Abstract
STAT3 and STAT5 (STAT3/5) proteins are crucial mediators of cytokine- or growth factor-induced cell survival and proliferation. These transcription factors are frequently overactivated in a variety of solid tumors and hematopoietic neoplasms and are targets of various oncogenes with tyrosine kinase activity. STAT3/5 proteins regulate expression of genes involved in survival and proliferation in the nucleus and interact with signaling pathways in the cytoplasm. Evidences for a cross-talk between STAT3/5 and oxidative metabolism have recently emerged. This review summarizes the current knowledge on the cross-regulation between STAT3/5 and oxidative metabolism in normal and cancer cells.
Keywords: STAT3, STAT5, ROS, oxidative stress, NOX, mitochondria, oncogene, cancer, leukemia
STAT3, STAT5A, and STAT5B, Three STAT Family Members with Redundant and Specific Oncogenic Properties
Signal transducer and activator of transcription (STAT) proteins are a seven-member family of cytoplasmic transcription factors that relay signals emanating from the cell surface cytokine and growth factors receptors to the nucleus. Upon ligand binding, cytoplasmic STAT proteins are activated through tyrosine phosphorylation, mainly by the JAK kinases. Activated STATs then dimerize and translocate into the nucleus where they bind to specific DNA elements and regulate transcription of target genes.1 STAT proteins control fundamental cellular processes, including survival, proliferation, differentiation, and immune responses.
It is now well-established that three of these family members, STAT3 and the closely related STAT5A and STAT5B, proteins are also important effectors of cellular transformation.2 Aberrant STAT3, STAT5A, and STAT5B signaling has been described in different solid tumors such as prostate, breast, colon, gliomas, head and neck cancer, melanoma, and in hematopoietic malignancies.2,3 Persistent activation of these transcription factors is frequently found in many tumor cells most probably as a consequence of deregulated tyrosine kinase activity. STAT5A/B and/or STAT3 are the common targets for different oncoproteins with tyrosine kinase activity like Tel-JAK2, JAK2V617F, Src, Bcr-Abl, Tel-Syk, NPM-ALK, Tel-PDGFR, and mutated forms of FLT3 and Kit receptors.2,4 Inhibition of STAT3/STAT5 signaling interrupts the transforming potential of these tyrosine kinases and the induction of cancer or leukemia in mouse models.5-13Evidences for a direct role of STAT3 or STAT5A/B in cell transformation were provided by the use of constitutively active variants.14,15 These proteins designated STAT3C, STAT51*6, or cS5F are able to transform cell lines or hematopoietic cells and to induce solid tumors or leukemias in mice.
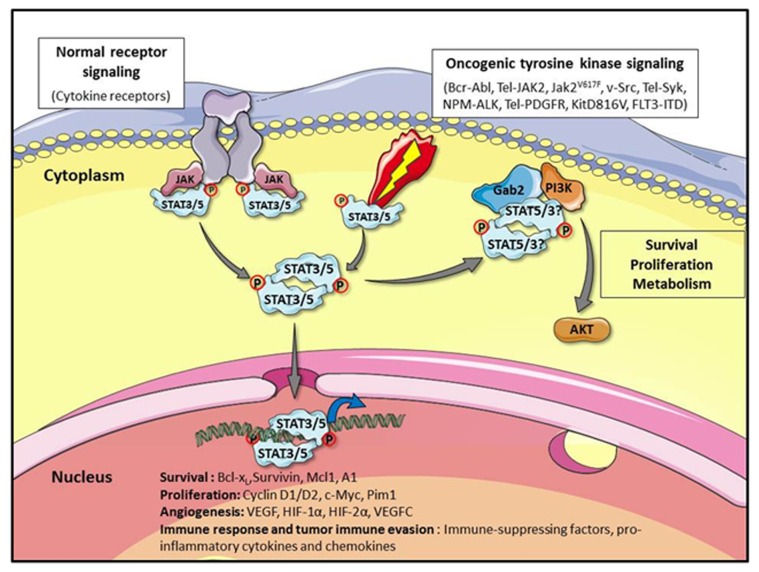
Tumor progression and metastasis involve the combined ability of cancer cells to resist apoptosis, to proliferate and to promote angiogenesis and tumor cell migration. STAT3 and/or STAT5a/b have been shown to participate in these processes. These proteins regulate indirectly or directly the expression of genes that are required for survival like the anti-apoptotic bcl-x, a1, mcl-1, and survivin genes or cell growth like c-myc, cyclin D1, and D2.2,4 STAT3 is a key regulator of VEGF expression which is critical in tumor angiogenesis while VEGF-C, another member of the VEGF family is dependent on STAT5 activity in prostate cancer.16-18 STAT3 and/or STAT5 have been shown to regulate expression of different cytokines, chemokines, pro-inflammatory mediators and integrins that play a role in tumor cell migration and immune surveillance evasion.2,18 Aberrant STAT3 and/or STAT5 signaling also promote epithelial–mesenchymal transition (EMT), a key event in the tumor invasion process, and STAT3 was shown to induce expression of TWIST1, a critical regulator of EMT.19,20 Beside their nuclear function, tyrosine phosphorylated STAT5 (P-Y-STAT5) proteins have an important role in the cytoplasm of cancer cells. P-Y-STAT5 has been detected in the cytoplasm of colorectal cancer and myeloid leukemic cells.21,22 We previously showed that P-Y-STAT5 proteins interact with the scaffolding adaptor Gab2 to activate the PI3-kinase/Akt pathway in myeloid leukemias.22,23Activation of this pathway is implicated in STAT5-dependent transformation of hematopoietic cells and induction of leukemia in mice.24 P-Y-STAT3 has also been detected in the cytoplasm of cancer cells and was shown to interact with Gab2 and the PI3-kinase25,26 (see Fig. 1).
