Abstract
The liz1+ gene of the fission yeast Schizosaccharomyces pombe was previously identified by complementation of a mutation that causes abnormal mitosis when ribonucleotide reductase is inhibited. Liz1 has similarity to transport proteins from Saccharomyces cerevisiae, but the potential substrate and its connection to the cell division cycle remain elusive. We report here that liz1+ encodes a plasma membrane-localized active transport protein for the vitamin pantothenate, the precursor of coenzyme A (CoA). Liz1 is required for pantothenate uptake at low extracellular concentrations. A lack of pantothenate uptake results in three phenotypes: (i) slow growth, (ii) delayed septation, and (iii) aberrant mitosis in the presence of hydroxyurea (HU). All three phenotypes are suppressed by high extracellular concentrations of pantothenate, where pantothenate uptake occurs by passive diffusion. liz1Δ mutants are viable because they can synthesize pantothenate from uracil as an endogenous source. The use of uracil for both pantothenate biosynthesis and deoxyribonucleotide generation provides an explanation for the aberrant mitosis in the presence of HU. HU blocks ribonucleotide reductase, and we propose that the accumulation of ribonucleotides reduces uracil biosynthesis by feedback inhibition of aspartate transcarbamoylase. Thus, the addition of HU to liz1Δ mutants results in a shortage of pantothenate. Because liz1Δ mutants show striking similarities to mutants with defects in fatty acid biosynthesis, we propose that the shortage of pantothenate compromises fatty acid synthesis, resulting in slow growth and mitotic defects.
The cell division cycle is a tightly controlled cascade of independent events. Biochemical pathways, known as cell cycle checkpoints, impose dependency by ensuring that the initiation of later events does not occur before earlier events have been completed. Chromosome replication is monitored by the intra-S-phase checkpoint, which coordinates DNA replication, and by the S-M checkpoint, which delays entry into mitosis until DNA replication is complete (reviewed in reference 5). Both checkpoint pathways have been extensively studied in the fission yeast Schizosaccharomyces pombe (6, 34). The inhibition of ribonucleotide reductase (RNR), the enzyme that converts ribonucleotides to deoxyribonucleotides, by the addition of hydroxyurea (HU) to the growth medium blocks chromosome replication. In wild-type cells, this activates the intra-S-phase checkpoint to prevent further progress through S phase and activates the S-M checkpoint to prevent mitosis. Genetic screens for HU sensitivity have been exploited to identify mutants that inappropriately responded to incomplete DNA replication (11). These mutations not only identified checkpoint proteins but also defined proteins that are required to survive arrest within S phase (3, 4, 26).
One mutant obtained in such a screen, the liz1− (lives if zapped 1) mutant, has an unusual phenotype for a HU-sensitive mutant: like wild-type cells, most of the liz1− cells appear to arrest mitosis following addition of HU. However, a proportion of liz1− mutants undergo a highly aberrant mitosis (28). Intriguingly, when HU was added to a culture of liz1− cells, passage through S phase was not a requirement for this defective mitosis to occur. In synchronized G2 cells, the first mitosis following HU addition was seen to be aberrant. In some of these cells, the new septum was seen to bisect an unseparated nucleus, a lethal event known as the “cut” phenotype (28). In less drastic cases, in the presence of HU, the liz1− cells completed nuclear division and formed a septum but failed to decondense their chromosomes. Thus, liz1− cells in G2 display a catastrophic mitosis when treated with HU. Conversely, when liz1− cells in G1 are treated with HU, they arrest in S phase and activate a normal S-M checkpoint, and mitosis is prevented.
To rule out the possibility that HU acted to inhibit an alternative process in liz1− cells, Moynihan and Enoch (28) introduced the cdc22-M45 mutation into a liz1 null mutant background. Cdc22 is the large subunit of RNR, and the cdc22-M45 mutation causes this to be inactive at the restrictive temperature. Inactivating RNR by a temperature shift resulted in phenotypes in liz1− cells that are the same as those observed when cells are treated with HU (28). This confirms that RNR activity is required for a normal mitosis in liz1− cells and that this phenotype is independent of passage through S phase.
While a cut cell phenotype is frequently observed in checkpoint mutants treated with HU, the phenotype of the liz1− mutant is fundamentally different. For example, when hus1− cells are treated with HU while they are in G2, they progress normally through the first mitosis and show no cuts. It is only when they enter the subsequent S phase and are unable to activate the S-M checkpoint that they then proceed into the second mitosis with catastrophic and lethal results. This contrasts with the liz1− mutant, which can activate a normal S-M checkpoint, but in which a significant proportion of cells are unable to successfully complete the previous mitosis (28). Thus, the conundrum is why a cell with replicated chromosomes should require RNR activity to complete mitosis.
The liz1+ gene was cloned and shown to encode a protein with similarity to the Saccharomyces cerevisiae allantoate permease family of transport proteins (28, 30). Although the substrate of Liz1 remained unidentified, the authors speculated that Liz1 might be involved in the transport of an intermediate in uracil biosynthesis (28). During our analysis of two S. cerevisiae transport proteins, Vht1p and Fen2p, we identified Liz1 as a closely related protein by database searches. Vht1p and Fen2p facilitate the uptake of biotin and pantothenate, respectively (36, 37). This similarity suggested either biotin or pantothenate as a possible substrate for the Liz1-dependent transport. In this report, we provide evidence that Liz1 functions in the uptake of pantothenate, transporting it across the plasma membrane. Since pantothenate is essential for coenzyme A (CoA) biosynthesis, the failure of liz1− mutants to complete nuclear separation in the presence of HU may be related to a defect in fatty acid synthesis. CoA is required for fatty acid synthesis, and it is well established that, in S. pombe, mutants in the fatty acid biosynthesis pathway exhibit a cut phenotype (31). This presumably reflects a high demand for fatty acids during nuclear division.
