Abstract
TSC1 acts coordinately with TSC2 in a complex to inhibit mTOR, an emerging therapeutic target and known promoter of cell growth and cell cycle progression. Perturbation of the mTOR pathway, through abnormal expression or function of pathway genes, could lead to tumorigenesis. TSC1 and TSC2 expression is reduced in invasive breast cancer as compared with normal mammary epithelium. Because single nucleotide polymorphisms (SNPs) in regulatory genes have been implicated in risk and age at diagnosis of breast cancers, systematic SNP association studies were performed on TSC1 and TSC2 SNPs for their associations with clinical features of breast cancer. TSC1 and TSC2 haplotypes were constructed from genotyping of multiple loci in both genes in healthy volunteers. SNPs were selected for further study using a bioinformatics approach based on SNP associations with drug response in NCI-60 cell lines and evidence of selection bias based on haplotype frequencies. Genotyping for five TSC1 and one TSC2 loci were performed on genomic DNA from 1,137 women with breast cancer. This study found that for TSC1 rs7874234, TT variant carriers had a 9-year later age at diagnosis of estrogen receptor positive (ER+), but not ER−, ductal carcinomas (P = 0.0049). No other SNP locus showed an association with age at diagnosis, nor any other breast cancer phenotype. TSC1 rs7874234 is hypothesized to be functional in ER+ breast cancer because the T allele, but not the C allele, may create an estrogen receptor element (ERE) site, resulting in increased TSC1 transcription and subsequent inhibition of mTOR.
Keywords: Breast cancer risk, SNP, TSC1/TSC2, Case study
Background
Breast cancer is one of the most frequently diagnosed cancers in women [1], where etiology is postulated to be polygenic [2]. Polymorphisms in genes contributing to tumorigenesis are less penetrant, but may account for the majority of breast cancers due to their higher frequency in the general population [2]. In fact, single nucleotide polymorphisms (SNPs) have been implicated in risk and age at diagnosis of breast cancers [3, 4]. One such polymorphism in MDM2 was found to lead to increased risk for earlier age at diagnosis of estrogen receptor (ER) positive breast cancers in women carrying the variant allele of the SNP [5].
Although breast cancers are also associated with mutations in the highly penetrant breast cancer susceptibility genes, BRCA1 and BRCA2, fewer than 10% of cases can be attributed to this etiology [2]. What is known based on BRCA1 and BRCA2 is that these familial syndromes are associated with particular breast cancer subtypes [6]. Breast cancer is a heterogeneous disease that is reflective of its molecular features and histology [7]. Whereas BRCA1 carriers frequently develop hormone receptor negative disease, the phenotype in BRCA2 carriers reflects the distribution observed for spontaneous breast cancer [6]. Therefore, other genetic contributors, including SNPs, may also be reflected in tumor heterogeneity.
Tuberous sclerosis is an autosomal dominant disorder that results from mutations in the tumor suppressor genes, TSC1 or TSC2 [8–10]. Tuberous Sclerosis is characterized by benign hamartomas that affect multiple systems including the brain, skin, heart, and kidneys [8–10]. Lymphangiomyomatosis, a manifestation of tuberous sclerosis seen in 30–40% of affected women, shares some of the same characteristics of breast cancers including the following: gender-dimorphism, estrogen-modulation, increased cell proliferation and migration [11–13]. TSC1 acts to stabilize TSC2, preventing its degradation and both function together as the TSC1/TSC2 heterodimer [8, 10]. This complex inhibits the G protein, Rheb, which acts upstream of the mammalian target of rapamycin (mTOR) kinase [10, 14]. The serine–threonine kinase mTOR plays a central role in the control of cell growth and proliferation through phosphorylation of its effector molecules, 4E-BP1 and S6K1 [15]. Activation of the pathway occurs in response to growth factors, amino acids, and nutrients, leading to mRNA translation and ribosome biogenesis [15]. TSC1/TSC2 complex when not functional, leads to uncontrolled mTOR activity causing uncontrolled cell growth and tumor formation. Rapamycin inhibits mTOR and has been used successfully to treat individuals with tuberous sclerosis [16]. Furthermore, analogs of rapamycin are currently being tested in breast and other cancer clinical trials [14].
Despite the implications highlighted by tuberous sclerosis and the key position of TSC1 and TSC2 in the mTOR pathway, few studies have examined the role of TSC1 and TSC2 in breast cancer [17]. We investigated a panel of 18 TSC1 and 14 TSC2 SNPs by first mapping their haplotypes and using bioinformatics approaches to further select potentially functional SNPs. Five germline TSC1 SNPs and one germline TSC2 SNP were evaluated from women with breast cancer and correlated with clinicopathologic characteristics. The data from these studies indicates that estrogen signaling may modulate the effect of a TSC1 SNP in age at diagnosis of breast cancer.