Figure 1. STAT3 and STAT5 signaling in normal and transformed cells. In normal cells, cytokines binding to their receptors results in the activation of JAK tyrosine kinases and STAT3 or STAT5 (STAT3/5). Tyrosine phosphorylated STAT3/5 proteins dimerize and translocate to the nucleus to regulate gene expression. Whereas STAT3/5 activation is tightly regulated in normal cells, the oncogenic activation of tyrosine kinases causes constitutive tyrosine phosphorylation of STAT3/5. This leads to permanent changes in the expression of genes that control cellular processes such as proliferation survival, angiogenesis, and immune response which are commonly disrupted in cancer. Constitutively active STAT5 and possibly STAT3 proteins also interact with the scaffolding adaptor Gab2 in the cytoplasm of transformed cells to activate the PI3-Kinase/Akt pathway, which plays an important role in survival, proliferation, and metabolism regulation. This figure was produced using Servier Medical Art: www.servier.com
ROS-Dependent Regulation of STAT Protein Activity in Normal and Cancer Cells
It is now well recognized that free radicals and reactive oxygen species (ROS) produced by O2 metabolism are important initiators and promoters of carcinogenesis and contribute to tumor progression. ROS, including superoxide (O2-), hydrogen peroxide (H2O2), and the hydroxyl free radical (HO-), are mainly generated by the mitochondrion, endoplasmic reticulum and the membrane bound nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) which is a multi-subunit enzyme consisting of a catalytic subunit gp91phox (also termed NOX2) and regulatory subunits, p67phox, p47phox, p22phox, and the small GTPase Rac1. Seven members of the NOX family have been identified (NOX1–5 and DUOX1–2) that differ in tissue distribution and regulatory mechanisms. ROS have been shown to influence cell-cycle progression, apoptosis, and growth factor signaling in a variety of cell types. Normal cells regulate the intracellular ROS content by balancing the ROS generation and scavenging systems. Oxidative stress is due to an excessive cellular ROS production and/or a deficiency in antioxidant defenses.27,28
Cytokines and growth factors activate intracellular regulation of redox processes through generation of ROS.29,30 In these cases, ROS may act as second messengers to regulate activities of redox-sensitive enzymes including phosphatases.31 Several protein tyrosine phosphatases are highly sensitive to oxidation because of a critical thiol group in the active site of the enzyme.32 ROS induce the reversible oxidation and inactivation of tyrosine phosphatases and increase the tyrosine phosphorylation of cellular proteins. Studies with antioxidant treatment of cytokine-stimulated cells and with H2O2 stimulation in cytokine deprived cells indicate that ROS may play roles in cytokine activation of JAK kinases as well as STAT3, STAT5, and other signaling proteins.33 Activation of STAT3 and/or STAT5 by cytokines or growth factors-induced ROS might involve NOX activity.34-37 In contrast, data from the literature have demonstrated that oxidative stress inhibits cytokine-induced JAK-STAT pathway.38,39 These discrepancies remain unclear but it is possible that in oxidative stress conditions, cytokines are not able to activate the JAK-STAT pathway. The inhibitory action of oxidative stress might also involve a direct oxidation of JAK and STAT proteins. Oxidation of cysteine residues in the catalytic domain of JAK2 inhibits the kinase activity of this molecule.40 Similarly, STAT3 is directly sensitive to intracellular oxidants. Formation of dimers, trimers and tetramers of STAT3 by disulfide-linked homodimerization and oligomerization has been evidenced in cells treated with H2O2.41,42 In this case, oxidation of conserved cysteine residues inhibits STAT3 DNA binding and transcriptional activity without affecting tyrosine phosphorylation. Formation of these multimers would be reversible and dependent on the intracellular redox potential. The higher the ROS levels, the more STAT3 proteins may be cross-linked into a redox tetramer. The precise function of these oxidized STAT3 complexes still remains unclear. STAT3 was shown to be present in mitochondria and contribute to the regulation of electron transport chain function. It has been proposed that oxidation of STAT3 may be regulated by mitochondrial peroxide, which could in turn modulate the association of STAT3 with mitochondria in a feedback mechanism to control mitochondrial respiration.43 Interestingly, expression of redox insensitive STAT3 cysteine mutant in breast cancer cells accelerated their proliferation but reduced their resistance to oxidative stress. STAT3 might then be involved in the coupling between redox homeostasis and cell proliferation.42 In addition to disulfide oligomerization, cysteine glutathionylation was shown to inhibit tyrosine phosphorylation of STAT3 in oxidative stress-induced cells.44 Specific oxidation of STAT5 in macrophages of aged mice has been reported. This oxidation interferes with cytokine-induced tyrosine phosphorylation of STAT5 via a mechanism that remains unknown.45
Oxidative stress has been found in many cancers, both in solid tumors and in several hematopoietic malignancies. There is evidence that tumor-derived ROS may promote proliferation, cell survival, migration and metastasis. These observations suggest that chronically increased endogenous ROS levels lead to adaptive changes that play pivotal roles in tumor progression.27,28,46 Moreover, chronic oxidative stress may cause DNA, protein, and/or lipid damage, leading to changes in chromosome instability, genetic mutations, and even drug resistance.47 Oncogenes can recapitulate most of these processes. Transformation by oncogenic tyrosine kinases is often accompanied by an increase of intracellular ROS which can in turn promote chromosomal instability. Elevated ROS levels have been detected in cell transformed by JAK2V617F, NPM-ALK, FLT3-ITD, c-Src, TEL-PDGFR, and Bcr-Abl oncogenes.47-52 The mechanisms by which oncogenic tyrosine kinases regulate the intracellular level of ROS are not fully determined, however, there may be important overlap.53 In Bcr-Abl-transformed cells, ROS are generated via distinct mechanisms involving the mitochondrial electron transport chain, the PI3K/mTOR pathway and the Rac/NOX protein complex. The origin of ROS associated with Src or FLT3-ITD transformation is also linked to activation of the Rac/NOX complex.27 Activation of NOX is restricted to specialized membrane areas in Src expressing cells.54 A large majority of ROS (H2O2) associated with FLT3-ITD in leukemic cells is localized in the endoplasmic reticulum and is produced by the small membrane bound component of the NOX complex, p22phox.36 In addition to the mechanisms of ROS generation described here, NPM-ALK was shown to induce ROS production by a pathway involving the arachidonic acid-metabolizing enzymes of the lipoxygenase (LOX) family. The use of specific LOX inhibitor or the anti-oxidant N-acetyl-cystein (NAC) demonstrated the importance of ROS in maintaining the ALK kinase active.49
Since ROS contribute to the regulation of redox sensitive proteins such as phosphatases, kinases and other signaling proteins, it is therefore not surprising that ROS production induced by oncogenic tyrosine kinases would affect indirectly the phosphorylation of STAT3 and STAT5. ROS seem to be important for the sustained activity of NPM-ALK and Src is a redox sensitive kinase.49,54 H2O2-dependent Src oxidation was shown to increase the catalytic activity of the kinase whereas NAC treatment had an inhibitory effect. Tyrosine phosphorylation of STAT3 and STAT5 would therefore be triggered as a consequence of ROS-mediated inhibition of tyrosine phosphatases and activation of tyrosine kinases. It will be important in the future to determine whether oxidation of STAT3 and/or STAT5 induced by ROS also exists in cells transformed by these oncogenic tyrosine kinases.