We also present data demonstrating that uracil serves as a precursor for CoA biosynthesis in the absence of pantothenate uptake in liz1− mutants. This metabolic link provides an explanation for why the inhibition of RNR leads to defects in normal mitotic progression in liz1− mutants. HU blocks RNR activity, and this is likely to cause an increase in CTP and UTP. Since CTP and UTP allosterically inhibit aspartate transcarbamoylase, an essential enzyme in the uracil biosynthetic pathway, the cellular uracil pool would be expected to drop, reducing pantothenate synthesis in liz1− mutant cells. liz1− mutant cells may thus experience a depletion of CoA in the presence of HU, which in turn may compromise the supply of fatty acids that are required for a normal mitosis.
MATERIALS AND METHODS
Yeast strains and media.
The S. pombe leu1-32 strain (17) served as a donor of genomic DNA for the amplification of liz1+ and pan6+ genes. S. pombe FY254 (ade6-M210 can1-1 leu1-32 ura4-D18 h−) (21) was used for all other experiments. The genotype of the hus1− mutant was hus1Δ::LEU2 ura4-D18 leu1-32 h− (19). Where indicated, cells were made ura4-positive by transformation with pREP4X (12). The pantothenate transport-deficient S. cerevisiae strain 711/1c (MATα ura3-52 trp1Δ1 fen2Δ::URA3) (1, 37) was used for the expression of the liz1+ gene.
S. cerevisiae and S. pombe cells were grown in synthetic minimal medium (0.67% yeast nitrogen base without amino acids [Difco], 2% d-glucose) that contained a fixed concentration of pantothenate (1.68 μM). Media for growth assays were prepared from an autoclaved solution containing 2% d-glucose and 2% Difco Bacto agar. Pantothenate-free yeast nitrogen base (final concentration, 0.67%), 50 mg of adenine/liter, and 50 mg of leucine/liter, as well as other compounds, were added from filter-sterilized stocks before pouring the plates. For growth assays, cells were washed and diluted in water to an A600 of 0.8, and four serial 10-fold dilutions were performed in a 96-well plate. Aliquots of the cells were transferred to plates by using a metal replicating device.
Plasmid constructs and gene disruptions.
The open reading frame of the liz1+ gene (SPBC2G2.01c) was amplified by PCR and ligated into the EcoRI site of pUC19 (pUC19LIZ1). liz1+ does not contain introns. For overproduction of Liz1 in S. pombe, the EcoRI fragment was introduced into SAP-E (38) to give plasmid pSAPLIZ1. For expression in S. cerevisiae, the EcoRI fragment was ligated into p424MET25 (29) to give plasmid pLIZ1mc.
To create a disrupted allele of liz1+, the ura4+ gene was subcloned from pREP4X (12) into the HindIII site of pIC20R (25), liberated with BglII and PstI, and ligated into the BclI and NsiI sites of pUC19LIZ1. In the resulting plasmid, nucleotides 194 to 1152 of the liz1+ open reading frame were replaced by ura4+, which was in antisense orientation relative to liz1+. The disrupted allele was liberated with EcoRI and transformed into S. pombe, and the cells were spread on plates containing 100 μM pantothenate.
To generate a knockout construct for pan6+ (SPAC5H10.08c), the gene was amplified by PCR and ligated into the single NotI site of a modified pUC19 vector. Next, the kanMX gene from pFA6a-kanMX4 (39) was introduced as a SalI-EcoRI restriction fragment into the SalI-MfeI sites within pan6+, eliminating 202 bp of the coding region. The final disruption construct was liberated with NotI and used to transform S. pombe cells. All gene disruptions were verified by PCR.
Transport assays and FACS and microscopic analyses.
The transport of pantothenate was determined at an initial outside concentration of 1.75 μM at pH 6.0 in 18 mM citric acid, 64 mM Na2HPO4, and 1% d-glucose, as previously described (37). DAPI (4′,6′-diamidino-2-phenylindole) was used to stain the DNA for scoring cut phenotypes, and calcofluor was used to stain the septa of dividing cells. At least 200 cells were counted for each time point. Fluorescence-activated cell sorter (FACS) analysis of S. pombe was performed as previously described (10) by using propidium iodide to stain the DNA.
RESULTS
liz1+ complements mutations in the S. cerevisiae FEN2 gene.
The genes encoding the S. cerevisiae plasma membrane transporters for pantothenate (FEN2) and biotin (VHT1) were recently identified (36, 37). The protein encoded by the S. pombe liz1+ gene is a homolog of both vitamin transporters. Based on direct protein comparisons and phylogenetic analyses of proteins from the allantoate transporter families of S. cerevisiae and S. pombe (35), Liz1 is most similar to the S. cerevisiae pantothenate transporter Fen2p.
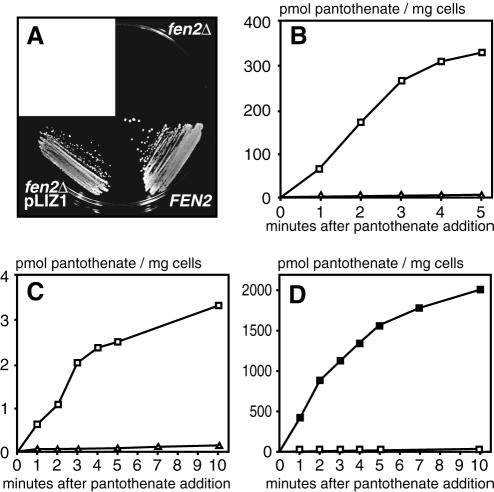
To test whether Liz1 functions as a pantothenate transporter, we expressed liz1+ in S. cerevisiae fen2Δ mutants (Fig. 1A). fen2Δ mutants were unable to grow on plates containing 1 μM pantothenate, whereas this concentration was sufficient for growth of S. cerevisiae wild-type cells (Fig. 1A). Heterologous expression of liz1+ restored the growth of fen2Δ cells in the presence of low pantothenate concentrations, strongly indicating that Liz1 acts as a pantothenate transporter. Consistent with this view, expression of liz1+ failed to sustain the growth of S. cerevisiae vht1Δ mutants defective in biotin transport (data not shown). Together with the recent cloning of the S. pombe biotin transporter gene vht1+ (35), these data identify pantothenate as the substrate for Liz1.
FIG. 1.
