Abstract
STAT2 is an essential transcription factor in type I IFN mediated anti-viral and anti-proliferative signaling. STAT2 function is regulated by tyrosine phosphorylation, which is the trigger for STAT-dimerization, subsequent nuclear translocation, and transcriptional activation of IFN stimulated genes. Evidence of additional STAT2 phosphorylation sites has emerged as well as novel roles for STAT2 separate from the classical ISGF3-signaling. This review aims to summarize knowledge of phosphorylation-mediated STAT2-regulation and future avenues of related STAT2 research.
Keywords: interferon, STAT2, phosphorylation, regulation, receptors
Introduction
The signal transducer and activator of transcription (STAT) protein family was discovered in the early 1990s and quickly regarded as elegant and straightforward cytoplasmic proteins that following activation by a ligand-receptor-kinase complex translocated to the nucleus and induced gene expression.1 STAT2 was defined as a co-factor only involved in type I IFN (IFN-α, -β, -τ, -ω) signaling, as compared with STAT1, which was found to be important in additional cytokine-induced signaling pathways, such as IFN-γ, IL-4, IL-6, and IL-27 to name a few examples.2 In the past decade numerous reports have hinted at STAT2 being involved in contexts other than the classical type I IFN signaling pathway. This, together with the discovery of several novel STAT2 phosphorylation sites, prompts a summary of where we are in our current understanding of STAT2 function and regulation. This review will shed light on the mechanisms, relevance, and recent developments regarding phosphorylation-dependent STAT2 regulation.
Overview of the JAK-STAT Pathway
STAT proteins are critical for transmitting information from many different transmembrane surface receptors, such as cytokine and hormone receptors, to the nucleus.3 The STAT family contains seven members (STAT1–6 including STAT5A and STAT5B) all of which share the same overall domain architecture: an N-terminal domain (ND), a coiled-coil domain (CCD), a DNA-binding domain (DBD), a linker domain (LD), a Src homology 2 (SH2) domain, and a transactivation domain (TAD) (Fig. 1). STATs are activated by phosphorylation of a conserved tyrosine performed by one or more receptor-associated tyrosine kinases from the Janus family of tyrosine kinases named JAK. Tyrosine phosphorylation results in reciprocal binding of one phosphorylated STAT to another, forming either a homodimer or a heterodimer. Dimer formation is the critical step for nuclear translocation since the nuclear localization signal, required for nuclear import, is formed by parts of dimerized STAT DBDs.4 In the nucleus, the STAT-dimer binds the promoter of genes and either induce or repress mRNA expression. In type I IFN signaling, type I IFN binds its cognate IFN receptor (IFNAR) that is pre-associated with receptor-associated kinases TYK2 and JAK1, which become activated through close proximity trans-phosphorylation (Fig. 2). This in turn causes TYK2 and JAK1 to phosphorylate the intracellular chains of the IFNARs that then serve as docking sites for STAT1 and STAT2. JAK1 and TYK2 phosphorylate STAT1 and STAT2, triggering their dimerization and association with the DNA-binding Interferon regulatory factor 9 (IRF9) resulting in the formation of the IFN stimulated gene factor 3 (ISGF3) complex. ISGF3 then binds to the promoters of IFN stimulated genes (ISGs) to activate gene expression.5 ISGF3 recognizes an IFN stimulated response element (ISRE) and its direct binding to DNA is mediated by STAT1 and IRF9, whereas STAT2 is responsible for recruiting transcriptional co-activators through its TAD.6,7 STAT1 is present as two isoforms, STAT1α and STAT1β.8 STAT1α is the full-length form, whereas STAT1β has a truncated and, therefore, non-functional TAD. The important TAD in ISGF3 is provided by STAT2 whereas STAT1α and STAT1β are interchangeable in ISGF3 signaling.6,9 STAT2-TAD has been shown to bind and recruit important transcriptional co-activators, such as p300/CBP, GCN5, DRIP150 and pp32.10-14 The biological significance of STAT2 in type I IFN signaling is apparent in STAT2 deficient mice.15 These mice are vulnerable to viral infection and host immune response is compromised. It is important to stress that mouse and human STAT2 are markedly divergent in the TAD. Mouse STAT2 contains 12 copies of a 24-nucleotide mini-satellite inserted into its TAD, in addition to having flanking non-conserved sequences, and yet it is still capable of activating transcription in human cells at comparable levels to human STAT2.11,16 The role of STAT2 in anti-viral signaling is further supported by a recent study in which a child lacking STAT2 had a history of disseminated vaccine-strain measles.17 The deficiency in STAT2, although not lethal, was caused by a homozygous mutation in an intron donor site that prevented correct STAT2 splicing. Further analysis of common anti-viral pathways pinpointed loss of STAT2 as the most likely cause for viral sensitivity.

Figure 1. Schematic of the domain architecture of STAT2 and location of identified phosphorylation sites. ND, N-terminal domain; CCD, coiled-coil domain; DBD, DNA-binding domain; LD, linker domain; SH2, Src homology 2 domain; TAD, transactivation domain.
