter in this region.
The results reported here for similar analyses of LmjF chr3 show that transcription of the coding strand was ∼8-fold higher than that of the noncoding region and that specific Pol II transcription initiates in the strand-switch region between LmjF3.0010 (gene 1) and LmjF3.0020 (gene 2), near the left telomere, and upstream of LmjF3.0980 (gene 98) at the right telomere (Fig. 1 and 2). In addition, there may be another Pol II TSS in the large intergenic region upstream of LmjF3.0680 (gene 68). Since we did not include fragments from all the genes on chr3, we cannot formally exclude the possibility that there are other TSSs in other areas of the chromosome. As for chr1, several TSSs were mapped in the strand-switch region between LmjF3.0010 and LmjF3.0020, the most 5′ of which were separated by only 247 bp (Fig. 3A). The region of initiation of transcription at the right telomere appears to be unidirectional (away from the telomere), but it also has multiple TSSs. There is no substantial sequence homology between the regions of transcription initiation identified on LmjF chr1 and chr3, and they do not contain any typical Pol II promoter elements. Although each region contains one or two G or C tracts, which can also found in the SL promoter region of chr2, similar-sized G or C tracts are also found in other intergenic regions throughout the genome. Thus, rigorously conserved sequence recognition sites do not appear to be required for Pol II transcription initiation in Leishmania.
Nuclear run-on and transient-transfection experiments (Fig. 1 and 6) indicated the presence of a Pol III promoter for the tRNA gene in the strand-switch region between the two large convergent gene clusters on chr3, although the drug inhibition studies were not totally conclusive (Fig. 4). Assays performed in the presence of 80 μM tagetitoxin, a concentration reported to inhibit Pol III in other trypanosomatids (8, 32), did not have any appreciable effect on the hybridization signal (data not shown) and the inhibition obtained with 160 μM tagetitoxin was only 52%. Others have also reported conflicting results when trying to identify the polymerase involved in transcription of other small RNA genes in trypanosomatids (12, 32, 43). The results obtained here could be explained by the fact the 339-nt tRNA-gene fragment contains 145 nt upstream of the 73-nt tRNA gene and thus this region is most likely transcribed by Pol II (Fig. 5). Hence, the top strand of the tRNA-gene region of chr3 appears to be transcribed by both Pol II and Pol III, while the bottom strand is transcribed only by Pol II. The noncoding (bottom) strand of the tRNA-gene cluster from chr23 is also transcribed by Pol II, but it appears that the coding strand may be transcribed by Pol III only (Fig. 4).
Comparison with tRNA sequences from other trypanosomatids and Saccharomyces cerevisiae indicated that the LmjF3.tRNALys.01 gene contains the two DNA elements, box A and box B (Fig. 5), typically located downstream of TSS in tRNA-gene promoters in eukaryotes (9, 34). A few other Pol III promoters have been characterized in trypanosomatids: in T. brucei the genes coding for the small nuclear RNAs (snRNAs) U2, U3, and U6, as well as the 7SL RNA, are transcribed by Pol III (8, 24). All these genes have a divergently oriented tRNA gene in their 5′-flanking region, and boxes A and B from the neighbor tRNA genes are essential for expression of the snRNAs and the 7SL RNA (24). The 7SL RNA gene from Leptomonas collosoma also requires a companion tRNA gene for efficient expression (3). In most cases, the snRNAs and 7SL RNA genes also require intragenic regulatory elements to achieve a good level of expression (3, 25). We have not yet ruled out the possibility that there is an snRNA gene adjacent to the LmjF3.tRNALys.01 gene, although none is apparent from the sequence.
