Abstract
The niiA (nitrite reductase) and niaD (nitrate reductase) genes of Aspergillus nidulans are subject to both induction by nitrate and repression by ammonium or glutamine. The intergenic region between these genes functions as a bidirectional promoter. In this region, nucleosomes are positioned under nonexpression conditions. On nitrate induction under derepressing conditions, total loss of positioning occurs. This is independent of transcription and of the NirA-specific transcription factor but absolutely dependent on the wide-domain GATA-binding AreA factor. We show here that a 3-amino-acid deletion in the basic carboxy-terminal sequence of the DNA-binding domain results in a protein with paradoxical properties. Its weak DNA binding is consistent with its loss-of-function phenotype on most nitrogen sources. However, it results in constitutive expression and superinducibility of niiA and niaD. Nucleosome loss of positioning is also constitutive. The mutation partially suppresses null mutations in the transcription factor NirA. AreA binds NirA in vitro, and the mutation does not affect this interaction. The in vivo methylation pattern of the promoter is drastically altered, suggesting the recruitment of one or more unknown transcription factors and/or a local distortion on the DNA double helix.
The transcriptional activation of the niiA (nitrite reductase) and niaD (nitrate reductase) genes of Aspergillus nidulans involves a 1.2-kb intergenic region. The activation process is strictly dependent on two transcription factors, NirA, a Zn binuclear cluster protein, and AreA, a GATA factor (7, 41, 48, 61) NirA is specific for the nitrate assimilation pathway, while AreA is involved in the transcriptional activation of scores of genes involved in the utilisation of different nitrogen sources (see references 4, 35, and 51 for reviews). Activation of transcription has an absolute requirement for both the presence of the specific inducer (nitrate) and the absence of repressing nitrogen metabolites (ammonium and glutamine [41]). The standard model is that AreA mediates general derepression in the absence of ammonium or glutamine while NirA mediates specific induction by nitrate (7, 39). Results presented here and in reference 41 make this simple model untenable. A very similar system of control is extant in Neurospora crassa, where close homologues of NirA and AreA operate (39), and probably in all filamentous ascomycetes able to assimilate nitrate (see, for example, references 18 and 19).
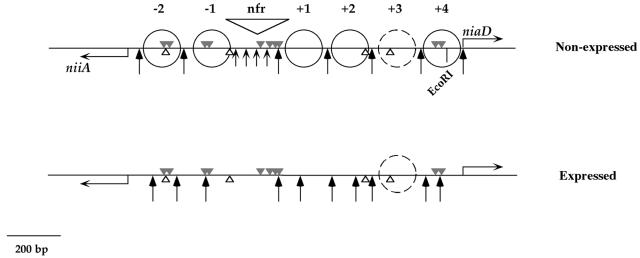
Transcriptional activation of the niiA-niaD bidirectional promoter is accompanied by a drastic chromatin restructuring, resulting in the loss of positioning of five or six nucleosomes. In the wild type, both the presence of nitrate and the absence of ammonium are necessary for this process. This remodeling is (with the possible exception of the loss of positioning of one nucleosome) independent of transcription and of NirA but strictly dependent on the GATA factor AreA (41). The chromatin structure of the promoter is schematized in Fig. 1. AreA is also necessary for in vivo binding of NirA to site 2, the only one that can be visualized by in vivo footprinting (42).
FIG. 1.
Nucleosome positioning in the niiA-niaD bidirectional promoter. Shown are the positions of nucleosomes in the niiA-niaD promoter under nonexpressed (noninduced-derepressed, noninduced-repressed, and induced-repressed) conditions (top) and expressed (induced-derepressed) conditions (bottom). The grey triangles above the line are the 10 AreA-binding sites, the white triangles below the line are the four NirA-binding sites. Positioned nucleosomes are indicated by full circles. Nucleosomes are numbered from the nucleosome-free region, + toward niaD and − toward niiA. There is a space between nucleosomes +2 and +4 for an additional positioned nucleosome (+3), but this is indicated by a dashed circle, since the absence of MNase cuts in this region does not allow us to determine whether this nucleosome is postioned. Long arrows indicate MNase cuts; short arrows indicate a smear in the nucleosome-free region. The latter is indicated by “nfr”. This figure summarizes data from a previous publication (41). Numbering of AreA and NirA sites throughout this article is from the left (niiA) to the right (niaD) of this figure.
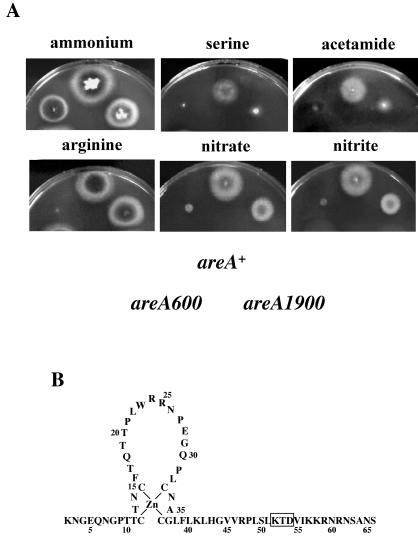
areA1900 is a 9-bp deletion mutation resulting in the loss of 3 amino acids in the basic region following the Zn finger in the DNA-binding domain (Fig. 2B). areA null mutants are unable to use all nitrogen sources except ammonium and glutamine (4, 35). areA1900 behaves as a total- or partial-loss-of-function mutation for the utilization of all nitrogen sources tested, with three exceptions, nitrate, nitrite, and arginine (examples are shown in Fig. 2A) (45, 63). areA1900 is a mutation resulting in a paradoxical phenotype. We show in this article that this mutation results, in spite of its weak binding to all extant DNA sites, in constitutive nucleosome loss of positioning in the niiA-niaD intergenic region and in partially constitutive activation of transcription and that these effects are correlated with a drastic alteration of the in vivo methylation protection pattern, which may reflect the recruitment of an uncharacterized transcription factor and/or local alterations in DNA structure.
FIG. 2.
In vivo phenotype of areA1900 strains. (A) Phenotypes of strains carrying different areA alleles on different nitrogen sources. The position of each strain in the plates is indicated below the growth tests. areA+ is the wild-type allele, and areA600 is a total-loss-of-function chain termination mutation (35). The plates contain A. nidulans minimal medium supplemented with different nitrogen sources as indicated; concentrations are those used in reference 4. (B) Schematic representation of the DNA-binding domain of the AreA protein. The 3 amino acid residues deleted in the AreA1900 protein are boxed.
MATERIALS AND METHODS
Strains, plasmids, and genetic techniques.
A biA1 A. nidulans strain was used as the wild type, argB2 inoB2 was used as the recipient strain for transformation with pTRAN3-1A derivatives, argB2 pabaA1 areA600 was used in crosses to obtain strains containing pTRAN3-1A in an areA600 background, and argB2 yA2 areA1900 was used in crosses to obtain strains containing pTRAN3-1A and derivatives in an areA1900 background. biA1 sB43 areA600, inoB2 areA1900, pabaA1 nirA637, yA2 biA1 pantoB100 prnB110 nirA514, inoB2 areA1900 nirA637, and inoB2 areA1900 nirA514 are the complete genotypes of the strains carrying areA or nirA mutations as described below. areA600 is a chain termination mutation in codon 646, eliminating the AreA DNA-binding domain (1, 35), areA1900 is a 9-bp deletion resulting in the loss of 3 amino acids (45) (see Fig. 2), nirA514 is a chain termination mutation in codon 82 (GGA to TGA) 12 residues carboxy terminal to the DNA-binding domain, and nirA637 is an in-phase 566-bp deletion eliminating 153 amino acids after the first methionine of the protein (41). Double mutants were obtained by standard genetic crosses.
biA1, inoB2, sb43, argB2, pantoB100, and pabaA1 are standard auxotrophic markers for biotin, inositol, thiosulfate, pantotenic acid, arginine, and p-aminobenzoic acid, respectively (16). prnB110 is a mutation in the major proline permease (5). Genetic techniques for A. nidulans were as described by Pontecorvo et al. (46).
pTRAN3-1A is a twin-reporter vector containing two divergently oriented reporter genes, encoding Escherichia coli β-glucuronidase (uidA) and E. coli β-galactosidase (lacZ) (47). pAN302 contains a 2.7-kb EcoRI-EcoRI fragment, comprising a 1,500-bp fragment of the niiA open reading frame and 1,214 bp of the niiA-niaD intergenic region (19, 33); this plasmid was used to obtain the HindIII-BglII 360-bp probe for indirect end labeling and the EcoRI-SalI 1.6-kb probe used in Northern blots for the niiA gene.
E. coli DH5α [F− endA1 hsdR17 (mk+ rk−) supE44 thi-1 recA1 gyrA96 relA1 ΔlacU169 (f80d-lacZΔM15)] was used for routine plasmid preparation, and LC137 (htpR lon supC tsx::Tn 10) was used for glutathione S-transferase (GST) fusion protein expression.
Isolation of A. nidulans strains carrying single copies of the various pTRAN3-1A derivatives.
