Abstract
The Crithidia fasciculata KAP2 and KAP3 proteins are closely related kinetoplast-specific histone-like DNA-binding proteins. The KAP2 and KAP3 genes are 46% identical and are arranged in tandem on the chromosomal DNA. Disruption of both alleles of either gene alone shows no detectable phenotype. However, replacement of both copies of the sequence encoding the entire KAP2 and KAP3 locus increases maxicircle mRNA levels two- to fourfold. These double-knockout cells are viable but grow extremely slowly, have reduced respiration and very abnormal cell morphologies, and accumulate numerous large vacuoles. The extreme phenotype of these mutant cells suggests an important role for the KAP2 and KAP3 proteins in mitochondrial metabolism and cell growth.
The kinetoplastid protozoa are among the earliest diverging eukaryotic organisms containing a mitochondrion. Their mitochondrial DNA, termed kinetoplast DNA (kDNA), is one of the most complex and unusual DNAs in nature (10, 16, 21). Most studies of kDNA have focused on protozoan parasites of the family Trypanosomatidae. The kDNA of the trypanosomatid Crithidia fasciculata is contained within a highly organized and condensed disk-shaped structure (∼1 μm in diameter and 0.4 μm thick) called the kinetoplast and consists of two types of circular DNA molecules, minicircles and maxicircles. These circular DNAs are topologically interlocked to form a single DNA network. Each minicircle in the network is interlocked with two to three other minicircles on average (8). The maxicircles in kinetoplastids usually range in size from 20 to 40 kb and encode a set of essential genes involved in the assembly of the respiratory chain and the mitochondrial ribosome. Unlike other eukaryotic mitochondrial transcripts, most maxicircle-encoded transcripts require the specific addition and deletion of many uridine residues to create a translatable mRNA (2, 11, 20). Minicircles range in size from 0.9 to 10 kb and encode guide RNAs, small RNAs that provide sequence information for editing of maxicircle transcripts (22). In C. fasciculata a single kDNA network contains 25 to 50 maxicircles and 5,000 to 10,000 minicircles. Each minicircle contains two replication origins located 180° apart on the 2.5-kb circular DNA (3, 4).
Until recently, the proteins directly involved in the nucleoprotein structure of the kDNA were unknown. Several histone H1-like proteins have been identified and isolated based on in vivo cross-linking to kDNA in C. fasciculata (24, 25). The genes encoding these proteins are contained within the nucleus, and their protein products are synthesized with a 9-amino-acid cleavable presequence, which appears to be necessary for mitochondrial import (13). These kinetoplast-associated proteins, or KAP proteins, are highly basic proteins rich in lysine and alanine residues and are all approximately 14 kDa in size. The KAP1 protein binds nonspecifically to kinetoplast minicircle DNA, whereas proteins KAP2, KAP3, and KAP4 all show preferential binding to a specific region of the minicircle DNA located halfway between the two replication origins and which has no known function (25).
We have initiated molecular genetic studies of the genes encoding the KAP proteins in an effort to identify the roles of these proteins in the structure and function of the kDNA. Disruption of both alleles encoding the KAP1 protein, the most basic of the KAP proteins, resulted in a dramatic rearrangement of the kinetoplast organization (17). Although the disk-shaped kinetoplast retained its shape and size in the mutant cells, the internal organization of the nucleoprotein fibers was significantly altered, implying an involvement of KAP1 protein in the organization of the kDNA.
The KAP2 and KAP3 proteins are closely related and share 46% identical residues. The genes encoding these proteins are present in tandem on the same chromosomal DNA, suggesting that this arrangement may have resulted from a gene duplication (14). Both proteins appear to localize exclusively to the kinetoplast, and both were found to rescue a chromosome condensation and segregation defect in an Escherichia coli strain carrying mutations in the genes encoding the subunits of the HU heterodimer, an abundant nonspecific DNA-binding protein with roles in replication, recombination, and chromosome segregation (25).
To gain further insight into the roles of the KAP2 and KAP3 proteins in the kinetoplast we have carried out disruptions of the genes encoding these proteins. Although cells with both alleles of either gene knocked out have no apparent phenotype, disruption of both alleles of both KAP2 and KAP3 increased the levels of several mitochondrial mRNAs, reduced respiration, and had a drastic effect on cell growth and morphology.
MATERIALS AND METHODS
Construction of targeting vectors.
To construct the targeting vector for replacement of the KAP2 and KAP3 locus, a 1-kb targeting sequence of the 5′ untranslated region of the KAP2 gene and a 0.7-kb targeting sequence consisting of the 3′ untranslated region of the KAP3 gene plus 238 nucleotides of KAP3 coding sequence were PCR amplified with oligonucleotide primers F98 (5′-GGAATTCCGATATCTCCTCACCTCTTCTGGTC 3′), F99 (5′-CGGGATCCCGCTCGAGCGGGTGTGGGTAAATAGCTG-3′), E66 (5′-CGGGATCCGCTGCGCGTGTCTCCTACCCTG-3′) and F85 (5′-GCTCTAGAGCGATATCTCTCCGCCACACAAGCTG-3′). The positions of the introduced restriction sites in oligonucleotide primers F98 (EcoRI and EcoRV), F99 (XhoI and BamHI), E66 (BamHI), and F85 (EcoRV and XbaI) are underlined. The PCR-amplified DNA fragments were digested with EcoRI, BamHI, and XbaI restriction enzymes and were agarose gel purified, and two DNA fragments (1.0 and 0.7 kb) were cloned into the EcoRI- and XbaI-digested pGEM11Zf(+) (Promega) to create the pANK1.7 plasmid.
