Abstract
Several genes are essential for Cryptococcus neoformans capsule synthesis, but their functions are unknown. We examined the localization of glucuronoxylomannan (GXM) in strain B-3501 and in cap59 mutants B-4131 and C536. Wild-type strain B-3501 showed a visible capsule by India ink staining and immunofluorescence with anticapsular monoclonal antibodies (MAbs) 12A1 and 18B7. B-4131, a mutant containing a missense mutation in CAP59, showed no capsule by India ink staining but revealed the presence of capsular polysaccharide on the cell surface by immunofluorescence. The cap59 gene deletion mutant (C536), however, did not show a capsule by either India ink staining or immunofluorescence. Analysis of cell lysates for GXM by enzyme-linked immunosorbent assay revealed GXM in C536 samples. Furthermore, the epitopes recognized by MAbs 12A1, 2D10, 13F1, and 18B7 were each detected in the cytoplasm of all strains by immunogold electron microscopy, although there were differences in location consistent with differences in epitope synthesis and/or transport. In addition, the cells of B-3501 and B-4131, but not those of the cap59 deletant, assimilated raffinose or urea. Hence, the missense mutation of CAP59 in B-4131 partially hampered the trafficking of GXM but allowed the secretion of enzymes involved in hydrolysis of raffinose or urea. Furthermore, the cell diameter and volume for strain C536 are higher than those for strain B-3501 or B-4131 and may suggest the accumulation of cellular material in the cytoplasm. Our results suggest that CAP59 is involved in capsule synthesis by participating in the process of GXM (polysaccharide) export.
Cryptococcus neoformans is the etiologic agent for cryptococcosis, a major opportunistic mycosis in patients with AIDS (23). Cryptococcosis usually is manifested clinically as a life-threatening meningoencephalitis (21, 23). C. neoformans polysaccharide capsule is considered to be the major virulence factor for this facultative intracellular pathogen (16, 17, 20). The polysaccharide capsule of C. neoformans is believed to contribute to virulence by being antiphagocytic, whereas shed capsular polysaccharide has been associated with a variety of deleterious effects that can affect the host immune response (2, 29). Capsule-deficient strains are avirulent for mice, and hypocapsular strains demonstrate attenuated virulence (17). In recent years genetic tools have been applied to dissect the capsular phenotype. Four genes essential for capsule formation, CAP59, CAP64, CAP60, and CAP10, have been identified by complementation studies of acapsular mutants (6-9). Nevertheless, the roles of these genes in capsule synthesis, assembly, and secretion remain obscure. Homologs of CAP10, CAP59, and CAP64 have been found in other nonencapsulated fungi, suggesting that these genes may be involved in processes other than capsule synthesis (1).
CAP59 was the first capsule gene found to be directly associated with the capsule phenotype and virulence (6), and it is present in all C. neoformans varieties (25). The gene was cloned by complementing an acapsular mutant, B-4131, with a genomic DNA library of C. neoformans B-3501, a reference strain of serotype D. This 1.9-kb CAP59 gene encodes a 458-amino-acid protein of unknown function and is located on chromosome 1. Functional analysis demonstrated that Cap59p contains a putative transmembrane domain at the N terminus that is required for its ability to complement the cap59 acapsular phenotype (10). B-4131 has a missense mutation at position 1345 leading to the change of a Gly to Ser (Table 1) (6). A Δcap59 strain, TYCC33 (C536), was generated by replacement of the wild-type allele with a disruption construct in a wild-type strain to demonstrate that the sequence of the CAP59 gene is involved in capsule formation (6). Both the B-4131 and TYCC33 strains were reported to be acapsular when analyzed by India ink staining and immunofluorescence (6) with capsule binding antibody E1 (12). A homolog of CAP59 known as CMT1 has recently been described to have alpha-1,3-mannosyltransferase activity. Deletion of the gene, however, resulted in a virulent strain with reduced capsule size (27), indicating that CAP59 and CMT1 have different functions in spite of the observed sequence homology.
TABLE 1.
Description of cap59 mutants
| Strain | Genetic description | Capsule |
|---|---|---|
| B-3501 | Wild type | + |
| B-4131 | Missense mutation in cap59 | +b |
| C536 | TYCC33 + pAUG/GUSa | − |
| C566 | F1 of TYCC33 | − |
TYCC33 is a cap59 deletant generated by homologous recombination. pAUG/GUS, empty vector.
Acapsular phenotype by India ink staining.
In recent years we have developed techniques for immunogold labeling of polysaccharide with monoclonal antibodies (MAbs) to glucuronoxylomannan (GXM) (15) that provide the opportunity for ultrastructural characterization of capsule mutants. Given that the function of CAP59 remains uncertain, we used several MAbs to analyze the phenotype of cap59 mutants with respect to intracellular and extracellular GXM localization. The results provide new insights into the function of CAP59 by linking this gene to capsule secretion.
(The data in this paper are from a thesis to be submitted by Javier Garcia-Rivera in partial fulfillment of the requirements for the degree of Doctor of Philosophy degree in the Sue Golding Graduate Division of Medical Science, Albert Einstein College of Medicine, Yeshiva University, Bronx, N.Y.)