The construction of mutant derivatives of expression vector pTRAN3-1A has been described previously (41). For the analysis of reporter gene expression in an areA1900 background, the transformants containing pTRAN3-1A or derivatives were crossed with an argB2 yA2 areA1900 strain by standard genetic techniques. Screening of the argB+ segregants carrying pTRAN3-1A and derivatives was carried out on plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (47) with nitrate as the sole nitrogen source.
Growth conditions.
A set of standard growth conditions was used for monitoring reporter gene expression, mRNA levels, promoter chromatin structures, and in vivo footprinting. A. nidulans strains carrying single copies of the various pTRAN3-1A derivatives were grown for 7 h at 37°C in minimal medium (46) with appropriate supplements plus 1.25 mM ammonium l-(+)-tartrate as the nitrogen source. For in vivo footprinting, mycelia were pregrown at 30°C for 14 h. This medium allows the growth of all strains, including areA600. The mycelia were then harvested by filtering through sterile Blutex tissue, washed with sterile water, and transferred to the same medium without any nitrogen source. Incubation was continued for 20 min in this medium, when the following nitrogen sources were added: 5 mM urea (noninduced-derepressed conditions [ND]), 10 mM NaNO3 (induced-derepressed conditions [ID]), 10 mM NaNO3 and 5 mM ammonium l-(+)-tartrate (induced-repressed conditions [IR]), or 5 mM ammonium l-(+)-tartrate (non induced-repressed conditions [NR]). Mycelia were incubated for an additional 3 h (reporter gene expression) or 2 h (Northern blots, chromatin determination, and in vivo footprints). The latter part of the growth protocol was always at 37°C.
Reporter gene expression.
The β-galactosidase and β-glucuronidase activities in mycelial extracts were determined as described by Punt et al. (47). The reporter activities are expressed as percentages of the activities of the pTRAN3-1A transformants (wild type for the intergenic region and all relevant regulatory genes) under ID conditions (NaNO3). Specific activities of 290 ± 30 nmol of p-nitrophenol · min−1 · mg of protein−1 for β-glucuronidase and 450 ± 15 nmol of o-nitrophenol · min−1 · mg of protein−1 for β-galactosidase were found in repeated assays with pTRAN3-1A transformants grown under these conditions. For all strains analyzed, means of three replicate assays are shown; standard errors were always lower than 15%.
Northern blots.
Mycelia of A. nidulans grown as described above were collected by filtration, washed with sterile water, and immediately frozen in liquid nitrogen. Total RNA was isolated from mycelia as described previously (15), glyoxal treated, and used in Northern blot analyses carried out by standard techniques. The probe for detecting niiA transcripts was an EcoRI-SalI 1.6-kb fragment from pAN302 (see above). An EcoRI 2.7-kb fragment containing the open reading frame of niaD was used for detection of these gene transcrips. An XbaI 2.7-kb fragment from pAN503 (20) was used as a probe for uapA transcripts. As a probe of the A. nidulans actin gene, we used a KpnI-SmaI 2-kb fragment from plasmid pSF5 (22).
Chromatin structure analysis.
Micrococcal nuclease MNase sensitivity analyses were performed by the indirect end-labeling technique as described for filamentous fungi by Gonzalez and Scazzocchio (27) on mycelial samples obtained as detailed above. MNase digestion profiles were obtained using 1 to 25 U of the enzyme per g of mycelium. After nuclease treatment, DNA was digested with HindIII and hybridized with a HindIII-BglII 360-bp probe as described previously (41).
Gel retardation assays and DNase I footprinting.
DNA restriction fragments were end labeled with a suitable α-32P-labeled deoxynucleoside triphosphate using Sequenase version 2.0 (USB, Cleveland, Ohio). Binding assays were performed in 20-μl reaction mixtures containing 2 ng of labeled DNA, 2 μg of poly(dI-dC), 25 mM Tris-HCl (pH 8.0), 100 mM KCl, 4 mM spermidine, 10% glycerol, and 9 to 300 ng of purified GST-AreA or GST-AreA1900 fusion proteins. After incubation at 25°C for 15 min, the reaction mixtures were run at 18 V/cm through a nondenaturing 6% polyacrylamide-10% glycerol gel in 0.25× Tris-borate-EDTA (TBE) buffer at 4°C. The gels were transferred to Whatman 3MM paper, dried, and subjected to autoradiography. For DNase I footprinting experiments, binding reactions were performed as described above and the mixtures were incubated with 2.5 mM CaCl2, 5 mM MgCl2 and 2 ng of DNase I (Boehringer, Manheim, Germany) for 1 min at 25°C. Digestions were stopped with 0.5 μmol of EDTA (pH 8.0), and the samples were immediately purified by acrylamide gel electrophoresis as in the gel retardation assays. The gels were autoradiographed, and the bound and free probes were excised from the gel and electroeluted onto DEAE membranes (Schleicher & Schuell, Dassel, Germany) and analyzed on a 6% polyacrylamide-urea sequencing gel. Nucleotide positions were identified by the Maxam-Gilbert reaction (40) for guanines run in parallel.
Construction of plasmids coding for and expression of the GST-AreA and GST-AreA1900 fusion proteins.
A BamHI fragment coding for 147 amino acids of AreA (amino acids 663 to 809) was amplified from the areA locus using appropriate primers (areA1 and areA2) each containing a BamHI restriction site not present in the areA sequence: areA1, 5′-C C A G G C G G A T C C A A G A A C G G A G A G C-3′; and areA2, 5′-CTGACGTTTAGGATCCACCTGTACC-3′. The amplified fragment was cloned into BamHI-restricted pGex4T. The absence of mutations in the PCR-amplified fragment was checked by sequencing. For the AreA1900-GST fusion protein, the same procedure was followed but the areA1900 locus was PCR amplified. E. coli LC137 was transformed with the GST-areA and GST-areA1900 plasmids. Expression and purification of the GST-AreA and GST-AreA1900 fusion proteins were performed by the method described by Smith and Johnson (57).
Construction of a plasmid coding for and expression of the NirA protein.
A full-length cDNA clone of nirA was constructed as follows. A NcoI-HaeII 314-bp fragment, obtained from the partial nirA cDNA clone cbs15J2 (12), and a HaeII-HindIII 633-bp fragment, PCR amplified from nirA locus, were ligated and cloned into pGex2T that was SmaI digested to give pGex2T-nirA(NcoI-HindIII). A 1.3-kb ApaI-EcoRI fragment was obtained from the partial nirA cDNA clone cbs 321 (12) and cloned into pGex2T-nirA(NcoI-HindIII) digested with the same enzymes. Finally, the remaining 1,089-bp ApaI fragment was obtained by reverse transcription-PCR from total A. nidulans RNA using the on2 primer (12) and the onJ9R primer (positions 4578 to 4556 according to Burger et al. [12]). This ApaI fragment was cloned into pGex2T-nirA(NcoI-HindIII) digested with the same enzyme to give pGex2T-nirA. The absence of mutations in the PCR- and reverse transcription-PCR-amplified fragments was checked by sequencing. A NcoI-EcoRI 3.2-kb fragment from pGex2T-nirA, containing the full-length nirA cDNA, was cloned into pET22-b(+) (Novagen) digested with the same enzymes to generate pNirA(1-892). This plasmid was digested with NcoI and NdeI, filled in, religated to eliminate some plasmid sequences coding for the pelB leader, and used in in vitro transcription and translation assays as described previously (61) for the synthesis of whole NirA protein.
Pull-down experiments.
A 3-μg portion of each of the purified AreA-GST, AreA1900-GST, and GST proteins was dialized against interaction buffer (buffer I) (10 mM Tris-HCl [pH 8], 150 mM NaCl, 0.3% NP-40, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluorider, 0.25% bovine serum albumin [BSA], 20 μM ZnCl2). These proteins were bound to glutathione-agarose resin equilibrated in buffer I (20 μl of 50% slurry). The beads were incubated for 2 h at 4°C with in vitro transcription-translation reaction mixtures containing [35S]methionine-labeled NirA protein, with occasional gentle mixing. After the unbound supernatant was removed, the beads were washed three times with 1 ml of buffer I and once with BSA-free buffer I. The beads were boiled in sodium dodecyl sulfate (SDS) buffer, and the samples were analyzed by SDS-polyacrylamide gel electrophoresis in a 10% polyacrylamide gel. The gel was stained with Coomassie blue and then transferred to Whatman 3MM paper, dried, and subjected to autoradiography.
In vivo footprinting assays.
To 18-ml aliquots of mycelia grown as described above, 2 ml of methylation buffer (300 mM MES [pH 6.2], 40 μl of dimethyl sulfate) was added, and the culture was incubated for 2 min on a rotary shaker. Mycelia grown as described above were collected by filtration, washed with sterile water, and immediately frozen in liquid nitrogen. DNA extraction and detection of in vivo protein-DNA interactions by radioactive ligation-mediated PCR were carried out as described by Wolschek et al. (66). The primers detecting the NirA-site 2 interaction and the protection of an uncharacterized site on the niiA coding strand were as follows: NIR II/1, 5′TGGCTAGAGCCGCTTGACGATAATG3′; NIR II/2, 5′GAGCCGGCGATAAGCATGATG-TTGGC3′; and NIR II/3, 5′CCGGCGATAAGCATGATGTTGGCGCTGTC3′. The primers detecting GATA site 5 occupation on the niaD coding strand were as follows: IGR-GATA5-6/1, 5′GGCTATTACTCACAGTCGATG3′; IGR-GATA5-6/2, 5′GGCAATTCCGATGACTCTCGATCG3′; and IGR-GATA5-6/1, 5′GGCAATTCCGATGACTCTCGATCGTGTC3′.
