Abstract
To identify new nonessential genes that affect genome integrity, we completed a screening for diploid mutant Saccharomyces cerevisiae strains that are sensitive to ionizing radiation (IR) and found 62 new genes that confer resistance. Along with those previously reported (Bennett et al., Nat. Genet. 29:426-434, 2001), these genes bring to 169 the total number of new IR resistance genes identified. Through the use of existing genetic and proteomic databases, many of these genes were found to interact in a damage response network with the transcription factor Ccr4, a core component of the CCR4-NOT and RNA polymerase-associated factor 1 (PAF1)-CDC73 transcription complexes. Deletions of individual members of these two complexes render cells sensitive to the lethal effects of IR as diploids, but not as haploids, indicating that the diploid G1 cell population is radiosensitive. Consistent with a role in G1, diploid ccr4Δ cells irradiated in G1 show enhanced lethality compared to cells exposed as a synchronous G2 population. In addition, a prolonged RAD9-dependent G1 arrest occurred following IR of ccr4Δ cells and CCR4 is a member of the RAD9 epistasis group, thus confirming a role for CCR4 in checkpoint control. Moreover, ccr4Δ cells that transit S phase in the presence of the replication inhibitor hydroxyurea (HU) undergo prolonged cell cycle arrest at G2 followed by cellular lysis. This S-phase replication defect is separate from that seen for rad52 mutants, since rad52Δ ccr4Δ cells show increased sensitivity to HU compared to rad52Δ or ccr4Δ mutants alone. These results indicate that cell cycle transition through G1 and S phases is CCR4 dependent following radiation or replication stress.
A failure to maintain genome stability following exposure to environmental agents that damage DNA is generally considered to be an early event in cancer progression. This is supported by observations that cancers (such as those of the breast and colon) are associated with defects in genes that normally maintain genomic integrity through DNA repair, recombination, and/or checkpoint functions (20, 38). Also, many physical and chemical agents that damage DNA, including ionizing radiation (IR), are carcinogens that induce a wide array of genome-destabilizing DNA lesions (62, 66). For IR, DNA double-strand breaks (DSBs) are thought to be the most biologically relevant lesion since their persistence appears to be the primary cause of genetic instability as well as lethality (7, 11). The inability to repair DSBs can lead to deletions, gross chromosomal rearrangements, and aneuploidy (18). To avoid the destabilizing effects of IR-induced DSBs, eukaryotes have evolved highly conserved DNA repair and checkpoint pathways that maintain genomic integrity through the accurate repair of DSB damage (41).
The yeast Saccharomyces cerevisiae has served as an important model organism for the identification of genetic controls associated with DNA repair and checkpoint functions. Most of the gene products involved in repair of DSBs in humans were first identified in yeast (58). The repair of DSBs in yeast primarily involves the RAD52 epistasis group of recombinational repair genes (26), while nonhomologous end joining appears to play only a minor role in the repair of IR-induced DSBs (42). Haploid yeast are extremely IR sensitive (IRS) in G1, since they lack a homologue for use as a template for the repair of IR-induced DSBs (15). Recombinational repair (using the newly replicated sister chromatid as a template) of DSBs in haploid cells can only occur in the S or G2 phase of the cell cycle. Conversely, diploid cells are very IR resistant (since recombinational repair can occur throughout the cell cycle using the homologous chromosome); however, mutations of RAD52 render diploid cells as sensitive to the killing effects of IR as haploid cells in G1 (59).