In higher eukaryotes, mRNA (Pol II) transcription termination is a complex process that depends on the presence of a functional poly(A) signal as well as other downstream signals and factors (30). The polyadenylation sites for LmjF3.0680 (gene 68) and LmjF3.0690 (gene 69) were mapped by RT-PCR. Three polyadenylation sites were localized 145, 155, and 336 bp downstream of the gene 68 stop codon, and two sites were mapped 368 and 440 bp downstream of the gene 69 stop codon (Fig. 5). Nuclear run-on analysis indicated that Pol II-mediated transcription of both large polycistronic gene clusters appears to terminate in the strand-switch region between these convergent clusters in the same vicinity as the Pol III-mediated initiation and termination for the tRNA gene. This conclusion was supported by RT-PCR experiments which identified a region within the tRNA gene where Pol II transcription terminates on the top strand. Interestingly, all the termination sites are located in the base-paired region of the anticodon arm of the tRNA. Transient-transfection experiments (Fig. 6) and RNase protection assays (data not shown) also support the role of the tRNA-gene region in transcription termination. In most tRNA genes, simple clusters of four or more T residues act as signals to terminate transcription (28). Our data indicate that the run of four Ts located immediately downstream of the LmjF3.tRNALys.01 gene is involved in Pol III transcription termination (Fig. 5). The T tract might also be involved in Pol II termination, in conjunction with other sequences such as the tRNA anticodon arm region. We have not yet determined the exact location of transcription termination on the bottom strand, although the nuclear run-on and RNase protection data suggest that it occurs within the tRNA-gene region. In Leishmania tarentolae, it has been shown that transcription of the SL-RNA gene (transcribed by Pol II) is terminated by the T tract located downstream of the gene (38). It is interesting that the region containing the LmjF3.tRNALys.01 gene was also able to terminate Pol I transcription in transient-transfection studies. Although Pol I does not transcribe chr3, the fact that the tRNA-gene region has the capacity to partially terminate (or attenuate) Pol I transcription supports the role of this region in termination of transcription. It is possible that the tRNA-gene region contains sequences that potentially promote the release of any RNA polymerase from the template DNA. Similar studies in T. brucei found that a 2-kb region in the PARP A transcription unit, was able to terminate Pol I transcription (from rRNA, PARP, or VSG promoters), but was unable to block transcription by Pol II (4).
In Saccharomyces cerevisiae, improper termination of two convergent genes resulted in the reduced expression of both genes by a mechanism known as transcriptional collision (29). The presence of a termination region between the two convergent polycistronic units on chr3 from LmjF suggests that Leishmania, like S. cerevisiae, might require separation of adjacent Pol II transcription units by proper termination signals to avoid transcriptional collision. It has been previously speculated that Pol III-gene regions might act as Pol II terminators in T. brucei, since Pol III genes have been found at the 3′ boundary of Pol II transcription units in several cases (17). This also appears to be the case in many (but not all) of the convergent strand-switch regions in LmjF (unpublished data).
The findings reported in this work significantly advance our understanding of transcription of protein-coding genes in trypanosomatids. Our data suggest that specific transcription generally starts in the strand-switch region between the first genes in two “divergent” gene clusters and terminates in the strand-switch region where the polycistronic clusters converge. The latter is often associated with Pol III transcription initiation and termination of tRNAs and/or other small RNAs. Since the trypanosomatid genome sequencing projects have revealed that most genes are organized into large clusters (22), the number of regions where Pol II transcription initiates in these organisms is low compared to other eukaryotes—probably only a few per chromosome.
Acknowledgments
We thank Aaron Leland for technical assistance.
This work was supported by PHS grant 1 U01 AI040599 to K.S., University of Washington Royalty Research Fund grant 95-3310 to P.J.M., and a postdoctoral fellowship from the International Training and Research in Emerging Infectious Diseases (ITREID) program to S.M.-C.
REFERENCES