RESULTS
Transcriptional phenotype of areA1900.
The in vivo experiments described in this article were carried out under four sets of standard growth conditions. While these are described exactly in Materials and Methods, it is important to keep in mind the conceptual differences among them: ND, noninduced-derepressed, a neutral nitrogen source (urea) is present in the growth medium; ID, induced-derepressed, nitrate is present in the growth medium; NR, noninduced-repressed, ammonium is present in the growth medium; IR, induced-repressed, both ammonium and nitrate are present in the growth medium.
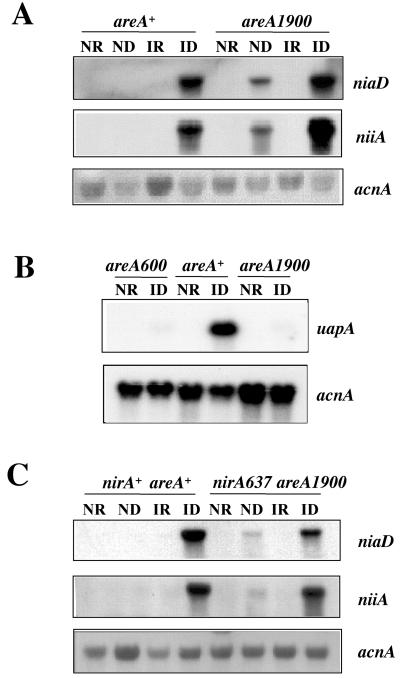
The areA1900 mutation results in strongly diminished growth on purines as nitrogen sources (63 and our unpublished results). In Fig. 3 we compare the effect of areA1900 on the transcription of the specific urate-xanthine transporter (28) with its effect on niiA-niaD expression. areA1900 behaves identically to the complete-loss-of-function mutation areA600 for the transcriptional activation of the uapA promoter. The uapA gene is strictly under the control of AreA and is as sensitive as niiA and niaD to nitrogen metabolite repression (28, 43, 50). The situation is radically different for the niiA and niaD mRNAs. areA1900 results in partial constitutivity of both transcripts and in overexpression in the presence of the inducer, nitrate. The repressibility by ammonium is not affected.
FIG. 3.
Transcriptional phenotype of the areA1900 mutant. (A) Steady-state levels of niaD and niiA mRNAs of the wild-type and areA1900 mycelia grown under the four different conditions described in Materials and Methods. The niiA and niaD mRNAs are undetectable in an areA600 strain. This was published previously (reference 41, Fig. 2). (B) Steady-state levels of uapA mRNA of wild-type, areA1900, and areA600 mycelia grown under NR and ID. Induction of uapA was done after pregrowing the mycelium for 7 h on 1.25 mM ammonium l-(+)-tartrate as the nitrogen source and then washing it with sterile water and transferring it for 2 h to 600 μM uric acid. (C) Steady-state levels of niaD and niiA mRNAs in wild-type and nirA637 areA1900 mycelia grown under the four different conditions described in Materials and Methods. Identical results were obtained with a nirA514 areA1900 double mutant. There are absolutely no detectable niiA and niaD mRNAs in nirA637 or nirA514 single mutants grown under any condition (reference 41, Fig. 2). The steady-state levels of the acnA gene (encoding actin) were monitored as a loading control in all the samples in all experiments.
To study quantitatively the effect of the areA1900 mutation, we used the bidirectional reporter vector described in previous papers (41, 47, 48). We had shown that the cluster of four AreA (Fig. 1)-binding sites (sites 5 to 8) contained in the nucleosome-free sequence accounts for more than 80% of the promoter transcriptional competence. We studied the effect of the areA1900 mutation both in the wild-type promoter and in mutant promoters carrying point mutations in one, some, or all of the four AreA-binding sites contained in the nucleosome-free region described previously (41). Table 1 confirms that areA1900 results in partial constitutivity and strong hyperinducibility of transcription of the wild-type promoter. The constitutive phenotype of areA1900 depends on the integrity of the four central GATA sites. Site 5 was shown to be the most important site in an areA+ strain (41); this is true in an areA1900 background for niiA but not for niaD expression under induced conditions. In this background, mutation of the four central GATA sites results in a clear reduction of induced expression, but the levels are still equal to (for niiA) or double (for niaD) those found in a areA+ strain carrying an intact intergenic region.
TABLE 1.
Reporter gene expression driven by the wild-type and mutant bidirectional promoters
| Host strain and plasmida | % β-Galactosidase (niiA) activityb
|
% β-Glucuronidase (niaD) activityb
|
||||||
|---|---|---|---|---|---|---|---|---|
| ND | ID | IR | NR | ND | ID | IR | NR | |
| areA+ | ||||||||
| pTRAN3-1A | 8.3 | 100 | 2.1 | 1.4 | 0.4 | 100 | 0.8 | 2.0 |
| pTRAN3-G5 | 2.0 | 43.6 | 2.9 | 1.8 | 3.1 | 55.0 | 2.0 | 4.0 |
| pTRAN3-G6 | 7.8 | 83.0 | 2.4 | 0.5 | 2.7 | 103.4 | 3.2 | 1.5 |
| pTRAN3-G5/6 | 1.8 | 40.7 | 3.3 | 1.9 | 1.3 | 38.9 | 0.7 | 0.0 |
| pTRAN3-G7/8 | 4.3 | 62.2 | 2.0 | 2.2 | 1.5 | 59.2 | 1.4 | 1.7 |
| pTRAN3-G5/6/7/8 | 1.7 | 18.9 | 3.3 | 0.6 | 1.3 | 16.5 | 0.7 | 0.8 |
| areA1900 | ||||||||
| pTRAN3-1A | 54.7 | 218.5 | 27.2 | 3.0 | 32.1 | 397.7 | 7.0 | 3.1 |
| pTRAN3-G5 | 22.1 | 131.9 | 14.5 | 1.1 | 24.6 | 338.2 | 12.7 | 7.3 |
| pTRAN3-G6 | 50.4 | 165.4 | 23.9 | 1.1 | 33.0 | 318.4 | 13.2 | 1.8 |
| pTRAN3-G5/6 | 17.1 | 119.1 | 19.0 | 2.2 | 11.9 | 237.0 | 9.4 | 0.0 |
| pTRAN3-G7/8 | 33.8 | 127.4 | 9.2 | 2.5 | 24.7 | 258.9 | 5.9 | 0.9 |
| pTRAN3-G5/6/7/8 | 4.9 | 112.4 | 5.2 | 2.5 | 1.9 | 224.8 | 2.1 | 0.6 |
| areA600 | ||||||||
| pTRAN3-1A | 1.6 | 6.2 | 2.5 | 1.1 | 0.7 | 0.7 | 0.3 | 0.3 |
The plasmids indicated are those used to introduce wild-type or mutated niiA-niaD intergenic regions driving the two reporter genes. pTRAN3-1A indicates a plasmid containing a wild-type intergenic region, and pTRAN3-G5, -G6, -G5/6, -G7/8, and G5/6/7/8, indicate plasmids containing niiA-niaD intergenic regions mutated as indicated in Materials and Methods for GATA sites 5, 6, 5 and 6, 7 and 8, and 5 to 8, respectively. The values obtained in areA+ and areA600 backgrounds have been published previously and are shown here for comparative purposes (41). The values reported are averages of three independent determinations with standard errors of <15%.
Enzyme activities are expressed as percentages of the activities obtained for the pTRAN3-1A transformant in an areA+ background, grown under ID conditions. All strains were pregrown with 1.25 mM ammonium l-(+)-tartrate as the nitrogen source, and after being harvested and washed as indicated in Materials and Methods, mycelia were transferred to the appropriate nitrogen sources. ND, 5 mM urea; ID, 10 mM NaNO3; IR, 10 mM NaNO3 and 5 mM ammonium l-(+)-tartrate; NR, 5 mM ammonium l-(+)-tartrate.
AreA1900 partially bypasses the function of NirA in transcriptional activation.
The areA1900 mutation was selected as a weak suppressor of the nirA1 mutation (63). We have investigated induction in double mutants of areA1900 and the two null mutations nirA637 and nirA514 with identical results. These mutations are respectively an in-phase deletion of 459 bp removing the whole DNA-binding domain and a chain termination mutation at codon 82 (41). Growth tests confirm that areA1900 is a weak suppressor of both complete-loss-of-function mutations in nirA (not shown). Northern blots (Fig. 3C [only areA1900 nirA637 is shown; identical results were obtained with areA1900 nirA514) show this suppression clearly. Under noninduced conditions, the double mutants show weak expression of both niiA and niaD. Surprisingly, this expression is clearly induced by nitrate. The double mutants are as sensitive to ammonium repression as is the areA1900 single mutant.