Liz1 is a plasma membrane transport protein for pantothenate. (A) Multicopy expression of liz1+ rescues the growth defect of S. cerevisiae fen2Δ mutants. Cells of the S. cerevisiae fen2Δ mutant strain 711/1c were transformed with an empty plasmid or with pLIZ1mc for multicopy expression of liz1+. The cells were resuspended in water (A600 = 0.1), and 10 μl of the suspension was streaked on a plate containing 1 μM pantothenate. A S. cerevisiae wild-type strain is shown for comparison. The plate was photographed after a 3-day incubation at 30°C. (B) Expression of liz1+ confers pantothenate uptake to S. cerevisiae fen2Δ cells. Transport of [14C]pantothenate was determined with fen2Δ cells harboring a control plasmid (▵) or with fen2Δ cells carrying liz1+ on a multicopy plasmid (□). (C) Pantothenate uptake into S. pombe cells after deletion of liz1+ (▵). A S. pombe wild-type strain carrying a control plasmid (□) is shown for comparison. (D) Pantothenate uptake in S. pombe cells after overexpression of liz1+ (▪) and in wild-type cells carrying a control plasmid (□).
liz1+ encodes a high-affinity proton pantothenate symporter.
The pantothenate transport activity of Liz1 was characterized in detail in S. cerevisiae fen2Δ mutants. Whereas the transport of [14C]pantothenate across the plasma membrane was negligible in control cells, liz1+-expressing cells showed a rapid uptake of pantothenate (Fig. 1B). Consistent with a proton symport mechanism, pantothenate uptake by Liz1 was stimulated by d-glucose and was strongly inhibited by protonophores (80% inhibition by 50 μM carbonyl cyanide m-chlorophenyl hydrazone; data not shown). Liz1-mediated pantothenate transport was maximal at an external pH of 6.0, at which pantothenate is negatively charged. Pantothenate transport was saturable and displayed an apparent Km of 2.75 μM. We found no reduction in pantothenate transport when a 50-fold molar excess of taurine, pantoyltaurine, β-alanine, or biotin was present during the uptake experiments (data not shown). Taken together, these experiments demonstrate that Liz1 acts as a specific high-affinity H+-pantothenate symporter.
To analyze the physiological function of liz1+ in S. pombe cells, we disrupted the open reading frame by the integration of the ura4+ gene. Although disruption of liz1+ was previously shown to cause partial uracil auxotrophy (28), we were able to recover ura4+ transformants that had the disruption construct correctly integrated. In agreement with a role in pantothenate uptake, disruption of the liz1+ gene in S. pombe cells strongly reduced pantothenate transport from 60 pmol of pantothenate h−1 × mg cell−1 in wild-type cells to 1.8 pmol of pantothenate h−1 × mg cell−1 for liz1Δ mutants (Fig. 1C). Overexpression of liz1+ increased pantothenate transport 430-fold over wild-type levels (26 mmol of pantothenate h−1 × mg cell−1 [Fig. 1D]). These data confirm that the physiological role of Liz1 is the uptake of pantothenate. Since pantothenate transport is almost completely lost upon disruption of liz1+ (Fig. 1C), Liz1 seems to be the only functional plasma membrane pantothenate transporter in S. pombe. Our data are consistent with the localization of green fluorescent protein-tagged Liz1 to the S. pombe plasma membrane (28).
Growth defects of liz1Δ mutants are rescued by high extracellular concentrations of pantothenate.
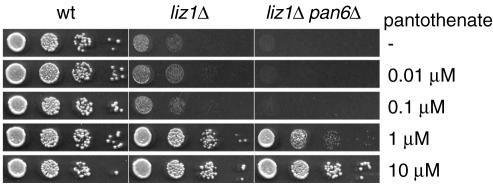
It is well established that the loss of plasma membrane transporters for various vitamins can be overcome by adding very high concentrations of the vitamin to the growth medium (22, 35-37). To test whether this was also true for S. pombe liz1Δ mutants, we analyzed growth in the presence of increasing concentrations of pantothenate (Fig. 2). Wild-type S. pombe cells were able to grow on any of the supplied pantothenate concentrations and were also capable of growing in the absence of pantothenate, indicating that they could synthesize pantothenate when it was absent. In contrast, the growth of liz1Δ cells was poor in the presence of pantothenate concentrations of 0.1 μM or below, but full growth was restored when pantothenate was present at concentrations of 1 μM or higher (Fig. 2). The fact that cells lacking Liz1-mediated pantothenate uptake were viable on pantothenate-free plates strongly implied that a pathway for pantothenate biosynthesis exists in S. pombe.
FIG. 2.
High concentrations of pantothenate complement the growth defects of liz1Δ cells. S. pombe wild-type cells (wt), carrying a ura4+ control plasmid, the liz1Δ::ura4+ mutant, and a liz1Δ::ura4+ pan6Δ::kanMX double mutant were grown in liquid culture, washed with water, and assayed for growth on plates containing the indicated concentrations of pantothenate. Growth was recorded after 3 days at 30°C.
To confirm the existence of such a pathway, we analyzed S. pombe cells lacking the pan6+ gene. The protein encoded by pan6+ is 60% identical to the Escherichia coli pantoate β-alanine ligase, an enzyme involved in pantothenate biosynthesis from β-alanine (27). As shown in Fig. 2, the deletion of pan6+ in the liz1Δ strain abolished all background growth at low pantothenate concentrations. This suggests that pantothenate is synthesized from β-alanine in the absence of exogenous pantothenate supplies. As expected, the addition of high pantothenate concentrations to the medium suppressed the growth defect of pan6Δ liz1Δ double mutants. Thus, S. pombe is capable of synthesizing pantothenate from an intracellular source, and this pathway is essential for the survival of liz1Δ mutants.
The cell cycle phenotypes of liz1Δ mutants are rescued by high extracellular concentrations of pantothenate.