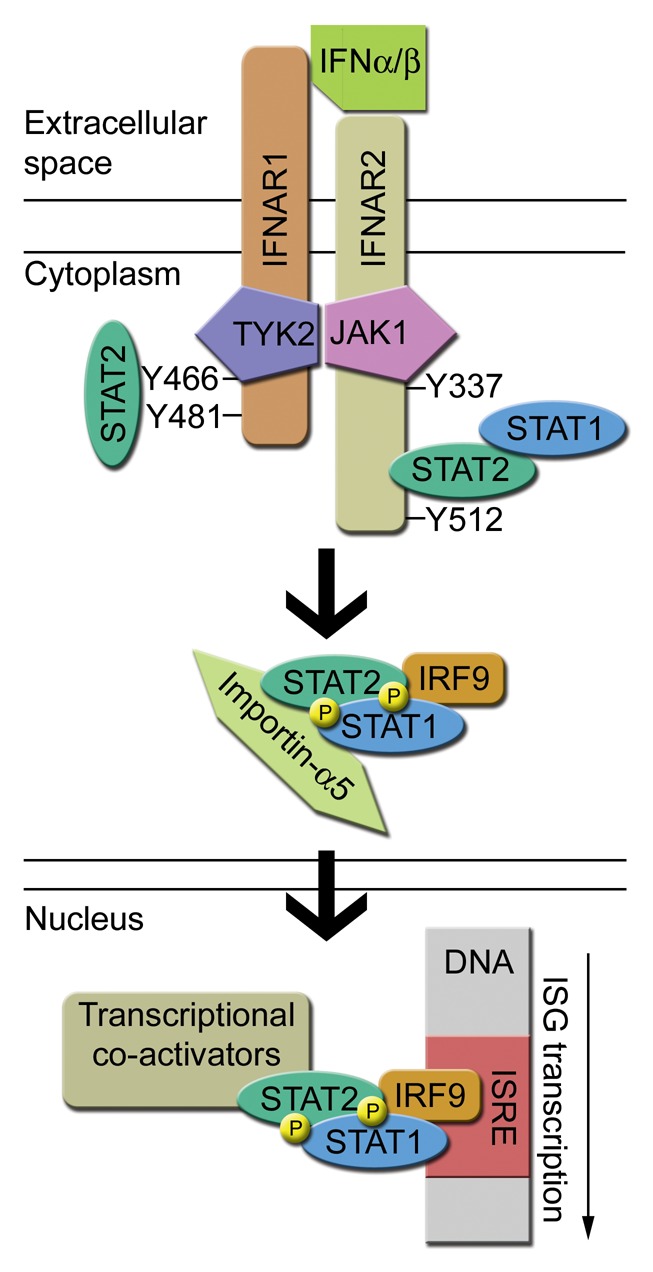
Figure 2. Illustration of type I interferon (IFN) signaling and ISGF3-mediated gene induction. Type I IFN binds IFNAR1 and IFNAR2 leading to transactivation of receptor-associated JAK kinases (JAK1 and TYK2). Several tyrosine residues on the intracellular chains of the receptor subunits are phosphorylated by JAKs, leading to recruitment and phosphorylation of STAT2 and STAT1. STAT2, STAT1, and IRF9 form the ISGF3 complex, which translocates to the nucleus by binding to importin-α5. In the nucleus, ISGF3 binds to the interferon-stimulated response element (ISRE) of interferon-stimulated genes (ISGs) and induce gene transcription with the help of additional co-activators recruited by STAT2.
Activation of STAT2
In 1988, Levy et al. identified the ISRE sequence and observed a cellular factor (named ISGF3) binding to an ISRE DNA-probe in response to IFN-α treatment.18 Subsequent studies uncovered that ISGF3 consisted of four pre-existing polypeptides. Three of these peptides, with the molecular size of 84, 91, and 113 kDa, were suggested to be from the same protein family and were later named STAT1β, STAT1α and STAT2, respectively.8,19 Reich and Pfeffer observed in 1990 that the broad-spectrum kinase inhibitor staurosporine prevented IFN-α induced ISGF3 formation and ISG expression, indicating a requirement of kinase activity for IFN-signaling.20 Phosphopeptide mapping confirmed that STAT2, as well as both forms of STAT1, were phosphorylated at tyrosine (Y)-690 (STAT2) and Y701 (STAT1) in response to IFN-α.21-24 At that same time, a panel of IFN unresponsive mutant cell lines derived from the human fibrosarcoma cell line 2fTGH were generated and became instrumental in the characterization of the type I IFN signaling pathway.25 Analysis of these non-responsive cell lines led to the identification of TYK2 and JAK1 as essential for STAT2 and STAT1 activation.26,27 The 2fTGH mutant cell line lacking STAT2 was named U6A and has been used in numerous studies to characterize STAT2 function. An early finding in these cells demonstrated that STAT1-activation was dependent on STAT2 in type I IFN signaling, but not in response to type II IFN (IFN-γ).28,29 This effect, however, is not common in all cell lines. We and others have observed that in STAT2-deficient cells other than U6A, STAT1 tyrosine phosphorylation is unaffected.30,31 Further studies in U6A cells have shown that the N-terminal part (1–315; includes ND and CCD) of STAT2 was critical for STAT1-activation most likely due to the pre-association of IFNAR2 with STAT2 in untreated cells.32 Together with the finding that STAT2 and STAT1 exist as dimers in resting cells, this suggested that a pre-initiation complex of IFNAR2:STAT2:STAT1 was critical for proper type I IFN signaling.33 Analysis of truncated and mutated versions of STAT2 in U6A cells shed several important regions and residues for proper STAT2-activation and gene induction (Table 1).6 Deletions from the STAT2 N-terminus disrupted tyrosine phosphorylation similarly to what was observed in the study by Li and colleagues.32 Mutation of a conserved arginine (R)-601 to lysine also abrogated STAT2 activation, thus highlighting the relevance of the SH2 domain for binding the phospho-tyrosine (pY) of the interacting partner.43 Our lab has shown that mutating Y631-STAT2 to phenylalanine, which is also close to the SH2-pY interface based on the crystal structure of STAT1, prolonged STAT1-activation and ISGF3-signaling due to a decrease in STAT1 dephosphorylation.35,44 Y631 was not found to be phosphorylated. In contrast, mutating proline 630-STAT2 to leucine produced the opposite phenotype; it reduced ISGF3-signaling due to impaired STAT2 tyrosine phosphorylation.36 The affinity of the pY for a SH2 domain is most often determined by one or more amino acids directly C-terminal of the pY.45 Amino acid residue R694-STAT2 was found to be important since R694 substitution to asparagine caused impaired STAT2 phosphorylation.6 C-terminal truncations of the TAD did not affect STAT2 tyrosine phosphorylation, and yet it dramatically affected induction of ISGs. The region between amino acids 800 and 831 was critical for ISRE-mediated ISG expression, and, interestingly, also for the induction of IRF1, a STAT1-driven ISG.