Yeast have efficient mechanisms for the detection and signaling of DNA damage that result in the transcriptional activation of damage-inducible genes (DIN) as well as the arrest of cells at specific points in the cell cycle (60, 71). Damage-induced cell cycle arrest is regulated by a large number of checkpoint genes that monitor DNA integrity in the G1/S, S, and G2/M phases of the cell cycle (24). In the presence of DSBs or replication stress, cells detect the damage and (through transducing pathways) signal an arrest of cell cycle progression. Most checkpoint genes do not participate directly in the repair of DSBs. Instead, their effects are indirect in that they allow additional time for recombinational repair to occur. Following damage-induced cell cycle arrest, another group of checkpoint-associated genes is required for cells to reenter or adapt back into the cell cycle (8, 10, 65). Defects in checkpoint adaptation result in prolonged cell cycle arrest following DNA damage. Prolonged cell cycle arrest can also occur when DNA damage persists due to a defect in a repair gene such as rad52, so care must be taken in describing a gene as an adaptation rather than a repair gene. Since loss of function in either checkpoint or adaptation genes can result in sensitivity to IR-induced damage, there appears to be an optimal time window during the cell cycle when repair must be completed and normal cell cycling must be resumed.
The availability of haploid and diploid yeast with a complete set of deletion mutations in nonessential genes has enabled a number of successful genome-wide screenings to be performed (5, 6, 8, 12-14, 16, 39, 56, 73). To identify new recombination or checkpoint genes that are required for the maintenance of genetic integrity following induction of DSBs, Bennett et al. previously examined 3,670 nonessential genes for the consequences of diploid homozygous mutations for growth and/or lethality following a single acute dose of IR (8). A total of 107 new genes that were required for radiation resistance were initially found. Many of these appear to affect replication, recombination, and checkpoint functions, and >50% share homology with human genes (including 17 implicated in cancer).
In this study, we report the completion of the genome-wide screening of nonessential genes and identify a total of 169 new genes that are required for radiation toleration. Many (35) of the new IR resistance genes interact genetically and/or physically in a network with the transcription factor Ccr4, which is a core component of the CCR4-NOT (CNOT) and RNA polymerase-associated factor 1-CDC73 (PAF) transcriptional complexes. We show that deletions of genes within the Ccr4 transcription complex render cells sensitive to the lethal effects of IR as diploids but not as haploids. Deletion of two core members (CCR4 and DHH1) of the CNOT complex does not directly affect recombination; instead, these mutants show reduced viability in G1 following IR due to a defect in G1 checkpoint transition. Moreover, ccr4 and rad9 mutants were found to be within the same checkpoint epistasis group and ccr4Δ cells demonstrate a prolonged IR-induced G1 arrest that is RAD9 dependent. Since ccr4Δ, pop2Δ, and dhh1Δ cells are also sensitive to the S-phase-specific agent hydroxyurea (HU), these results suggest that (following checkpoint arrest in G1) CNOT functions to promote cell cycle transition from G1 into S phase with effects that also extend into S phase. Furthermore, ccr4Δ cells that transit S phase in the presence of HU show prolonged arrest as large budded cells followed by cellular lysis, suggesting a replication defect. The synthetic slow growth and hypersensitivity to HU exhibited by rad52Δ ccr4Δ cells further suggests an S-phase replication defect in ccr4Δ cells that is RAD52 independent.
MATERIALS AND METHODS
Yeast strains and gamma-ray screening.
Deletions of individual nonessential genes (or open reading frames [ORFs]) were performed with MATa (BY4741) and MATα (BY4742) haploid S. cerevisiae strains as part of the Saccharomyces Gene Deletion Project. The diploid deletion strains (1,076 mutants) were purchased in 96-well microtiter dishes from Research Genetics (release II). Strains were screened for radiation and chemical sensitivity as previously described (8). Sensitivity to doxorubicin (dissolved in dimethyl sulfoxide; 20 mg/ml was then added to warm yeast extract-peptone-dextrose [YPD] agar) was determined at a final concentration of 50 μg/ml. YPD plates were immediately irradiated with 80 krads of gamma irradiation from a 137Cs source (J.L. Sheppard & Assoc., San Fernando, Calif.) at a dose rate of 2.4 krad/min or 60 J of UV light/m2 (dose rate of 1 J/m2/s). These plates (along with unirradiated control plates) were examined after 24 and 48 h of growth at 30°C. Putative gamma-ray-sensitive mutants were confirmed by (i) plating serial dilutions of the strains grown for 48 h at 30°C to YPD and again exposing them to 80 krads and (ii) using survival curve analysis as previously described (8). Briefly, following 3 to 5 days of growth at 30°C, relative survival levels were determined as the ratio of viable CFU levels on gamma-irradiated versus unirradiated plates. Haploid deletion strains used to derive the IRS diploids were also obtained from Research Genetics and individually examined for sensitivity to IR by survival curve analysis of logarithmically growing cultures.