MATERIALS AND METHODS
Strains.
C. neoformans strains B-3501, B-4131, and C536 were each grown in YNB (yeast nitrogen base without amino acids)-2% glucose in a rotatory shaker (150 rpm) until late log to early stationary phase at 30°C. A description of the strains used in this study is provided in Table 1.
Cell size and volume.
Three-day-old cells were suspended in India ink and imaged at a magnification of ×100 by using an Olympus AX70 instrument with a RETIGA 1300 (QImaging, Burnaby, Canada). Cell volume was calculated by using the equation (4/3)πr3 after determining the diameter of the cell body (cell wall to cell wall), excluding the capsule. A total of 150 cells were counted for each strain, and the results were analyzed by the t test with the software package in Excel (Microsoft Corporation, Redmond, Wash.).
Immunofluorescence.
Cells from 2-day-old cultures were harvested by centrifugation, washed three times with phosphate-buffered saline (PBS), and suspended to a density of approximately 106 cells/ml in blocking solution (2% bovine serum albumin and 0.5% goat serum in PBS) for 30 min at 37°C. Cell-associated GXM was detected by indirect immunofluorescence with MAbs 12A1 (μκ) and 18B7 (γ1κ) at 10 μg/ml. After MAb binding, the cells were washed three times with PBS and incubated with fluorescein isothiocyanate-conjugated goat anti-mouse (GAM) immunoglobulin M (IgM) or GAM IgG1 (Fisher) at 1 μg/ml for 30 min at 37°C. Cells were washed and suspended in a solution of n-propylgalactate-glycerol-PBS. Fluorescence was observed and digitally photographed by using an Olympus AX70 with a RETIGA 1300. No fluorescence was detected with isotype-matched control MAbs 5C11 (μκ) and ricin 45 (γ1κ), which have specificity for Mycobacterium tuberculosis arabinomannan (24) and ricin toxin, respectively. Figures were prepared by using Photoshop 7.0 (Adobe System Incorporated, San Jose, Calif.).
Capture ELISA for GXM.
Cultures from C. neoformans were grown for 3 days. Cells were collected by centrifugation and washed three times with sterile PBS. Approximately 2 × 109 to 6 × 109 cells were suspended in 5 ml of PBS and then homogenized with 0.5-mm-diameter zirconium-silica glass beads (Biospec, Bartlesville, Okla.) by using a glass bead beater (Biospec) for 4 min to ensure complete lysis. Cell debris was removed by centrifugation at 3,900 × g for 10 min at room temperature with a VSMC-13 centrifuge (Shelton Scientific, Shelton, Conn.). The supernatant was collected and treated with 30 μg of DNase I (Roche, Indianapolis, Ind.) per ml for 1 h at 37°C, followed by treatment with proteinase K (1 mg/ml) overnight at 37°C. The proteinase K was deactivated by boiling the samples for 25 min. GXM was precipitated by adding sodium acetate (10%, wt/vol) and 2.5 volumes of ethanol and incubated overnight at room temperature. The precipitate was suspended in 300 μl of PBS. Volumes of 100 μl were then analyzed for GXM by using a capture enzyme-linked immunosorbent assay (ELISA). A 96-well polystyrene plate (Corning Glass Works, Corning, N.Y.) was coated with 5 μg of GAM IgM (Fisher Scientific, Fairlawn, N.J.) per ml for 1 h and then blocked with 1% bovine serum albumin. In the first step, the solutions containing GXM were placed in the microtiter well for antigen capture by MAb 2D10 (μκ) (10 μg/ml) by incubating samples overnight at 4°C. The plates were then washed five times with a solution of Tris-buffered saline-0.1% Tween 20, followed by the detection of captured GXM with the IgG1 MAb 18B7 (2 μg/ml) for 1 h. Binding of 18B7 was detected with 1 μg of alkaline phosphatase-conjugated GAM IgG1 (Fisher) per ml for 1 h. The ELISA was developed with p-nitrophenyl phosphate disodium hexahydrate (Pierce, Rockford, Ill.) for 1 h, and the absorbance was measured at 405 nm with a Multiscan MS (Labsystem, Helsinki, Finland). Values in the figures represent the averages of two measurements after subtraction of background. The experiment was repeated several times with similar results.
Immunogold labeling.
Three-day-old cells grown in YNB-2% glucose medium were washed with PBS twice and fixed with 4% formaldehyde-1% glutaraldehyde for 3 h. Postfixed cells were treated with 1% osmium tetroxide followed by 1% uranyl acetate, dehydrated through a graded series of ethanol solutions, and embedded in Spurrs resin (Electron Microscopy Science, Fort Washington, Pa.). Ultrathin sections of 70 to 80 nm were prepared in nickel grids. The grids were incubated in 10% H2O2 for 10 min and washed with PBS, followed by etching in a saturated solution of sodium periodate for 10 min and washes with PBS. Grids were then blocked with a solution of 5% goat serum for 1 h and incubated overnight at 4° with 5 μg of MAbs 12A1 (μκ), 2D10 (μκ), 13F1 (μκ), 18B7 (γ1κ), 5C11 (μκ), and ricin 45 (γ1κ) per ml. After MAb binding, the grids were washed with 5% goat serum-0.1% gelatin (Sigma Chemical Co., St. Louis, Mo.)-0.01% Tween 20-PBS. Samples were washed and then incubated with GAM IgM-biotin (Fisher) or GAM IgG1-biotin (Fisher) for 1 h at room temperature. Streptavidin conjugated to 10-nm-diameter gold particles (Ted Pella, Redding, Calif.) was added and left for 2 h at room temperature. Finally, the grids were washed and fixed with 2% glutaraldehyde, followed by additional washes with PBS and double-distilled water and 10% uranyl staining. The immunogold labeling was observed with a JEOL 100 CXII instrument at 80 kV and quantified. t test analysis was performed with Microsoft Excel.