The specific phenotype found in areA1900 strains does not derive from privileged binding to any specific GATA sites.
areA mutations in the DNA-binding domain that differentially affect the expression of a number of genes have been described. Genetic, footprinting, modeling and nuclear magnetic resonance studies have shown that this is due to differential binding to different GATA sites within the HGATAR consensus (6, 31, 50, 59, 60, 65).
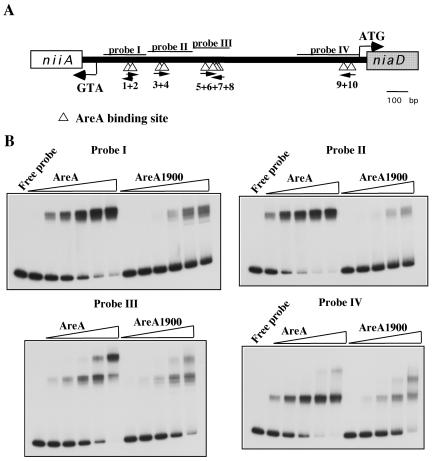
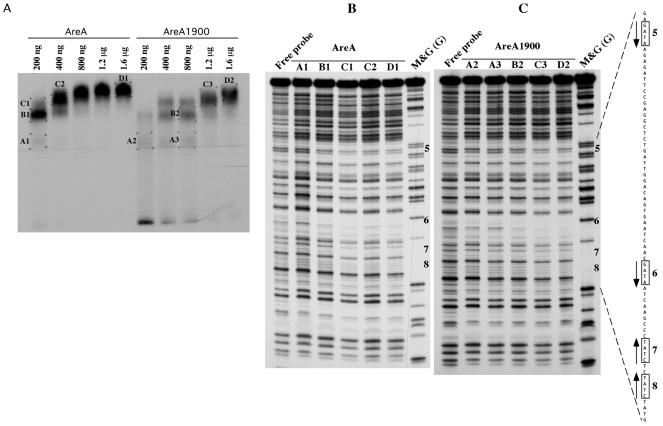
The results in the previous section could be accounted for if AreA1900 resulted in increased affinity for some specific AreA sites in the niiA-niaD promoter. Privileged binding to one or more of the central GATA sites (sites 5 to 8) could account for the constitutive phenotype of the areA1900 mutant. We have therefore studied, in vitro by electrophoretic mobility shift assay, the binding of an AreA-GST fusion protein to probes covering the entire niiA-niaD intergenic region. AreA1900 has a reduced affinity for all the probes (Fig. 4), including the one carrying the four central sites. We have repeated the electrophoretic mobility shift assays with the probe containing sites 5 to 8 with an AreA DNA-binding domain cleaved from the GST protein, with qualitatively identical results (not shown). We have also carried out DNase I footprinting of the four central sites; the pattern of protection found at the appropriate concentrations of protein (about four times higher for AreA1900) is identical for the wild-type AreA and AreA1900. One example of this is shown in Fig. 5.
FIG. 4.
AreA and AreA1900 binding to different probes from the niiA-niaD intergenic region. (A) Schematic representation of the niiA-niaD intergenic region. The triangles below the line indicate the 10 AreA-binding sites. The positions of the four probes used in the EMSA study are shown. (B) Gel retardation experiments with the four probes containing the AreA binding sites. 32P-labeled probes were incubated with different amounts (9 to 300 ng) of AreA or AreA1900 proteins. From left to right in each panel, 4.5, 9, 18, 75, 150, and 300 ng of protein were used. For probes II, III, and IV, the 4.5-ng point was omitted.
FIG. 5.
DNase I protection of the niiA-niaD promoter region containing AreA-binding sites 5 to 8. A relevant section of the sequence of the niaD coding strand is shown to the right of panel C. The probe used is probe III (Fig. 4) terminally labeled in the niaD coding strand. (A) Preparative gel retardation assay from which the DNA-protein complexes were excised (see Materials and Methods). The amounts of GST-AreA or GST-AreA1900 used in each assay are indicated. The different complexes analyzed by DNase I protection are labeled (A1, A2, etc.), and these labels are also indicated in the corresponding lane of panels B and C. (B) DNase I protection by the AreA protein. Lanes: free probe, DNase I digestion carried out on nonprotected probes; M&G (G), guanine Maxam and Gilbert reaction. (C) DNase I protection by the AreA1900 protein. Lanes are as in panel B.
Only one AreA-binding site can be revealed by in vivo methylation protection; this is AreA site 5, the most important for transcriptional activation in the wild type (41). No protection or, in some experiments, extremely weak protection of site 5 can be seen in the areA1900 mutant strain (data not shown). This is in line with the diminished affinity demonstrated in vitro, but the effect of the mutation of site 5 on the expression of reporter genes driven by the bidirectional promoter (Table 1) shows that AreA1900 must nevertheless bind to this site in vivo.
The wild-type and mutated AreA proteins bind equally well NirA in vitro.
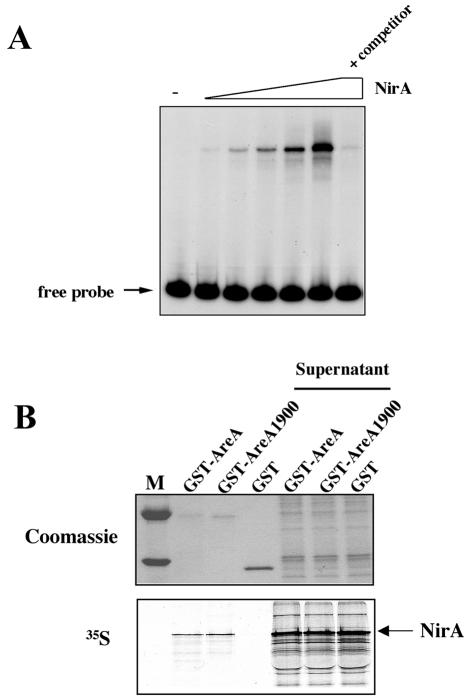
It has been shown by Feng and Marzluf that the DNA-binding domains of the close homologues of AreA (Nit2) and NirA (Nit4) in N. crassa interact in vitro and also in vivo in double-hybrid assays (21). The basic region following the Nit2 Zn finger is involved in this interaction. AreA is essential for NirA to bind to site 2 in vivo in response to nitrate induction, which may imply a direct interaction in vivo (see below [42]). The phenotype of the areA1900 mutant could be partially explained if the mutation resulted in an increased affinity for NirA. We show below that AreA and NirA interact in vitro. In Fig. 6 we show the interaction of a whole NirA protein with the DNA-binding domain of AreA. The wild-type and mutant AreA proteins behave identically. There is an excess of labeled NirA protein in the supernatant, and thus any increase in affinity of AreA1900 would have been detected in this experiment.
FIG. 6.
In vitro interaction of the full-length NirA protein with the DNA-binding domain of AreA. (A) Gel retardation experiment with the full-length in vitro-synthesized NirA protein. Aliquots of the in vitro transcription-translation reaction mixture (0.5 to 8 μl) containing the whole NirA protein were incubated with a 239-bp ClaI-BanI 32P-labeled DNA fragment containing the NirA binding site 2, used as a probe. In the lane labeled +competitor, 8 μl of reaction mixture was incubated with the labeled probe and a 60-fold excess of cold probe. This panel establishes the DNA in vitro binding activity of the full-length NirA protein synthesized in vitro and is a necessary control for the experiment shown in panel B. (B) Analysis of the NirA-AreA specific interaction by pull-down experiments. In vitro-synthesized 35S-labeled NirA protein was incubated with GST-AreA, GST-AreA1900, or GST (negative control) proteins immobilized on a glutathione-agarose resin, as described in Materials and Methods. Specifically bound 35S-labeled NirA protein was detected by autoradiography. After the molecular mass marker lane (M), the first three lanes correspond to the bound samples and the fourth to sixth lanes to the nonbound supernatant samples.
AreA1900 results in constitutive nucleosome rearrangement.
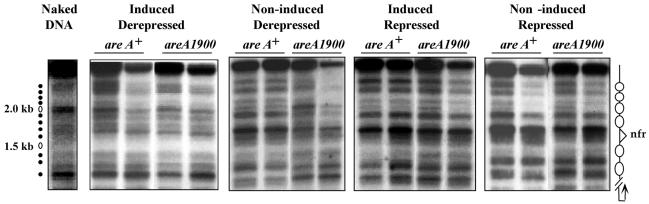
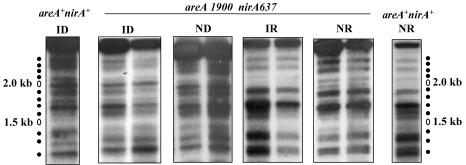
When grown on a neutral nitrogen source, urea, in the absence of both induction and repression, the wild type has a pattern of complete nucleosomal positioning that is virtually identical to that found under fully repressed conditions (41). At variance with the wild type, a strain carrying the areA1900 mutation showed an identical MNase digestion pattern under noninduced and induced conditions. This open pattern was identical to that of the wild-type strain grown under induced conditions (fig. 7). All nucleosome positioning was lost (as with the wild type, this experiment did not establish the fate of the putative nucleosome +3, since no MNase-sensitive sites were present in the stretch of DNA presumably occupied by this nucleosome). Thus, the areA1900 mutation results in constitutivity of both transcriptional activation and nucleosome positional loss. Repression affects transcription and nucleosomal positioning equally. We further investigated the chromatin structure of a promoter of nirA637 and nirA514 strains in an areA1900 background (shown only for nirA637 in Fig. 8; identical results were obtained with nirA514). The results with nirA637 and nirA514 show that neither the NirA DNA-binding domain nor the sequences carboxy-terminal to it are necessary for the constitutive nucleosome rearrangement occurring in the presence of an AreA1900 transcriptional activator (Fig. 8).