Diploid double-deletion strains were constructed as follows. A haploid MATα rad9Δ::URA3 deletion strain was constructed by transforming plasmid pRR330 (cut with SalI and EcoRI) into BY4742. Putative deletions were identified by enhanced sensitivity of Ura+ transformants to UV and gamma irradiation. Successful deletion of RAD9 was confirmed by PCR using genomic template DNA obtained from an isolated Ura+ colony and the appropriate RAD9 flaking primer and an internal URA3 primer (sequences available upon request). The rad9Δ strain was mated to either a MATa dhh1Δ::G418R or a MATa ccr4Δ::G418R strain constructed in the isogenic BY4741 background (Research Genetics). The heterozygote diploids were selected on synthetic complete (SC) glucose-uracil plates containing G418 (200 μg/ml). Through the use of standard genetic techniques, the heterozygotes were sporulated and 4 spore asci were dissected to obtain haploid rad9Δ dhh1Δ and rad9Δ ccr4Δ segregants of each mating type. MATa and MATα haploid double-deletion strains were mated, and diploids were visually identified by zygote formation during mating. Diploidy of the double-deletion strains was confirmed using appropriate mating type tester strains. The rad52Δ ccr4Δ diploid strain was constructed in a similar manner by crossing a MATα rad52Δ::LEU2 disruption in BY4742 (obtained from K. Lewis) to the MATa ccr4Δ::G418R strain described above and selecting the heterozygous diploid on SC glucose medium lacking leucine (SC GLU−LEU) and containing G418. Sporulation, selection of haploid double-mutant segregants, and construction of the diploid double mutant were similar to the procedures described above. The rad6Δ ccr4Δ diploid strain was also prepared in a manner similar to that used for the rad52Δ ccr4Δ diploid strain. Initially, we created a haploid MATα rad6::LEU2 deletion by transforming BY4742 with the deletion plasmid pDG315 (obtained from W. Xiao) cut with BamHI and HindIII. Successful deletion of RAD6 was confirmed by PCR using genomic template DNA obtained from an isolated Leu+ colony, the appropriate RAD6 flaking primer, and an internal LEU2 primer (sequences available upon request). The resulting rad6Δ::LEU2 MATα strain was also shown to be sensitive to radiation. This rad6 strain was mated with the MATa ccr4Δ::G418R strain described above, and heterozygote diploids were selected on SC GLU−LEU containing G418. Sporulation, selection of haploid double-mutant segregants, and construction of the diploid double mutant were similar to the procedures described above. The ccr4Δ his3Δ1 diploid strain was obtained by mating haploid ccr4Δ::G418R Ura+ his− or ccr4Δ::G418R Leu+ his− segregants from the sporulations of diploid rad9/RAD9 ccr4/CCR4 or rad52/RAD52 ccr4/CCR4 heterozygotes.
Targeted recombination at his3Δ1.
Cells were grown to logarithmic phase in YPD liquid culture and then transformed (as described previously) with 200 ng of pRS315 and 1 μg of a partial HIS3 PCR fragment that spanned the his3Δ1 deletion (8). PCR amplification of HIS3 produced a 729-bp fragment with an overlap of 225 bp 5′ and 317 bp 3′ of the his3Δ1 deletion. A functional HIS3 gene could only occur by targeted integration of the amplified PCR fragment into the genomic his3Δ1 sequences following transformation. Targeted integration efficiencies were determined by calculating the ratio of the colony-forming abilities of wild-type (WT) and deletion strains on SC medium lacking histidine. Ratios were then corrected for the relative transformation efficiency of circular plasmid DNA (pRS315; LEU2-selectable marker on SC GLU-LEU).