Carbon source utilization by and urease activity of CAP59 and cap59 strains.
B-3501 (MATα), B-4131 (MATα cap59), TYCC33 (MATa ura5 ade2 Δcap59::ADE2), and C566 (MATα Δcap59::ADE2; F1 progeny) strains were compared for their ability to utilize carbon sources by using an API 20C AUX instrument according to the suggestions of the manufacturer (bioMerieux, Hazelwood, Mo.), with a supplement of uracil (22.4 mg/liter). The acapsular MATα prototrophic strain, C566, was isolated by crossing TYCC33 with the wild type. Two methods were used to test their urease activities. A loopful of cells were streaked on Christiansen's urea agar slant (19) and incubated at 30°C for 24 h, and the color change from yellow to pink was monitored periodically. The rapid urease broth method used was as described by Kwon-Chung et al. (22).
RESULTS
Phenotypic cell analysis.
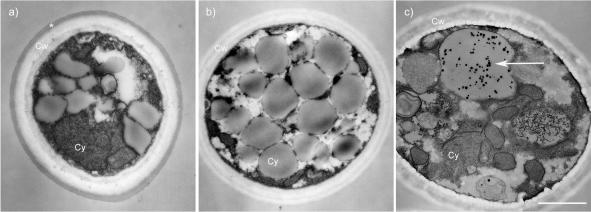
In an effort to gain insight into the function of CAP59, we analyzed the phenotypes of B-3501, B-4131, and C536. The cell diameter was measured and used to calculate the cell volume [(4/3)πr3] for strains B-3501, B-4131, and C536 (Table 2). Reference strain B-3501 had a diameter of 4.5 ± 0.8 μm and a volume of 51.5 ± 29.9 μm3. The cap59 deletant, C536, exhibited increases in diameter (to 5.0 ± 0.9 μm) and volume (to 73.3 ± 42.8 μm3) that were statistically significant (P > 0.05). Both the diameter (4.8 ± 0.5 μm) and volume (59.7 ± 18.8 μm3) of B-4131 were between those of B-3501 and C536 but were significantly different from those of both strains. In addition, we scrutinized the electron micrographs for phenotypic differences between the strains. In the cap59 deletant C536, we observed the presence of globose electron-dense particles inside vacuoles (Fig. 1).
TABLE 2.
Cell diameter and volume
| Strain | Diameter (μm) | Vol (μm3)a |
|---|---|---|
| B3501 | 4.5 ± 0.8 | 51.5 ± 29.9 |
| B4131 | 4.8 ± 0.5 | 59.7 ± 18.8* |
| C536 | 5.0 ± 0.9 | 73.3 ± 42.8** |
*, P = 0.0047; **, P = 0.0000 (n = 150).
FIG. 1.
Phenotypic differences in ultrastructures of strains B-3501 (a), B-4131 (b), and C536 (c). Globose electron-dense inclusion were observed in 0 of 6, 0 of 6, and 8 of 11 cells of strains B3501, B4131, and C536, respectively. *, capsule; Cw, cell wall; Cy, cytoplasm. The arrow points to electron-dense inclusions found in vacuoles of C536. Bar, 1 μm.
Capsule expression in strain B-3501 and cap59 mutants.
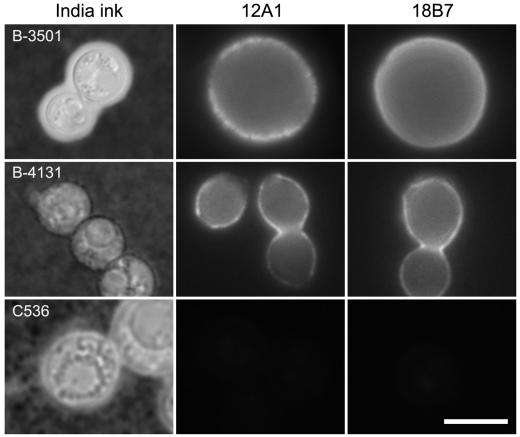
To study the contribution of CAP59 to the capsule phenotype of C. neoformans, the parental strain B-3501 and the cap59 mutants were analyzed for the presence of a capsule by India ink and immunofluorescence staining. As anticipated, only B-3501 cells had an area of particle exclusion surrounding the cell, which is characteristic of the C. neoformans capsule as visualized with India ink staining (Fig. 2). Mutants B-4131 and C536 had no evidence of capsule by India ink staining when grown in YNB-2% glucose (Fig. 2). Indirect immunofluorescence with MAbs 12A1 and 18B7 revealed a thin ring of fluorescence for B-4131 despite the absence of capsule by India ink staining (Fig. 2). Hence, B-4131 had capsular material on the cell surface, but the capsule was too thin to be visible by India ink preparation. The localization of fluorescence in B-4131 was closer to the cell wall than that in B-3501, presumably reflecting differences in capsule size between these strains. In contrast, no capsule was detected in the C536 strain by any of the methods used, suggesting differences between the phenotypes caused by the missense mutation and the deletion of CAP59.