Table 1. Summary of mutations that alter STAT2 signaling and function.
| Phenotype | STAT2 mutation | Domain | References |
|---|---|---|---|
| Prolonged Y690 phosphorylation |
S287A |
CCD |
34 |
| Y631F |
SH2 |
35 |
|
| Reduced or no Y690 phosphorylation |
S287D |
CCD |
34 |
| R601L |
SH2 |
6 |
|
| P630L |
SH2 |
36 |
|
| Y690F |
C-terminus of SH2 domain |
28 |
|
| R694N |
C-terminus of SH2 domain |
6 |
|
| Reduced or no nuclear translocation |
R374A + K375A |
DBD |
37 |
| R409A + K415A |
DBD |
37 |
|
| Reduced or no nuclear export |
L737A + L741A |
TAD |
38 |
| L740A + L741A |
TAD |
39 |
|
| L745A |
TAD |
39 |
|
| L751A |
TAD |
39 |
|
| Reduced DNA-binding |
V453I + V454I |
DBD |
40 |
| Reduced transcriptional activity |
K390R |
DBD |
41 |
| Abolished STAT4 activation | Y833F | TAD | 42 |
Type I IFN Receptors in STAT2 Activation
The type I IFN receptor is formed by two receptor chains, IFNAR1 and IFNAR2. Both chains interact with IFN to form a ternary complex (IFN:IFNAR1:IFNAR2), with IFNAR2 binding the ligand with higher affinity compared with IFNAR1.46 The close proximity between IFNAR1 and IFNAR2 after ligand binding facilitates reciprocal trans-phosphorylation and activation of the two receptor-associated kinases, TYK2 and JAK1. Activated TYK2 (bound to IFNAR1), and JAK1 (bound to IFNAR2) phosphorylate tyrosine residues on the receptor chains that act as docking sites for the SH2-domain of STAT2.47-49 As STAT2 binds to the receptor chain, it becomes phosphorylated on Y690 just C-terminal of the SH2-domain. The exact sites on the intracellular chains of IFNARs that STAT2 binds to are partially known. Y337 and Y512 of IFNAR2 are seemingly important because mutation of all tyrosines in IFNAR2 to phenylalanine (F) abrogated STAT2-activation.50 Restoring F337 or F512 back to tyrosine rescued the non-functional phenotype. STAT2 was also shown to constitutively bind a stretch of acidic residues located at amino acids 435–438 of IFNAR2.32,51,52 This pre-association is dependent on the ND of STAT2 when examined in 2fTGH cells. The importance of this interaction is unclear because truncation of IFNAR2 or mutating amino acids 435–438, thereby disrupting the STAT2-IFNAR2 interaction, minimally affected STAT2 activation by IFN-α. The intracellular region of IFNAR2 when overexpressed has been shown to be cleaved in a γ-secretase dependent manner in response to type I IFN, phorbol 12-myristate 13-acetate and EGF.53 The proteolytically released intracellular domain (ICD) modulated the transcriptional response in several GAL4 reporter assays.53,54 Binding of IFNAR2-ICD to STAT2 was necessary for the transcriptional effect of the ICD, as well as the presence of an intact STAT2 TAD. Because these observations were made using exogenously expressed IFNAR2, it is unknown if endogenous IFNAR2 is cleaved during IFN signaling or to what extent IFNAR2-ICD complements type I IFN signaling.
In vitro studies have pinpointed IFNAR1 Y466 and Y481 to be phosphorylated by TYK2, and that STAT2 binds to a phosphopeptide consisting of Y466 and surrounding residues in a SH2-dependent manner.32,47,55 STAT2 also bound pY481, but this interaction was weaker when compared with pY466. Treatment of permeabilized HeLa cells with IFNAR1 Y466 phosphopeptide prevented IFN-α induced STAT2 and STAT1 activation. In a separate study, however, Y466F or Y481F mutations did not abolish STAT2 activation, and neither did deleting IFNAR2-404-462, but co-expressing the two altered IFNARs led to loss of STAT2 phosphorylation.51 Some studies have questioned the in vivo significance of pY466, and to a lesser extent pY481, for STAT2 activation. These two tyrosines are found in human, but not in mouse IFNAR1, and the only two conserved tyrosines, Y518 and Y529 in mouse IFNAR1 (Y527 and Y538 in human IFNAR1), were dispensable since mouse IFNAR1 truncated at amino acid 511 remained fully capable of activating STAT2.56 The critical region in mouse IFNAR1 was confined to a section between residues 471 and 511, which is devoid of tyrosines indicating a different mechanism for STAT2-activation in mouse. This is likely given that the TAD domains of mouse and human STAT2 are 39% identical and were found to bind overlapping but also distinct proteins.16 The study of JAK1 and TYK2, and the role of each individual kinase in STAT2 tyrosine phosphorylation, has been difficult to discern due to the interdependence one kinase has on the other for activation.57 Nonetheless, knockdown or inhibition of either JAK1 or TYK2 severely decreases tyrosine phosphorylation of STAT2.9,26 The final step of STAT-dimer formation is the release from the receptor-chain and binding to another STAT through SH2-pY interaction. The exact mechanism that triggers this release from the receptor chain is not known, but pY-STAT2 was reported to not associate with phosphorylated Y466 of IFNAR1, whereas unphosphorylated STAT2 did.47 Presently, it is unknown if this is due to pY-STAT2 already binding to another STAT with higher affinity or an intrinsic change of the STAT2 SH2-domain in response to the intramolecular pY.
Deacetylation and acetylation events are important in type I IFN signaling. Early studies using the histone deacetylase inhibitor Trichostatin A demonstrated that increased global acetylation prevented proper ISGF3 function and reduced nuclear translocation of STAT2 in response to Newcastle disease virus infection.58 In contrast, a study by Tang et al. concluded that acetylation of ISGF3 was required for proper ISGF3-dependent signaling.41 The histone acetylase CREB-binding protein was recruited to IFNAR2 in response to IFN-α resulting in the acetylation of IFNAR2, IRF9, and STAT2 on several critical lysines (K) including K399-IFNAR2, K81-IRF9, and K390-STAT2. Mutation of any of these lysines reduced ISRE reporter gene response. Acetylation of K390-STAT2 was suggested to be important for proper formation of the active STAT2:STAT1 dimer.