Methods
Subjects
The cohort consisted of 1,137 consecutively enrolled patients, invited to participate in this prospective study from 2004-present, through the Stacy Goldstein Breast Cancer Center at The Cancer Institute of New Jersey (CINJ). Over 95% of eligible individuals gave consent for participation. A history of biopsy-proven breast cancer was verified by pathology records and confirmed on review by our institutional breast pathologist. Fewer than 5% of cases were not available for review and for those pathological features were based on pathology reports from other institutions. Lobular carcinoma in situ (LCIS) was excluded. Negative estrogen receptor (ER) staining was defined as<10%. BRCA1/2 testing was performed where clinically indicated and patients with known BRCA1/2 mutations were then excluded from age at diagnosis analysis due to potential confounding bias. Investigations were performed with prior approval by the University of Medicine and Dentistry of New Jersey Institutional Review Board.
Determining the haplotype structure
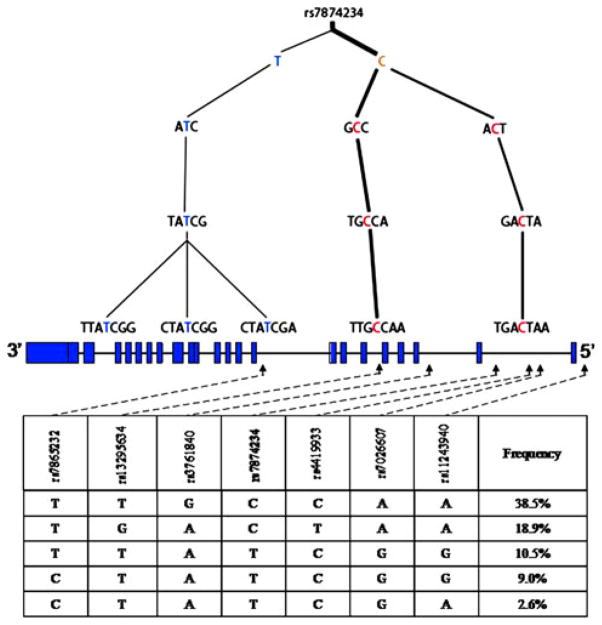
A list of 18-tagged SNPs in TSC1 and 14-tagged SNPs in TSC2 (Table 1) was generated based on HapMap data, representing the minimum number of SNPs necessary to complete the haplotype for TSC1 and TSC2. The SNP tagging formalism employed an information-theoretic definition of haplotype diversity, and the optimal tag SNPs were chosen using a greedy procedure [18] that minimized the haplotypic uncertainty (unpublished). This SNP tagging algorithm was designed independently of Haploview Tagger. It outperforms Tagger giving similar but better results, requiring slightly fewer SNPs to tag the haplotypes. Genomic DNA isolated from lymphoblastoid cell lines (LCLs) was genotyped to determine frequency of haplotypes (demonstrated for one locus in each gene; Fig. 1 and Online Resource 1). LCLs were obtained from Coriell Institute for Medical research (Camden, NJ, USA) and represent DNA from healthy individuals. For both TSC1 and TSC2, only haplotypes occurring at greater than 2% are depicted.
Table 1.
TSC1 and TSC2 SNPs used to generate haplotype structure
| Gene | rs# | Location | Allele |
|---|---|---|---|
| TSC1 | 3761840 | Intron 2 | C/T |
| 11243929 | Intron 19 | C/T | |
| 2809243 | Exon 22, 3′UTR | A/G | |
| 739441 | Exon 22, 3′UTR | C/T | |
| 2519757 | Intron 16 | C/T | |
| 739442 | Exon 22, 3′UTR | A/G | |
| 11243940 | 5′end (not in gene) | A/G | |
| 10491534 | 3′UTR | C/T | |
| 2809244 | 3′UTR | A/C | |
| 7026607 | Intron 1 | A/G | |
| 1076160 | Intron 19 | C/T | |
| 7874234 | Intron 1 | C/T | |
| 7865232 | Intron 8 | C/T | |
| 6597584 | Intron 19 | A/C | |
| 13295634 | Intron 5 | G/T | |
| 7870151 | Intron 13 | A/C | |
| 4419933 | Intron 1 | C/T | |
| 1073123 | Exon 10, missense mutation | A/G | |
| TSC2 | 2074969 | Intron 11 | A/G |
| 17654678 | Intron 15 | G/T | |
| 30259 | Intron 32 | C/T | |
| 13335638 | Intron 37 | C/T | |
| 8063461 | Intron 16 | A/G | |
| 2074968 | Intron 10 | C/G | |
| 2073636 | Intron 5 | A/G | |
| 8050755 | Intron 16 | C/T | |
| 13331451 | Intron 22 | C/T | |
| 7190284 | Intron 16 | A/C | |
| 1800720 | Intron 5 | C/T | |
| 12325450 | Intron 20 | C/T | |
| 13337626 | Exon 23 | C/T | |
| 8047396 | Intron 26 | C/G |
UTR untranslated region
Fig. 1.