FIG. 2.
India ink preparation and immunofluorescence after staining with MAbs 12A1 and 18B7 of strains B-3501, B-4131, and C536. Bar, 5 μm.
Presence of GXM in B-3501 and cap59 mutants.
Given that the absence of a capsule does not necessarily indicate the absence of polysaccharide synthesis, we proceeded to investigate the presence of GXM in cell lysates of B-3501 and the cap59 mutants by using a capture ELISA (Fig. 3). Since B-3501 and B-4131 have extracellular capsule, the amounts of GXM in the cell lysates of these strains were expected to be higher than that in C536. Similar amounts of GXM were detected in the concentrated cell lysates of B-3501 and B-4131. Despite the absence of extracellular capsular material in C536 as determined by immunofluorescence, small quantities of GXM were detected by capture ELISA.
FIG. 3.
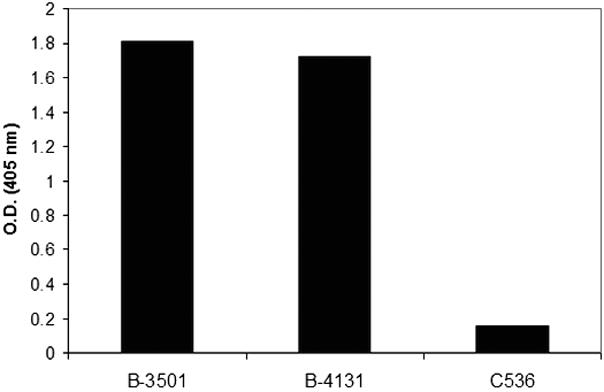
Analysis of the presence of GXM in cell lysates of B-3501, B-4131, and C536 as determined by capture ELISA. Cell lysates from B-3501 and B-4131 samples include capsular material. O.D., optical density.
Ultrastructural localization of GXM in B-3501 and cap59 mutants.
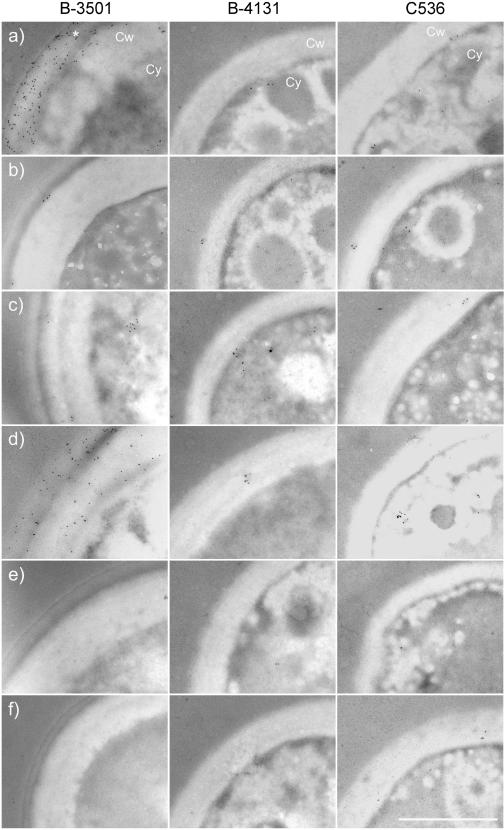
Immunogold electron microscopy was used to ascertain the location of GXM in B-3501 and the cap59 mutants. The presence of cytoplasmic GXM was determined by immunogold labeling with MAbs 12A1, 2D10, 13F1, and 18B7. Labeling of IgM MAb 2D10 in B-3501 resulted in a prominent staining of the cell wall and capsule fraction (Fig. 4a; Table 3). Strong binding intensity was also observed for IgG1 MAb 18B7 in the B-3501 cell wall-capsule region (Fig. 4d; Table 3). A significant reduction of 2D10 and 18B7 labeling was observed in the cell wall-capsule regions of the mutant strains B-4131 and C536, although labeling with 18B7 in the cytoplasm of C536 was significantly higher than in that of B-3501 or B-4131 (Table 3). However, the immunogold pattern obtained with the IgM MAbs (12A1 and 13F1) was different, and more discrete (Fig. 4b and c). Immunogold deposition resulting from MAb 12A1 staining was observed primarily in the cell wall and capsule in a cluster pattern. Examination of images for the immunogold staining with MAb 12A1 revealed that the position of the gold particle clusters varied in the cytoplasm and cell wall, consistent with a vesicular transfer mechanism (Fig. 5). Some clusters were observed in the cytoplasm (Fig. 5a). In contrast, immunogold deposition resulting from MAb 13F1 binding was found primarily as relatively rare single particles in the cytoplasm and cell wall that occasionally congregated into clusters. The intensity of the immunogold staining was much greater for MAbs 2D10 and 18B7 than for MAbs 13F1 and 12A1.