Zymocin production and killer eclipse assay.
WT and deletion strains were exposed (using a dilution plating technique described above) to zymocin on plates. Briefly, cells were grown for 2 days in liquid YPD (filter sterilized) in 96-well plates and serial fivefold dilutions were made in water. Cells (∼2 μl of each dilution) were then transferred to YPD and YPD-zymocin plates using a 48-pin replica-plating device. YPD plates containing zymocin were made by growing Kluyveromyces lactis strain AWJ137 on filter-sterilized liquid YPD for 2 days at room temperature. Briefly, 2 parts of a sterile YPD filtrate of conditioned medium from the 48-h culture of the K. lactis strain were mixed with 1 part of 3× agar made in fresh 1× YPD. Plates were immediately poured and allowed to solidify at room temperature. The killer eclipse assay using the K. lactis strains AWJ137 (zymocin producing) and NK40 (zymocin nonproducing) was performed on YPD plates as previously described (40).
Irradiation of synchronized cells.
Logarithmically growing cells (∼107 cells/ml) were exposed to benomyl (a 10 mg/ml solution of benomyl dissolved in dimethyl sulfoxide was added to cells in 5 ml of YPD to give a final concentration of 40 μg/ml) for a total of 4 h with vigorous shaking at 30°C. Exposure to benomyl by this method resulted in the arrest of ∼90% of logarithmically growing cells in G2, with no decrease in survival. Arrested cells were pelleted by low-speed centrifugation and irradiated (80 krads) following suspension of cells in water containing benomyl (40 μg/ml) as described above. Unirradiated and irradiated benomyl-arrested cells were diluted in water and plated to YPD as described above. Cells arrested by benomyl were released from the block by resuspending pelleted cells in liquid YPD (no benomyl) and grown at 30°C with vigorous shaking for 45 min. This release was asynchronous such that ∼50% of cells entered into G1 before the onset of S phase (i.e., in previous experiments newly budded cells were observed at 1 h following resuspension of benomyl arrested cells in fresh YPD). Following release from the block (after 45 min of YPD growth), cells were pelleted, irradiated (80 krads) in water, and plated to YPD as described above.
Checkpoint analysis.
Position in the cell cycle can be morphologically distinguished in yeast (unbudded cells are in G1; the beginning of S phase is marked by bud emergence; G2 cells are large budded). To examine the checkpoint transition from single (G1) cells into budded cells and microcolonies, logarithmically growing cells were plated to YPD, YPD-HU (200 mM), or YPD followed by exposure to 8 krads of IR. The time of transition from G1 to S phase was determined by marking the positions of cell fields (60 to 150 cells) from each strain and repeatedly photographing the same cells at hourly intervals with a Singer MSM dissecting microscope as previously described (8). Alternatively, single G1 cells were plated and repositioned into a grid pattern within one field of view. Cells were monitored hourly to determine the precise transition times for G1 to S phase (single cells to small budded cells) and G2 to M phase (large budded cells to microcolonies of 3 or more cells).
RESULTS
Genome-wide screening reveals 169 new genes that are required for toleration of gamma-ray damage in diploid yeast. We completed the genome-wide screening of the yeast diploid deletion strain collection for sensitivity to a single acute dose (80 krads) of IR (reference 8 and this study). As previously described (8), sensitivity in this screening system may be determined by decreased survival and/or slower postirradiation growth rate compared to that of the WT strain. The survival response is ascertainable only with additional tests (see below). The first study examined 3,670 genes. This study completes the genome-wide screening of nonessential genes. The remaining gene deletion mutants (1,076) were screened for sensitivity to IR and a number of other DNA-damaging agents, including UV light, methyl methanesulfonate (MMS), HU, bleomycin, camptothecin, and doxorubicin (Table 1; see Table S1 in the supplemental material).