Haplotype structure and gene map of TSC1 for Caucasians The haplotype structure of TSC1 determined from analysis of LCLs is shown, along with the gene structure, haplotype block for rs7874234, and relevant haplotype frequencies. Below the haplotype trees, is the gene structure of TSC1; boxes indicate exons with intervening spaces representing intronic sequence. Below the gene structure is the haplotype block and corresponding haplotype frequencies. Dashed lines and arrows indicate the SNP position within these blocks and location on the corresponding gene map above. The haplotype structure and corresponding haplotype frequencies represent the SNPs immediately adjacent, either right or left of the reference loci, rs7874234
Candidate TSC1 and TSC2 SNPs for Clinical Study
Candidate SNP selection for further study in the breast cancer cohort was achieved by two different approaches. The first approach searched for SNPs manifesting signatures of natural selection in the pattern of genotype correlations with nearby SNPs. To identify genetic variants that deviate from the standard assumptions of selective neutrality, a previously described and publicly available map of recent positive selection of the human genome (Haplotter, http://hg-wen.uchicago.edu/selection/haplotter.htm) was utilized. In Haplotter, recent positive selection is determined using a haplotype-based approach that looks for enrichment of the classic signal for strong directional selection using the phase II data of the HapMap. Haplotter utilizes a test statistic called the integrated haplotype score (iHS), which is a measure that includes the degree of haplotype homozygosity around a given SNP [19]. Highly positive and negative his scores denote SNPs that harbor higher haplotype homozygosity, compared with other SNPs with similar allele frequencies in the genome. Using this methodology, SNPs in TSC1 (rs7874234, rs1076160, rs1073123, and rs3761840) and SNPs in TSC2 (rs13335638) were identified.
The second approach was based on associations between the genotypes and the response of tumor derived cell lines to standard chemotherapeutic agents (Online Resource 2) [20, 21]. The mutational status of p53, the genotypes of 109,687 SNPs (Affymetrix 125K chip), and the GI50 data for the NCI60 cell panel of tumor derived cell lines was obtained from the NCI/NIH Developmental Therapeutics Program web site, http://www.dtp.nci.nih.gov. A univariate test was undertaken for 132 standard agents to evaluate allelic differences in the GI50s. Specifically, the average log GI50 [X = − log10(GI50)] for cells for each of the three genotypes of a given locus (AA, Aa, and aa) were calculated for cells either wild-type or mutant for p53. Subsequently, the probability (P value) was computed that just by chance the difference for the following groupings either was equal to or larger than the actual measurement:(a) Xa – XAA or (b) Xaa – XA, or (c) XAA – Xa, or (d) XA – Xaa, or (e) [Xaa – XaA and XaA – XAA], and (f)[XAA – XaA and XaA – Xaa]. These probabilities were estimated using a permutation test (106 permutations) that preserved the allele or genotype group sizes but permuted the samples among the groups. Results P < 0.05 were considered significant and P < 0.1 marginally significant. A multiple hypothesis test was performed for allelic differences in the GI50s across the entire panel of drugs. A Fisher’s exact test to compute the statistical significance of observing h univariate hits for a SNP on a total of D = 132 drugs, given that overall H significant hits are observed after testing S reference SNPs on the D drugs. All 109,687 Affymetrix genotyped SNPs were chosen as a reference set. Using this methodology, TSC1 rs2809243 and TSC2 rs13335638 were identified (Online resource 2).
Therefore a total of five SNPs in TSC1 and one SNP in TSC2 were further analyzed for associations with breast cancer phenotypes, i.e. associations of age at diagnosis and breast cancer subtype (ductal versus lobular), ER status (ER-positive versus ER-negative) and menopausal status as well as recurrence.