To generate the targeting pANKBSD and pANKHYG constructs, 2.7-kb SalI-BglII and 3.4-kb SalI-BglII gel-purified DNA fragments containing blasticidin S (BSD) and hygromycin (hygromycin B phosphotransferase [HYG]) selectable markers from plasmids pGL437B (5) and pX63HYG (9) were cloned into XhoI-BamHI-digested pANK1.7 plasmid. Coding sequences for the drug resistance markers are flanked by Leishmania major 5′ and 3′ sequences from the dihydrofolate reductase-thymidylate synthase (DHFR-TS) gene for expression of the BSD and HYG drug resistance markers. Plasmids pAKBSD and pANKHYG were digested with EcoRV, and released targeting fragments were agarose gel purified. Twenty to twenty-five micrograms of the first targeting DNA construct containing the BSD drug resistance cassette was electroporated into wild-type C. fasciculata cells as described previously (19). The single kap2/3+/− mutant cells of C. fasciculata were identified by Southern blot analysis of individual clones and were then transformed with the second targeting DNA construct containing the HYG selectable marker.
Southern blot analysis.
Genomic DNAs of C. fasciculata from wild-type, single kap2/3+/− mutant, and double kap2/3−/− mutant cells were isolated using a Wizard Genomic DNA Isolation kit as recommended by the supplier (Promega, Madison, Wis.). Total genomic DNA (2 μg) was digested with XhoI, separated on a 0.8% agarose gel, transferred to a nylon membrane (MagnaGraph; Micron Separations, Inc.) by vacuum blotting, and cross-linked to the membrane by UV irradiation. To generate a 1.7-kb DNA probe for Southern blot analysis, the pANK1.7 plasmid was digested with EcoRV, and 25 ng of gel-purified DNA fragment was labeled using Random Primer Labeling Beads (Amersham Bioscience, Piscataway, N.J.). The membrane was hybridized at 62.5°C overnight and washed three times in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 65°C for 20 min. The membrane was exposed to X-ray film with two screens for 12 h.
Western blot analysis.
C. fasciculata wild-type, single kap2/3+/−, and double kap2/3−/− mutant cells were harvested at 12,000 × g for 5 min. The cell pellets were washed three times with 1 ml of phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4), resuspended in PBS, mixed with 2× sample loading buffer (1× sample loading buffer is 25 mM Tris-HCl, 2% SDS, 8% glycerol, 2.5% 2-mercaptoethanol, and 0.0125% bromphenol blue [pH 6.8]), and boiled at 100°C for 4 to 5 min. Total lysates of 2 × 106 cells were separated by SDS-12.5% polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane with transfer buffer [10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS), 20% methanol, 10 mM NaOH] at 450 mA for 25 min. The membrane was blocked in Tris-buffered saline (TBS) (10 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20, pH 8.0) for 20 min, then was blocked with 5% (wt/vol) nonfat dry milk-5% (vol/vol) goat serum in TBS (blocking buffer) for 1 h, and then was incubated with either affinity-purified KAP2 or KAP3 antibodies (25) in blocking buffer (diluted 1:25 in blocking buffer). KAP2 and KAP3 protein bands were visualized using peroxidase conjugated anti-rabbit immunoglobulin G (IgG) secondary antibodies in blocking buffer (diluted 1:16,000 in blocking buffer) and the SuperSignal West Pico Chemiluminescent system (Pierce).
Analysis of oxygen consumption.
Logarithmically growing C. fasciculata wild-type cells (1.53 ×107 cells/ml) and kap2/3−/− double-mutant cells (1.27 ×106 cells/ml) were harvested, washed, and resuspended in 3 ml of fresh brain heart infusion medium (Difco) at concentrations of 1.5 × 107 and 1.3 × 107 cells/ml, respectively. Oxygen uptake was determined polarographically with a Clark type oxygen electrode (YSI model 53; Yellow Springs Instrument Co., Inc.) in a total volume of 3 ml at 28°C. The electrode was calibrated with air-saturated brain heart infusion medium at 28°C.
Northern blot analysis.