- 1.Adeniyi-Jones, S., P. H. Romeo, and M. Zasloff. 1984. Generation of long read-through transcripts in vivo and in vitro by deletion of 3′ termination and processing sequences in the human tRNAimet gene. Nucleic Acids Res. 12:1101-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben Amar, M. F., D. Jefferies, A. Pays, N. Bakalara, G. Kendall, and E. Pays. 1991. The actin gene promoter of Trypanosoma brucei. Nucleic Acids Res. 19:5857-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Shlomo, H., A. Levitan, O. Beja, and S. Michaeli. 1997. The trypanosomatid Leptomonas collosoma 7SL RNA gene. Analysis of elements controlling its expression. Nucleic Acids Res. 25:4977-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berberof, M., A. Pays, S. Lips, P. Tebabi, and E. Pays. 1996. Characterization of a transcription terminator of the procyclin PARP A unit of Trypanosoma brucei. Mol. Cell. Biol. 16:914-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton, C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coquelet, H., M. Steinert, and E. Pays. 1991. Ultraviolet irradiation inhibits RNA decay and modifies ribosomal RNA processing in Trypanosoma brucei. Mol. Biochem. Parasitol. 44:33-42. [DOI] [PubMed] [Google Scholar]
- 7.Downey, N., and J. E. Donelson. 1999. Search for promoters for the GARP and rRNA genes of Trypanosoma congolense. Mol. Biochem. Parasitol. 104:25-38. [DOI] [PubMed] [Google Scholar]
- 8.Fantoni, A., A. O. Dare, and C. Tschudi. 1994. RNA polymerase III-mediated transcription of the trypanosome U2 small nuclear RNA gene is controlled by both intragenic and extragenic regulatory elements. Mol. Cell. Biol. 14:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galli, G., H. Hofstetter, and M. L. Birnstiel. 1981. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 294:626-631. [DOI] [PubMed] [Google Scholar]
- 10.Gilinger, G., and V. Bellofatto. 2001. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 29:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, S. V., D. Jefferies, and J. D. Barry. 1996. A promoter directing α-amanitin-sensitive transcription of GARP, the major surface antigen of insect stage Trypanosoma congolense. Nucleic Acids Res. 24:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grondal, E. J. M., R. Evers, K. Kosubek, and A. W. C. A. Cornelissen. 1989. Characterization of the RNA polymerases of Trypanosoma brucei: trypanosomal mRNAs are composed of transcripts derived from both RNA polymerase II and III. EMBO J. 8:3383-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, P. J., J. M. Kooter, and P. Borst. 1987. Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell 51:273-281. [DOI] [PubMed] [Google Scholar]
- 14.Kapler, G. M., K. Zhang, and S. M. Beverley. 1990. Nuclease mapping and DNA sequence analysis of transcripts from the dihydrofolate reductase-thymidylate synthase (R) region of Leishmania major. Nucleic Acids Res. 18:6399-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, M. G. S. 1996. An RNA polymerase II promoter in the hsp70 locus of Trypanosoma brucei. Mol. Cell. Biol. 16:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo, H., G. Gilinger, D. Mukherjee, and V. Bellofatto. 1999. Transcription initiation at the TATA-less spliced leader RNA gene promoter requires at least two DNA-binding proteins and a tripartite architecture that includes an initiator element. J. Biol. Chem. 274:31947-31954. [DOI] [PubMed] [Google Scholar]
- 17.Marchetti, M. A., C. Tschudi, E. Silva, and E. Ullu. 1998. Physical and transcriptional analysis of the Trypanosoma brucei genome reveals a typical eukaryotic arrangement with close interspersion of RNA polymerase II- and III-transcribed genes. Nucleic Acids Res. 26:3591-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Calvillo, S., S. M. Sunkin, S. Yan, M. Fox, K. Stuart, and P. J. Myler. 2001. Genomic organization and functional characterization of the Leishmania major Friedlin ribosomal RNA gene locus. Mol. Biochem. Parasitol. 116:147-157. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Calvillo, S., S. Yan, D. Nguyen, M. Fox, K. D. Stuart, and P. J. Myler. 2003. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell 11:1291-1299. [DOI] [PubMed] [Google Scholar]
- 20.McAndrew, M., S. Graham, C. Hartmann, and C. Clayton. 1998. Testing promoter activity in the trypanosome genome: isolation of a metacyclic-type VSG promoter, and unexpected insights into RNA polymerase II transcription. Exp. Parasitol. 90:65-76. [DOI] [PubMed] [Google Scholar]
- 21.Myler, P. J., L. Audleman, T. deVos, G. Hixson, P. Kiser, C. Lemley, C. Magness, E. Rickell, E. Sisk, S. Sunkin, S. Swartzell, T. Westlake, P. Bastien, G. Fu, A. Ivens, and K. Stuart. 1999. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl. Acad. Sci. USA 96:2902-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myler, P. J., E. Sisk, P. D. McDonagh, S. Martinez-Calvillo, A. Schnaufer, S. M. Sunkin, S. Yan, R. Madhubala, Ivens, A., and K. Stuart. 2000. Genomic organization and gene function in Leishmania. Biochem. Soc. Trans. 28:527-531. [DOI] [PubMed] [Google Scholar]