FIG. 7.
Chromatin structure of the niiA-niaD intergenic region in the wild-type and areA1900 strains. The MNase digestion patterns of wild-type (areA+) and areA1900 strains grown under all conditions previously described are shown. The pattern for the wild-type strain under all conditions has already been published, (41) and is included here as a necessary control. Increasing quantities of enzyme were used as described in Materials and Methods. Growth conditions are as described in the legend to Fig. 3. To the left of the figure, the position of markers corresponding to the 100-bp ladder from Pharmacia is shown. To the right of the figure, the nucleosome structure deduced from the MNase digest is shown. The samples corresponding to the same growth conditions for both strains were run side by side. Note the typical nonpositionated pattern shown by the areA1900 strain grown under noninduced-derepressed conditions.
FIG. 8.
Chromatin structure of the niiA-niaD intergenic region in an areA1900 nirA637 strain. To the left and right of the figure, the MNase digestion patterns of the same region in a wild-type strain (areA+ nirA+) under conditions of repression (presence of ammonium) and induction (presence of nitrate) are also shown. Increasing quantities of enzyme were used as described in Materials and Methods. Growth conditions are as described in the legend to Fig. 3. On both sides of the figure, the positions of markers corresponding to the 100-bp ladder from Pharmacia is shown. Note the typical nonpositionated pattern shown by the areA1900 nirA637 strain grown under ND conditions (compare with the ID lane and the relevant lanes of Fig. 7). Identical results were obtained with a nirA514 areA1900 double mutant.
The AreA1900 protein does not recruit NirA constitutively in vivo but results in a drastically altered methylation protection pattern.
If an AreA1900 protein could recruit NirA constitutively, its phenotype in a nirA+ background could be easily explained, at the levels both of transcriptional activation of the promoter and of chromatin restructuring. Nevertheless, this proposed mechanism fails to explain the ability of the areA1900 mutation to suppress null mutations in nirA. This could be explained if, as an alternative or in addition to constitutive recruitment of nirA, AreA1900 recruited an extraneous transcription factor that is not involved normally in the transcriptional activation of the niiA-niaD promoter.
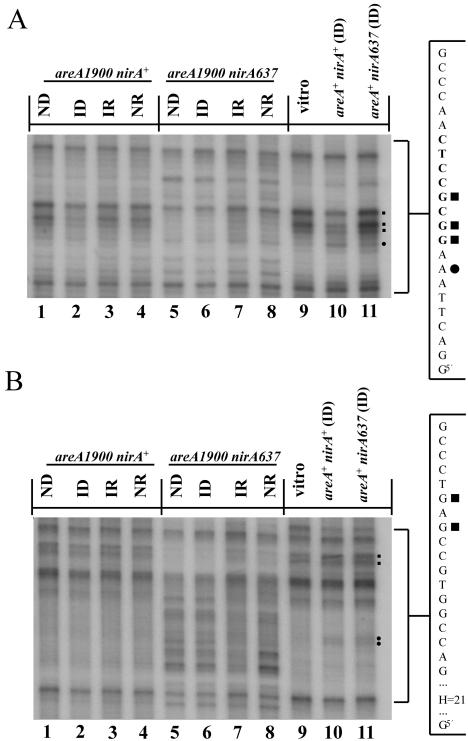
Only the most important site of NirA transcriptional activation (NirA site 2) (48) can be visualized by in vivo methylation protection. In vivo occupancy of NirA site 2 is strictly dependent in the wild type on induction by nitrate and on the presence of an active AreA protein (42). Figure 9A shows that in an areA1900 mutant, NirA binding in vivo to site 2 is strictly dependent on the presence of nitrate. As in the wild type, binding did not take place in the presence of ammonium. In Fig. 9A we show the pattern of protection of NirA site 2 in an areA1900 background. Three guanines (in the niiA coding strand) were protected in the wild-type (lane 10, partial protection seen in this experiment [indicated by squares to the right of the panel]) and areA1900 (lane 2) strains only when the strains were grown under induced-derepressed conditions. Under these conditions, a hypersensitive adenine, which is also seen in vitro (42, 61) was equally visible in the areA+ and areA1900 strains (lanes 10 and 2 [indicated by a dot to the right of the panel]). The pattern of protection seen in the niaD coding strand in the areA1900 mutant is again identical to that seen in areA+ (42). Protection of three guanines is seen only under conditions of induction and is lost in presence of ammonium (not shown). The only additional feature seen in areA1900 strains is the protection of a single guanine in the niaD coding strand 9 bp downstream of AGATAA (GATA site 5). This is seen under all physiological conditions (data not shown). No protection in this region is seen either for the AreA or AreA1900 proteins in the in vitro DNase I protection experiments. Thus, the recruitment of NirA to site 2 proceeds identically in areA+ and areA1900 backgrounds.
FIG. 9.
In vivo footprinting of NirA site 2 and adjacent sequences. (A) In vivo methylation protection of NirA binding site 2. The niiA coding strand from the niiA-niaD intergenic region of the areA1900 single mutant and the areA1900 nirA637 double mutant are compared with wild-type strain (areA+ nirA+) and the nirA loss-of-function mutant (nirA637), both grown under inducing conditions. A relevant section of the sequence of the niiA coding strand is shown at the right of the figure. The sequence highlighted in bold type corresponds to the asymmetric NirA-binding sequence (61). Filled squares indicate the protected guanines within the NirA-binding site. A dot indicates the position of a hypersensitive adenine seen in the NirA-DNA complex both in vitro (61) and in vivo (42). “Vitro” indicates the protein-free control DNA. Strains are grown under the standard conditions described in the text. Identical results were obtained with a nirA514 areA1900 double mutant. (B) In vivo methylation pattern of an unknown target. In vivo methylation protection and the hypersensitivity pattern of a so far uncharacterized sequence at position −439 from the niiA start codon is shown. Shown in the figure is the niiA coding strand of the areA1900 single mutant and the areA1900 nirA637 double mutant compared with the wild-type strain (areA+ nirA+) and the nirA loss-of-function mutant (nirA637), both grown under inducing conditions. The two guanines which are protected in the nirA637 areA1900 double mutant are indicated by squares to the right of the panel. The hypersensitive region comprises 21 nucleotides (H=21), including adenine, thymine, and cytosine residues. Strains were grown under standard conditions. A relevant section of the sequence of the niiA coding strand is shown at the right of the figure. The hypersensitive region corresponds to the sequence 5′ActTCCcAaaTaTCaTCatTc3′, where the hypersensitive bases are in capital letters. The two adenines which are hypersensitive in the wild-type and nirA637 strains but not in the nirA63 areA1900 double mutant are indicated by dots to the right of the figure. “Vitro” indicates the protein-free control DNA. Identical results were obtained with a nirA514 areA1900 double mutant.
The most striking results are found in the nirA− areA1900 double mutants. All experiments were carried out with both areA1900 nirA637 and areA1900 nirA514, with identical results. We have shown previously (42) that no methylation protection is seen in this region in null nirA− mutants under any conditions (fig. 9A, lane 11, here shown only under ID conditions). In the nirA− areA1900 double mutants, there were a number of unexpected changes in the niiA coding strand. (i) Two guanines of the NirA site 2, which are strongly protected in the in vivo footprinting experiments in nirA+ areA+ and nirA+ areA1900 strains only under induced conditions (lane 2), are protected under all conditions in the double mutant (lanes 5 to 8). Three guanines corresponding to NirA site 2 in the niaD coding strand are also protected under all conditions (data not shown). (ii) Two guanines between NirA site 2 and AreA site 5 (46 bp from NirA site 2 toward niaD) on the niiA coding strand are protected constitutively in this mutant (Fig. 9B, lanes 5 to 8 [indicated by squares to the right of the panel]). No protection of this stretch of sequence is seen on the niaD coding strand (data not shown). (iii) Strikingly, immediately adjacent to the protected sequence, a region of 21 bp is strongly hypersensitive to methylation under all growth conditions (lanes 5 to 8). The sequence is 5′ActTCCcAaaTaTCaTCatTc 3′, where the sensitive bases are indicated in capital letters. The niaD coding strand is, in this region, identical in the areA1900 and areA+ strains (data not shown). Note also that two adenines in this region which are sensitive to methylation in the wild-type and nirA− strains are not exposed to methylation in both the areA1900 and the nirA− areA1900 strains (indicated by dots to the right of the panel).
DISCUSSION
Roles of AreA and NirA in niiA-niaD expression.