STAT-Dependent Regulation of ROS Production and Oxidative Metabolism in Normal and Cancer Cells
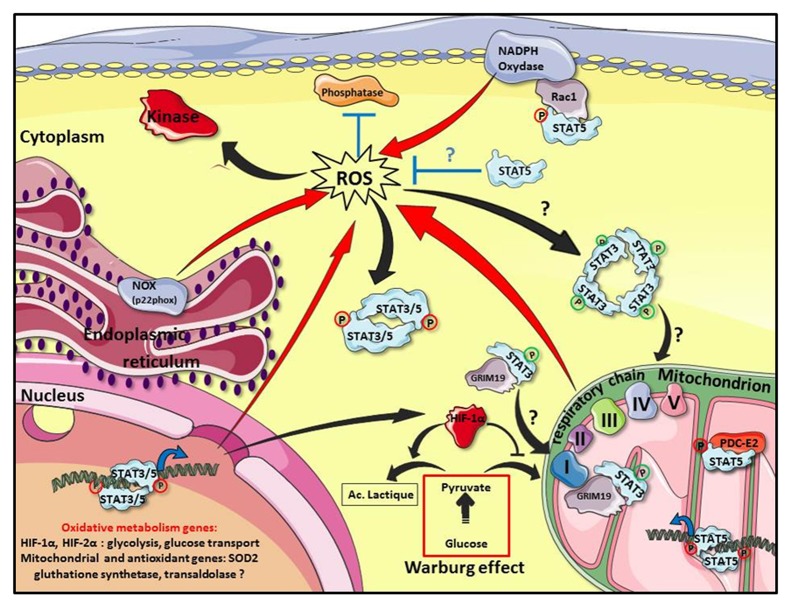
The past 5 years highlighted the emerging roles of STAT3 and STAT5 in the regulation of ROS production and oxidative metabolism in normal and cancer cells. The starting point of this story came with the discovery of a non-canonical role of STAT3 in mitochondria that did not require the DNA binding domain or the tyrosine phosphorylation.55 STAT3 was shown to localize in mitochondria of pro-B cells and to regulate the electron transport chain (ETC) activity, the ROS producing “enzymatic machinery” in mitochondria. Other reports indicate that GRIM19, a component of the ETC complex 1 and a previously identified binding partner of STAT3, is necessary for STAT3 uptake in mitochondria.56,57 Phosphorylation of the serine 727 residue present in the carboxyl-terminal region of STAT3 seems to be important for GRIM19-mediated recruitment of STAT3 and mitochondrial function (see Fig. 2). More recently, a mitochondrial dysfunction associated with high ROS levels has been reported in hematopoietic stem cells from Stat3−/− mice suggesting that STAT3 is essential for normal mitochondrial activity in hematopoietic cells.58 Fascinatingly, malignant transformation of mouse embryo fibroblasts by activated Ras oncogene also requires mitochondrial STAT3. Serine (ser727) but not tyrosine (tyr705) phosphorylation of STAT3 is crucial for Ras-mediated cellular transformation and mitochondrial function.59 Mitochondrial STAT3 upregulates ETC activity in Ras-transformed cells. Paradoxically, lactate dehydrogenase activity was also increased in these cells indicating a shift to aerobic glycolysis. Cancer cells exhibit a distinct metabolic shift from oxidative phosphorylation in the mitochondrion to aerobic glycolysis in the cytoplasm, known as the Warburg effect.60 In an elegant work, Demaria et al. demonstrated that oncogenic STAT3 proteins induced an aerobic glycolysis in primary fibroblasts and in STAT3-dependent tumor cell lines.61 This shift toward aerobic glycolysis is dependent on hypoxia inducible factor-1α (HIF-1α) upregulation that is partly due to STAT3-dependent transcription (see Fig. 2).62 HIF-1α is a transcription factor that is primarily regulated by cellular oxygen levels but also by oncogenes or growth factors.63 HIF-1α is known to regulate expression of genes involved in glycolysis and glucose transport. The increased glycolysis observed in STAT3-transformed cells is accompanied by a downregulation of mitochondrial respiration which is caused by a STAT3-mediated decrease in mitochondrial protein expression leading to reduced levels of ETC complexes.61 Conversely, inhibition of STAT3 expression and activity in normal and/or cancer cells is often accompanied by increased ROS levels and a mitochondrial dysfunction.58,64,65
Figure 2. Oxidative metabolism and STAT3/STAT5 in cancer cells. In cancer cells, regulation of ROS levels is tightly controlled by the mitochondrion, endoplasmic reticulum (p22phox), the membrane bound NADPH oxidase (NOX) and by the transcriptional regulation of genes that might affect oxidative metabolism (red arrows). ROS indirectly influence STAT3 and STAT5 (STAT3/5) signaling by inhibiting phosphatases and activating kinases (black arrows). An excessive ROS production may also induce oxidation of STAT3 by disulphide-linked oligomerization to modulate STAT3 and mitochondrial activities. Conversely, STAT3 and STAT5 proteins regulate ROS levels in transformed cells by distinct mechanisms. In mitochondria, serine phosphorylated STAT3 (ser727) (green circle) regulates the electron transport chain (ETC, respiratory chain) activity. Translocation of STAT3 to mitochondrion is dependent on GRIM19, a binding partner of STAT3 and a component of the ETC complex 1. Tyrosine phosphorylated STAT5 (red circle) interacts with PDC-E2, a component of the pyruvate dehydrogenase complex and the mitochondrial genome. In the nucleus, tyrosine phosphorylated STAT3 induces HIF-1α expression to promote an aerobic glycolysis in the cytoplasm of cancer cells and downregulates mitochondrial activity by repressing expression of unidentified mitochondrial genes. Activated STAT3 may also regulate expression of antioxidant genes such as SOD2. Tyrosine phosphorylated STAT5 upregulates HIF-2α expression which has been associated with increased glycolysis. In the cytoplasm, tyrosine phosphorylated STAT5 induces the production of ROS via its binding to Rac1 and activation of NADPH oxidase. In contrast, unphosphorylated STAT5 has a protective effect against excessive ROS levels in pre-B leukemic cells by directly or indirectly regulating expression of proteins involved in oxidative metabolism. This figure was produced using Servier Medical Art: www.servier.com