STAT2 Serine/Threonine Phosphorylation
Serine and threonine phosphorylations are responsible for 90% and 10% of all eukaryotic phosphorylation events; respectively, with tyrosine phosphorylation being much rarer at around 0.05%.59 Serine phosphorylation is also involved in modulating the function of STATs. The main site for STAT serine phosphorylation, except for STAT2 and STAT6, is a conserved serine flanked by two prolines in the TAD. This site is usually situated 20–30 residues C-terminal of the pY. For STAT1, STAT3, STAT4, and STAT5B this post-translational modification (PTM) can increase the transcriptional activity of these STATs.60-63 In the case of STAT3, phosphorylation of Serine (S)727 can also decrease tyrosine phosphorylation.60,64,65 S727-STAT1 phosphorylation is necessary for STAT1 homodimers to induce a strong transcriptional response during IFN signaling. However, pS727-STAT1 is seemingly not important for ISGF3-mediated ISG induction since STAT1β, which has a truncated TAD with a C-terminus lacking S727, is interchangeable with STAT1α for gene expression.9 This raises the question of whether STAT2, as the TAD provider, is modified by additional phosphorylation events to potentiate or suppress gene expression. Several studies have hinted at or shown that STAT2 is serine or threonine phosphorylated. While mapping IFN-induced STAT2 phosphorylation, serine-phosphorylated STAT2 was detected in IFN-α stimulated cells.22 In a recent study by Ng et al. STAT2 was “phospho-shifted” as a consequence of exogenous expression of Inhibitor of nuclear factor κ-B Kinase subunit epsilon (IKKε, a serine/threonine kinase), but not by catalytically inactive IKKε.66 In most recent reports, STAT2 was found phosphorylated at S595 in a phosphoproteomic screen designed to identify phosphorylation events in Jurkat cells responding to T-cell receptor activation.67 S595, however, was not validated to be phosphorylated uniquely by T-cell receptor activation indicating the possibility that this site could be constitutively modified or phosphorylated in response to another signaling pathway. Furthermore, a phosphoproteomic analysis, as a part of the Chromosome-Centric Human Proteome Project, found threonine (T)-800 of STAT2 to be phosphorylated in human colorectal cancer tissue.68 T597 and Y833 have also been identified as phosphorylation sites from curated information (www.phosphosite.org).69 In a study investigating STAT4-activation in response to type I IFN in human T-cells, the recruitment of STAT4 to the IFNAR complex was found to be STAT2-dependent.42 The STAT4-interacting region of STAT2 was mapped to amino acids 811–851, with Y833 being a critical residue.29 The phosphorylation-status of Y833 was not assessed, but modification of Y833 may be important in STAT2-STAT4 interaction. No biological data on the importance or function of phosphorylated T597 or T800 are available at the moment.
We recently analyzed STAT2 by mass spectrometry in an attempt to find additional regulatory PTMs. Serine 287 was identified as phosphorylated in response to IFN-α treatment.34 Sequence alignment revealed no conservation of S287 between human and mouse STAT2. U6A-cells reconstituted with S287A-STAT2 were found more responsive to the antiproliferative and antiviral effects of IFN-α. Biochemical analysis further revealed that Y690-phosphorylation of S287A-STAT2 was prolonged and ISG-induction was enhanced after an overnight treatment with IFN-α compared with WT-STAT2. The phosphomimetic mutant, S287D-STAT2, had the opposite effect of S287A. Expression of S287D-STAT2 reduced IFN-α-induced pY690-STAT2 and drastically decreased nuclear translocated STAT2 and consequently ISG induction. S287D-STAT2 also poorly protected cells against vesicular stomatitis virus infection. Because an antibody recognizing pS287-STAT2 is not available, the dynamics of S287 phosphorylation remain unknown at the moment. Comparison of WT-STAT2 to S287A-STAT2 and S287D-STAT2 indicates that the majority of WT-STAT2 molecules are most likely not phosphorylated on S287 since the biological and biochemical difference between WT and S287A were less than between WT and S287D. Inhibitory phosphorylation events are not unprecedented in the STAT family with the existence of such events in STAT5A, STAT5B, and STAT6.63,70,71 The analogous site to S287-STAT2 in STAT1 is T288. This STAT1 site is found mutated to alanine in some patients with chronic mucocutaneous candidiasis.72 Cells expressing this mutation had protracted responses to cytokines that depend on STAT1 for signaling. T288A-STAT1 activation was prolonged due to defects in STAT1 tyrosine dephosphorylation, and believed to be the result of the mutation restricting the STAT1-dimer to adopt a favorable conformation for dephosphorylation. The similarities between S287A-STAT2 and T288A-STAT1 suggest that phosphorylation of S287 accelerates dephosphorylation of STAT2 thus contributing to termination of JAK-STAT signaling.
Phosphorylation that negatively affects intermolecular binding between STATs is not rare. Two interesting studies from the Maniatis lab have shown that STAT1 can be phosphorylated at S708 by IKKε.66,73 Loss of IKKε reduced the expression of a subset of ISGs. The affected genes lacked a suggested STAT2-binding motif upstream of the consensus ISRE, thereby making the ISRE a lower-affinity binding site for ISGF3. Through several in vitro biochemical experiments, phosphorylation of S708-STAT1 was shown to decrease STAT1 homodimer formation due to interference of the pY701-SH2 interaction. However, formation of the STAT2:STAT1 heterodimer was not affected leading to increased ISGF3 assembly in the presence of active IKKε and higher expression of ISGs with low-affinity ISREs. There is no analogous site to S708-STAT1 in STAT2.