Isolation and Northern blot analysis of total RNA were performed as described previously (1). Briefly, total RNA was isolated from wild-type cells in the logarithmic (2 × 107 cells/ml) and stationary (2 × 108 to 3 × 108 cells/ml) phases and kap2/3−/− double-mutant cells in the logarithmic (2 × 106 cells/ml), early stationary (6 × 106 cells/ml), and stationary (2 × 107 cells/ml) phases. Total RNA (10 μg) was separated on 1.2% agarose-formaldehyde gels and blotted to nylon membranes (Hybond-XL; Amersham Pharmacia Biotech). The DNA fragments encoding mitochondrial proteins (ND7, Cox3, Cyb, ATPase 6, and Cox2) and nuclear encoded proteins (ISP and Cyt-c1) were PCR amplified with gene-specific oligonucleotide primers using the purified kDNA or genomic DNA of C. fasciculata as a template, respectively. The oligonucleotide sequences and size of PCR amplified DNA fragments are shown in Table 1. The gel-purified DNA fragments were labeled using Random Primer Labeling Beads (Amersham Bioscience), purified on a Sephadex G-50 column (Amersham Pharmacia Biotech), hybridized at 42°C for 15 to 17 h, and washed twice with 2.0× SSC-0.1% SDS at 60°C and then twice with 0.1× SSC-0.1% SDS at 60°C for 15 min each time. In contrast, the Northern blot membrane hybridized with ATPase 6 DNA probe was washed once with 2.0× SSC-0.1% at 30°C and at 50°C, and once with 0.1× SSC-0.1% at 45°C. The membrane was subsequently stripped and rehybridized with the labeled DNA fragments shown in Table 1. Northern blots were quantitated using a Molecular Dynamics PhosphorImager and ImageQuant software and then also exposed to X-ray film with two intensifying screens.
TABLE 1.
Oligonucleotide primers used in PCR amplification of probe sequences for kinetoplast and nuclear mRNAs
| Gene | Primer | Primer sequence (direction) | DNA fragment size (bp) |
|---|---|---|---|
| ND 7 | K13 | 5′-AACATCCTGCAGCTCATG-3′ (forward) | 373 |
| K14 | 5′-AACATCCTGCAGCTATG-3′ (reverse) | ||
| Cox3 | J42 | 5′-ATAGACGGAGGGGTTGAG-3′ (forward) | 839 |
| J43 | 5′-TTATGCACATAAATACAC-3′ (reverse) | ||
| Cyb | K40 | 5′-AAAGCGGAGAAAGAAGAAAAG-3′ (forward) | 1,000 |
| K41 | 5′-TCTAAACGACAAACTACTAGC-3′ (reverse) | ||
| ATPase 6 | K15 | 5′-CTTATGTATGTTTTTGTG-3′ (forward) | 517 |
| K16 | 5′-AAAGTCCATTATGAGCTG-3′ (reverse) | ||
| Cox2 | J44 | 5′-ATGGCGTTTATATTATCG-3′ (forward) | 573 |
| J45 | 5′-TGCATTGACCGTATACTG-3′ (reverse) | ||
| ISP | K11 | 5′-TCTTCTGTCTCGGGTCTG-3′ (forward) | 198 |
| K12 | 5′-GGTGGTGCCCTCGAGCTG-3′ (reverse) | ||
| Cyt-c1 | J46 | 5′-ATGGCGGGAAAGAAGGCG-3′ (forward) | 360 |
| J47 | 5′-CTGCAGATCCGGCGGCTC-3′ (reverse) |
Phase-contrast and immunofluorescence microscopy.
Immunofluorescence microscopy was performed essentially as described previously (1). Briefly, logarithmic cells were harvested by centrifugation (at 1,000 × g for 5 min), washed twice in an equal volume of PBS, and resuspended in PBS to 4 × 107 cells/ml. Cells were applied to microscope slides coated with 0.1% poly-l-lysine (Sigma-Aldrich) and incubated in a humid chamber for 20 to 30 min to allow cells to adhere. Slides were then washed in PBS, fixed in 4% paraformaldehyde in PBS for 10 min, and quenched by addition of glycine to a final concentration of 0.1 M. The fixed cells were washed two times in PBS for 5 min, permeabilized in PBST (PBS plus 0.1% Triton X-100) for 10 min at room temperature, and then washed three times in PBS. Slides were blocked in PBS plus 20% goat serum in a humid chamber at 37°C for 30 to 45 min, then incubated with a rabbit anti-RPA serum (diluted 1:500 in PBS plus 20% goat serum) in a humid chamber at 37°C for 1 h 30 min, and then washed three times in PBS for 2 to 3 min. Slides were incubated with Alexa fluor 568-labeled goat anti-rabbit IgG secondary antibodies (Molecular Probes) and washed three times in PBS for 2 to 3 min. Finally, the cells were incubated with 4′,6′-diamidino-2-phenylindole (DAPI) (0.2 μg/ml) for 3 min, rinsed with PBS, drained, and mounted in Vectashield (Vector Laboratories, Inc., Burlingame, Calif.). Cell images were captured using an Axioscope 2 microscope (Carl Zeiss, Jena, Germany), and the images were processed with Adobe PhotoShop (Adobe System, Mountain, Calif.).
Electron microscopy.
Cells were harvested, washed in PBS, and again pelleted, and the pellet was carefully resuspended in the fixative (2.5% glutaraldehyde [Sigma] in 0.1 M phosphate buffer), incubated at 4°C overnight, and embedded in Epon-Araldite (Polysciences). Thin sections stained with uranyl acetate and lead citrate were examined in a JEOL JEM 1010 electron microscope.