FIG. 4.
Intracellular localization of GXM epitopes recognized by MAbs 2D10 (a), 12A1 (b), 13F1 (c), 18B7 (d), 5C11 (e), and ricin 45 (f) in strains B-3501, B-4131, and C536 as determined by immunogold labeling. *, capsule; Cw, cell wall; Cy, and cytoplasm. Bar, 1 μm.
TABLE 3.
Quantification of gold particles in the cytoplasm and cell wall-capsule of C. neoformans cells as determined by immunogold labeling
| Area of cell | Strain | Labelinga with MAb (no. of gold particles):
|
|||||
|---|---|---|---|---|---|---|---|
| 2D10 | 12A1 | 13F1 | 18B7 | 5C11 | Ricin 45 | ||
| Cytoplasm | B-3501 | 10.8 ± 8.5* | 1.6 ± 1.5 | 5.0 ± 4.4* | 1.6 ± 2.6 | 1.3 ± 1.8 | 0.8 ± 0.8 |
| B-4131 | 6.6 ± 6.0 | 1.2 ± 1.3 | 16.8 ± 6.3 | 0.0 ± 0.0 | 1.2 ± 2.2 | 0.8 ± 1.3 | |
| C536 | 19.8 ± 13.3* | 2.0 ± 2.3 | 5.0 ± 3.9 | 12.8 ± 4.9** | 0.6 ± 0.9 | 1.2 ± 2.2 | |
| Cell wall-capsule | B-3501 | 621.4 ± 401.0* | 7.8 ± 4.1* | 11.6 ± 3.6* | 309.2 ± 239.4 | 1.3 ± 2.2 | 0.2 ± 0.4 |
| B-4131 | 0.2 ± 0.4** | 2.0 ± 1.9 | 5.8 ± 1.6 | 3.8 ± 1.9** | 0.4 ± 0.9 | 0.0 ± 0.0 | |
| C536 | 5.0 ± 3.5** | 5.3 ± 1.3 | 9.0 ± 4.4 | 2.4 ± 2.5** | 1.8 ± 2.2 | 2.0 ± 2.0 | |
*, significant compared to 5C11; **, significant compared to B-3501.
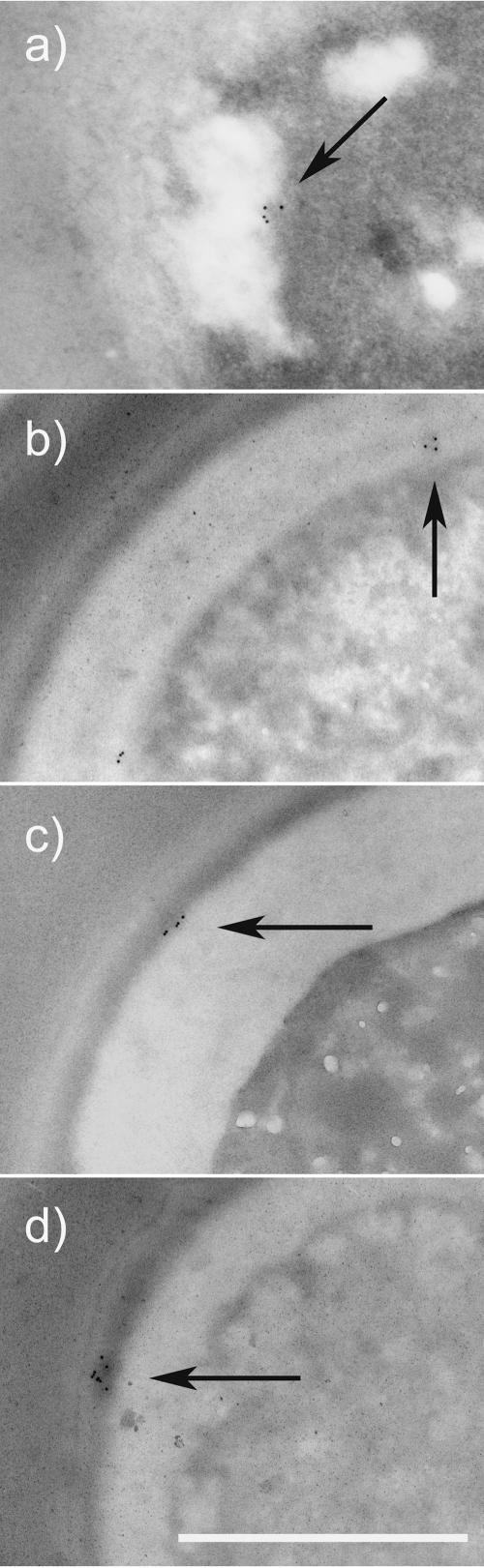
FIG. 5.
Localization of the 12A1 epitope (arrows) throughout the cell cytoplasm (a), cell wall (b and c), and capsule (d). Bar, 1 μm.