The accepted model of nitrogen regulation in the ascomycetes is that a GATA factor (AreA in A. nidulans) is active in the absence of ammonium (or glutamine) and thus mediates derepression and that for each pathway a specific transcriptional activator mediates the specific induction signal (7, 53). Thus, in A. nidulans, UaY would mediate uric acid induction of the purine degradation pathway (54, 62), PrnA would mediate the induction of the proline utilization pathway (14, 25, 56), AmdR would mediate the induction by β-alanine of two unrelated pathways (3, 30), and HxnR would mediate the induction by 6-hydroxynicotinate of the nicotinate utilization pathway (55; R. Fernández, A. Cultrone, and C. Scazzocchio, unpublished results). Similar patterns of regulation have been found in S. cerevisiae and N. crassa (38, 39). In the nitrate pathway, NirA would mediate nitrate induction and AreA would be independently inactivated by ammonium (or glutamine). One should expect constitutive mutations (those where transcriptional activation occurs in the absence of nitrate) to map in nirA and derepressed mutations (those where transcriptional activation occurs in the presence of ammonium) to map in areA. These mutations were described long ago (4, 17, 35, 44, 45). Derepressed mutations mapping in nirA have also been described (12, 49, 64). In this article, we describe the fourth possible class, a mutation constitutive for the expression of niiA and niaD and mapping in areA. These last two classes of mutations strongly suggest that NirA and AreA act as complex in the niiA-niaD promoter, and we show in this article that they are able to interact in vitro. The areA1900 mutation does not alter this interaction.
Platt et al. described areA1900 as a derepressed mutant (45), while our results show clearly that it is not. Platt et al. observed partial derepression (repressed value, 28% of the fully induced level for nitrate reductase compared with 13% for the wild type). We saw some derepression (Table 1) when we used reporter enzyme assays (a maximum of 12% of the fully induced value for nitrite reductase compared with 2% for the wild type), but we saw no derepression at all in Northern blots. The Northern blot analyses were repeated several times, always with identical results. We consider the results of Northern blot analyses to be a more reliable and definitive indication of transcriptional repression. Enzyme assays, whether of the physiological enzymes or those encoded by reporter genes, are affected by the accumulation of protein, which is usually far more stable than the cognate mRNAs. This effect is magnified by the different, less stringent growth protocol used by Platt et al. (45).
Does induction require NirA?
Results presented in this article show that the simple and satisfying scheme presented above is incorrect or at least incomplete for the nitrate assimilation pathway. NirA cannot mediate the nucleosomal loss of positioning elicited by the nitrate signal, since this occurs in null nirA mutants (41). We show here that NirA could not be the sole transducer of the nitrate induction signal. Nitrate induction occurs in areA1900 nirA− double mutants, where no active NirA protein is present. However, we have not shown rigorously that nitrate induction in areA1900 nirA− double mutants occurs at the level of transcription. We have measured the niiA and niaD mRNA steady states, and thus the apparent nitrate-dependent induction in these mutants may result from an increase in mRNA stability. Wherever this nitrate signal acts, it does not require an active NirA protein. If the nirA-independent signal acts at the level of mRNA stability, it follows that there must be two topographically distinct nitrate-dependent signals, one acting at the level of chromatin and resulting in nucleosome loss of positioning and the other acting at the level of mRNA.
AreA and nitrate are necessary for in vivo occupancy of NirA binding site 2 (42). If AreA were the transducer of the nitrate signal, all results could be interpreted in a unified scheme. A model coherent with the experimental results would be one where NirA is essential as a transcriptional activator in an areA+ strain and is recruited to the niiA-niaD promoter by AreA, which, to be active in this capacity, requires both the presence of nitrate and the absence of ammonium. Nothing excludes the possibility that both AreA and NirA could respond to nitrate; alternatively, a third protein could be the nitrate receptor transducing the nitrate signal to both AreA and NirA and/or mediating mRNA stability. The only model excluded by the data presented here and by Muro-Pastor et al. (41) is one where NirA is the exclusive receptor of the nitrate signal.
A model where AreA is the transducer of the specific induction signal cannot possibly account for the transcriptional activation of all nitrogen utilization pathways. It has been shown that Zn finger structures can mediate a host of protein-protein interactions (37). The AreA Zn finger could interact directly with many binuclear Zn clusters of very similar structure. However, it is highly unlikely that the AreA protein could recognize coinducers as different as nitrate, uric acid (52, 62), β-alanine (3, 30), proline (56), 6-hydroxynicotinate (55), or arginine (8), to cite a few specific inducers of pathways requiring a pathway-specific transcription factor acting together with AreA. The nitrate assimilation pathway is possibly an interesting exception, and the general scheme could remain valid for most and perhaps all other nitrogen assimilation pathways.
AreA1900 is or can recruit a transcriptional activator.
The results obtained with the areA1900 nirA− double mutants show that AreA1900 can act as or recruit a transcriptional activator in the absence of NirA. Since the mutation does not affect any of the putative activation domains of AreA (13) but promotes a constitutive chromatin rearrangement, the most economical model would be one in which the chromatin rearrangement is the only prerequisite for the recruitment of the transcriptional machinery. This model is untenable, since in an areA wild-type background NirA is essential for transcriptional activation but not for nucleosomal loss of positioning (41). Thus, nucleosomal loss of positioning may be necessary for transcriptional activation, but it is surely not sufficient. The minimal interpretation of the results is that AreA1900, but not the wild-type AreA protein, is able to substitute for NirA in recruiting the transcriptional machinery to the niiA-niaD promoter.
A mutation with paradoxical properties.
AreA1900 is a partial-loss-of-function mutation that is paradoxically constitutive (but not derepressed) for the expression of the niiA-niaD promoter. At variance with other areA specificity mutations reported previously (50), the privileged transcriptional activation of the niiA-niaD promoter is not correlated with better binding of the mutant protein to specific HGATAR sites. On the contrary, the deletion in the basic region of the transcriptional activator decreases, in vitro, the affinity of AreA for all GATA sites present in the niiA-niaD promoter. For site 5, this has been confirmed in vivo. That binding is necessary for the activity of AreA1900 is shown for the four central GATA sites 5 to 8 by individual and collective mutation of the sites. Quantitative data of the reporter constructs show that the constitutive level of expression is lost (90% reduction) in a strain mutated for GATA sites 5 to 8 but that the induced level is only partially affected (50% reduction) by these mutations. Thus, the AreA1900 protein must bind to the critical central sites in order to recruit the transcriptional machinery in the absence of inducer. In the presence of inducer, binding to the central sites contributes to the overall overexpression, but about 50% of this activity must by accounted by binding to other GATA sites present in the intergenic region.
All data lead to the conclusion that AreA1900 is able to promote nucleosome loss of postioning and transcriptional activation constitutively in spite of its weaker binding to all GATA sites. The inescapable conclusion is that the mutation changes the interaction of AreA with the transcriptional and chromatin rearrangement machinery and that this change is specific for the niiA-niaD promoter. Data presented in Results show that these novel interactions do not require NirA.
In an areA1900 nirA+ background, the methylation protection pattern in the region of NirA site 2 does not differ from that seen in the wild type. These results could be accounted for by a simple model: AreA1900 would be able to activate transcription independently from NirA and nucleosome loss of postionining independently from nitrate, but the latter metabolite, by promoting NirA recruitment, would substantially increase transcriptional activation. The results with the areA1900 nirA− double mutants are very surprising and cannot be accommodated by this simple model. In the double mutants, sequences overlapping the Nir2 site are protected, independently of growth conditions, including in the presence of ammonium. If this protection is due to an unknown transcription factor recruited by AreA1900, this effect shows some surprising peculiarities. (i) It is found only in nirA null mutants. Since NirA is bound to site 2 only under induced-derepressed conditions and the new pattern of protection is not seen in nirA+ strains under any conditions, we have to postulate that in wild-type strains NirA is able to prevent the binding of this unknown factor even when not yet bound to site 2. While formally this effect could occur anywhere in the cell, it is plausible to suppose that it occurs at the chromatin level. (ii) It is also seen under conditions of repression. The lower affinity of AreA1900 for DNA results in loss of in vivo protection of AreA site 5 under all conditions; hence, we cannot know under which conditions Are1900 is bound to this site. In the wild-type strain, repression does not result in loss of AreA binding to GATA site 5 (41).
Other changes are seen in the double mutant in this region. Two protected guanines toward niaD are visible under all conditions. This may indicate the binding of an unknown factor or a local modification of the DNA in this region. The most striking modification is an extended area of hypersensitivity, which includes a row of pyrimidines. This implies that these are methylated at N-3, which in turn is only possible if there is a local melting of the double helix. Methylation of N-3 of cytosine by diethyl sulfate has been reported previously (2, 23) and has been shown in vitro to result from a melting of the double helix (2). N-3 methylation of thymines should be equally possible, but to our knowledge it has never been reported previously. How AreA1900 is able to promote directly or indirectly this effect and how NirA is able to prevent it could only be a matter of speculation at the present stage of this work. The most economical hypothesis is that the mutant AreA protein promotes the binding of an unknown protein to NirA site 2 only in the absence of NirA and that this, in turn, would result in a melting of the double helix about 40 bp away.