RESULTS
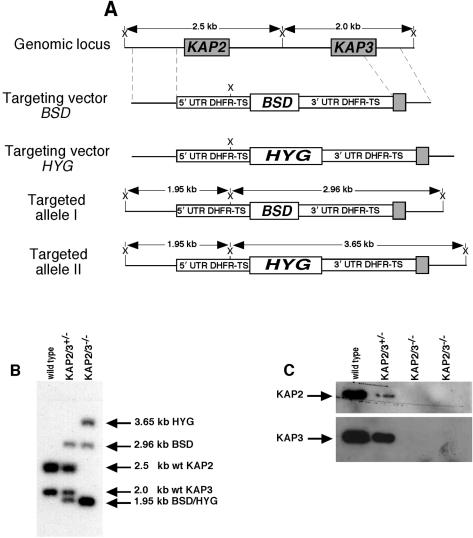
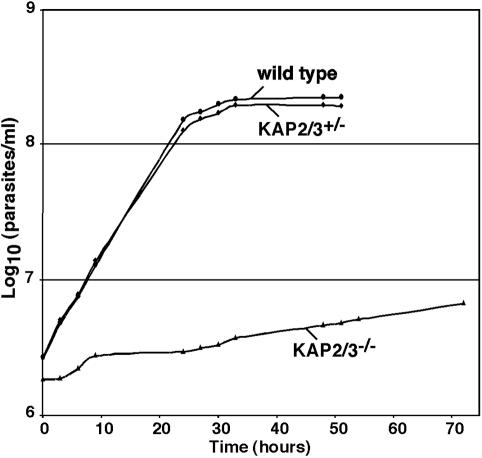
In our effort to reveal the function of the KAP proteins we have initiated gene replacement studies of each of the genes encoding these histone-like proteins. Since C. fasciculata is a diploid organism we have attempted to replace both alleles of each gene in order to observe a possible phenotype. In the case of the KAP1 gene we found that the double-mutant cells were viable but had drastically altered kDNA morphology (17). In contrast, we have not observed any phenotype for double mutants of either the KAP2 gene or the KAP3 gene (not shown). These cells have growth rates and morphologies similar to those of wild-type cells. Because of the close relationship of the KAP2 and KAP3 genes, we have investigated the possibility that the KAP2 and KAP3 proteins are functionally redundant by replacing the entire KAP2/3 locus on both chromosomes as outlined in Fig. 1A. BSD and HYG drug resistance cassettes derived from the pGL437B and pX63HYG vectors were used to sequentially replace the KAP2/3 locus on each chromosome. Figure 1B shows a Southern blot of cellular DNA from cells in which one or both loci were replaced. Loss of expression of the KAP2 and KAP3 proteins was confirmed by Western blotting as shown in Fig. 1C. The heterozygous strain (kap2/3+/−) expresses reduced levels of both KAP2 and KAP3 proteins, with a greater reduction of the KAP2 protein level. Several such isolates have been found to grow at the same rate as wild-type cells (Fig. 2) and have a normal appearance as observed by phase-contrast microscopy. Mutants in which both loci have been replaced are devoid of both KAP2 and KAP3 proteins (Fig. 1C) and grow extremely slowly (Fig. 2). The double-replacement mutant cells become stationary at an approximately 20-fold-lower cell density than that of wild-type cells, and frequent dilution of these cultures is required to maintain their growth.
FIG. 1.
Targeted disruption of the KAP2/KAP3 locus of C. fasciculata. (A) Schematic representations of the KAP2 and KAP3 genomic locus, targeting vectors, and targeted alleles. Diagrams indicate the size differences between the wild type, the kap2/3+/− single-disrupted alleles, and the kap2/3−/− double-disrupted alleles and show the positions of XhoI restriction sites (X). The gene-targeting vectors contain the BSD or HYG selectable markers flanked by the L. major 5′ and 3′ flanking regions of the DHFR-TS gene for expression of the drug resistance markers. UTR, untranslated region. (B) Southern blot of XhoI digests of C. fasciculata DNA from wild-type cells and from kap2/3+/− cells in which one allele of KAP2 and KAP3 was disrupted by insertion of the BSD cassette or kap2/3−/− cells in which the remaining alleles were disrupted by insertion of the hygromycin cassette (BSD/HYG). (C) Absence of KAP2 and KAP3 proteins in the kap2/3−/− double-mutant cells. Cell lysates of 2 × 106 wild-type, kap2/3+/− single-mutant, and kap2/3−/− double-mutant cells were analyzed by Western blotting with either affinity-purified KAP2 or KAP3 antibodies.
FIG. 2.
C. fasciculata kap2/3−/− double-mutant cells grow significantly more slowly than wild-type and kap2/3+/− single-mutant cells. Growth curves of wild-type, kap2/3+/− single-mutant, and kap2/3−/− double-mutant cells were determined using a Coulter Counter Z1. Numbers of cells of wild-type (circles), kap2/3+/− single-mutant (diamonds), and kap2/3−/− double-mutant (triangles) cells are indicated.