Enumeration of gold particles in electron micrographs was used to obtain a quantitative measure of the differences in immunogold staining (Table 3). For MAbs 2D10 and 18B7, the number of gold particles in the cell wall and capsule of strain B-3501 was significantly greater than those for the mutant strains, whereas for MAbs 12A1 and 13F1, the differences were not significant. However, when gold particles in the cytoplasm were enumerated, greater numbers were observed for MAbs 2D10 and 18B7 in C536 than in B-3501 (wild type) or B-4131 (hypocapsular), consistent with accumulation of polysaccharide containing these epitopes in the cap59 deletion strain.
Utilization of oligosaccharides by and urease activity of CAP59 and cap59 strains.
Since Cap59p appeared to be involved in the secretion of GXM, we investigated whether it was also involved in secretion of hydrolytic enzymes. For Saccharomyces cerevisiae to assimilate di-, tri-, or oligosaccharides, such as sucrose, raffinose, or starch, as sole carbon sources, extracellular enzymes must first hydrolyze them to monosaccharides (18). The carbon utilization patterns of the various strains were analyzed with the API 20C AUX system, which can test a variety of carbohydrates as sole carbon sources. Carbon utilization patterns were similar among these strains, except that cap59 deletants (TYCC33 and its F1 progeny C566) failed to use raffinose as a carbon source (Table 4). Raffinose is a trisaccharide composed of galactose, glucose, and fructose. The trisaccharide is hydrolyzed to monosaccharide by combination of invertase and melibiase (18). Since strains B-3501 and B-4131 utilized raffinose but the cap59 deletants did not, it is likely that the secretion of a hydrolytic enzyme that breaks down raffinose is hampered in cap59 deletants. Urease is also a secreted enzyme which breaks down urea and releases ammonia. Only cap59 deletants showed negative results on Christiansen's urea agar for 24 h. In the rapid urease broth test, the cap59 deletant and its acapsular F1 progeny showed negligible reactions, while B-3501 and B-4131 were strongly positive in urease activity within an hour (Table 4). These results also support the notion that Cap59p is involved in secretory function.
TABLE 4.
Utilization of raffinose by and urease activity of CAP59 and cap59 strains
| Strain | Raffinose utilization | Urease activity |
|---|---|---|
| B-3501 | + | + |
| B-4131 | + | + |
| TYCC33 | − | − |
| C566 | − | − |
DISCUSSION
Mutations in CAP59 are known to eliminate the capsular phenotype as detected by India ink preparation, but the function of the CAP59 gene is essentially unknown. In this study we have reexamined the capsular phenotype in C. neoformans cap59 mutants by using MAbs in combination with light, fluorescence, and electron microscopy and a sensitive capture ELISA for the detection of intracellular capsular polysaccharide. Our goal was to compare strains and gain insight into the function of CAP59 and the mechanisms of capsule assembly.
First, the cell diameter was measured and used to calculate the cell volume for strains B3501, B-4131, and C536. Significant differences in cell diameter and volume were observed among strains B-3501, B,-4131 and C536, with deletion of CAP59 resulting in larger cells. Interestingly, B-4131 exhibited an intermediate phenotype between those of B-3501 and C536. Second, we analyzed electron micrographs for evidence of phenotypic differences and observed the presence of electron-dense inclusions in C536 located almost exclusively in vacuolar structures. Hence, disruption of CAP59 affected other features of the cell phenotype in addition to eliminating the capsule. The nature of the inclusions remains unknown, and they may be the result of cell components that precipitate intracellularly because they are not secreted.
As expected from earlier studies (6, 10), strains B-4131 and C536 lacked a capsule when the cells were examined in India ink suspensions. However, when strain B-4131 was studied by indirect immunofluorescence with MAbs 12A1 and 18B7, a thin external coating of polysaccharide was apparent. Previously, strain B-4131 had been analyzed by indirect immunofluorescence with MAb E1, and that study had revealed no evidence for capsule or capsular polysaccharide (6). However, MAb E1 was generated by using serotype A C. neoformans capsular polysaccharide and has specificity primarily for serotype A strains. In fact, MAb E1 binds poorly to strains of serotypes B and D (14), and its specificity for serotype A can be used for typing strains (13). We repeated the analysis of B-4131 with MAb E1 and again confirmed the prior negative results (unpublished data). Since B-4131 is from a serotype D background, the most likely explanation for the negative results with MAb E1 is the weak reactivity of this reagent for serotype D polysaccharide. In contrast, MAb 18B7 binds strongly to cells of all serotypes (4). For strain C536, both India ink and immunofluorescence studies were negative, indicating differences between these two cap59 mutants. This result may be explained by the known genetic differences between strains B-4131 and C536. Strain B-4131 has a missense mutation in the cap59 gene nucleotide 1345 resulting in a Gly to Ser change, whereas C536 is a Δcap59 strain containing empty vector (6).
These two strains were further differentiated by their biochemical properties. In contrast to B-3501 and B-4131, C536 failed to assimilate raffinose and degrade urea, both of which are hydrolyzed by secreted enzymes. Hence, it is conceivable that the missense mutation in B-4131 results in a partially functional protein with lower secretory activity, whereas gene deletion of CAP59 has a pleiotropic effect on secretion that translates into the absence of capsule.