All GATA factors bind DNA in a very similar way. There is, however, a difference in structure between the chicken GATA-1 and AreA-DNA complexes. The carboxy-terminal tail of the DNA-binding domain of GATA-1 lies in the minor groove, while that of AreA is parallel to the phosphate backbone (59), and the latter has been invoked as a possible mechanism of nucleosome destabilization (11). The effects of AreA1900 could be explained if the mutational change leads to a protein which not only would destabilize the nucleosomes once bound but also could interact with DNA in such a way as to promote the local melting of the double helix. While the former cannot be excluded, the latter seems impossible since local melting occurs 72 bp from the nearer AreA binding site. Modeling of the AreA1900-DNA complex on the AreA-DNA complex (59, 60) does not provide any obvious explanation for the decrease in affinity of AreA1900 to its cognate HGATAR sites, nor does it suggest that it may interact in the minor groove in a way reminiscent of GATA-1 (results not shown).
AreA1900 discriminates between two classes of AreA-dependent promoters.
Vertebrate GATA factors are involved in local chromatin alterations (10, 58), but it has not been shown that they are involved in loss of nucleosome postioning in vivo. The lysines at the carboxy-terminal end of the vertebrate GATA factor DNA-binding domain can be acetylated by enzymes known to act as histone acetylases. The DNA-binding domain of both chicken and murine GATA-1 and murine GATA-3 can be acetylated by p300/CBP in vitro and in vivo (9, 29, 67). Murine GATA-1 is involved in establishing the histone acetylation patterns of the β-globin locus (32, 34, 36). We can speculate that acetylation of histones may be the missing link between AreA and its role in nucleosome loss of depositioning in the niiA-niaD promoter. Whatever the mechanism involved, the phenotype of areA1900 highlights the fact that AreA fulfills different functions in different promoters. For growth on most nitrogen sources, areA1900 behaves like a total-loss-of-function (uapA, Fig. 3) or partial-loss-of-function mutation. This is in line with its considerably weaker DNA binding to all GATA sites tested. We can assume that in the promoters involved in the utilization of such nitrogen sources, AreA behaves just as a transcription factor acting synergistically (uapA [28, 43]) or additively (prnD-prnB [26]) with the pathway-specific transcription factors and hence that loss of binding results in a loss-of-function phenotype. areA1900 behaves as a loss of function mutation for the utilisation of proline (our unpublished results, 63). We have analyzed in detail the chromatin rearrangements in the bidirectional promoter prnD-prnB (proline oxidase and proline transporter, respectively). In contrast to what happens in niaD-niiA, AreA is irrelevant to the massive nucleosome rearrangement that occurs in this promoter on induction, which necessitates only the specific transcription factor PrnA (24). In niiA-niaD, AreA must have different and novel interactions with the transcriptional and chromatin rearrangement machinery such that the partial loss of DNA binding occurring in areA1900 is bypassed by a gain-of-function mutation in these other functions. Presumably the same must be true for one or more promoters involved in the regulation of the utilization of arginine as a nitrogen source. It is tempting to speculate that in these promoters AreA interacts with the chromatin rearrangement machinery whether in others it does not.
Acknowledgments
We thank Eric Quiniou for invaluable help with the modeling of the AreA1900 DNA-binding domain and Rachid Rahmouni for helpful discussion. H. N. Arst, Jr., is thanked for providing the areA1900 strain and for communicating unpublished results.
M.I.M.-P. was supported successively by a postdoctoral fellowship of the Ministerio de Educación y Ciencia of the Spanish Government, EC fellowship Bio-CT94-8102, and a fellowship from the Foundation pour la Recherche Médicale. A.R. was supported by EU contract BIO4-CT96-0535 and a fellowship from the Fondation pour la Recherche Médicale. Work at Orsay was supported by the Université Paris-Sud, CNRS, the IUF, and EU contract BIO4-CT96-0535. Work at Vienna was supported by START Program grant. Y114-MOB from the Austrian Science Fund (FWF) to J.S.
REFERENCES
- 1.Al-Taho, N. M., H. M. Sealy-Lewis, and C. Scazzocchio. 1984. Suppressible alleles in a wide domain regulatory gene in Aspergillus nidulans. Curr. Genet. 8:245-251. [DOI] [PubMed] [Google Scholar]
- 2.Amouyal, M., and H. Buc. 1987. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J. Mol. Biol. 195:795-808. [DOI] [PubMed] [Google Scholar]
- 3.Arst, H. N., Jr. 1976. Integrator gene in Aspergillus nidulans. Nature 262:231-234. [DOI] [PubMed] [Google Scholar]
- 4.Arst, H. N., Jr., and D. J. Cove. 1973. Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 126:111-141. [DOI] [PubMed] [Google Scholar]
- 5.Arst, H. N., Jr., S. A. Jones, and C. R. Bailey. 1981. A method for the selection of deletion mutations in the l-proline catabolism gene cluster of Aspergillus nidulans. Genet. Res. 38:171-195. [DOI] [PubMed] [Google Scholar]
- 6.Arst, H. N., Jr., and C. Scazzocchio. 1975. Initiator constitutive mutation with an ′up-promoter' effect in Aspergillus nidulans. Nature 254:31-34. [DOI] [PubMed] [Google Scholar]
- 7.Arst, H. N., Jr., and C. Scazzocchio. 1985. Formal genetics and molecular biology of the controlof gene expression in Aspergillus nidulans, p. 309-343. In L. L. Lasure (ed.), Gene manipulations in fungi. Academic Press, Inc., Orlando, Fla.
- 8.Bartnik, E., and P. Weglenski. 1974. Regulation of arginine catabolism in Aspergillus nidulans. Nature 250:590-592. [DOI] [PubMed] [Google Scholar]
- 9.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed] [Google Scholar]
- 10.Boyes, J., and G. Felsenfeld. 1996. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 15:2496-2507. [PMC free article] [PubMed] [Google Scholar]
- 11.Boyes, J., J. Omichinski, D. Clark, M. Pikaart, and G. Felsenfeld. 1998. Perturbation of nucleosome structure by the erythroid transcription factor GATA-1. J. Mol. Biol. 279:529-544. [DOI] [PubMed] [Google Scholar]
- 12.Burger, G., J. Strauss, C. Scazzocchio, and B. F. Lang. 1991. nirA, the pathway-specific regulatory gene of nitrate assimilation in Aspergillus nidulans, encodes a putative GAL4-type zinc finger protein and contains four introns in highly conserved regions. Mol. Cell. Biol. 11:5746-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caddick, M. X. 1996. Nitrogen regulation in mycelial fungi, p. 349-368. In R. Brambl and G. A. Marzluf (ed.), The Mycota, 2nd ed., vol. III. Biochemistry and molecular biology. Springer-Verlag, Berlin, Germany.
- 14.Cazelle, B., A. Pokorska, E. Hull, P. M. Green, G. Stanway, and C. Scazzocchio. 1998. Sequence, exon-intron organization, transcription and mutational analysis of prnA, the gene encoding the transcriptional activator of the prn gene cluster in Aspergillus nidulans. Mol. Microbiol. 28:355-370. (Erratum, 31:1283, 1999.) [DOI] [PubMed]
- 15.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 16.Clutterbuck, A. J. 1994. Linkage map and locus list. Prog. Ind. Microbiol. 29:791-824. [PubMed] [Google Scholar]
- 17.Cove, D. J., and J. A. Pateman. 1969. Autoregulation of the synthesis of nitrate reductase in Aspergillus nidulans. J. Bacteriol. 97:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daboussi, M. J., T. Langin, F. Deschamps, Y. Brygoo, C. Scazzocchio, and G. Burger. 1991. Heterologous expression of the Aspergillus nidulans regulatory gene nirA in Fusarium oxysporum. Gene 109:155-160. [DOI] [PubMed] [Google Scholar]
- 19.de Lamotte-Malardier, L. 1991. Utilisation des gènes de la voie d'assimilation du nitrate chez Aspergillus nidulans pour la mise au point de la transformation chez Fusarium oxysporum et l'étude de leur régulation chez A. nidulans. Ph.D. thesis. Université de Paris-Sud, Orsay, France.