An important observation that arises from these different studies is the ability of STAT3 to upregulate or downregulate ETC activity. Tyrosine phosphorylation that discriminates between nuclear and mitochondrial STAT3 is probably essential in this apparent contradictory effect. The downregulation of ETC activity is accompanied by a decreased ROS accumulation in cells transformed by constitutively active STAT3 suggesting that STAT3 protects cells from apoptosis by preventing an overproduction of ROS. STAT3-dependent transcriptional repression of nuclear encoded mitochondrial genes has been suggested to explain the downregulation of ETC activity and the decreased ROS levels.61 Alternatively, STAT3 might also upregulate expression of antioxidant genes such as SOD2 (superoxide dismutase) which could in turn contribute to reduce ROS levels.66 In contrast to STAT3, translocation of STAT5 to mitochondria requires tyrosine phosphorylation.67 Mitochondrial localization of tyrosine phosphorylated STAT5 was observed in IL-2-stimulated T cells or leukemic T cells expressing constitutively active STAT5. In mitochondria, STAT5 interacts with the protein E2, a component of the pyruvate dehydrogenase complex (PDC-E2). It also binds to the D-loop regulatory region of mitochondrial DNA suggesting that STAT5 might be involved in the regulation of mitochondrial genome (see Fig. 2). The mitochondrial localization of STAT5 coincides with the metabolic shift to aerobic glycolysis observed in cytokine-stimulated and leukemic T cells. Interestingly, HIF-2α, an HIF isoform closely related to HIF-1α, was identified as a STAT5 target gene in hematopoietic stem cell (HSC).68 Downregulation of HIF-2α expression reduced STAT5-induced HSC expansion as well as progenitor and stem cell frequencies. Glucose uptake was also enhanced in STAT5-expressing HSC and HIF-2α was shown to be required for STAT5-induced upregulation of genes associated with glucose metabolism. STAT5-mediated glucose uptake was also observed in T cells.69
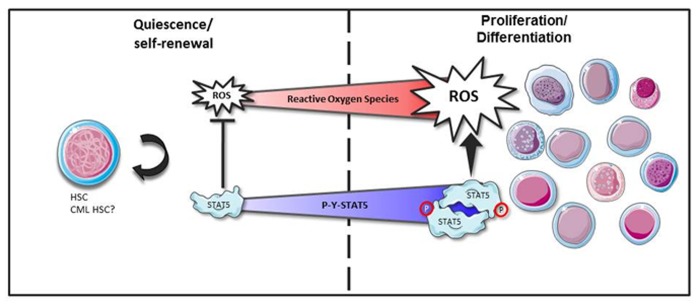
Oncogenic tyrosine kinases FLT3-ITD in acute myeloid leukemia (AML) cells and BCR-Abl in chronic myeloid leukemia (CML) cells have been shown to induce the production of ROS which can in turn increase DNA double-strand breaks and repair errors promoting genomic instability and mutagenesis. In both cases, increased ROS levels appear to be produced via STAT5 signaling.70,71 Evidences for a direct effect of STAT5 on ROS production was observed in human fibroblasts expressing the constitutively active STAT51*6 mutant.72 We also recently showed that STAT51*6-induced transformation of hematopoietic cells is accompanied by an increase of ROS levels (unpublished data). In FLT3-ITD expressing AML cells, tyrosine phosphorylated STAT5 interacts with the small GTPase protein Rac1 to regulate NOX activity (see Fig. 2).70 ROS production in AML cells is reduced by treatment with FLT3-ITD or NOX inhibitors indicating that ROS production is FLT3-ITD and NOX dependent. Moreover, FLT3-ITD inhibition abrogated tyrosine phosphorylation of STAT5, decreased Rac1 activity and binding to NOX2. As previously mentioned, a large majority of H2O2 produced by the p22phox was found in the endoplasmic reticulum of FLT3-ITD expressing cells. Inhibition of FLT3-ITD blocked the production of ROS in the endoplasmic reticulum without affecting mitochondrial ROS levels. Importantly, downregulation of p22phox dramatically reduced the production of ROS and STAT5 signaling in these transformed cells suggesting the existence of a feed forward loop in which p22phox-derived ROS maintain STAT5 signaling to increase ROS levels via the Rac1/NOX complex activity.36 The mechanisms implicated in STAT5-induced ROS levels in Bcr-Abl-transformed cells still remain enigmatic. DNA binding, transcriptional activity and oligomer formation are required for STAT5-mediated ROS production indicating that STAT5 regulates expression of genes implicated in the oxidative metabolism.71 NOX4 was a good candidate because it was an important source of increased ROS levels in Bcr-Abl expressing cells and was shown to be regulated by STAT5 in the liver.73,74 However, the STAT5-dependent expression of NOX4 was not observed in Bcr-Abl-transformed cells. STAT5-mediated ROS production induced by Bcr-Abl is also independent of the mitochondrial respiratory chain and it seems then to be unlikely that STAT5 regulates expression of ETC components. In contrast to these data, analysis of STAT5A and STAT5B functions in hematopoietic stem/progenitors cells (HS/P cells) from healthy donors and CML patients showed that STAT5A plays an oxidative stress protective role.75 In this study, the authors used a RNAi-based strategy to specifically downregulate expression of STAT5A, STAT5B, or both. Remarkably, they showed that specific attenuation of STAT5A but not STAT5B expression is sufficient to enhance basal oxidative stress and DNA damage of normal and CML HS/P cells. Basal ROS production following downregulation of STAT5A is partly dependent on NOX activity but not on mitochondrial function. Importantly, expression of STAT5A and a transactivation-deleted STAT5AΔ749 mutant rescues these activities suggesting that STAT5A expression protects HSC from an overproduction of ROS via a non-canonical mechanism. These data are reminiscent of our recent published work in which we showed a similar protective role of STAT5A in pre-B leukemic cells.76 Inhibition of STAT5A activity or expression in these cells was sufficient to induce a basal oxidative stress and apoptosis. We used a proteomic approach to identify proteins that were differentially regulated in