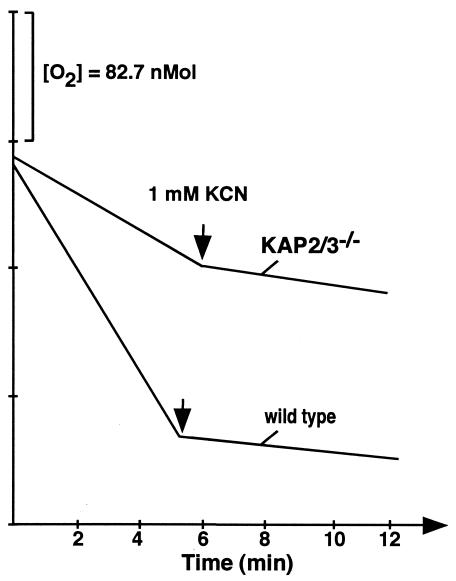
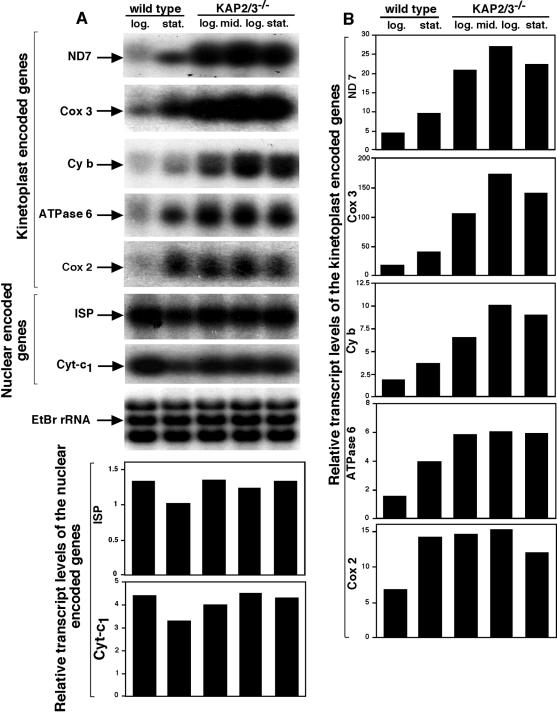
To evaluate possible defects in mitochondrial function we have examined the respiration of both the mutant and wild-type cells. The double-mutant cells were found to have a significantly reduced cyanide-sensitive respiration (Fig. 3). Further evidence of altered mitochondrial function is indicated in Northern blot analysis of several mitochondrial mRNAs (Fig. 4). The relative levels of mRNAs encoding maxicircle genes that code for ND7, Cox2, Cyb, ATPase 6, and Cox3 in wild-type cells all increased approximately twofold in stationary-phase cells relative to the levels in log-phase cells. Surprisingly, the levels of these transcripts in the double-mutant cells were found to be two- to fourfold higher than those of the stationary-phase wild-type cells, even at low cell densities of the mutant cells. In contrast, nucleus-encoded transcripts encoding the mitochondrial proteins Cyt-c1 and ISP did not show similar increases in the mutant cells.
FIG. 3.
Respiration of C. fasciculata wild-type and kap2/3−/− double-mutant cells. Oxygen consumption of logarithmically growing C. fasciculata wild-type and kap2/3−/− double-mutant cells was determined polarographically. Cyanide was added directly to cells at a final concentration of 1 mM at the time indicated. C. fasciculata wild-type and kap2/3−/− double-mutant cells consumed oxygen at rates of 25 and 9.8 nmol/min per 107 cells, respectively.
FIG. 4.
Increased levels of maxicircle transcripts in kap2/3−/− mutant cells. (A) Northern blot analysis of mitochondrial and nuclear transcripts. Total RNA was isolated from wild-type cells in logarithmic (log.) or stationary (stat.) phase and kap2/3−/− mutant cells in logarithmic (log.), middle logarithmic (mid.log.), and stationary (stat.) phase. Total RNA (10 μg) was fractionated on a 1.2% agarose-formaldehyde gel, stained with ethidium bromide, and transferred to a nylon membrane. The membrane was hybridized to the ND 7 DNA probe and was stripped and rehybridized sequentially to mitochondrial Cox3, Cyb, ATPase 6, Cox2, and nuclear ISP, Cyt-c1 DNA probes. rRNA stained with ethidium bromide was used as a loading control (EtBr rRNA). (B) PhosphorImager quantitation of expression levels of several mitochondrial and two nuclear transcripts.
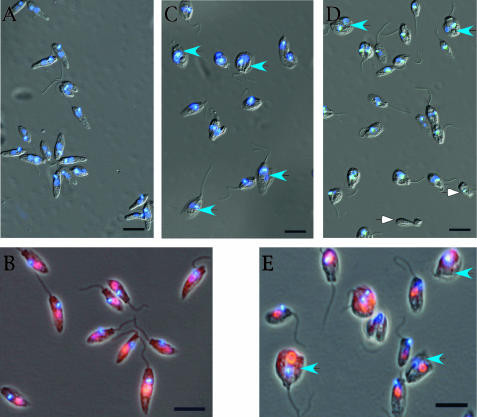
Microscopic examination of the double-mutant cells revealed very abnormal morphologies of the cells. Mutant cells stained with DAPI are shown in Fig. 5C and D. Many of the mutant cells appear to be blocked at a late stage of division, and a small percentage of the cells either lack a nucleus or lack both a nucleus and a kinetoplast. In contrast, the wild-type cells are much more uniform in size and shape (Fig. 5A). Since the nucleus generally fluoresces much less brightly than the kinetoplast in DAPI-stained C. fasciculata cells, we have also examined wild-type and mutant cells by additionally immunostaining the cells with antibodies against the nuclear protein RPA1 (6) (Fig. 5B and E). These results again appear to reflect an inhibition of cell division and the accumulation of cells with abnormal morphologies.