- 23.Myung, K. S., J. K. Beetham, M. E. Wilson, and J. E. Donelson. 2002. Comparison of the post-transcriptional regulation of the mRNAs for the surface proteins PSA (GP46) and MSP (GP63) of Leishmania chagasi. J. Biol. Chem. 277:16489-16497. [DOI] [PubMed] [Google Scholar]
- 24.Nakaar, V., A. O. Dare, D. Hong, E. Ullu, and C. Tschudi. 1994. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol. Cell. Biol. 14:6736-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakaar, V., A. Gunzl, E. Ullu, and C. Tschudi. 1997. Structure of the Trypanosoma brucei U6 snRNA gene promoter. Mol. Biochem. Parasitol. 88:13-23. [DOI] [PubMed] [Google Scholar]
- 26.Nakaar, V., C. Tschudi, and E. Ullu. 1995. An unusual liaison: small nuclear and cytoplasmic RNA genes team up with tRNA genes in trypanosomatid protozoa. Parasitol. Today 11:225-228. [Google Scholar]
- 27.Parsons, M., R. G. Nelson, K. P. Watkins, and N. Agabian. 1984. Trypanosome mRNAs share a common 5′ spliced leader sequence. Cell 38:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paule, M. R., and R. J. White. 2000. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 28:1283-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prescott, E. M., and N. J. Proudfoot. 2002. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. USA 99:8796-8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 31.Rudenko, G., P. A. Blundell, A. Dirks-Mulder, R. Kieft, and P. Borst. 1995. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell 83:547-553. [DOI] [PubMed] [Google Scholar]
- 32.Saito, R. M., M. G. Elgort, and D. A. Campbell. 1994. A conserved upstream element is essential for transcription of the Leishmania tarentolae mini-exon gene. EMBO J. 13:5460-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauerbier, W., and K. Hercules. 1978. Gene and transcription unit mapping by radiation effects. Annu. Rev. Genet. 12:329-363. [DOI] [PubMed] [Google Scholar]
- 34.Sharp, S., D. DeFranco, T. Dingermann, P. Farrell, and D. Soll. 1981. Internal control regions for transcription of eukaryotic tRNA genes. Proc. Natl. Acad. Sci. USA 78:6657-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, J. J., and R. Lainson. 1987. Ecology and epidemiology: New World, p. 291-361. In W. Peters and R. Killick-Kendrick (ed.), The leishmaniases in biology and medicine, vol. I. Academic Press, London, United Kingdom.
- 36.Steinberg, T. H., D. E. Mathews, R. D. Durbin, and R. R. Burgess. 1990. Tagetitoxin: a new inhibitor of eukaryotic transcription by RNA polymerase III. J. Biol. Chem. 265:499-505. [PubMed] [Google Scholar]
- 37.Stuart, K., T. E. Allen, S. Heidmann, and S. D. Seiwert. 1997. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 61:105-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sturm, N. R., M. C. Yu, and D. A. Campbell. 1999. Transcription termination and 3′-end processing of the spliced leader RNA in kinetoplastids. Mol. Cell. Biol. 19:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swindle, J., and A. Tait. 1996. Trypanosomatid genetics, p. 6-34. In D. F. Smith and M. Parsons (ed.), Molecular biology of parasitic protozoa. Oxford University Press, Oxford, United Kingdom.
- 40.Wincker, P., C. Ravel, C. Blaineau, M. Pages, Y. Jauffret, J. Dedet, P. Bastien, and J. P. Dedet. 1996. The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 24:1688-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong, A. K. C., M. A. Curotto de Lafaille, and D. F. Wirth. 1994. Identification of a cis-acting gene regulatory element from the lemdr1 locus of Leishmania enriettii. J. Biol. Chem. 269:26497-26502. [PubMed] [Google Scholar]
- 42.Worthey, E., S. Martinez-Calvillo, A. Schnaufer, G. Aggarwal, J. Cawthra, G. Fazelinia, G. Fu, M. Hassebrock, G. Hixson, A. C. Ivens, P. Kiser, F. Marsolini, E. Rickell, R. Salavati, E. Sisk, S. M. Sunkin, K. D. Stuart, and P. J. Myler. 2003. Leishmania major chromosome 3 contains two long “convergent” polycistronic gene clusters separated by a tRNA gene. Nucleic Acids Res. 31:4201-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, Y., L. Liu, C. Lopez-Estrano, and S. Michaeli. 2001. Expression studies on clustered trypanosomatid box C/D small nucleolar RNAs. J. Biol. Chem. 276:14289-14298. [DOI] [PubMed] [Google Scholar]
- 44.Yan, S., M. J. Lodes, M. Fox, P. J. Myler, and K. Stuart. 1999. Characterization of the Leishmania donovani ribosomal RNA promoter. Mol. Biochem. Parasitol. 103:197-210. [DOI] [PubMed] [Google Scholar]
- 45.Yan, S., S. Martinez-Calvillo, A. Schnaufer, P. J. Myler, and K. Stuart. 2002. A low background inducible promoter system in Leishmania donovani. Mol. Biochem. Parasitol. 119:217-223. [DOI] [PubMed] [Google Scholar]
- 46.Zomerdijk, J. C. B. M., R. Kieft, P. G. Shiels, and P. Borst. 1991. Alpha-amanitin-resistant transcription units in trypanosomes: a comparison of promoter sequences for a VSG gene expression site and for the ribosomal RNA genes. Nucleic Acids Res. 19:5153-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]