cells expressing or not a dominant negative form of STAT5A and showed that 40% of the identified proteins were related to the oxidative metabolism and 14% to the stress-related heat shock proteins. Some of these proteins have been shown to control the levels of ROS and might regulate apoptosis as well. Among these proteins, we identified the glutathione synthetase (GSS) and transaldolase, which are enzymes involved in the generation of glutathione and NADPH, respectively. Expression of both proteins was downregulated in cells expressing the dominant negative form of STAT5A. In contrast to these results, we recently found that STAT5 induced the production of ROS in hematopoietic cells transformed by the constitutively active STAT51*6 mutant or in Bcr-Abl expressing cells, consistent with recently published work.71 This opposite behavior of STAT5 in the regulation of ROS production might be related to the levels of tyrosine phosphorylation as mentioned for STAT3. In pre-B cells in which STAT5 exerts a protective role against oxidative stress, tyrosine phosphorylation is undetectable while STAT5 is persistently tyrosine phosphorylated in STAT51*6 or Bcr-Abl expressing cells. This might explain the differences observed between the apparent “antioxidant” effect of STAT5 in CML HS/P cells which have a low proliferating rate and the “oxidant” effect of STAT5 in CML cell lines with high proliferating rates. It would be interesting to properly analyze levels of tyrosine phosphorylated STAT5 in CML HSC and their progeny. STAT5 is now well-recognized as an important regulator of HSC function.77 Maintenance, expansion and differentiation of HSC are dependent on STAT5 protein levels.78 Similarly, ROS appear to control HSC behavior. Low ROS levels favor quiescence and maintenance of HSC while high ROS levels are found in proliferating and differentiating HS/P cells.79 STAT5 could play a dual role in HSC. As proposed in Figure 3, non-tyrosine phosphorylated STAT5 would protect HSC from a ROS overproduction helping them to stay in a quiescent or a self-renewal state. The rise of tyrosine phosphorylated STAT5 would favor their proliferation by inducing ROS production. In a similar vein, quiescence, self-renewal and proliferation of CML HSC might be also dependent on the degree of STAT5 tyrosine phosphorylation and expression.
Figure 3. Hypothetical model of STAT5-dependent regulation of ROS in HSC. ROS levels are critical in regulating the balance between HSC self-renewal and proliferation/differentiation. Low ROS levels favor quiescence and/or self-renewal of HSC while high ROS levels promote their proliferation and differentiation. The balance between quiescence/self-renewal and proliferation/differentiation is also dependent on STAT5 expression and/or activation levels. In the proposed model, non-tyrosine phosphorylated STAT5 might contribute to the protection of HSC against a ROS overproduction, keeping them in a quiescent or self-renewal state. An increase of ROS levels through cytokine receptors signaling would facilitate tyrosine phosphorylation of STAT5. High tyrosine phosphorylated STAT5 levels would in turn modulate ROS production to promote proliferation and differentiation. Quiescence and self-renewal of CML HSC might also be dependent on ROS and tyrosine phosphorylated STAT5 levels. This figure was produced using Servier Medical Art: www.servier.com
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/25764
References
- 1.Leaman DW, Leung S, Li X, Stark GR. Regulation of STAT-dependent pathways by growth factors and cytokines. FASEB J. 1996;10:1578–88. [PubMed] [Google Scholar]
- 2.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 3.Benekli M, Baer MR, Baumann H, Wetzler M. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101:2940–54. doi: 10.1182/blood-2002-04-1204. [DOI] [PubMed] [Google Scholar]
- 4.Ferbeyre G, Moriggl R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochim Biophys Acta. 2011;1815:104–14. doi: 10.1016/j.bbcan.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11:623–9. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 6.Harir N, Boudot C, Friedbichler K, Sonneck K, Kondo R, Martin-Lannerée S, et al. Oncogenic Kit controls neoplastic mast cell growth through a Stat5/PI3-kinase signaling cascade. Blood. 2008;112:2463–73. doi: 10.1182/blood-2007-09-115477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson EA, Walker SR, Xiang M, Weisberg E, Bar-Natan M, Barrett R, et al. The STAT5 Inhibitor Pimozide Displays Efficacy in Models of Acute Myelogenous Leukemia Driven by FLT3 Mutations. Genes Cancer. 2012;3:503–11. doi: 10.1177/1947601912466555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR, et al. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol Cell. 2000;6:693–704. doi: 10.1016/S1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 10.Yan D, Hutchison RE, Mohi G. Critical requirement for Stat5 in a mouse model of polycythemia vera. Blood. 2012;119:3539–49. doi: 10.1182/blood-2011-03-345215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cain JA, Xiang Z, O’Neal J, Kreisel F, Colson A, Luo H, et al. Myeloproliferative disease induced by TEL-PDGFRB displays dynamic range sensitivity to Stat5 gene dosage. Blood. 