FIG. 5.
Abnormal cell size and morphology of cells lacking the KAP2 and KAP3 proteins. Combined phase-contrast and immunofluorescence microscopy of wild-type and kap2/3−/− mutant cells of C. fasciculata. Wild-type (A and B) and kap2/3−/− mutant (C to E) C. fasciculata cells were fixed and stained with the DNA intercalating dye DAPI to identify the nucleus and kinetoplast. To further identify the nucleus (B and E) the cells were incubated with a rabbit antiserum against C. fasciculata RPA1, a nuclear replication protein, and Alexa fluor 568-labeled goat anti-rabbit IgG secondary antibody. Wild-type cells (A and B) show normal morphologies, whereas kap2/3−/− mutant cells (C to E) show many large cells with abnormal morphologies. Blue arrowheads show cells of abnormal size. White arrowheads show a cell body and a cell lacking a nucleus. Scale bar, 10 μm.
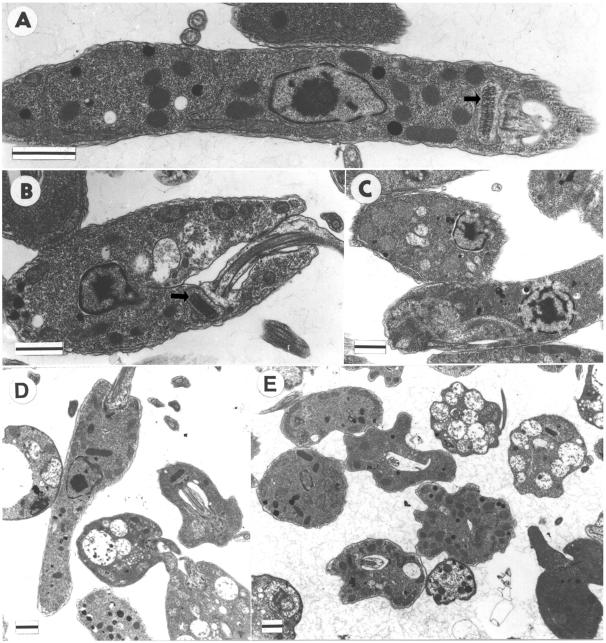
Electron microscopic examination of thin sections of wild-type and mutant cells shows the abnormal morphologies of the double-mutant cells in greater detail. Figure 6A shows a typical wild-type cell. Cells in which both alleles of only the KAP2 gene (Fig. 6B) or of only the KAP3 gene (Fig. 6C) have been knocked out show one to three large vacuoles on average but otherwise appear similar in morphology to wild-type cells. Wild-type cells generally show one to two smaller vacuoles. In striking contrast, cells in which the entire KAP2/3 locus has been replaced on both chromosomes accumulate numerous large vacuoles and display a wide range of morphologies (Fig. 6D and E). However, kinetoplasts in the latter mutant cells appear to have morphologies and positioning relative to the base of the flagellum similar to that of wild-type cells. Many of the cells are packed with large vacuoles while others have only a few such vacuoles. The double-mutant cells also frequently display numerous invaginations in the cell surface.
FIG. 6.
Ultrastructural analysis of C. fasciculata wild-type and kap2−/−, kap3−/−, and kap2/3−/− mutant cells. Shown are electron micrographs of wild-type cells (A) and cells lacking KAP2 (B), KAP3 (C), or both KAP2 and KAP3 (D and E) proteins. (A and B) Longitudinal sections of the kinetoplast disk (indicated by arrows). Scale bar, 1 μm.
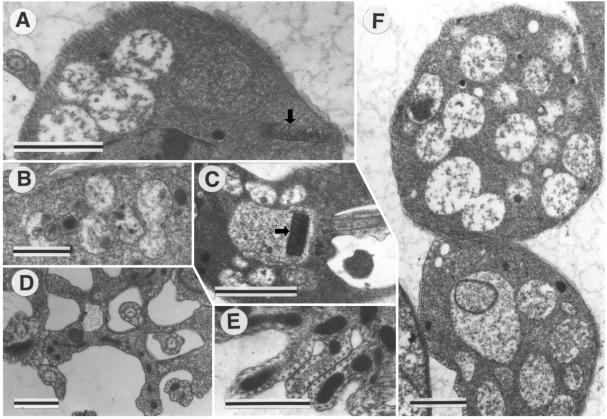
Examples of the range of unusual morphologies in the kap2/3−/− double replacement mutants are shown in Fig. 7. Kinetoplasts shown in Fig. 7A and C appear similar to those in wild-type cells in size and morphology. Figure 7B and F show vacuoles that appear to be fusing. Generally these numerous vacuoles are relatively electron translucent, although Fig. 7A also shows vacuoles that are much more electron opaque. Figure 7D and E show the folding and invagination of the cell surface. The section in Fig. 7D appears to represent a transverse section through the flagellar pockets of a dividing cell showing the two daughter flagella. The corset of closely spaced subpellicular microtubules is clearly visible in Fig. 7D to F. The lower cell in Fig. 7F appears to have a vacuole within a vacuole.
FIG. 7.