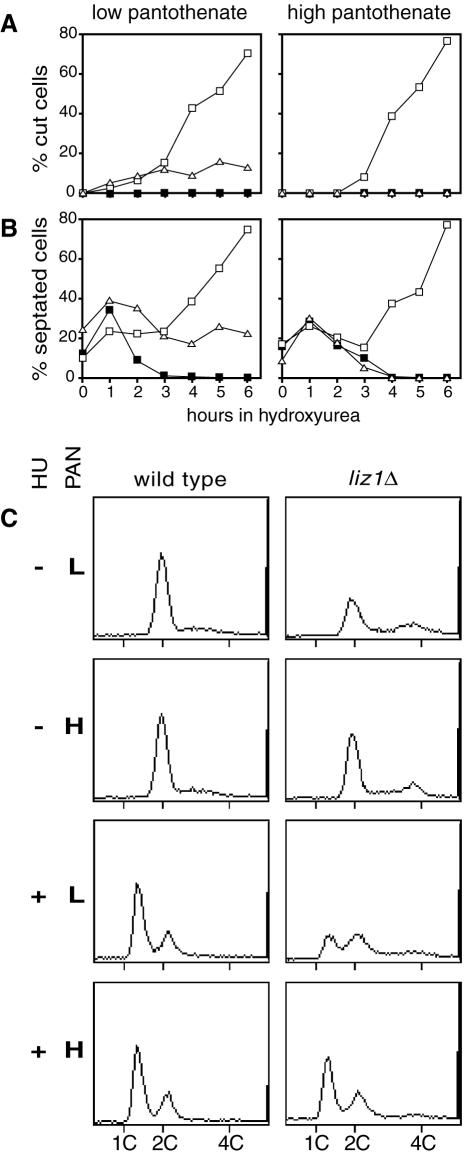
Our data and that of Moynihan and Enoch (28) indicate that the HU sensitivity of liz1Δ mutants is caused by the absence of pantothenate uptake. To confirm this, we compared liz1Δ, hus1Δ, and wild-type cells for their responses to HU. The experiments were performed in medium containing a low concentration of pantothenate (left panels of Fig. 3A and B) or upon the addition of 1 mM pantothenate, which supplies the vitamin in the absence of Liz1 (right panels of Fig. 3A and B).
FIG. 3.
Supplementation of pantothenate corrects the cell cycle phenotypes of S. pombe liz1Δ mutants. S. pombe cells were grown in media containing a low (1.68 μM) or high (1 mM) concentration of pantothenate (PAN), freshly diluted into the same media, and 10 mM HU was added. Cells were stained with calcofluor to visualize septa and with DAPI to visualize nuclei. At least 200 cells were examined for each time point. All strains analyzed were prototrophic for uracil and were grown in media lacking uracil. ▪, wild type; ▵, liz1Δ; □, hus1Δ. In panel A, the percentages of cells showing the cut phenotype are given, and in panel B, the percentages of cells showing septa (i.e., cells in S phase) are given. Inpanel C, the DNA content of asynchronous wild-type or liz1Δ cells was determined by FACS analysis. Samples were obtained after growth in media with low (L) or high (H) concentrations of pantothenate, as described above. Where indicated, the cells were treated for 2 h with 10 mM HU. Approximately 2,000 cells were fixed, stained with propidium iodide, and analyzed.
At low pantothenate concentrations, the inhibition of RNR by HU triggered an increase in the number of liz1− mutant cells undergoing a catastrophic mitosis (cut phenotype). This increase was completely suppressed by the presence of high pantothenate concentrations, suggesting that pantothenate uptake is indeed required for normal mitotic progression in the presence of HU. Unlike liz1Δ cells, hus1Δ cells entered a catastrophic mitosis independently of the pantothenate concentration in the medium (Fig. 3A). The different behavior of hus1Δ and liz1Δ cells is consistent with the function of Hus1 in the replication checkpoint pathway and supports the previous observation that the replication checkpoint functions normally in liz1− cells (28).
A careful analysis of the kinetics of cell cycle progression in these experiments suggested to us that septation proceeded more slowly in liz1− cells. When HU was added to asynchronous wild-type cell cultures, the septation index dropped from about 15 to 0%, reflecting the fact that the cells have stopped cell cycle progression. In contrast, when HU was added to an asynchronous culture of checkpoint-deficient hus1Δ cells, they failed to arrest the cell cycle, and septated cells accumulated from approximately 15 to over 70%, because the septum cuts through the nucleus leading to cell death (the terminal phenotype). Interestingly, in media containing a low pantothenate concentration, logarithmically growing cultures of liz1− cells possess a twofold higher content of septated cells prior to treatment with HU, indicating that septation proceeds more slowly in these mutants. Consistent with the interpretation that liz1− cells spend an increased length of time septating, the septation index of liz1Δ mutants decreased more slowly in the presence of HU than in wild-type cells and leveled off at around 20% at later time points (Fig. 3B), reflecting the terminal phenotype. All the septation-related phenotypes were suppressed to wild-type levels by the addition of high pantothenate concentrations to the growth medium, demonstrating a direct link to pantothenate import.
We next performed FACS analysis to determine the DNA content of wild-type cells and liz1Δ mutants (Fig. 3C). In the absence of HU, wild-type cells possessed a 2C DNA content. This is expected for an asynchronous S. pombe culture. In contrast to wild-type cells, the liz1Δ culture contained cells with a 4C DNA content (Fig. 3C). This 4C peak is consistent with the idea that the lack of pantothenate slows septation. Because septation is slow, the daughter cells remain attached for longer. During this time they each undergo S phase and reach a 2C DNA content. The fact that two 2C cells remain attached thus explains the appearance of a 4C peak in FACS analysis. As predicted, we also observed that high panothenate supplementation increased the 2C peak of liz1Δ cultures at the expense of the 4C peak (Fig. 3C). Upon the addition of HU, liz1Δ cells started to arrest in G1, with a 1C DNA content consistent with a functional checkpoint pathway (Fig. 3C). In contrast to wild-type cells, liz1Δ mutant cultures took longer to exhibit a peak of cells with 1C DNA content by FACS analysis. This is again consistent with a delay in cell cycle progression during septation, since two 1C cells joined by a septum will migrate as a 2C particle in this analysis. High concentrations of pantothenate restored normal arrest kinetics, supporting the model that pantothenate is required not only for a normal mitosis in the presence of HU (27) but also for normal progression of septation (Fig. 3C).