Genotyping
Genomic DNA was extracted from 1 ml of peripheral blood, obtained through venipuncture, using a spin column-based method according to the manufacturer’s protocol (QIAGEN). Genotyping for TSC1 and TSC2 SNPs was performed using Taqman assays on the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). Briefly, reactions were performed using 5–10 ng genomic DNA in 10 μl volume. For TSC1 rs7874234, rs1076160, rs2809243, rs3761840, and rs1073123 and for TSC2 rs13335638, PCR cycling conditions were 50°C for 2 min, 95°C for 10 min, followed by 45 cycles of 92°C for 15 s and 60°C for 1 min. Conditions were modified to optimize reactions for the following several loci: TSC1 rs11243940, conditions were identical except only 40 cycles (instead of 45 cycles) were performed; for TSC1 rs739442 and rs10491534, conditions were identical except the final annealing temperature was 58 °C.
Statistical Analysis
A permutation test was performed to determine the statistical significance of differences in mean age at diagnosis between different genotype groups (e.g., wild-type homozygote or heterozygote vs. variant homozygote). This permutation test was chosen because it is non-parametric, with the assumption that all genotype groups, or categories, are equivalent and making no assumptions about the age of diagnosis distribution. Fisher’s exact test was used to determine the statistical significance of the association between categorical values for each genotype group. The odds ratio and 95% confidence interval were then computed using a Bayesian estimate for the odds ratio posterior distribution.
Results
Analysis in LCLs and haplotype generation
Using genotype data generated by the Hapmap project, the minimal set of TSC1 and TSC2 SNPs sufficient to reconstruct their haplotypes was determined. These 18-tagged SNPs in TSC1 and 14-tagged SNPs in TSC2 were analyzed in LCLs, obtained from healthy individuals, and used to generate haplotype frequencies and are demonstrated for two loci (Fig. 1 and Online Resource 1). For TSC1 rs7874234, since genotype frequencies were similar between Caucasian and African American populations (Table 3), only the haplotype tree for the Caucasian population is shown (Fig. 1). However, since genotype frequencies differed between the Caucasian and African American populations, haplotype trees for both populations were generated for TSC2 rs13335638 (Online Resource 1).
Table 3.
Genotype Frequencies for relevant TSC1 and TSC2 SNPs for all populations in the CINJ breast cancer cohort
| SNP | Genotype | Caucasian, n (%) | African American, n (%) | Asian, n (%) | Hispanic, n (%) |
|---|---|---|---|---|---|
| TSC1 rs7874234 | CC | 503 (59.5) | 35 (57.4) | 54 (75.0) | 45 (58.4) |
| CT | 306 (36.2) | 20 (32.8) | 17 (23.6) | 26 (33.8) | |
| TT | 36 (4.3) | 6 (9.8) | 1 (1.4) | 6 (7.8) | |
| TSC2 rs13335638 | CC | 30 (3.5) | 18 (29.0) | 0 (0) | 7 (9.0) |
| CT | 266 (31.0) | 26 (41.9) | 5 (7.1) | 19 (24.4) | |
| TT | 563 (65.5) | 18 (29.0) | 65 (92.9) | 52 (66.7) |
The major TSC1 haplotypes generated, with rs7874234 as the reference locus, accounted for 38.5, 18.9, 10.5, 9, and 2.6% of the genotypes in the Caucasian population (Fig. 1) and was similar in African Americans (Table 3). The C allele was more prevalent and represented 57.4% of the haplotypes. The major TSC2 haplotypes generated, with rs13335638 as the reference locus, accounted for 63.0%, 7.6%, 6.1%, 3.2% and 3.0% of the genotypes in Caucasians, as depicted in Online Resource 1. In the Caucasian population, the major allele was the T allele for this TSC2 SNP. Haplotype trees for both Caucasian and African American populations are shown for TSC2 (Online Resource 1). For African Americans, there was more heterogeneity, and the major haplotypes occurred at frequencies of 24.0, 22.2, 16.2, 11.0, 93, 5.1, and 2.0% (with six haplotypes occurring at 2.0%). In the African American population, both the C and the T allele are represented almost equally, with one C allele haplotype accounting for 22.2% of the genotypes, and one T allele haplotype accounting for 24.0% of the genotypes.
Demographics and SNP Frequencies in the Breast Cancer Cohort
The demographics for the breast cancer cohort are depicted in Table 2. The data shows that the majority of women were Caucasian and the majority of cancers were ductal in origin. The average age at diagnosis was 51.5 years, with patients ranging from 19–89 years of age. Nearly 75% of all breast cancers were ER positive.
Table 2.