TABLE 1.
| UV | Bleo | MMS | HU | Camp | Doxo | Functional grouping(s) | Gamma-ray-sensitive gene-ORF deletion(s) (n = 65) |
|---|---|---|---|---|---|---|---|
| S | S | S | S | S | R | Bud site selection | BUD32 |
| S | S | S | S | R | S | Recombination, transcription | RLR1, NOT4, NOT5, SRB5, MDM20 |
| S | S | S | S | R | R | Cytoskeleton, sporulation | SRV2 |
| S | S | S | R | R | S | Transcription, DNA recombination | YAF9, SLX8 |
| S | S | S | R | R | R | Transcription | BDF1 |
| S | S | R | S | R | S | α-1,6-Mannosyltransferase | OCH1 |
| S | S | R | R | R | S | Trehelose synthesis | TPS1 |
| S | S | R | R | R | R | Checkpoint | DDC1, EAP1 |
| S | R | S | R | S | S | ? | YJL184Wd |
| S | R | R | R | R | S | mRNA processing | LSM7 |
| S | R | R | R | R | R | Checkpoint | RAD24 |
| R | S | S | S | S | S | 60S ribosomal protein subunit | RPL31A |
| R | S | S | S | R | S | Vacuolar organization-biogenesis | FAB1, YGL218W |
| R | S | S | R | S | S | ? | YBR100W |
| R | S | S | R | S | R | Recombination | RAD59 |
| R | S | S | R | R | S | DNA repair, mitochondrial | SAE2, MDM10 |
| R | S | R | S | R | S | ? (Diverse functions) | ATP4, PLC1, VPS33, MAP1, YDJ1d |
| R | S | R | R | R | S | Transcription, protein synthesis | GLO3, ADA2, GCN5, RPL34B, TIF4631, YDL041W, YDR532C, YBR077C |
| R | S | R | R | R | R | Checkpoint, DNA repair | RDH54, PSO2, SCO1, YML036W |
| R | R | S | S | R | S | Elongated bud morphologyc | YJL075C |
| R | R | S | R | S | S | DNA repair | MMS4 |
| R | R | S | R | R | R | Chromatin | NAT1, YLR358C |
| R | R | R | S | R | S | Transcription | SPT20, TUP1 |
| R | R | R | S | R | R | Actin cytoskeleton | ARP8, ARP5 |
| R | R | R | R | R | S | Chromatin, mitochondrial, transcription | ASM4, HMO1, MBP1, ATP2, MRP10, DEG1, ADE12, BMH1, MDJ1, YGR272C |
| R | R | R | R | R | R | ? (Diverse functions) | DOT1, RIM1, BRE1, TPS2, YDR417C, YNL080C |
Approximately 1,100 diploid deletion strains were simultaneously screened for sensitivity to seven physical and chemical agents. Relative sensitivity levels were determined by dilution plating, and some strains were more sensitive than others (see Table S1 in the supplemental material). Strains sensitive to IR are listed (cross-sensitivity to other agents is indicated). Several of the genes have multiple functions. Eight genes (indicated with boldface characters) have been previously characterized as participating in DNA repair or checkpoint functions.
UV, 60 J/m2 (sensitivity defined as described above); Bleo, 4 ug of bleomycin/ml (sensitivity defined as described above); MMS, 2 mM methyl methane sulfonate (sensitivity defined as described above); HU, 100 mM hydroxyurea (sensitivity defined as described above); Camp, 10 ug of camptothecin/ml in 25 mM HEPES buffer, pH 7.2 (sensitivity defined as described above); Doxo, 50 ug of doxorubicin (sensitivity defined as described above).
APQ13 enhanced apical growth detected by quantitative analyses (Yoshikazu Ohya, personal communication).
Poor survival upon refrigeration at 4°C on YPD plates; unable to be grown from frozen stocks at Research Genetics.