2007;109:3906–14. doi: 10.1182/blood-2006-07-036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–52. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walz C, Ahmed W, Lazarides K, Betancur M, Patel N, Hennighausen L, et al. Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and JAK2(V617F) in mice. Blood. 2012;119:3550–60. doi: 10.1182/blood-2011-12-397554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 15.Moriggl R, Sexl V, Kenner L, Duntsch C, Stangl K, Gingras S, et al. Stat5 tetramer formation is associated with leukemogenesis. Cancer Cell. 2005;7:87–99. doi: 10.1016/j.ccr.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–8. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 17.Jung JE, Lee HG, Cho IH, Chung DH, Yoon SH, Yang YM, et al. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. FASEB J. 2005;19:1296–8. doi: 10.1096/fj.04-3099fje. [DOI] [PubMed] [Google Scholar]
- 18.Gu L, Vogiatzi P, Puhr M, Dagvadorj A, Lutz J, Ryder A, et al. Stat5 promotes metastatic behavior of human prostate cancer cells in vitro and in vivo. Endocr Relat Cancer. 2010;17:481–93. doi: 10.1677/ERC-09-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho KH, Jeong KJ, Shin SC, Kang J, Park CG, Lee HY. STAT3 mediates TGF-β1-induced TWIST1 expression and prostate cancer invasion. Cancer Lett. 2013;336:167–73. doi: 10.1016/j.canlet.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 20.Lee TK, Man K, Poon RT, Lo CM, Yuen AP, Ng IO, et al. Signal transducers and activators of transcription 5b activation enhances hepatocellular carcinoma aggressiveness through induction of epithelial-mesenchymal transition. Cancer Res. 2006;66:9948–56. doi: 10.1158/0008-5472.CAN-06-1092. [DOI] [PubMed] [Google Scholar]
- 21.Xiong H, Su WY, Liang QC, Zhang ZG, Chen HM, Du W, et al. Inhibition of STAT5 induces G1 cell cycle arrest and reduces tumor cell invasion in human colorectal cancer cells. Lab Invest. 2009;89:717–25. doi: 10.1038/labinvest.2009.11. [DOI] [PubMed] [Google Scholar]
- 22.Harir N, Pecquet C, Kerenyi M, Sonneck K, Kovacic B, Nyga R, et al. Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood. 2007;109:1678–86. doi: 10.1182/blood-2006-01-029918. [DOI] [PubMed] [Google Scholar]
- 23.Nyga R, Pecquet C, Harir N, Gu H, Dhennin-Duthille I, Régnier A, et al. Activated STAT5 proteins induce activation of the PI 3-kinase/Akt and Ras/MAPK pathways via the Gab2 scaffolding adapter. Biochem J. 2005;390:359–66. doi: 10.1042/BJ20041523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G, Miskimen KL, Wang Z, Xie XY, Tse W, Gouilleux F, et al. Effective targeting of STAT5-mediated survival in myeloproliferative neoplasms using ABT-737 combined with rapamycin. Leukemia. 2010;24:1397–405. doi: 10.1038/leu.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni S, Zhao C, Feng GS, Paulson RF, Correll PH. A novel Stat3 binding motif in Gab2 mediates transformation of primary hematopoietic cells by the Stk/Ron receptor tyrosine kinase in response to Friend virus infection. Mol Cell Biol. 2007;27:3708–15. doi: 10.1128/MCB.01838-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeffer LM, Mullersman JE, Pfeffer SR, Murti A, Shi W, Yang CH. STAT3 as an adapter to couple phosphatidylinositol 3-kinase to the IFNAR1 chain of the type I interferon receptor. Science. 1997;276:1418–20. doi: 10.1126/science.276.5317.1418. [DOI] [PubMed] [Google Scholar]
- 27.Hole PS, Darley RL, Tonks A. Do reactive oxygen species play a role in myeloid leukemias? Blood. 2011;117:5816–26. doi: 10.1182/blood-2011-01-326025. [DOI] [PubMed] [Google Scholar]
- 28.Ogasawara MA, Zhang H. Redox regulation and its emerging roles in stem cells and stem-like cancer cells. Antioxid Redox Signal. 2009;11:1107–22. doi: 10.1089/ars.2008.2308. [DOI] [PubMed] [Google Scholar]
- 29.Sattler M, Winkler T, Verma S, Byrne CH, Shrikhande G, Salgia R, et al. Hematopoietic growth factors signal through the formation of reactive oxygen species. Blood. 1999;93:2928–35. [PubMed] [Google Scholar]
- 30.Iiyama M, Kakihana K, Kurosu T, Miura O. Reactive oxygen species generated by hematopoietic cytokines play roles in activation of receptor-mediated signaling and in cell cycle progression. Cell Signal. 2006;18:174–82. doi: 10.1016/j.cellsig.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–28. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- 32.Salmeen A, Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid Redox Signal. 2005;7:560–77. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 33.Simon AR, Rai U, Fanburg BL, Cochran BH. Activation of the JAK-STAT pathway by reactive oxygen species. Am J Physiol. 1998;275:C1640–52. doi: 10.1152/ajpcell.1998.275.6.C1640. [DOI] [PubMed] [Google Scholar]