Demographics of the breast cancer cohort
| n | % | |
|---|---|---|
| Ethnicity | ||
| African American | 63 | 5.6 |
| Asian | 72 | 6.4 |
| Caucasian | 869 | 77.5 |
| Hispanic | 78 | 7 |
| Other | 39 | 3.5 |
| Type of Breast Cancer | ||
| Ductal | 807 | 83.9 |
| Lobular | 94 | 9.8 |
| Other | 61 | 6.3 |
| Age at Diagnosis, years | ||
| Mean | 51.5 | |
| Median | 50 | |
| Range | 19–89 | |
| ER Status | ||
| Neg | 235 | 25.4 |
| Pos | 692 | 74.6 |
Population-specific genotype frequencies were observed for TSC1 rs7874234 and TSC2 rs13335638 (Table 3). TSC1 genotype frequencies did not deviate from Hardy–Weinberg equilibrium (HWE) for any of the races in this cohort. TSC1 rs7874234 genotype frequencies were similar between the populations depicted, except for the Asian population. TSC2 rs13335638 genotypes were in HWE for all populations except the Hispanic subset. Both Asian and Hispanic populations represent a small number of individuals and heterogeneity in area of participant origin. In Caucasians, the TT genotype was observed in 65.5% of the population. However, in the African American population, both CC and TT genotypes were equally prevalent (each accounting for 29% of the population).
Association between TSC1 and TSC2 SNPs and breast cancer phenotypes
Six SNPs across TSC1 and TSC2 were evaluated for associations with breast cancer phenotypes (Table 4). For all comparisons, ancestral homozygotes versus variant homozygotes were used to determine odds ratios and P values. Associations for TSC1 and TSC2 SNPs with breast cancer subtype (ductal versus lobular), ER status (ER-positive versus ER-negative) and menopausal status at diagnosis were evaluated but no significant associations were found. A trend was observed for an association between ER status and TSC1 rs1073123 genotype, though it did not reach significance. Likewise, for rs7874234, the mean age at diagnosis between ancestral and variant homozygotes differed by 3.4 years, but this did not reach significance.
Table 4.
Association between TSC SNPs and breast cancer phenotypes
| SNP | Ductal versus lobular
|
ER− versus ER+
|
Pre- versus post- menopausal diagnosis
|
Δ Mean age at diagnosisa | P value | |||
|---|---|---|---|---|---|---|---|---|
| OR | P value | OR | P value | OR | P value | |||
| TSC1 rs7874234 CC vs. TT | 0.21 (0.03–1.53) | 0.09 | 0.73 (0.38–1.42) | 0.36 | 1.52 (0.68–3.41) | 0.31 | +3.36 | 0.06 |
| TSC1 rs1076160 CC vs. TT | 0.85 (0.51–1.42) | 0.53 | 1.01 (0.66–1.53) | 0.98 | 1.02 (0.66–1.58) | 0.91 | −0.14 | 0.44 |
| TSC1 rs2809243 AA vs. GG | 1.50 (0.82–2.76) | 0.19 | 1.09 (0.69–1.73) | 0.71 | 0.8 (0.50–1.28) | 0.34 | −1.3 | 0.12 |
| TSC1 rs1073123 AA vs. GG | 0.70 (0.09–5.50) | 0.74 | 0.39 (0.14–1.08) | 0.06 | 0.72 (0.22–2.38) | 0.58 | +0.75 | 0.41 |
| TSC2 rs13335638 TT vs. CC | 1 (0.38–2.63) | 1 | 1.23 (0.61–2.46) | 0.56 | 1.22 (0.54–2.79) | 0.63 | −1.18 | 0.47 |
Values reflect analysis of entire breast cancer cohort
All SNP loci were evaluated for association with age at diagnosis. One TSC1 SNP, rs7874234, showed a significant association with age at diagnosis as depicted in Fig. 2. To reduce the heterogeneity in the study population and potential confounders, samples were stratified into groups with the same cancer subtype and ethnic background. In a case-only analysis of rs7874234, for Caucasian women who had ER+ ductal carcinomas, CC, CT, and TT carriers had an average age at diagnosis of 52, 55 years and 61 years, respectively. Homozygous variant TT carriers had a 9-year later age at diagnosis of ER+ ductal carcinomas as compared with CC carriers (P = 0.0049). When comparing all three genotypes, the curve for heterozygotes fell in between the TT and CC curves, and associated with a 3-year later age at diagnosis than CC carriers and a 6-year earlier age at diagnosis than TT carriers (P = 0.00036). When stratifying the analysis by post-menopausal status, post-menopausal women with ER+ ductal carcinomas showed a similar pattern of genotypes for age at onset as was observed in Fig. 2 (data not shown). Specifically, an additive effect was observed in the analysis of post-menopausal women with ER+ ductal carcinomas. CC, CT, and TT carriers were diagnosed at 61, 64, and 68 years of age, respectively (P = 0.00046). None of the other five SNPs showed significant associations with age of diagnosis by any stratification.