Among the members of this collection, we have identified a further 65 diploid deletion strains that are IRS. These were confirmed to be IRS by replica plating serially diluted stationary-phase cells to YPD plates and exposing these to 80 krads as previously described (8). Of these, 57 have not been previously associated with sensitivity to DNA damage (Table 1; see Table S1 in the supplemental material). Therefore, 164 new genes (57 in this study plus 107 previously described) that are required for the toleration of IR damage have been identified. In addition, we identified in the combined screenings all 31 of the expected, well-characterized recombination, checkpoint, and postreplication repair genes (such as RAD52, RAD9, and RAD6) that are required for radiation resistance. Five gene deletion strains (pop2, dbf2, not3, paf1, and elm1) were not detected as radiation sensitive in the initial screening. On the basis of their described physical or genetic network interactions with identified IRS gene deletions, however, these mutants were predicted to be IRS. Subsequent retesting confirmed them to be IRS. In this genome-wide screening, therefore, 4.2% (200/4,746) of the nonessential yeast genes were found to contribute to the toleration of IR damage.
Of the 169 new genes, 131 correspond to genes for which a function or genetic role has been suggested on the basis of experimental evidence (see Saccharomyces Genome Database [SGD]; http://www.yeastgenome.org/). Most (90%) of these deletion mutants show cross-sensitivity to one or more of the damaging agents described above (Table 1) (8). On the basis of the cross-sensitivities to other DNA-damaging agents, we can group these new IR resistance genes into 24 functional groupings, of which 6 contain previously identified DNA damage or checkpoint repair genes (Table 1). As in our previous study, many genes can also be grouped on the basis of shared functions such as transcription or protein synthesis (Table 1; see Table S1 in the supplemental material). When screened for sensitivity to other DNA-damaging agents, some of the new IRS deletions show cross-sensitivity profiles similar to those of known recombinational repair or checkpoint genes (Table 1). With the exception of the RAD59 deletion mutant (see Table S1 in the supplemental material), strains lacking members of the RAD52 group of recombinational repair genes were cross-sensitive to each of the DNA-damaging agents tested.
Genetic and physical relationships among newly identified IR resistance genes identify a novel damage response network.
Using literature annotated in the SGD, the Yeast Proteome Database (https://www.incyte.com/proteome/YPD/), the Munich Information Center for Protein Sequences (http://mips.gsf.de/), and the General Repository for Interaction Datasets (http://biodata.mshri.on.ca:80/grid/servlet/Index) plus recently published data describing large interactive genetic and proteomic networks, we have identified genetic and/or physical interactions among the genes and gene products required for the toleration of IR damage. The criteria for these interactions include epistasis analysis, synthetic lethal interactions, two-hybrid analysis, and mass spectrometry of immunoprecipitated protein complexes. This has allowed us to create networks that overlay our functional genomic screening with genomic and proteomic interaction maps. Using this approach, we have identified a new damage response network (as described for Fig. 1) that directly links three separate well-characterized transcriptional complexes through their interactions with CCR4. These transcription complexes include the CNOT complex (seven genes: CCR4, DHH1, POP2, NOT3, NOT4, NOT5, and DBF2), the PAF complex (four genes: CCR4, PAF1, HPR1, and RTF1) and the SRB transcription complex (SRB5 and ANC1). Furthermore, RLR1 (THO2) interacts with HPR1 in the THO complex that is required for transcriptional elongation. Individual deletions of these genes render cells IRS as diploids.