- 34.Jay DB, Papaharalambus CA, Seidel-Rogol B, Dikalova AE, Lassègue B, Griendling KK. Nox5 mediates PDGF-induced proliferation in human aortic smooth muscle cells. Free Radic Biol Med. 2008;45:329–35. doi: 10.1016/j.freeradbiomed.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shatynski KE, Chen H, Kwon J, Williams MS. Decreased STAT5 phosphorylation and GATA-3 expression in NOX2-deficient T cells: role in T helper development. Eur J Immunol. 2012;42:3202–11. doi: 10.1002/eji.201242659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woolley JF, Naughton R, Stanicka J, Gough DR, Bhatt L, Dickinson BC, et al. H2O2 production downstream of FLT3 is mediated by p22phox in the endoplasmic reticulum and is required for STAT5 signalling. PLoS One. 2012;7:e34050. doi: 10.1371/journal.pone.0034050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon S, Woo SU, Kang JH, Kim K, Kwon MH, Park S, et al. STAT3 transcriptional factor activated by reactive oxygen species induces IL6 in starvation-induced autophagy of cancer cells. Autophagy. 2010;6:1125–38. doi: 10.4161/auto.6.8.13547. [DOI] [PubMed] [Google Scholar]
- 38.Kaur N, Lu B, Monroe RK, Ward SM, Halvorsen SW. Inducers of oxidative stress block ciliary neurotrophic factor activation of Jak/STAT signaling in neurons. J Neurochem. 2005;92:1521–30. doi: 10.1111/j.1471-4159.2004.02990.x. [DOI] [PubMed] [Google Scholar]
- 39.Di Bona D, Cippitelli M, Fionda C, Cammà C, Licata A, Santoni A, et al. Oxidative stress inhibits IFN-alpha-induced antiviral gene expression by blocking the JAK-STAT pathway. J Hepatol. 2006;45:271–9. doi: 10.1016/j.jhep.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 40.Smith JK, Patil CN, Patlolla S, Gunter BW, Booz GW, Duhé RJ. Identification of a redox-sensitive switch within the JAK2 catalytic domain. Free Radic Biol Med. 2012;52:1101–10. doi: 10.1016/j.freeradbiomed.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Shaw PEA. A STAT3 dimer formed by inter-chain disulphide bridging during oxidative stress. Biochem Biophys Res Commun. 2004;322:1005–11. doi: 10.1016/j.bbrc.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Cheung SH, Evans EL, Shaw PE. Modulation of gene expression and tumor cell growth by redox modification of STAT3. Cancer Res. 2010;70:8222–32. doi: 10.1158/0008-5472.CAN-10-0894. [DOI] [PubMed] [Google Scholar]
- 43.Shaw PE. Could STAT3 provide a link between respiration and cell cycle progression? Cell Cycle. 2010;9:4294–6. doi: 10.4161/cc.9.21.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie Y, Kole S, Precht P, Pazin MJ, Bernier M. S-glutathionylation impairs signal transducer and activator of transcription 3 activation and signaling. Endocrinology. 2009;150:1122–31. doi: 10.1210/en.2008-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sebastián C, Herrero C, Serra M, Lloberas J, Blasco MA, Celada A. Telomere shortening and oxidative stress in aged macrophages results in impaired STAT5a phosphorylation. J Immunol. 2009;183:2356–64. doi: 10.4049/jimmunol.0901131. [DOI] [PubMed] [Google Scholar]
- 46.Shi X, Zhang Y, Zheng J, Pan J. Reactive oxygen species in cancer stem cells. Antioxid Redox Signal. 2012;16:1215–28. doi: 10.1089/ars.2012.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sallmyr A, Fan J, Rassool FV. Genomic instability in myeloid malignancies: increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Lett. 2008;270:1–9. doi: 10.1016/j.canlet.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 48.Marty C, Lacout C, Droin N, Le Couédic JP, Ribrag V, Solary E, et al. A role for reactive oxygen species in JAK2(V617F) myeloproliferative neoplasm progression. Leukemia. 2013 doi: 10.1038/leu.2013.102. In press. [DOI] [PubMed] [Google Scholar]
- 49.Thornber K, Colomba A, Ceccato L, Delsol G, Payrastre B, Gaits-Iacovoni F. Reactive oxygen species and lipoxygenases regulate the oncogenicity of NPM-ALK-positive anaplastic large cell lymphomas. Oncogene. 2009;28:2690–6. doi: 10.1038/onc.2009.125. [DOI] [PubMed] [Google Scholar]
- 50.Gianni D, Bohl B, Courtneidge SA, Bokoch GM. The involvement of the tyrosine kinase c-Src in the regulation of reactive oxygen species generation mediated by NADPH oxidase-1. Mol Biol Cell. 2008;19:2984–94. doi: 10.1091/mbc.E08-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slupianek A, Hoser G, Majsterek I, Bronisz A, Malecki M, Blasiak J, et al. Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G(2)/M phase, and protection from apoptosis. Mol Cell Biol. 2002;22:4189–201. doi: 10.1128/MCB.22.12.4189-4201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koptyra M, Cramer K, Slupianek A, Richardson C, Skorski T. BCR/ABL promotes accumulation of chromosomal aberrations induced by oxidative and genotoxic stress. Leukemia. 2008;22:1969–72. doi: 10.1038/leu.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reddy MM, Fernandes MS, Salgia R, Levine RL, Griffin JD, Sattler M. NADPH oxidases regulate cell growth and migration in myeloid cells transformed by oncogenic tyrosine kinases. Leukemia. 2011;25:281–9. doi: 10.1038/leu.2010.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giannoni E, Taddei ML, Chiarugi P. Src redox regulation: again in the front line. Free Radic Biol Med. 2010;49:516–27. doi: 10.1016/j.freeradbiomed.2010.04.025. [DOI] [PubMed] [Google Scholar]