The growth defects of S. pombe in the absence of pantothenate can be overcome with uracil, ureidopropionate, and β-alanine.
To this point, we have demonstrated that a lack of pantothenate uptake correlates with a slow septation rate and an increase in the number of cut cells upon inhibition of RNR. In addition, we have provided evidence that pantothenate can be synthesized from β-alanine in the absence of pantothenate transport. We wondered which metabolic pathway allowed S. pombe cells to produce β-alanine and if this knowledge might provide us with an explanation for the impact of pantothenate on the cell division cycle and HU sensitivity of liz1Δ mutants. For S. cerevisiae, it is known that β-alanine is an intermediate in the degradation of polyamines and pantothenate biosynthesis is driven by spermine degradation (40). In E. coli, β-alanine is derived from decarboxylation of l-aspartate (8). In Saccharomyces kluyveri, β-alanine production is driven by the degradation of uracil, and two of the gene products involved in this pathway have been identified (14, 16).
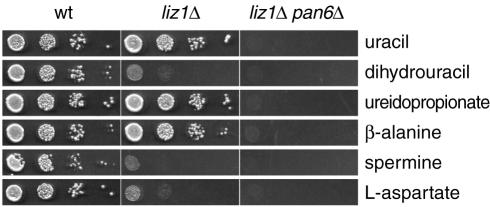
By growing liz1Δ cells in high concentrations of potential β-alanine precursors, we tested which potential pathway might operate to produce β-alanine in S. pombe. Out of the three potential precursors tested (spermine, l-aspartate, and uracil), only uracil equaled β-alanine in promoting the growth of the liz1Δ strain (Fig. 4). Stimulation of growth by uracil strictly depended on the pan6+ gene, indicating that uracil is the endogenous source of pantothenate in S. pombe cells. We also tested dihydrouracil and ureidopropionate, two known β-alanine precursors in S. kluyveri (15). Whereas dihydrouracil was not effective in supporting the growth of liz1Δ cells, ureidopropionate was very effective and its stimulatory effect was also dependent on pan6+ (Fig. 4).
FIG. 4.
S. pombe liz1Δ mutants are complemented by uracil, β-alanine, and ureidopropionate. Wild-type S. pombe cells or cells with deletions in liz1+ or in liz1+ and pan6+ were spotted on plates containing no pantothenate. All cells tested here were ura-positive. The indicated compounds were present at a concentration of 1 mM. Growth was recorded after 3 days at 30°C.
Together, these data strongly indicate that S. pombe cells synthesize pantothenate from uracil via β-alanine. This opens the possibility that the competition for uracil by two essential pathways, the production of nucleotides and the synthesis of pantothenate, is responsible for the cell cycle defects observed in liz1Δ mutants.
DISCUSSION
Liz1 is the S. pombe transporter for pantothenate.
In a previous study (37), the S. cerevisiae gene coding for the plasma membrane pantothenate transporter Fen2p was identified. In this report, we characterize the S. pombe gene liz1+ that is homologous to FEN2. Moynihan and Enoch (28) isolated liz1+ as a mutation that affected cell cycle progression through mitosis when RNR was inhibited. Our complementation experiments and growth assays as well as pantothenate uptake experiments unequivocally establish that Liz1 functions in the transport of pantothenate. Pantothenate transport by Liz1 is saturable, stimulated by d-glucose, and sensitive to uncouplers of the transmembrane proton gradient, strongly suggesting a proton symport mechanism. Taken together, the activities of S. pombe Liz1 and S. cerevisiae Fen2p are highly similar.
Lack of pantothenate uptake affects the cell division cycle.
The characterization of Liz1 as a pantothenate transporter was surprising because a connection between pantothenate transport and specific cell cycle events has not been revealed by previous studies. Here we demonstrate that the lack of pantothenate uptake is indeed responsible for all of the reported phenotypes of liz1Δ mutants and provide a rational framework to explain how this occurs.
The phenotypes of liz1Δ cells include a high percentage of septated cells in the absence of HU (Fig. 3B) and the inability to separate the nucleus (cut phenotype) in the presence of HU (Fig. 3A and reference 27). Both of these phenotypes are reversed when pantothenate is supplemented in concentrations that allow uptake in the absence of Liz1 activity. This demonstrates that the lack of pantothenate transport is responsible for these cell cycle perturbations.
This raises the question of how pantothenate influences septation and mitosis. Pantothenate is an essential metabolite and functions as a building block for the synthesis of CoA. CoA and metabolites derived from it have multiple cellular roles. Fatty acid biosynthesis and elongation depend on malonyl-CoA, which is produced from acetyl-CoA in a reaction catalyzed by acetyl-CoA carboxylase. We have noticed that S. pombe mutants defective in key enzymes required for fatty acid biosynthesis display phenotypes that are related to those observed for liz1Δ cells. These mutants, cut6− and lsd1−, encode acetyl-CoA carboxylase and the α subunit of fatty acid synthase, respectively (31).
cut6− and lsd1− mutants are temperature sensitive and, at the restrictive temperature, produce daughter nuclei of unequal size accompanied by a high degree of chromatin condensation in the smaller daughter nucleus (31). Moreover, lsd1− mutants were shown to complete cytokinesis (septation) in these circumstances, despite the fact that nuclear division has not been completed. This resulted in cut cells, similar to the phenotype described for liz1− mutants. These data strongly suggest that fatty acid metabolism plays a central role in nuclear division and chromatin condensation and that fatty acid metabolism is compromised in liz1Δ cells. Fatty acids are structural components of phospholipids. Thus, cut6−, lsd1−, and liz1− mutants may be defective in the biosynthesis of phospholipids. In S. cerevisiae, the activities of acetyl-CoA carboxylase (encoded by ACC1) and of other proteins involved in lipid metabolism are increased in G1 as a prelude to DNA synthesis (7). Moreover, net phospholipid accumulation occurs specifically during S phase (reviewed in reference 18). Thus, mutations in cut6+, lsd1+, or liz1+ may be deleterious because they interfere with the increased demand for phospholipids, which are required for nuclear division and septation.