Range of abnormal morphologies in kap2/3−/− double-mutant cells. (A to F) Electron micrographs of cells lacking both KAP2 and KAP3 proteins show numerous large vacuoles and cell wall invaginations in the mutant cells. (A and C) Kinetoplasts are indicated by arrows. Scale bar, 1 μm.
DISCUSSION
DNA-binding proteins play central roles in repair, replication, recombination, transcription, and nucleoprotein organization. Often a single such protein will have roles in two or more of these processes leading to complex phenotypes of cells in which the protein is mutated or absent. DNA-binding proteins have also been identified in the mitochondria of a variety of eukaryotic organisms ranging from yeast to humans (18). These proteins are encoded in the cell nucleus and are posttranslationally imported into the mitochondrion. In the case of C. fasciculata we were surprised to find little, if any, effect of the loss of either the histone-like KAP2 or KAP3 DNA-binding protein alone. The close relationship of their amino acid sequences (14) suggested that the proteins might be capable of substituting for one another and the strong phenotype resulting from replacing both copies of the locus encoding KAP2 and KAP3 proteins supports this interpretation.
The most striking effects of the loss of the KAP2 and KAP3 proteins are the extremely slow growth, the highly abnormal cell morphologies, and the accumulation of numerous large vacuoles. The frequent invaginations of the cell surface observed in these mutants could potentially result from repeated attempts at cell division in the absence of nuclear and kinetoplast duplication. Aberrant cytokinesis in some cells might result in cell bodies lacking nuclear DNA and/or kDNA such as those observed here. The origin and composition of the accumulated vacuoles are unknown. Many of the mutant cells have numerous large vacuoles while others have none. Since these cells are clonal, the heterogeneity in the population may represent an unequal distribution of essential cellular components at cell division. The cells with large numbers of vacuoles may possibly represent dying or dead cells. Isolation of these vacuoles and determination of their protein contents will be important for ultimately revealing their origin. However, the extremely slow growth, the reduced respiration, the abnormal morphologies, and the formation of vacuoles by the mutant cells are likely to be indirect effects of the loss of the KAP2 and KAP3 proteins. While not providing insight into the mechanistic basis for these defects, these results imply an important role for the KAP2 and KAP3 proteins in normal cell growth and in mitochondrial function.
The increased expression of maxicircle mRNAs but not that of nucleus-encoded mitochondrial proteins suggests a possible direct effect of the loss of the KAP2 and KAP3 proteins on maxicircle gene expression. The KAP2 and KAP3 proteins were isolated initially based on chemical cross-linking to kDNA in vivo (24). Although recombinant forms of these proteins show preferential binding to a region of minicircle DNA with no known function (25), the highly basic nature of the KAP2 and KAP3 proteins (pIs of 10.7 and 11.2, respectively) and their localization exclusively within the kinetoplast make it likely that these proteins also bind to maxicircle DNA and might therefore affect transcription of maxicircle genes. While the loss of KAP2 and KAP3 proteins might also affect RNA editing or message stability, we consider these possibilities less likely. The ratio of edited to unedited maxicircle transcripts observed in poisoned primer extension experiments (not shown) are similar in mutant and wild-type cells and it is unclear how the loss of these basic proteins could increase the stability of maxicircle transcripts.
Expression of either the KAP2 or KAP3 protein in an E. coli strain having a mutation in the gene encoding the HU protein rescued a defect in chromosome condensation and segregation (25). Although the size and morphology of the kinetoplast in the double-mutant cells appear indistinguishable from those of wild-type cells, the KAP2 and KAP3 proteins could have a subtle effect on the kDNA organization affecting expression of the maxicircle genes. We note that small histone-like proteins in prokaryotes are known to be involved in transcriptional repression. Several such proteins have been identified in association with the bacterial nucleoid and, like KAP2 and KAP3, have no DNA consensus motif and bind DNA cooperatively. The E. coli HU protein appears to repress transcription through its effect on coiling of the chromosomal DNA (15). A similar protein (H-NS), also a major constituent of the E. coli nucleoid, is implicated in the compact organization of the chromosome and in transcriptional repression (23). The bacteriophage DNA-binding protein p6 is functionally related to these prokaryotic histone-like proteins and plays a role in repression through effects on local DNA topology (7). The recent identification and cloning of the gene encoding a kinetoplast RNA polymerase (12) should now permit an investigation of the possible effect of the KAP2 and KAP3 proteins on transcription of maxicircle templates by the purified polymerase.
Acknowledgments
We thank Kent Hill for valuable comments on the manuscript and for advice on and use of his fluorescence microscope and Martina Nebohácová and Larry Simpson for assistance with and use of their oxygen electrode.
This research was supported by NIH grant AI45536 (to D.S.R.) and grant IAA5022302 (to J.L.) from the Grant Agency of the Czech Academy of Sciences.