- 55.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–7. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shulga N, Pastorino JG. GRIM-19-mediated translocation of STAT3 to mitochondria is necessary for TNF-induced necroptosis. J Cell Sci. 2012;125:2995–3003. doi: 10.1242/jcs.103093. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Zhang J, Yang J, Roy SK, Tininini S, Hu J, Bromberg JF, et al. The cell death regulator GRIM-19 is an inhibitor of signal transducer and activator of transcription 3. Proc Natl Acad Sci U S A. 2003;100:9342–7. doi: 10.1073/pnas.1633516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mantel C, Messina-Graham S, Moh A, Cooper S, Hangoc G, Fu XY, et al. Mouse hematopoietic cell-targeted STAT3 deletion: stem/progenitor cell defects, mitochondrial dysfunction, ROS overproduction, and a rapid aging-like phenotype. Blood. 2012;120:2589–99. doi: 10.1182/blood-2012-01-404004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–6. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Solaini G, Sgarbi G, Baracca A. Oxidative phosphorylation in cancer cells. Biochim Biophys Acta. 2011;1807:534–42. doi: 10.1016/j.bbabio.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 61.Demaria M, Giorgi C, Lebiedzinska M, Esposito G, D’Angeli L, Bartoli A, et al. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging (Albany NY) 2010;2:823–42. doi: 10.18632/aging.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu G, Briggs J, Deng J, Ma Y, Lee H, Kortylewski M, et al. Signal transducer and activator of transcription 3 is required for hypoxia-inducible factor-1alpha RNA expression in both tumor cells and tumor-associated myeloid cells. Mol Cancer Res. 2008;6:1099–105. doi: 10.1158/1541-7786.MCR-07-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patiar S, Harris AL. Role of hypoxia-inducible factor-1alpha as a cancer therapy target. Endocr Relat Cancer. 2006;13(Suppl 1):S61–75. doi: 10.1677/erc.1.01290. [DOI] [PubMed] [Google Scholar]
- 64.Du W, Hong J, Wang YC, Zhang YJ, Wang P, Su WY, et al. Inhibition of JAK2/STAT3 signalling induces colorectal cancer cell apoptosis via mitochondrial pathway. J Cell Mol Med. 2012;16:1878–88. doi: 10.1111/j.1582-4934.2011.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu X, Guo W, Wu S, Wang L, Wang J, Dai B, et al. Antitumor activity of a novel STAT3 inhibitor and redox modulator in non-small cell lung cancer cells. Biochem Pharmacol. 2012;83:1456–64. doi: 10.1016/j.bcp.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jung JE, Kim GS, Narasimhan P, Song YS, Chan PH. Regulation of Mn-superoxide dismutase activity and neuroprotection by STAT3 in mice after cerebral ischemia. J Neurosci. 2009;29:7003–14. doi: 10.1523/JNEUROSCI.1110-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chueh FY, Leong KF, Yu CL. Mitochondrial translocation of signal transducer and activator of transcription 5 (STAT5) in leukemic T cells and cytokine-stimulated cells. Biochem Biophys Res Commun. 2010;402:778–83. doi: 10.1016/j.bbrc.2010.10.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fatrai S, Wierenga AT, Daenen SM, Vellenga E, Schuringa JJ. Identification of HIF2alpha as an important STAT5 target gene in human hematopoietic stem cells. Blood. 2011;117:3320–30. doi: 10.1182/blood-2010-08-303669. [DOI] [PubMed] [Google Scholar]
- 69.Wofford JA, Wieman HL, Jacobs SR, Zhao Y, Rathmell JC. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood. 2008;111:2101–11. doi: 10.1182/blood-2007-06-096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sallmyr A, Fan J, Datta K, Kim KT, Grosu D, Shapiro P, et al. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111:3173–82. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- 71.Warsch W, Grundschober E, Berger A, Gille L, Cerny-Reiterer S, Tigan AS, et al. STAT5 triggers BCR-ABL1 mutation by mediating ROS production in chronic myeloid leukaemia. Oncotarget. 2012;3:1669–87. doi: 10.18632/oncotarget.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–8. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naughton R, Quiney C, Turner SD, Cotter TG. Bcr-Abl-mediated redox regulation of the PI3K/AKT pathway. Leukemia. 2009;23:1432–40. doi: 10.1038/leu.2009.49. [DOI] [PubMed] [Google Scholar]
- 74.Yu JH, Zhu BM, Riedlinger G, Kang K, Hennighausen L. The liver-specific tumor suppressor STAT5 controls expression of the reactive oxygen species-generating enzyme NOX4 and the proapoptotic proteins PUMA and BIM in mice. Hepatology. 2012;56:2375–86. doi: 10.1002/hep.25900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casetti L, Martin-Lannerée S, Najjar I, Plo I, Augé S, Roy L, et al. Differential contributions of STAT5A and STAT5B to stress protection and tyrosine kinase inhibitor resistance of chronic myeloid leukemia stem/progenitor cells. Cancer Res. 2013;73:2052–8. doi: 10.1158/0008-5472.CAN-12-3955. [DOI] [PubMed] [Google Scholar]
- 76.Cholez E, Debuysscher V, Bourgeais J, Boudot C, Leprince J, Tron F, et al. Evidence for a protective role of the STAT5 transcription factor against oxidative stress in human leukemic pre-B cells. Leukemia. 2012;26:2390–7. doi: 10.1038/leu.2012.112. [DOI] [PubMed] [Google Scholar]
- 77.Bunting KD. STAT5 signaling in normal and pathologic hematopoiesis. Front Biosci. 2007;12:2807–20. doi: 10.2741/2274. [DOI] [PubMed] [Google Scholar]
- 78.Wierenga AT, Vellenga E, Schuringa JJ. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol Cell Biol. 2008;28:6668–80. doi: 10.1128/MCB.01025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Urao N, Ushio-Fukai M. Redox regulation of stem/progenitor cells and bone marrow niche. Free Radic Biol Med. 2013;54:26–39. doi: 10.1016/j.freeradbiomed.2012.10.532. [DOI] [PMC free article] [PubMed] [Google Scholar]