Fig. 2.
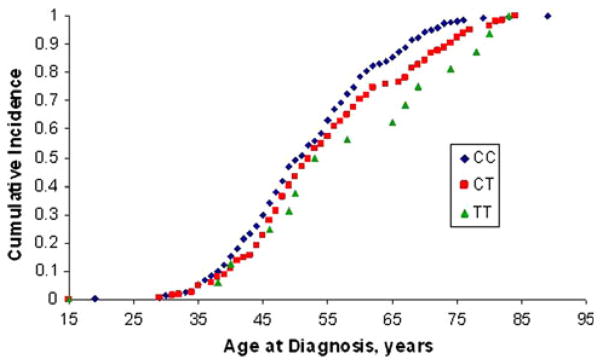
TT genotype of TSC1 rs7874234 associates with a later age at diagnosis of ER+ ductal carcinomas in Caucasian women cumulative incidence as a function of age at diagnosis was evaluated for TSC1 rs7874234 to determine genotype-specific effects. The breast cancer population demonstrates ductal ER+ carcinomas in Caucasian women. CC carriers are depicted by filled diamonds, CT carriers by filled squares, and TT carriers by filled triangles
Discussion
Due to the ubiquitous role of TSC1 and TSC2 in the mTOR pathway and the pathway in tumor biology [22], a systematic evaluation of clinical associations with SNPs in TSC1 and TSC2 was undertaken. Analysis of TSC1 SNP rs7874234 in this breast cancer cohort showed that in Caucasian women, variant homozygote TT carriers developed ductal ER+ breast carcinomas on average 9 years later than CC carriers. Furthermore, the intermediate age of diagnosis of heterozygotes in comparison to either homozygote indicates an additive effect. The largest differential appears in the postmenopausal age group (assuming the average age of menopause in the US is 51 years of age). No other significant associations were observed for other TSC SNP loci and other clinico-pathologic variables, including ER status, breast cancer subtype and menopausal status.
The effect observed with TSC1 rs7874234 indicates a deleterious effect with the CC genotype and that the TT genotype confers protection against earlier development of ER+ ductal carcinomas. Alternatively, a later age at diagnosis for TT carriers could also mean that tumors in TT carriers are slower-growing and take longer to reach a threshold for detection than CC carriers. In silico analysis showed an estrogen receptor element (ERE) within the flanking sequence of rs7874234. Although there is high variability in ERE sequences, comparison of known EREs in other human genes, revealed sequence homology between “GTTAG” in TSC1 with the ERE identified for Human calbindin-D9k [23]. However, this homology only exists for the T allele. This is suggestive that different alleles in rs7874234 may affect ER binding.
Taken together, these findings support the possibility that the T allele may mediate estrogen-specific effects in risk towards later onset breast cancer. It is hypothesized that the T allele allows for ER to bind to this ERE in TSC1, activating TSC1 transcription and increased inhibition of mTOR, delaying breast cancer in TT carriers. The C allele, however, does not allow for ER binding, and therefore there is no increased inhibition of mTOR, furthering earlier tumorigenesis in CC carriers.
The post-menopausal effect observed fits nicely in our hypothesized mode of action for the T allele: if the T allele increases binding affinity of activated ER and hence leads to increased TSC1 transcription, this would make TSC1 in carriers more sensitive to the low levels of circulating estrogens in post-menopausal women. In premenopausal women, because the circulating levels of estrogens are much higher, ER binding to the TSC1 ERE is not limiting. Furthermore, effects of estrogens on other cell pathways may be more critical in premenopausal women [24]. Other SNP association studies have demonstrated similar post-menopausal effects [25–27]. A recent study of SNPs in folate and alcohol metabolic pathway genes and breast cancer risk showed an association with a SNP in 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (MTRR) and increased risk for breast cancer in postmenopausal but not premenopausal women [25]. Another study investigated the role of SNPs in ER-alpha and other estrogen-metabolizing genes and breast cancer risk in Chinese women and found that postmenopausal but not premenopausal breast cancer risk was associated with a heterozygous CYP17 genotype [26].
One advantage of using this study cohort is the availability of extensive clinico-pathologic information that was collected for all study participants. Analysis was enhanced by the ability to reduce heterogeneity within the study cohort. SNP selection which was supported by bioinformatic analysis reduced the number of SNPs to be analyzed, making this a cost-effective approach. This analysis was stratified by race, while other subgroup analysis was limited due to small numbers.