REFERENCES
- 1.Avliyakulov, N. K., J. C. Hines, and D. S. Ray. 2003. Sequence elements in both the intergenic space and the 3′ untranslated region of the Crithidia fasciculata KAP3 gene are required for cell cycle regulation of KAP3 mRNA. Eukaryot. Cell 2:671-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benne, R., J. Van den Burg, J. P. Brakenhoff, P. Sloof, J. H. Van Boom, and M. C. Tromp. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46:819-826. [DOI] [PubMed] [Google Scholar]
- 3.Birkenmeyer, L., and D. S. Ray. 1986. Replication of kinetoplast DNA in isolated kinetoplasts from Crithidia fasciculata. J. Biol. Chem. 261:2362-2368. [PubMed] [Google Scholar]
- 4.Birkenmeyer, L., H. Sugisaki, and D. S. Ray. 1987. Structural characterization of site-specific discontinuities associated with replication origins of minicircle DNA from Crithidia fasciculata. J. Biol. Chem. 262:2384-2392. [PubMed] [Google Scholar]
- 5.Brooks, D. R., R. McCulloch, G. H. Coombs, and J. C. Mottram. 2000. Stable transformation of trypanosomatids through targeted chromosomal integration of the selectable marker gene encoding blasticidin S deaminase. FEMS Microbiol. Lett. 186:287-291. [DOI] [PubMed] [Google Scholar]
- 6.Brown, G. W., T. E. Melendy, and D. S. Ray. 1992. Conservation of structure and function of DNA replication protein A in the trypanosomatid Crithidia fasciculata. Proc. Natl. Acad. Sci. USA 89:10227-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camacho, A., and M. Salas. 2001. Mechanism for the switch of phi29 DNA early to late transcription by regulatory protein p4 and histone-like protein p6. EMBO J. 20:6060-6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., C. A. Rauch, J. H. White, P. T. Englund, and N. R. Cozzarelli. 1995. The topology of the kinetoplast DNA network. Cell 80:61-69. [DOI] [PubMed] [Google Scholar]
- 9.Cruz, A., C. M. Coburn, and S. M. Beverley. 1991. Double targeted gene replacement for creating null mutants. Proc. Natl. Acad. Sci. USA 88:7170-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Englund, P. T. 1981. Kinetoplast DNA. Biochem. Physiol. Protozoa 4:333-383. [Google Scholar]
- 11.Feagin, J. E., J. M. Abraham, and K. Stuart. 1988. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 53:413-422. [DOI] [PubMed] [Google Scholar]
- 12.Grams, J., J. C. Morris, M. E. Drew, Z. Wang, P. T. Englund, and S. L. Hajduk. 2002. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J. Biol. Chem. 277:16952-16959. [DOI] [PubMed] [Google Scholar]
- 13.Hausler, T., Y. D. Stierhof, J. Blattner, and C. Clayton. 1997. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur. J. Cell Biol. 73:240-251. [PubMed] [Google Scholar]
- 14.Hines, J. C., and D. S. Ray. 1997. Tandem arrangement of two genes encoding kinetoplast-associated H1 histone-like proteins. Mol. Biochem. Parasitol. 89:41-49. [DOI] [PubMed] [Google Scholar]
- 15.Lewis, D. E., M. Geanacopoulos, and S. Adhya. 1999. Role of HU and DNA supercoiling in transcription repression: specialized nucleoprotein repression complex at gal promoters in Escherichia coli. Mol. Microbiol. 31:451-461. [DOI] [PubMed] [Google Scholar]
- 16.Lukeš, J., D. L. Guilbride, J. Votypka, A. Zikova, R. Benne, and P. T. Englund. 2002. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell 1:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukeš, J., J. C. Hines, C. J. Evans, N. K. Avliyakulov, V. P. Prabhu, J. Chen, and D. S. Ray. 2001. Disruption of the Crithidia fasciculata KAP1 gene results in structural rearrangement of the kinetoplast disc. Mol. Biochem. Parasitol. 117:179-186. [DOI] [PubMed] [Google Scholar]
- 18.Maier, D., C. L. Farr, B. Poeck, A. Alahari, M. Vogel, S. Fischer, L. S. Kaguni, and S. Schneuwly. 2001. Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol. Biol. Cell 12:821-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasion, S. G., G. W. Brown, L. M. Brown, and D. S. Ray. 1994. Periodic expression of nuclear and mitochondrial DNA replication genes during the trypanosomatid cell cycle. J. Cell Sci. 107:3515-3520. [DOI] [PubMed] [Google Scholar]
- 20.Shaw, J. M., J. E. Feagin, K. Stuart, and L. Simpson. 1988. Editing of kinetoplastid mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell 53:401-411. [DOI] [PubMed] [Google Scholar]
- 21.Stuart, K. 1983. Kinetoplast DNA, mitochondrial DNA with a difference. Mol. Biochem. Parasitol. 9:93-104. [DOI] [PubMed] [Google Scholar]
- 22.Sturm, N. R., and L. Simpson. 1990. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell 61:879-884. [DOI] [PubMed] [Google Scholar]
- 23.Ueguchi, C., and T. Mizuno. 1993. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 12:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu, C., and D. S. Ray. 1993. Isolation of proteins associated with kinetoplast DNA networks in vivo. Proc. Natl. Acad. Sci. USA 90:1786-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu, C. W., J. C. Hines, M. L. Engel, D. G. Russell, and D. S. Ray. 1996. Nucleus-encoded histone H1-like proteins are associated with kinetoplast DNA in the trypanosomatid Crithidia fasciculata. Mol. Cell. Biol. 16:564-576. [DOI] [PMC free article] [PubMed] [Google Scholar]