Alternatively, mutations in cut6+, lsd1+, or liz1+ may affect the biosynthesis of a specific class of lipids. In S. cerevisiae, fatty acid biosynthesis has a 17-fold higher affinity for malonyl-CoA than does fatty acid elongation (9). Thus, a shortage of malonyl-CoA may predominantly affect the elongation of fatty acids. Evidence in favor of this model comes from conditional S. cerevisiae acc1 mutants. These mutants possess highly aberrant nuclear envelopes, which display expansions of the intermembrane space and accumulation of vesicles between the two membranes (32). Moreover, the acc1 mutants develop large nuclei that do not enter the daughter cells during mitosis (2). The defect in these mutants has been assigned to a reduced abundance of C26 fatty acids on sphingolipids (2, 32). The fact that supplementation of C16 fatty acids does not rescue the growth defect of liz1− mutants (data not shown) may indicate that it is indeed the lack of very-long-chain fatty acids that slows the growth in liz1− mutants. This model, however, cannot be tested directly because exogenous very-long-chain fatty acids are not readily taken up or activated (33).
A metabolic link between uracil catabolism and pantothenate biosynthesis.
Whereas mutations in cut6+ or lsd1+ have severe effects on the nuclear morphology by themselves, liz1Δ mutants display their most drastic phenotypes only when RNR is inhibited (Fig. 3 and reference 28). This indicates that there are metabolic links between the two unrelated processes, pantothenate uptake and reduction of ribonucleotides.
Our experiments led to the discovery that S. pombe cells are capable of deriving pantothenate from the breakdown of uracil (Fig. 2 and 4). This pathway of pantothenate biosynthesis depends on the presence of the pan6+ gene (Fig. 2 and 4), and thus proceeds via β-alanine. We find that uracil, ureidopropionate, and β-alanine are able to substitute for pantothenate in S. pombe (Fig. 4). Ureidopropionate and β-alanine are part of a catabolic pathway in S. kluyveri that provides β-alanine (15). Two enzymes of this pathway have been identified: 5,6-dihydropyrimidine amidohydrolase (encoded by PYD2 [14]) and β-alanine synthase (encoded by PYD3 [16]). S. pombe lacks an obvious ortholog of PYD2, consistent with our finding that dihydrouracil is not able to substitute for pantothenate (Fig. 4). S. pombe also has no ortholog of PYD3, but our experiments showed that the cells were able to grow in the presence of ureidopropionate (Fig. 4). It is not clear if ureidopropionate is converted to β-alanine by spontaneous hydrolysis (8) or if the identification of the S. pombe β-alanine synthase is prevented by the sequence diversity known to occur in eukaryotic β-alanine synthases (16, 24).
Nonetheless, our data support the existence of a novel metabolic pathway in S. pombe that leads from uracil to β-alanine and does not have dihydrouracil as an intermediate. This pathway may not be unique to S. pombe. When LaRue and Spencer (20) analyzed 123 yeast species for their ability to utilize different pyrimidines as their sole source of nitrogen, they found that 69 strains were able to grow on uracil. Of these, 22 were unable to utilize dihydrouracil, demonstrating that in these strains dihydrouracil was not an intermediate of uracil catabolism.
Although the precise pathway of uracil degradation in S. pombe remains to be established, our data clearly show that uracil is a precursor of pantothenate. Thus, when the plasma membrane pantothenate transporter Liz1 is defective, uracil biosynthesis has to satisfy the demand of two essential processes: CoA synthesis and deoxyribonucleotide production via RNR (Fig. 5). S. pombe cells are unable to synthesize sufficient uracil to support both pathways (Fig. 2 and 4). We speculate that the concentrations of CTP and UTP increase in the presence of HU, which blocks RNR activity. This may lead to a feedback inhibition of aspartate transcarbamoylase, the committed step in the synthesis of pyrimidines which is known to be allosterically regulated by CTP and UTP (13, 23, 41). Thus, HU may lead to a reduction of de novo production of uracil (Fig. 5) and a further drop in the CoA concentrations in liz1Δ cells.
FIG. 5.
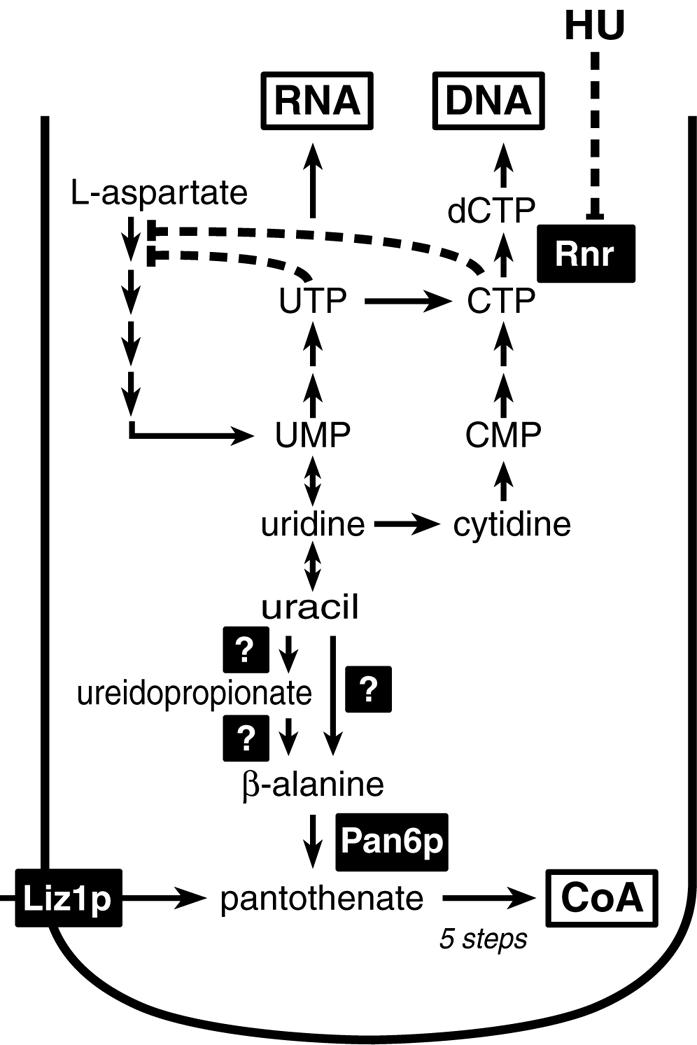
A model illustrating uracil biosynthesis, generation of nucleotides, and degradation of uracil to β-alanine for pantothenate and CoA biosynthesis. The model depicts the roles of uracil as a precursor of nucleotides for RNA and DNA synthesis and as a precursor of CoA. It is not known if ureidopropionate is a catabolite of uracil, but both uracil and ureidopropionate lead to the formation of β-alanine, which is then converted to pantothenate by the action of Pan6. CoA is synthesized from pantothenate in five enzymatic reactions. The deletion of liz1+ abolishes pantothenate uptake and diverts more uracil to the synthesis of CoA. In the presence of HU, RNR is inhibited (dashed line) and CTP and UTP accumulate. Both CTP and UTP are likely to block uracil biosynthesis (two dashed lines) of aspartate transcarbamoylase, the first enzyme in the synthesis of pyrimidines from l-aspartate, by feedback inhibition.