There is evidence that the capsule components of C. neoformans are synthesized in the cytoplasm of C. neoformans (15). This, combined with evidence that CAP59 was involved in secretion, led us to investigate whether acapsular strains had accumulation of GXM in their cytoplasm. The presence of cytoplasmic GXM was determined by a sensitive capture ELISA that can detect nanogram quantities of polysaccharide (5). The presence of GXM was analyzed after concentration of cell lysates. No differences were observed between B-3501 and B-4131, despite the fact that B-4131 is hypocapsular, suggesting saturation of the capture ELISA. Interestingly, GXM was detected in C536 samples, although in significantly smaller amounts. In experiments in which the lysates were not concentrated, the amount of GXM in B-4131 was smaller than that in B-3501 but larger than that in C536 (unpublished data). Pretreatment of B-4131 with dimethyl sulfoxide to remove extracellular GXM, followed by breakage of cells and concentration of lysates, gave results similar to those for C536, suggesting that the amount of GXM in the cytoplasm is relatively small (unpublished data). The presence of GXM in the cytoplasm and capsule as detected by immunofluorescence indicates that cap59 cells make GXM. Hence, the capsular defect in C536 is consistent with a function for CAP59 in trafficking of GXM to the extracellular space. In strain B-4131, the replacement of glycine with serine in the cap59 product yields a protein with weaker functional activity.
In the past, attempts to detect cytoplasmic GXM in acapsular strains have failed (28). In 2001, Feldmesser et al. demonstrated the presence of cytoplasmic GXM in Cap67 with MAbs 2H1, 12A1, and 13F1 by immunogold labeling (15). This discrepancy could reflect the differences in the epitopes recognized by the MAbs used or in the methods used in the preparation of the samples. Our analysis of strain B-3501 and the cap59 mutants revealed the presence of cytoplasmic GXM in all of the strains, including C536. This result implies that the capsule defect in these mutants does not involve a defect in the synthesis of GXM but rather that the problem is in the trafficking of GXM to the extracellular space for capsule assembly.
The gold deposition patterns formed by the binding of MAbs 12A1 and 13F1 to strain B-3501 and its variants were similar to those previously reported for another encapsulated serotype D strain (15). The observation that immunogold labeling with MAb 12A1 localizes to cytoplasmic and cell wall clusters provides strong support for the suggestion that the capsular polysaccharide is synthesized in the cytoplasm and exported to the exterior of the cell in secretory vesicles that traverse the cell wall (15, 26). MAbs 2D10 and 18B7 labeled epitopes found through out the cell wall and capsule, suggesting that the epitope recognized by these MAbs was widely distributed in the cells and in great abundance. The amount of cell wall-capsule labeling was significantly reduced in B-4131 and C536, consistent with the absence of a capsular structure. Interestingly, there was a significant increase in labeling with MAb 18B7 in the cytoplasm of C536, consistent with an accumulation of a GXM constituent recognized by this MAb. The observation that the MAbs used in this study displayed differences in cellular binding patterns is important because it establishes that different epitopes of GXM are found in different locations in the cell. Since the epitopes recognized by these MAbs must reflect structurally different polysaccharide components, this finding suggests that GXM is assembled from components made in geographically distinct areas of the cell. Hence, different components or precursors of GXM may traffic by different cellular pathways.
Our results are also relevant for the association of the capsular phenotype with virulence in C. neoformans. The capsule was conclusively established to be a virulence factor by the demonstration that acapsular mutants were avirulent (3) and that virulence could be restored by genetic complementation (6-9). Furthermore, the association of the capsule with virulence was supported by extensive experimental work that demonstrated mechanisms by which the capsule and soluble capsular polysaccharide could mediate deleterious effects on immune function (29). Our finding of the involvement of Cap59p in secretion of GXM underscores the importance of secretory pathways for expression of the virulence phenotype in C. neoformans. In this regard, we note that C536 and C566 were urease negative and that urease has been associated with virulence (11). Since secretion includes capsule material and possibly other molecules, our results suggest the importance of evaluating other acapsular mutants for secretion defects. It also implies a need for generating acapsular variants that result from genetic lesions in capsule biosynthesis to assess the role of capsule in virulence.
In summary, C. neoformans strains lacking CAP59 or expressing mutated CAP59 manifested pleiotropic effects that included the lack of the capsular phenotype, larger cell size, vacuolar inclusions, the inability to utilize certain disaccharides, and markedly reduced or no urease activity. Since B-3501 and B-4131 have markedly different capsule size but similar rates of urease activity and degradation of raffinose, the missense mutation in B-4131 may not significantly affect the trafficking of these enzymes. Only when the CAP59 gene was deleted were the urease activity and hydrolysis of raffinose hampered.
The most straightforward interpretation of our findings is that CAP59 is essential for the extracellular trafficking and/or secretion of several compounds, including GXM. However, the mechanism by which CAP59 mediates this function remains unknown.
Acknowledgments
We thank Clemenia Cayetano, Leslie Gunther, and Frank Macaluso from the Einstein College of Medicine Analytical Imaging Facility for technical assistance with electron microscopy.