Unresolved Areas of STAT2 Function
The identification of several phosphorylated residues on STAT2 raises the question if the gaps in our knowledge of STAT2 could be explained by additional post-translational events. The role of STAT2 as a critical unit in ISGF3 is well established. However, multiple studies indicate a larger role for STAT2 in gene regulation beyond ISFG3. Several STAT2 complexes have been identified in vitro, including STAT2:STAT1 heterodimers without IRF9, STAT2 homodimers and STAT2:STAT3 heterodimers.7,74-76 STAT2:STAT1 and STAT2 homodimers could bind a version of gamma-activated sequence, the consensus sequence that several STAT-dimers bind to including STAT1, in gel-shift assays, but with much lower affinity than STAT1 homodimers. Careful examination of the DBD of STAT2 revealed two conserved valines (V453 and V454), previously shown to be essential in STAT5 for DNA-binding, and also found to be important in STAT2-dependent IFN-α induced growth inhibition and viral protection.40 A STAT2:STAT1 complex, not containing IRF9 and relying on both STAT2 and STAT1 for DNA-binding, was suggested to be the complex affected by the DNA-binding deficient STAT2 mutant, but no direct evidence for this was found. The STAT2 DNA-binding deficient mutant was subsequently used for DNA microarray analysis. A small subset of ISGs (19 genes) were identified as dependent on a functional STAT2-DBD for induction in response to IFN-α.77
STAT1-Independent STAT2 Function
The in vivo importance of alternative STAT2-containing dimers is not very clear, but several studies indicate that STAT2-dependent, ISGF3-independent factors are involved in type I IFN signaling. A study on STAT1-deficient mice showed that STAT2 supported the expression of ISGs.78 In studies evaluating DNA-binding and gene activation in spleen and bone marrow-derived macrophages, STAT2 was found associated with ISG promoters (Oas1a, Oas1b, and Irf7) without STAT1; albeit weakly, and mRNA expression of several ISGs remained intact with no STAT1 present.78 STAT1-independent induction of ISGs in mouse splenocytes has also been observed by Zimmerer and colleagues.79 APOBEC3G (A3G), a cytidine deaminase important in restricting retroviral replication, was induced in response to IFN-α by STAT2 and IRF9, in the absence of STAT1.80 Similarly to what was found with pS708-STAT1, phosphorylation (or any other PTM) could potentially direct the formation or stability of a specific STAT2-complex. STAT2 has not only been associated with the activation of IFN-target genes. Mutated p53 (p53-mut) can provide a gain-of-function phenotype in cancer cells, such as enhanced drug resistance, proliferation and cell motility. Some p53-mut phenotypes have been connected to an increase in NF-κB2 expression.81 Although p53-mut did not bind the NFκB2 promoter, STAT2 and CBP were found to be enriched at the promoter in p53-mut H1299 cells compared with cells expressing WT p53. STAT2 has also been implicated in myogenic differentiation. JAK2, STAT2, and STAT3 were all important for differentiation of primary myocytes and C12C2 cells, a skeletal muscle satellite-cell-derived line, into multinucleated myotubes.82 The exact mechanism of action was not described, but STAT2 expression was important for reducing HGF expression and increasing IGF2 expression, two events required for myocyte differentiation.
A Role for Non-Tyrosine Phosphorylated STAT2
Several papers have uncovered novel roles for non-tyrosine-phosphorylated STATs. One of the first studies demonstrated a role for S727-STAT1, but not Y701-STAT1, in supporting the expression of caspase-1, -2, and -3.83 In a more recent study, STAT3 was detected in mitochondria with a role in oxidative phosphorylation.84 The function of STAT3 in this context was dependent on S727 instead of Y705 phosphorylation. The effect of increased levels of STAT1 protein as a result of extended IFN-treatment was studied by Cheon and Stark.85 They concluded that non-tyrosine-phosphorylated STAT1 prolonged ISG expression of a subset of genes, primarily immune regulatory ISGs. The exact mechanism, however, remains unknown. A role for upregulated STAT2 has also been suggested in IFN-γ signaling. An ISGF3-like complex formed by non-tyrosine phosphorylated STAT2, pY701-STAT1, and IRF9 assembled after 24 h of IFN-γ treatment, and induced a subset of ISGs.86 Furthermore, non-tyrosine-phosphorylated STAT2 was detected to be associated with a number of ISG promoters, prior to and/or after IFN-α treatment in a chromatin immunoprecipitation sequencing assay.87 Most recently, our lab, in collaboration with the lab of Graham Foster, made the observation that NFκB-mediated expression of pro-inflammatory cytokines required STAT2. Phosphorylation of Y690-STAT2 was dispensable because reconstitution of Y690F-STAT2 in STAT2 deficient immortalized mouse macrophages restored NFκB transcriptional activity.88
In conclusion, all of these observations depict a more complex picture of STAT2 function and ISG-regulation than was initially thought when STAT2 was first studied. The future of STAT2 signaling research will most likely develop alongside general transcriptional control and epigenetics research. The core of STAT2 signaling is the transcriptional regulation of genes, of which we have much more left to discover. Multimeric transcriptional complexes with interchangeable parts, repressor complexes, and cross-talk between several different promoter elements all influence and dynamically regulate gene transcription. Add to that the complex language of chromatin remodeling in the form of histone acetylation, methylation and phosphorylation, and you have the assembly of one of the most complicated and dynamic processes in nature. Therefore, identifying additional PTMs might, in the end, give us new insights into the different aspects of STAT2 function; and when deregulated, whether they are implicated in human diseases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by a grant from the National Cancer Institute (RO1CA140499 to AM Gamero).