Summary.
All three phenotypes of liz1Δ cells, slow growth, delayed septation, and aberrant mitosis in the presence of HU, are reversed by the addition of high pantothenate concentrations to the growth medium. Since pantothenate is required for CoA synthesis, the lack of Liz1 is expected to reduce the cellular CoA pool, rendering cells dependent on CoA synthesis from endogenous sources, a pathway requiring uracil degradation. The use of uracil for both CoA generation and deoxyribonucleotide synthesis by RNR provides the explanation for the appearance of aberrant mitosis in presence of HU. HU blocks RNR activity, and the increase in RNR substrates slows uracil production. As a result, the CoA pool will drop below a critical threshold, resulting in a shortage of fatty acids during mitosis. The pantothenate transporter Liz1 thus provides a surprising example that shows how substrate transport across the plasma membrane can influence cellular events in many as yet unexpected ways.
Acknowledgments
The experiments shown in Fig. 1 were initiated and performed by J.S. in the lab of N.S. in Erlangen, Germany. T.C. performed the experiments shown in Fig. 3 in the lab of A.M.C. in Sussex, United Kingdom. We thank Sabine Strahl (Universität Regensburg, Germany) for providing S. pombe strains and plasmids. W. Tanner (Universität Regensburg), Eckhard Schweizer and Uschi Hoja (Universität Erlangen-Nürnberg, Germany), Sepp Kohlwein (Universität Graz, Austria), and W. Hunter White (Eli Lilly Corp., Greenfield, Ill.) are acknowledged for many helpful discussions. Excellent technical assistance was provided by Manuela Reich, Petra Schitko, and Sabine Laberer.
This work was supported by the DFG (grants SA382/4, STO434-1/2, and SFB521-C7), the MRC (grant G0001129), and by a grant from the Bavarian Ministry for Science, Research and Arts.
REFERENCES
- 1.Agostoni Carbone, M. L., L. Panzeri, M. Muzi Falconi, C. Carcano, P. Plevani, and G. Lucchini. 1992. Nucleotide sequence of 9.2 kb left of CRY1 on yeast chromosome III from strain AB972: evidence for a Ty insertion and functional analysis of open reading frame YCR28. Yeast 8:805-812. [DOI] [PubMed] [Google Scholar]
- 2.Al-Feel, W., J. C. DeMar, and S. J. Wakil. 2003. A Saccharomyces cerevisiae mutant strain defective in acetyl-CoA carboxylase arrests at the G2/M phase of the cell cycle. Proc. Natl. Acad. Sci. USA 100:3095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.al-Khodairy, F., T. Enoch, I. M. Hagan, and A. M. Carr. 1995. The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J. Cell Sci. 108:475-486. [DOI] [PubMed] [Google Scholar]
- 4.al-Khodairy, F., E. Fotou, K. S. Sheldrick, D. J. Griffiths, A. R. Lehmann, and A. M. Carr. 1994. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell 5:147-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr, A. M. 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair (Amsterdam) 1:983-994. [DOI] [PubMed] [Google Scholar]
- 6.Caspari, T., and A. M. Carr. 1999. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie 81:173-181. [DOI] [PubMed] [Google Scholar]
- 7.Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway, L. Wodicka, T. G. Wolfsberg, A. E. Gabrielian, D. Landsman, D. J. Lockhart, and R. W. Davis. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2:65-73. [DOI] [PubMed] [Google Scholar]
- 8.Cronan, J. E., Jr. 1980. β-Alanine synthesis in Escherichia coli. J. Bacteriol. 141:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dittrich, F., D. Zajonc, K. Huhne, U. Hoja, A. Ekici, E. Greiner, H. Klein, J. Hofmann, J. J. Bessoule, P. Sperling, and E. Schweizer. 1998. Fatty acid elongation in yeast-biochemical characteristics of the enzyme system and isolation of elongation-defective mutants. Eur. J. Biochem. 252:477-485. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, R. J., and A. M. Carr. 1997. Analysis of radiation-sensitive mutants of fission yeast. Methods Enzymol. 283:471-494. [DOI] [PubMed] [Google Scholar]
- 11.Enoch, T., A. M. Carr, and P. Nurse. 1992. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 6:2035-2046. [DOI] [PubMed] [Google Scholar]
- 12.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebhart, J. C., and A. C. Pardee. 1962. The enzymology of control by feedback inhibition. J. Biol. Chem. 237:891-896. [PubMed] [Google Scholar]
- 14.Gojkovic, Z., K. Jahnke, K. D. Schnackerz, and J. Piskur. 2000. PYD2 encodes 5,6-dihydropyrimidine amidohydrolase, which participates in a novel fungal catabolic pathway. J. Mol. Biol. 295:1073-1087. [DOI] [PubMed] [Google Scholar]
- 15.Gojkovic, Z., S. Paracchini, and J. Piskur. 1998. A new model organism for studying the catabolism of pyrimidines and purines. Adv. Exp. Med. Biol. 431:475-479. [DOI] [PubMed] [Google Scholar]
- 16.Gojkovic, Z., M. P. Sandrini, and J. Piskur. 2001. Eukaryotic beta-alanine synthases are functionally related but have a high degree of structural diversity. Genetics 158:999-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe, p. 395-446. In R. C. King (ed.), Handbook of genetics. Plenum Press, New York, N.Y.