Further study in a larger cohort would allow the detection of associations in other subgroups. For example, analysis of this SNP in the African American population showed no significant association with age at diagnosis of ductal ER+ breast carcinomas; however, this may be due to the small number of African American patients with a TT genotype. Furthermore, defining the molecular mechanism of SNP functionality would further support the association observed for rs7874234. TSC1 has not been previously shown to be regulated by estrogen, and while informatics gave evidence for a plausible explanation for rs7874234 functionality, the possibility exists that another SNP, not rs7874234, could be the functional SNP. Therefore, the effect observed could be due to other SNP(s) in the haplotype represented by rs7874234. As a case-only study, a limitation of this study is that it only evaluated one aspect of risk, i.e., age at diagnosis, whereas overall risk for development of breast cancer would provide further clinical utility in potentially identifying individuals at risk for the disease and knowing the optimal time for screening and prevention.
In this cohort, no association was found between any TSC SNPs and recurrence (data not shown). Although analysis of drug response in the NCI-60 cell lines demonstrated genotype-specific effects for another TSC1 SNP, rs2809243, these results were dependent on p53 status (Online Resource 2). In a p53 mutant background, the genotypes for rs2809243 significantly differed in their growth inhibition to several different chemotherapeutic agents(P = 4.7 × 10−7). However, the heat maps showed a divergent effect between a p53 mutant versus p53 wild-type background. For example, the TT genotype associated with a better response to alkylating agents in a p53 mutant background, while the CC genotype associated with better response to these same agents in a p53 wild-type background, indicating an opposing effect between p53 mutant and p53 wild-type backgrounds for the same class of drugs (Online Resource 2). Overall, the T allele associated with better response in the p53 setting, while the C allele correlated with better response in a mutant p53 background. Since upwards of 30% of tumors harbor p53 mutations in breast cancer [28], the absence of a recurrence-phenotype in the breast cancer cohort, where p53 status is unknown, may not reflect a true lack of association.
In summary, we found that the TSC1 rs7874234 variant associated with delayed age at diagnosis of ER-positive ductal carcinomas in Caucasian women. The observed findings are intriguing and bear confirmation in other breast cancer populations.
Acknowledgments
We would like to thank all of the study participants at the Stacy Goldstein Breast Cancer Center at The Cancer Institute of New Jersey, as well as the researchers at the CINJ Tissue Analytic Services who provided support for sample collection, storage and distribution, Joseph Miktus and Jacqueline Harris. This work was supported by the Breast Cancer Research Foundation (Hirshfield/ Levine), Ohl Foundation (Toppmeyer), New Jersey Commission on Cancer Research (Vazquez), and the Ruth Estrin Goldberg Memorial for Cancer Research (Hirshfield).
Abbreviations
- TSC1
Tuberous sclerosis 1
- TSC2
Tuberous sclerosis 2
- SNP
Single nucleotide polymorphism
- ER
Estrogen receptor
- ERE
Estrogen receptor element
- mTOR
Mammalian target of rapamycin
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-010-1062-1) contains supplementary material, which is available to authorized users.
Contributor Information
Madhura S. Mehta, Department of Medicine, Division of Medical Oncology, The Cancer Institute of New Jersey, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08901, USA
Alexei Vazquez, Department of Radiation Oncology, The Cancer Institute of New Jersey, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08901, USA.
Diptee A. Kulkarni, Department of Medicine, Division of Medical Oncology, The Cancer Institute of New Jersey, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08901, USA
John E. Kerrigan, Department of Bioinformatics, The Cancer Institute of New Jersey, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08901, USA
Gurinder Atwal, Quantitative Biology, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA.
Shoichi Metsugi, Simons Center for Systems Biology, Institute for Advanced Study, Princeton, NJ 08540, USA.