Glossary
Abbreviations:
- STAT
signal transducer and activator of transcription
- IFN
interferon
- ISGF3
interferon-stimulated gene factor 3
- CCD
coiled-coil domain
- DBD
DNA-binding domain
- LD
linker domain
- SH2
Src homology 2
- TAD
transactivation domain
- IRF9
interferon regulatory factor 9
- ISG
interferon stimulated gene
- IFNAR
interferon alpha/beta receptor
- ISRE
interferon stimulated response element
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/25790
References
- 1.Darnell JE., Jr. STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Ramana CV, Chatterjee-Kishore M, Nguyen H, Stark GR. Complex roles of Stat1 in regulating gene expression. Oncogene. 2000;19:2619–27. doi: 10.1038/sj.onc.1203525. [DOI] [PubMed] [Google Scholar]
- 3.Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 4.Meyer T, Vinkemeier U. Nucleocytoplasmic shuttling of STAT transcription factors. Eur J Biochem. 2004;271:4606–12. doi: 10.1111/j.1432-1033.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Moczygemba M, Gutch MJ, French DL, Reich NC. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J Biol Chem. 1997;272:20070–6. doi: 10.1074/jbc.272.32.20070. [DOI] [PubMed] [Google Scholar]
- 6.Qureshi SA, Leung S, Kerr IM, Stark GR, Darnell JE., Jr. Function of Stat2 protein in transcriptional activation by alpha interferon. Mol Cell Biol. 1996;16:288–93. doi: 10.1128/mcb.16.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bluyssen HA, Levy DE. Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J Biol Chem. 1997;272:4600–5. doi: 10.1074/jbc.272.7.4600. [DOI] [PubMed] [Google Scholar]
- 8.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci U S A. 1992;89:7836–9. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell JE, Jr., et al. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 1993;12:4221–8. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, et al. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–7. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 11.Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J Biol Chem. 1999;274:25343–9. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 12.Paulson M, Press C, Smith E, Tanese N, Levy DE. IFN-Stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat Cell Biol. 2002;4:140–7. doi: 10.1038/ncb747. [DOI] [PubMed] [Google Scholar]
- 13.Lau JF, Nusinzon I, Burakov D, Freedman LP, Horvath CM. Role of metazoan mediator proteins in interferon-responsive transcription. Mol Cell Biol. 2003;23:620–8. doi: 10.1128/MCB.23.2.620-628.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadota S, Nagata K. pp32, an INHAT component, is a transcription machinery recruiter for maximal induction of IFN-stimulated genes. J Cell Sci. 2011;124:892–9. doi: 10.1242/jcs.078253. [DOI] [PubMed] [Google Scholar]
- 15.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/S1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 16.Park C, Lecomte MJ, Schindler C. Murine Stat2 is uncharacteristically divergent. Nucleic Acids Res. 1999;27:4191–9. doi: 10.1093/nar/27.21.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hambleton S, Goodbourn S, Young DF, Dickinson P, Mohamad SM, Valappil M, et al. STAT2 deficiency and susceptibility to viral illness in humans. Proc Natl Acad Sci U S A. 2013;110:3053–8. doi: 10.1073/pnas.1220098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr. Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–93. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 19.Fu XY, Schindler C, Improta T, Aebersold R, Darnell JE., Jr. The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci U S A. 1992;89:7840–3. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reich NC, Pfeffer LM. Evidence for involvement of protein kinase C in the cellular response to interferon alpha. Proc Natl Acad Sci U S A. 1990;87:8761–5. doi: 10.1073/pnas.87.22.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu XY. A transcription factor with SH2 and SH3 domains is directly activated by an interferon alpha-induced cytoplasmic protein tyrosine kinase(s) Cell. 1992;70:323–35. doi: 10.1016/0092-8674(92)90106-M. [DOI] [PubMed] [Google Scholar]
- 22.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–13. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 23.Improta T, Schindler C, Horvath CM, Kerr IM, Stark GR, Darnell JE., Jr. Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc Natl Acad Sci U S A. 1994;91:4776–80. doi: 10.1073/pnas.91.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–6. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 25.Pellegrini S, John J, Shearer M, Kerr IM, Stark GR. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol Cell Biol. 1989;9:4605–12. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell. 1992;70:313–22. doi: 10.1016/0092-8674(92)90105-L. [DOI] [PubMed] [Google Scholar]
- 27.Müller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, et al. The protein tyrosine kinase JAK1 complements defects in interferon-alpha/beta and -gamma signal transduction. Nature. 1993;366:129–35. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 28.Leung S, Qureshi SA, Kerr IM, Darnell JE, Jr., Stark GR. Role of STAT2 in the alpha interferon signaling pathway. Mol Cell Biol. 1995;15:1312–7. doi: 10.1128/mcb.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farrar JD, Smith JD, Murphy TL, Leung S, Stark GR, Murphy KM. Selective loss of type I interferon-induced STAT4 activation caused by a minisatellite insertion in mouse Stat2. Nat Immunol. 2000;1:65–9. doi: 10.1038/76932. [DOI] [PubMed] [Google Scholar]
- 30.Romero-Weaver AL, Wang HW, Steen HC, Scarzello AJ, Hall VL, Sheikh F, et al. Resistance to IFN-alpha-induced apoptosis is linked to a loss of STAT2. Mol Cancer Res. 2010;8:80–92. doi: 10.1158/1541-7786.MCR-08-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Z, Fan M, Kim JG, Eckerle D, Lothstein L, Wei L, et al. Interferon-resistant Daudi cell line with a Stat2 defect is resistant to apoptosis induced by chemotherapeutic agents. J Biol Chem. 2009;284:27808–15. doi: 10.1074/jbc.M109.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Leung S, Kerr IM, Stark GR. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol Cell Biol. 1997;17:2048–56. doi: 10.1128/mcb.17.4.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stancato LF, David M, Carter-Su C, Larner AC, Pratt WB. Preassociation of STAT1 with STAT2 and STAT3 in separate signalling complexes prior to cytokine stimulation. J Biol Chem. 1996;271:4134–7. doi: 10.1074/jbc.271.8.4134. [DOI] [PubMed] [Google Scholar]