- 18.Jackowski, S. 1996. Cell cycle regulation of membrane phospholipid metabolism. J. Biol. Chem. 271:20219-20222. [DOI] [PubMed] [Google Scholar]
- 19.Kostrub, C. F., F. al-Khodairy, H. Ghazizadeh, A. M. Carr, and T. Enoch. 1997. Molecular analysis of hus1+, a fission yeast gene required for S-M and DNA damage checkpoints. Mol. Gen. Genet. 254:389-399. [DOI] [PubMed] [Google Scholar]
- 20.LaRue, T. A., and J. F. Spencer. 1968. The utilization of purines and pyrimidines by yeasts. Can. J. Microbiol. 14:79-86. [DOI] [PubMed] [Google Scholar]
- 21.Liang, D. T., J. A. Hodson, and S. L. Forsburg. 1999. Reduced dosage of a single fission yeast MCM protein causes genetic instability and S phase delay. J. Cell Sci. 112:559-567. [DOI] [PubMed] [Google Scholar]
- 22.Llorente, B., and B. Dujon. 2000. Transcriptional regulation of the Saccharomyces cerevisiae DAL5 gene family and identification of the high affinity nicotinic acid permease TNA1 (YGR260w). FEBS Lett. 475:237-241. [DOI] [PubMed] [Google Scholar]
- 23.Lollier, M., L. Jaquet, T. Nedeva, F. Lacroute, S. Potier, and J. L. Souciet. 1995. As in Saccharomyces cerevisiae, aspartate transcarbamoylase is assembled on a multifunctional protein including a dihydroorotase-like cryptic domain in Schizosaccharomyces pombe. Curr. Genet. 28:138-149. [DOI] [PubMed] [Google Scholar]
- 24.Lundgren, S., Z. Gojkovic, J. Piskur, and D. Dobritzsch. 2003. Yeast beta-alanine synthase shares a structural scaffold and origin with dizinc-dependent exopeptidases. J. Biol. Chem. 278:51851-51862. [DOI] [PubMed] [Google Scholar]
- 25.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-485. [DOI] [PubMed] [Google Scholar]
- 26.McFarlane, R. J., A. M. Carr, and C. Price. 1997. Characterisation of the Schizosaccharomyces pombe rad4/cut5 mutant phenotypes: dissection of DNA replication and G2 checkpoint control function. Mol. Gen. Genet. 255:332-340. [DOI] [PubMed] [Google Scholar]
- 27.Merkel, W. K., and B. P. Nichols. 1996. Characterization and sequence of the Escherichia coli panBCD gene cluster. FEMS Microbiol. Lett. 143:247-252. [DOI] [PubMed] [Google Scholar]
- 28.Moynihan, E. B., and T. Enoch. 1999. Liz1p, a novel fission yeast membrane protein, is required for normal cell division when ribonucleotide reductase is inhibited. Mol. Biol. Cell 10:245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mumberg, D., R. Müller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelissen, B., R. De Wachter, and A. Goffeau. 1997. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 21:113-134. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh, S., K. Takahashi, K. Nabeshima, Y. Yamashita, Y. Nakaseko, A. Hirata, and M. Yanagida. 1996. Aberrant mitosis in fission yeast mutants defective in fatty acid synthetase and acetyl CoA carboxylase. J. Cell Biol. 134:949-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneiter, R., M. Hitomi, A. S. Ivessa, E. V. Fasch, S. D. Kohlwein, and A. M. Tartakoff. 1996. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol. Cell. Biol. 16:7161-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneiter, R., and S. D. Kohlwein. 1997. Organelle structure, function, and inheritance in yeast: a role for fatty acid synthesis? Cell 88:431-434. [DOI] [PubMed] [Google Scholar]
- 34.Stewart, E., and T. Enoch. 1996. S-phase and DNA-damage checkpoints: a tale of two yeasts. Curr. Opin. Cell Biol. 8:781-787. [DOI] [PubMed] [Google Scholar]
- 35.Stolz, J. 2003. Isolation and characterization of the plasma membrane biotin transporter from Schizosaccharomyces pombe. Yeast 20:221-231. [DOI] [PubMed] [Google Scholar]
- 36.Stolz, J., U. Hoja, S. Meier, N. Sauer, and E. Schweizer. 1999. Identification of the plasma membrane H+-biotin symporter of Saccharomyces cerevisiae by rescue of a fatty acid-auxotrophic mutant. J. Biol. Chem. 274:18741-18746. [DOI] [PubMed] [Google Scholar]
- 37.Stolz, J., and N. Sauer. 1999. The fenpropimorph resistance gene FEN2 from Saccharomyces cerevisiae encodes a plasma membrane H+-pantothenate symporter. J. Biol. Chem. 274:18747-18752. [DOI] [PubMed] [Google Scholar]
- 38.Truernit, E., J. Schmid, P. Epple, J. Illig, and N. Sauer. 1996. The sink-specific and stress-regulated Arabidopsis STP4 gene: enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 8:2169-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 40.White, W. H., P. L. Gunyuzlu, and J. H. Toyn. 2001. Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of beta-alanine production from spermine. J. Biol. Chem. 276:10794-10800. [DOI] [PubMed] [Google Scholar]
- 41.Wild, J. R., S. J. Loughrey-Chen, and T. S. Corder. 1989. In the presence of CTP, UTP becomes an allosteric inhibitor of aspartate transcarbamolyase. Proc. Natl. Acad. Sci. USA 86:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]