Deborah L. Toppmeyer, Department of Medicine, Division of Medical Oncology, The Cancer Institute of New Jersey, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08901, USA
Arnold J. Levine, Department of Pediatrics, The Cancer Institute of New Jersey, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08901, USA. Simons Center for Systems Biology, Institute for Advanced Study, Princeton, NJ 08540, USA
Kim M. Hirshfield, Email: hirshfie@umdnj.edu, Department of Medicine, Division of Medical Oncology, The Cancer Institute of New Jersey, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08901, USA
References
- 1.Key TJ, Verkasalo PK, Banks E. Epidemiology of breast cancer. Lancet Oncol. 2001;2(3):133–140. doi: 10.1016/S1470-2045(00)00254-0. [DOI] [PubMed] [Google Scholar]
- 2.Bennett IC, Gattas M, Teh BT. The genetic basis of breast cancer and its clinical implications. Aust N Z J Surg. 1999;69(2):95–105. doi: 10.1046/j.1440-1622.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 3.Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet. 2008;9:321–345. doi: 10.1146/annurev.genom.9.081307.164339. [DOI] [PubMed] [Google Scholar]
- 4.Dunning AM, et al. A systematic review of genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1999;8(10):843–854. [PubMed] [Google Scholar]
- 5.Bond GL, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66(10):5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 6.Rubinstein WS. Hereditary breast cancer: pathobiology, clinical translation, and potential for targeted cancer therapeutics. Fam Cancer. 2008;7(1):83–89. doi: 10.1007/s10689-007-9147-7. [DOI] [PubMed] [Google Scholar]
- 7.Martin M. Molecular biology of breast cancer. Clin Transl Oncol. 2006;8(1):7–14. doi: 10.1007/s12094-006-0089-6. [DOI] [PubMed] [Google Scholar]
- 8.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412(2):179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries PJ, Howe CJ. The tuberous sclerosis complex proteins—a GRIPP on cognition and neurodevelopment. Trends Mol Med. 2007;13(8):319–326. doi: 10.1016/j.molmed.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Orlova KA, Crino PB. The tuberous sclerosis complex. Ann N Y Acad Sci. 2010;1184:87–105. doi: 10.1111/j.1749-6632.2009.05117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu JJ, et al. Estrogen promotes the survival and pulmonary metastasis of tuberin-null cells. Proc Natl Acad Sci USA. 2009;106(8):2635–2640. doi: 10.1073/pnas.0810790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.York B, et al. Cross-talk between tuberin, calmodulin, and estrogen signaling pathways. FASEB J. 2005;19(9):1202–1204. doi: 10.1096/fj.04-3142fje. [DOI] [PubMed] [Google Scholar]
- 13.Sampson JR. Therapeutic targeting of mTOR in tuberous sclerosis. Biochem Soc Trans. 2009;37(Pt 1):259–264. doi: 10.1042/BST0370259. [DOI] [PubMed] [Google Scholar]
- 14.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27(13):2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 16.Franz DN, et al. Rapamycin causes regression of astrocytomas in tuberous sclerosis complex. Ann Neurol. 2006;59(3):490–498. doi: 10.1002/ana.20784. [DOI] [PubMed] [Google Scholar]
- 17.Jiang WG, et al. Tuberin and hamartin are aberrantly expressed and linked to clinical outcome in human breast cancer: the role of promoter methylation of TSC genes. Eur J Cancer. 2005;41(11):1628–1636. doi: 10.1016/j.ejca.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 18.Feo TA, Resende MGC. Greedy randomized adaptive search procedures. J Glob Optim. 1995;6(2):109–133. [Google Scholar]
- 19.Mizuno H, et al. Fine-scale detection of population-specific linkage disequilibrium using haplotype entropy in the human genome. BMC Genet. 2010;11(1):27. doi: 10.1186/1471-2156-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazquez A, et al. Chemosensitivity profiles identify polymorphisms in the p53 network genes 14-3-3tau and CD44 that affect sarcoma incidence and survival. Cancer Res. 2010;70(1):172–180. doi: 10.1158/0008-5472.CAN-09-2218. [DOI] [PubMed] [Google Scholar]
- 21.Vazquez A, et al. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7(12):979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 22.Feng Z, et al. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102(23):8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucl Acids Res. 2001;29(14):2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerry DJ, Dunphy KA, Hagen MJ. Estrogens, regulation of p53 and breast cancer risk: a balancing act. Cell Mol Life Sci. 2010;67(7):1017–1023. doi: 10.1007/s00018-009-0244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sangrajrang S, et al. Genetic polymorphisms of estrogen metabolizing enzyme and breast cancer risk in Thai women. Int J Cancer. 2009;125(4):837–843. doi: 10.1002/ijc.24434. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, et al. Association of genetic polymorphisms of ER-alpha and the estradiol-synthesizing enzyme genes CYP17 and CYP19 with breast cancer risk in Chinese women. Breast Cancer Res Treat. 2009;114(2):327–338. doi: 10.1007/s10549-008-9998-0. [DOI] [PubMed] [Google Scholar]
- 27.Setiawan VW, et al. Two estrogen-related variants in CYP19A1 and endometrial cancer risk: a pooled analysis in the Epidemiology of Endometrial Cancer Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18(1):242–247. doi: 10.1158/1055-9965.EPI-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertheau P, et al. TP53 status and response to chemotherapy in breast cancer. Pathobiology. 2008;75(2):132–139. doi: 10.1159/000123851. [DOI] [PubMed] [Google Scholar]