- 34.Steen HC, Nogusa S, Thapa RJ, Basagoudanavar SH, Gill AL, Merali S, et al. Identification of STAT2 serine 287 as a novel regulatory phosphorylation site in type I interferon-induced cellular responses. J Biol Chem. 2013;288:747–58. doi: 10.1074/jbc.M112.402529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scarzello AJ, Romero-Weaver AL, Maher SG, Veenstra TD, Zhou M, Qin A, et al. A Mutation in the SH2 domain of STAT2 prolongs tyrosine phosphorylation of STAT1 and promotes type I IFN-induced apoptosis. Mol Biol Cell. 2007;18:2455–62. doi: 10.1091/mbc.E06-09-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamero AM, Sakamoto S, Montenegro J, Larner AC. Identification of a novel conserved motif in the STAT family that is required for tyrosine phosphorylation. J Biol Chem. 2004;279:12379–85. doi: 10.1074/jbc.M310787200. [DOI] [PubMed] [Google Scholar]
- 37.Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J Biol Chem. 2001;276:16447–55. doi: 10.1074/jbc.M008821200. [DOI] [PubMed] [Google Scholar]
- 38.Frahm T, Hauser H, Köster M. IFN-type-I-mediated signaling is regulated by modulation of STAT2 nuclear export. J Cell Sci. 2006;119:1092–104. doi: 10.1242/jcs.02822. [DOI] [PubMed] [Google Scholar]
- 39.Banninger G, Reich NC. STAT2 nuclear trafficking. J Biol Chem. 2004;279:39199–206. doi: 10.1074/jbc.M400815200. [DOI] [PubMed] [Google Scholar]
- 40.Brierley MM, Fish EN. Functional relevance of the conserved DNA-binding domain of STAT2. J Biol Chem. 2005;280:13029–36. doi: 10.1074/jbc.M500426200. [DOI] [PubMed] [Google Scholar]
- 41.Tang X, Gao J-S, Guan YJ, McLane KE, Yuan Z-L, Ramratnam B, et al. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 42.Farrar JD, Smith JD, Murphy TL, Murphy KM. Recruitment of Stat4 to the human interferon-alpha/beta receptor requires activated Stat2. J Biol Chem. 2000;275:2693–7. doi: 10.1074/jbc.275.4.2693. [DOI] [PubMed] [Google Scholar]
- 43.Waksman G, Kominos D, Robertson SC, Pant N, Baltimore D, Birge RB, et al. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature. 1992;358:646–53. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr., Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–39. doi: 10.1016/S0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 45.Liu BA, Engelmann BW, Nash PD. The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction. FEBS Lett. 2012;586:2597–605. doi: 10.1016/j.febslet.2012.04.054. [DOI] [PubMed] [Google Scholar]
- 46.Piehler J, Thomas C, Garcia KC, Schreiber G. Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol Rev. 2012;250:317–34. doi: 10.1111/imr.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan H, Krishnan K, Greenlund AC, Gupta S, Lim JT, Schreiber RD, et al. Phosphorylated interferon-alpha receptor 1 subunit (IFNaR1) acts as a docking site for the latent form of the 113 kDa STAT2 protein. EMBO J. 1996;15:1064–74. [PMC free article] [PubMed] [Google Scholar]
- 48.Colamonici OR, Domanski P, Krolewski JJ, Fu XY, Reich NC, Pfeffer LM, et al. Interferon alpha (IFN alpha) signaling in cells expressing the variant form of the type I IFN receptor. J Biol Chem. 1994;269:5660–5. [PubMed] [Google Scholar]
- 49.Domanski P, Fish E, Nadeau OW, Witte M, Platanias LC, Yan H, et al. A region of the beta subunit of the interferon alpha receptor different from box 1 interacts with Jak1 and is sufficient to activate the Jak-Stat pathway and induce an antiviral state. J Biol Chem. 1997;272:26388–93. doi: 10.1074/jbc.272.42.26388. [DOI] [PubMed] [Google Scholar]
- 50.Wagner TC, Velichko S, Vogel D, Rani MR, Leung S, Ransohoff RM, et al. Interferon signaling is dependent on specific tyrosines located within the intracellular domain of IFNAR2c. Expression of IFNAR2c tyrosine mutants in U5A cells. J Biol Chem. 2002;277:1493–9. doi: 10.1074/jbc.M108928200. [DOI] [PubMed] [Google Scholar]
- 51.Nadeau OW, Domanski P, Usacheva A, Uddin S, Platanias LC, Pitha P, et al. The proximal tyrosines of the cytoplasmic domain of the beta chain of the type I interferon receptor are essential for signal transducer and activator of transcription (Stat) 2 activation. Evidence that two Stat2 sites are required to reach a threshold of interferon alpha-induced Stat2 tyrosine phosphorylation that allows normal formation of interferon-stimulated gene factor 3. J Biol Chem. 1999;274:4045–52. doi: 10.1074/jbc.274.7.4045. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen VP, Saleh AZ, Arch AE, Yan H, Piazza F, Kim J, et al. Stat2 binding to the interferon-alpha receptor 2 subunit is not required for interferon-alpha signaling. J Biol Chem. 2002;277:9713–21. doi: 10.1074/jbc.M111161200. [DOI] [PubMed] [Google Scholar]
- 53.Saleh AZ, Fang AT, Arch AE, Neupane D, El Fiky A, Krolewski JJ. Regulated proteolysis of the IFNaR2 subunit of the interferon-alpha receptor. Oncogene. 2004;23:7076–86. doi: 10.1038/sj.onc.1207955. [DOI] [PubMed] [Google Scholar]
- 54.El Fiky A, Arch AE, Krolewski JJ. Intracellular domain of the IFNaR2 interferon receptor subunit mediates transcription via Stat2. J Cell Physiol. 2005;204:567–73. doi: 10.1002/jcp.20305. [DOI] [PubMed] [Google Scholar]
- 55.Krishnan K, Yan H, Lim JT, Krolewski JJ. Dimerization of a chimeric CD4-interferon-alpha receptor reconstitutes the signaling events preceding STAT phosphorylation. Oncogene. 1996;13:125–33. [PubMed] [Google Scholar]
- 56.Zhao W, Lee C, Piganis R, Plumlee C, de Weerd N, Hertzog PJ, et al. A conserved IFN-alpha receptor tyrosine motif directs the biological response to type I IFNs. J Immunol. 2008;180:5483–9. doi: 10.4049/jimmunol.180.8.5483. [DOI] [PubMed] [Google Scholar]
- 57.Krishnan K, Pine R, Krolewski JJ. Kinase-deficient forms of Jak1 and Tyk2 inhibit interferon alpha signaling in a dominant manner. Eur J Biochem. 1997;247:298–305. doi: 10.1111/j.1432-1033.1997.00298.x. [DOI] [PubMed] [Google Scholar]