- 20.Diallinas, G., and C. Scazzocchio. 1989. A gene coding for the uric acid-xanthine permease of Aspergillus nidulans: inactivational cloning, characterization, and sequence of a cis-acting mutation. Genetics 122:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng, B., and G. A. Marzluf. 1998. Interaction between major nitrogen regulatory protein NIT2 and pathway-specific regulatory factor NIT4 is required for their synergistic activation of gene expression in Neurospora crassa. Mol. Cell. Biol. 18:3983-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fidel, S., J. H. Doonan, and N. R. Morris. 1988. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a gamma-actin. Gene 70:283-293. [DOI] [PubMed] [Google Scholar]
- 23.Gallagher, R. C., and E. H. Blackburn. 1998. A promoter region mutation affecting replication of the Tetrahymena ribosomal DNA minichromosome. Mol. Cell. Biol. 18:3021-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García, I., R. González, D. Gómez, and C. Scazzocchio. 2004. Chromatin rearrangements in the prnD-prnB promoter: dependence on transcription factors. Eukaryot. Cell 3:144-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gómez, D., B. Cubero, G. Cecchetto, and C. Scazzocchio. 2002. PrnA, a Zn2Cys6 activator with a unique DNA recognition mode, requires inducer for in vivo binding. Mol. Microbiol. 44:585-597. [DOI] [PubMed] [Google Scholar]
- 26.Gómez, D., I. García, C. Scazzocchio, and B. Cubero. 2003. Multiple GATA sites: protein binding and physiological relevance for the regulation of the proline transporter gene of Aspergillus nidulans. Mol. Microbiol. 50:277-289. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez, R., and C. Scazzocchio. 1997. A rapid method for chromatin structure analysis in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 25:3955-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorfinkiel, L., G. Diallinas, and C. Scazzocchio. 1993. Sequence and regulation of the uapA gene encoding a uric acid-xanthine permease in the fungus Aspergillus nidulans. J. Biol. Chem. 268:23376-23381. [PubMed] [Google Scholar]
- 29.Hung, H. L., J. Lau, A. Y. Kim, M. J. Weiss, and G. A. Blobel. 1999. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol. 19:3496-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hynes, M. J. 1978. Multiple independent control mechanisms affecting the acetamidase of Aspergillus nidulans. Mol. Gen. Genet. 161:59-65. [DOI] [PubMed] [Google Scholar]
- 31.Hynes, M. J. 1975. Studies on the role of the areA gene in the regulation of nitrogen catabolism in Aspergillus nidulans. Aust. J. Biol. Sci. 28:301-313. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 33.Johnstone, I. L., P. C. McCabe, P. Greaves, S. J. Gurr, G. E. Cole, M. A. Brow, S. E. Unkles, A. J. Clutterbuck, J. R. Kinghorn, and M. A. Innis. 1990. Isolation and characterisation of the crnA-niiA-niaD gene cluster for nitrate assimilation in Aspergillus nidulans. Gene 90:181-192. [DOI] [PubMed] [Google Scholar]
- 34.Kiekhaefer, C. M., J. A. Grass, K. D. Johnson, M. E. Boyer, and E. H. Bresnick. 2002. Hematopoietic-specific activators establish an overlapping pattern of histone acetylation and methylation within a mammalian chromatin domain. Proc. Natl. Acad. Sci. USA 99:14309-14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kudla, B., M. X. Caddick, T. Langdon, N. M. Martinez-Rossi, C. F. Bennett, S. Sibley, R. W. Davies, and H. N. Arst, Jr. 1990. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 9:1355-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letting, D. L., C. Rakowski, M. J. Weiss, and G. A. Blobel. 2003. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 23:1334-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackay, J. P., and M. Crossley. 1998. Zinc fingers are sticking together. Trends Biochem. Sci. 23:1-4. [DOI] [PubMed] [Google Scholar]
- 38.Magasanik, B., and C. A. Kaiser. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1-18. [DOI] [PubMed] [Google Scholar]
- 39.Marzluf, G. A. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 41.Muro-Pastor, M. I., R. Gonzalez, J. Strauss, F. Narendja, and C. Scazzocchio. 1999. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J. 18:1584-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narendja, F., S. Goller, M. Wolschek, and J. Strauss. 2002. Nitrate and the GATA factor AreA are necessary for in vivo binding of NirA, the pathway-specific transcriptional activator of Aspergillus nidulans. Mol. Microbiol. 44:573-583. [DOI] [PubMed] [Google Scholar]
- 43.Oestreicher, N., and C. Scazzocchio. 1995. A single amino acid change in a pathway-specific transcription factor results in differing degrees of constitutivity, hyperinducibility and derepression of several structural genes. J. Mol. Biol. 249:693-699. [DOI] [PubMed] [Google Scholar]
- 44.Pateman, J. A., and D. J. Cove. 1967. Regulation of nitrate reduction in Aspergillus nidulans. Nature 215:1234-1237. [DOI] [PubMed] [Google Scholar]
- 45.Platt, A., T. Langdon, H. Arst, D. Kirk, D. Tollervey, J. M. Sanchez, and M. X. Caddick. 1996. Nitrogen metabolite signalling involves the C-terminus and the GATA domain of the Aspergillus transcription factor AREA and the 3′ untranslated region of its mRNA. EMBO J. 15:2791-2801. [PMC free article] [PubMed] [Google Scholar]
- 46.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 47.Punt, P. J., P. A. Greaves, A. Kuyvenhoven, J. C. van Deutekom, J. R. Kinghorn, P. H. Pouwels, and C. A. van den Hondel. 1991. A twin-reporter vector for simultaneous analysis of expression signals of divergently transcribed, contiguous genes in filamentous fungi. Gene 104:119-122. [DOI] [PubMed] [Google Scholar]
- 48.Punt, P. J., J. Strauss, R. Smit, J. R. Kinghorn, C. A. van den Hondel, and C. Scazzocchio. 1995. The intergenic region between the divergently transcribed niiA and niaD genes of Aspergillus nidulans contains multiple NirA binding sites which act bidirectionally. Mol. Cell. Biol. 15:5688-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rand, K. N., and H. N. Arst, Jr. 1978. Mutations in nirA gene of Aspergillus nidulans and nitrogen metabolism. Nature 272:732-734. [DOI] [PubMed] [Google Scholar]
- 50.Ravagnani, A., L. Gorfinkiel, T. Langdon, G. Diallinas, E. Adjadj, S. Demais, D. Gorton, H. N. Arst, Jr., and C. Scazzocchio. 1997. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter- specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 16:3974-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scazzocchio, C. 2000. The fungal GATA factors. Curr. Opin. Microbiol. 3:126-131. [DOI] [PubMed] [Google Scholar]
- 52.Scazzocchio, C. 1994. The purine degradation pathway, genetics, biochemistry and regulation. Prog. Ind. Microbiol. 29:221-257. [PubMed] [Google Scholar]
- 53.Scazzocchio, C., and H. N. Arst. 1989. Regulation of nitrate assimilation in Aspergillus nidulans, p. 299-313. In J. R. Kinghorn (ed.), Molecular and genetic aspects of nitrate assimilation. Oxford Science Publications, Oxford, United Kingdom.
- 54.Scazzocchio, C., N. Sdrin, and G. Ong. 1982. Positive regulation in a eukaryote, a study of the uaY gene of Aspergillus nidulans. I. Characterization of alleles, dominance and complementation studies, and a fine structure map of the uaY-oxpA cluster. Genetics 100:185-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sealy-Lewis, H. M., D. Lycan, and C. Scazzocchio. 1979. Product induction of purine hydroxylase II in Aspergillus nidulans. Mol. Gen. Genet. 174:105-106. [DOI] [PubMed] [Google Scholar]
- 56.Sharma, K. K., and H. N. Arst, Jr. 1985. The product of the regulatory gene of the proline catabolism gene cluster of Aspergillus nidulans is a positive-acting protein. Curr. Genet. 9:299-304. [DOI] [PubMed] [Google Scholar]
- 57.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 58.Stamatoyannopoulos, J. A., A. Goodwin, T. Joyce, and C. H. Lowrey. 1995. NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human beta-globin locus control region. EMBO J. 14:106-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Starich, M. R., M. Wikstrom, H. N. Arst, Jr., G. M. Clore, and A. M. Gronenborn. 1998. The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors. J. Mol. Biol. 277:605-620. [DOI] [PubMed] [Google Scholar]
- 60.Starich, M. R., M. Wikstrom, S. Schumacher, H. N. Arst, Jr., A. M. Gronenborn, and G. M. Clore. 1998. The solution structure of the Leu22→Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity. J. Mol. Biol. 277:621-634. [DOI] [PubMed] [Google Scholar]
- 61.Strauss, J., M. I. Muro-Pastor, and C. Scazzocchio. 1998. The regulator of nitrate assimilation in ascomycetes is a dimer which binds a nonrepeated, asymmetrical sequence. Mol. Cell. Biol. 18:1339-13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suarez, T., M. V. de Queiroz, N. Oestreicher, and C. Scazzocchio. 1995. The sequence and binding specificity of UaY, the specific regulator of the purine utilization pathway in Aspergillus nidulans, suggest an evolutionary relationship with the PPR1 protein of Saccharomyces cerevisiae. EMBO J. 14:1453-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tollervey, D. 1981. Aspects of nitrogen metabolic regulation in Aspergillus nidulans. Ph.D. thesis. University of Cambridge, Cambridge, United Kingdom.
- 64.Tollervey, D., and H. N. Arst. 1981. Mutations to constitutivity and dere-pression are separate and separable in a regulatory gene of Aspergillus nidulans. Curr. Genet. 4:63-68. [DOI] [PubMed] [Google Scholar]
- 65.Wilson, R. A., and H. N. Arst, Jr. 1998. Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “streetwise” GATA family of transcription factors. Microbiol. Mol. Biol. Rev. 62:586-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolschek, M. F., F. Narendja, J. Karlseder, C. P. Kubicek, C. Scazzocchio, and J. Strauss. 1998. In situ detection of protein-DNA interactions in filamentous fungi by in vivo footprinting. Nucleic Acids Res. 26:3862-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamagata, T., K. Mitani, H. Oda, T. Suzuki, H. Honda, T. Asai, K. Maki, T. Nakamoto, and H. Hirai. 2000. Acetylation of GATA-3 affects T-cell survival and homing to secondary lymphoid organs. EMBO J. 19:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]