J. Garcia-Rivera is supported by Ruth L. Kirschstein National Research Service Award GM64100. A. Casadevall is supported by National Institutes of Health Awards AI33774, AI13342, and HL59842.
REFERENCES
- 1.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchanan, K. L., and J. W. Murphy. 1998. What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 4:71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulmer, G. S., M. D. Sans, and C. M. Gunn. 1967. Cryptococcus neoformans. I. Nonencapsulated mutants. J. Bacteriol. 94:1475-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall, A., W. Cleare, M. Feldmesser, A. Glatman-Freedman, D. L. Goldman, T. R. Kozel, N. Lendvai, J. Mukherjee, L. A. Pirofski, J. Rivera, A. L. Rosas, M. D. Scharff, P. Valadon, K. Westin, and Z. Zhong. 1998. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 42:1437-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall, A., J. Mukherjee, and M. D. Scharff. 1992. Monoclonal antibody based ELISAs for cryptococcal polysaccharide. J. Immunol. Methods 154:27-35. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. C., and K. J. Kwon-Chung. 1998. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 66:2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. C., and K. J. Kwon-Chung. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. C., B. L. Wickes, and K. J. Kwon-Chung. 1995. Further analysis of the CAP59 locus of Cryptococcus neoformans: structure defined by forced expression and description of a new ribosomal protein-encoding gene. Gene 167:179-183. [DOI] [PubMed] [Google Scholar]
- 11.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dromer, F., J. Charreire, A. Contrepois, C. Carbon, and P. Yeni. 1987. Protection of mice against experimental cryptococcosis by anti-Cryptococcus neoformans monoclonal antibody. Infect. Immun. 55:749-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dromer, F., E. Gueho, O. Ronin, and B. Dupont. 1993. Serotyping of Cryptococcus neoformans by using a monoclonal antibody specific for capsular polysaccharide. J. Clin. Microbiol. 31:359-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dromer, F., J. Salamero, A. Contrepois, C. Carbon, and P. Yeni. 1987. Production, characterization, and antibody specificity of a mouse monoclonal antibody reactive with Cryptococcus neoformans capsular polysaccharide. Infect. Immun. 55:742-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldmesser, M., Y. Kress, and A. Casadevall. 2001. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 147:2355-2365. [DOI] [PubMed] [Google Scholar]
- 16.Feldmesser, M., S. Tucker, and A. Casadevall. 2001. Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol. 9:273-278. [DOI] [PubMed] [Google Scholar]
- 17.Fromtling, R. A., H. J. Shadomy, and E. S. Jacobson. 1982. Decreased virulence in stable, acapsular mutants of Cryptococcus neoformans. Mycopathologia 79:23-29. [DOI] [PubMed] [Google Scholar]
- 18.Johnston, M., and M. Carlson. 1992. Regulation of carbon and phosphate utilization, vol. II. Gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology. Lea & Febiger, Philadelphia, Pa.
- 20.Kwon-Chung, K. J., and J. C. Rhodes. 1986. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect. Immun. 51:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon-Chung, K. J., T. C. Sorrell, F. Dromer, E. Fung, and S. M. Levitz. 2000. Cryptococcosis: clinical and biological aspects. Med. Mycol. 38:205-213. [PubMed] [Google Scholar]
- 22.Kwon-Chung, K. J., B. L. Wickes, J. L. Booth, H. S. Vishniac, and J. E. Bennett. 1987. Urease inhibition by EDTA in the two varieties of Cryptococcus neoformans. Infect. Immun. 55:1751-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navoa, J. A., S. Laal, L. A. Pirofski, G. R. McLean, Z. Dai, J. B. Robbins, R. Schneerson, A. Casadevall, and A. Glatman-Freedman. 2003. Specificity and diversity of antibodies to Mycobacterium tuberculosis arabinomannan. Clin. Diagn. Lab. Immunol. 10:88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petter, R., B. S. Kang, T. Boekhout, B. J. Davis, and K. J. Kwon-Chung. 2001. A survey of heterobasidiomycetous yeasts for the presence of the genes homologous to virulence factors of Filobasidiella neoformans, CNLAC1 and CAP59. Microbiology 147:2029-2036. [DOI] [PubMed] [Google Scholar]
- 26.Sakaguchi, N., T. Baba, M. Fukuzawa, and S. Ohno. 1993. Ultrastructural study of Cryptococcus neoformans by quick-freezing and deep-etching method. Mycopathologia 121:133-141. [DOI] [PubMed] [Google Scholar]
- 27.Sommer, U., H. Liu, and T. L. Doering. 2003. An alpha-1,3-mannosyltransferase of Cryptococcus neoformans. J. Biol. Chem. 21:21. [DOI] [PubMed] [Google Scholar]
- 28.Todaro-Luck, F., E. H. White, E. Reiss, and R. Cherniak. 1989. Immunoelectronmicroscopic characterization of monoclonal antibodies (MAbs) against Cryptococcus neoformans. Mol. Cell. Probes 3:345-361. [DOI] [PubMed] [Google Scholar]
- 29.Vecchiarelli, A. 2000. Immunoregulation by capsular components of Cryptococcus neoformans. Med. Mycol. 38:407-417. [DOI] [PubMed] [Google Scholar]