- 58.Génin P, Morin P, Civas A. Impairment of interferon-induced IRF-7 gene expression due to inhibition of ISGF3 formation by trichostatin A. J Virol. 2003;77:7113–9. doi: 10.1128/JVI.77.12.7113-7119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tichy A, Salovska B, Rehulka P, Klimentova J, Vavrova J, Stulik J, et al. Phosphoproteomics: searching for a needle in a haystack. J Proteomics. 2011;74:2786–97. doi: 10.1016/j.jprot.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 60.Wen Z, Zhong Z, Darnell JE., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–50. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 61.Zhu X, Wen Z, Xu LZ, Darnell JE., Jr. Stat1 serine phosphorylation occurs independently of tyrosine phosphorylation and requires an activated Jak2 kinase. Mol Cell Biol. 1997;17:6618–23. doi: 10.1128/mcb.17.11.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, et al. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc Natl Acad Sci U S A. 2002;99:12281–6. doi: 10.1073/pnas.182618999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park SH, Yamashita H, Rui H, Waxman DJ. Serine phosphorylation of GH-activated signal transducer and activator of transcription 5a (STAT5a) and STAT5b: impact on STAT5 transcriptional activity. Mol Endocrinol. 2001;15:2157–71. doi: 10.1210/me.15.12.2157. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, Blenis J, Li HC, Schindler C, Chen-Kiang S. Requirement of serine phosphorylation for formation of STAT-promoter complexes. Science. 1995;267:1990–4. doi: 10.1126/science.7701321. [DOI] [PubMed] [Google Scholar]
- 65.Chung J, Uchida E, Grammer TC, Blenis J. STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol Cell Biol. 1997;17:6508–16. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng SL, Friedman BA, Schmid S, Gertz J, Myers RM, Tenoever BR, et al. IκB kinase epsilon (IKK(epsilon)) regulates the balance between type I and type II interferon responses. Proc Natl Acad Sci U S A. 2011;108:21170–5. doi: 10.1073/pnas.1119137109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 68.Shiromizu T, Adachi J, Watanabe S, Murakami T, Kuga T, Muraoka S, et al. Identification of Missing Proteins in the neXtProt Database and Unregistered Phosphopeptides in the PhosphoSitePlus Database As Part of the Chromosome-Centric Human Proteome Project. J Proteome Res. 2013;12:2414–21. doi: 10.1021/pr300825v. [DOI] [PubMed] [Google Scholar]
- 69.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40(Database issue):D261–70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beuvink I, Hess D, Flotow H, Hofsteenge J, Groner B, Hynes NE. Stat5a serine phosphorylation. Serine 779 is constitutively phosphorylated in the mammary gland, and serine 725 phosphorylation influences prolactin-stimulated in vitro DNA binding activity. J Biol Chem. 2000;275:10247–55. doi: 10.1074/jbc.275.14.10247. [DOI] [PubMed] [Google Scholar]
- 71.Maiti NR, Sharma P, Harbor PC, Haque SJ. Serine phosphorylation of Stat6 negatively controls its DNA-binding function. J Interferon Cytokine Res. 2005;25:553–63. doi: 10.1089/jir.2005.25.553. [DOI] [PubMed] [Google Scholar]
- 72.Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208:1635–48. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tenoever BR, Ng SL, Chua MA, McWhirter SM, García-Sastre A, Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–8. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 74.Ghislain JJ, Fish EN. Application of genomic DNA affinity chromatography identifies multiple interferon-α-regulated Stat2 complexes. J Biol Chem. 1996;271:12408–13. doi: 10.1074/jbc.271.21.12408. [DOI] [PubMed] [Google Scholar]
- 75.Li X, Leung S, Qureshi S, Darnell JE, Jr., Stark GR. Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-alpha. J Biol Chem. 1996;271:5790–4. doi: 10.1074/jbc.271.10.5790. [DOI] [PubMed] [Google Scholar]
- 76.Ghislain JJ, Wong T, Nguyen M, Fish EN. The interferon-inducible Stat2:Stat1 heterodimer preferentially binds in vitro to a consensus element found in the promoters of a subset of interferon-stimulated genes. J Interferon Cytokine Res. 2001;21:379–88. doi: 10.1089/107999001750277853. [DOI] [PubMed] [Google Scholar]
- 77.Brierley MM, Marchington KL, Jurisica I, Fish EN. Identification of GAS-dependent interferon-sensitive target genes whose transcription is STAT2-dependent but ISGF3-independent. FEBS J. 2006;273:1569–81. doi: 10.1111/j.1742-4658.2006.05176.x. [DOI] [PubMed] [Google Scholar]
- 78.Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zimmerer JM, Lesinski GB, Radmacher MD, Ruppert A, Carson WE., 3rd STAT1-dependent and STAT1-independent gene expression in murine immune cells following stimulation with interferon-alpha. Cancer Immunol Immunother. 2007;56:1845–52. doi: 10.1007/s00262-007-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sarkis PT, Ying S, Xu R, Yu XF. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J Immunol. 2006;177:4530–40. doi: 10.4049/jimmunol.177.7.4530. [DOI] [PubMed] [Google Scholar]
- 81.Vaughan CA, Singh S, Windle B, Sankala HM, Graves PR, Andrew Yeudall W, et al. p53 mutants induce transcription of NF-κB2 in H1299 cells through CBP and STAT binding on the NF-κB2 promoter and gain of function activity. Arch Biochem Biophys. 2012;518:79–88. doi: 10.1016/j.abb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang K, Wang C, Xiao F, Wang H, Wu Z. JAK2/STAT2/STAT3 are required for myogenic differentiation. J Biol Chem. 2008;283:34029–36. doi: 10.1074/jbc.M803012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar A, Commane M, Flickinger TW, Horvath CM, Stark GR. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–2. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 84.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–7. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A. 2009;106:9373–8. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morrow AN, Schmeisser H, Tsuno T, Zoon KC. A novel role for IFN-stimulated gene factor 3II in IFN-γ signaling and induction of antiviral activity in human cells. J Immunol. 2011;186:1685–93. doi: 10.4049/jimmunol.1001359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Testoni B, Völlenkle C, Guerrieri F, Gerbal-Chaloin S, Blandino G, Levrero M. Chromatin dynamics of gene activation and repression in response to interferon alpha (IFN(alpha)) reveal new roles for phosphorylated and unphosphorylated forms of the transcription factor STAT2. J Biol Chem. 2011;286:20217–27. doi: 10.1074/jbc.M111.231068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alazawi W, Heath H, Waters JA, Woodfin A, O’Brien AJ, Scarzello AJ, et al. Stat2 loss leads to cytokine-independent, cell-mediated lethality in LPS-induced sepsis. Proc Natl Acad Sci U S A. 2013;110:8656–61. doi: 10.1073/pnas.1221652110. [DOI] [PMC free article] [PubMed] [Google Scholar]