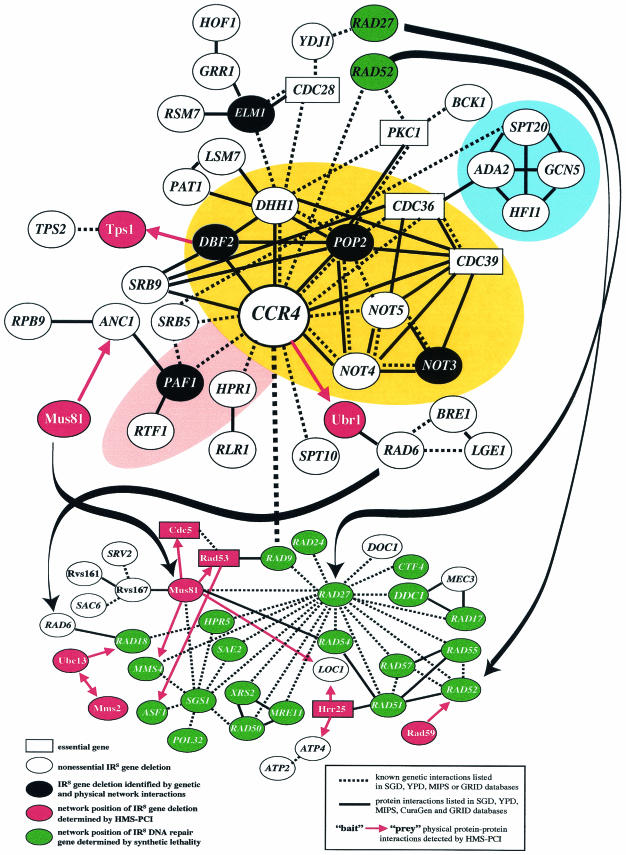
FIG.1.
Interaction of the CCR4 damage response network with known repair and checkpoint genes. Nonessential, IRS gene deletions are depicted by an oval enclosing the gene name. Genes or their protein products that interact either genetically (dotted lines) or physically (solid black lines without arrows or red lines with arrows) in the network are shown. Critical essential genes that have described roles in recombination and checkpoint functions or link nonessential genes within the CCR4 network have been indicated with a rectangular box enclosing the gene name. Ccr4 is a core member of two separate transcription complexes. IRS members of the CNOT complex are highlighted in yellow, while members of the PAF complex are highlighted in pink. CCR4 appears to be the core of this network, since it shows the largest number (13) of genetic and physical interactions with other radiation resistance genes. Members of the SAGA transcriptional complex that confer radiation resistance have been highlighted in blue. On the basis of their network interactions, gene deletions not initially identified in the primary screening as IRS (indicated with an oval containing white characters on a black background) were subsequently identified (using dilution pronging or survival curve analysis) as radiation sensitive (see Fig. 2). Genetic and physical interactions were determined using the following databases: SGD, the Yeast Proteome Database, the Munich Information Center for Protein Sequences, the Pathcalling yeast interaction database at CuraGen Corporation, and the General Repository for Interaction Datasets. Network positions of the genes indicated with white characters on a red background have been determined by high-throughput mass-spectrometric protein complex identification (HMS-PCI). Red arrows indicate the direction (from the bait protein to the prey) of the interaction (33). Network positions of the genes indicated with white characters on a green background have been determined from a systematic examination of synthetic lethal interactions (67). Some proteins (Ydj1 and Atp2) identified by HMS-PCI but omitted due to high frequencies of interaction have been included in this figure on the basis of supporting data indicating genetic interactions with other known IR resistance genes. The CCR4 network (upper grouping) interacts with other established repair networks or pathways (lower grouping of genes) through at least four intermediary IR resistance genes (MUS81, RAD6, RAD27, and RAD52 [indicated with black lines with arrows]) that serve as a linkage between the two networks. The genetic linkage of CCR4 with RAD9 was determined by epistasis analysis in this study.
Members of the SAGA complex (SPT20, ADA2, GCN5, and HFI1) which are required for transcriptional activation of a subset of RNA polymerase (Pol) II-dependent genes are linked to the CNOT complex through interactions with the essential gene CDC36 (NOT2). The CCR4 gene product appears to play a central role in the toleration of IR damage, since it interacts with at least 13 other nonessential genes (including RAD9) whose absence confers IRS (this study; see below and Fig. 1). Another 23 radiation resistance genes are indirectly linked to CCR4 (through pathways that connect to the 13 genes that directly interact with CCR4) (Fig. 1). Because CCR4 has the largest number of genetic and/or physical associations among the members of our combined collection of newly identified damage toleration genes, we have collectively named these genes the CCR4 damage response network.
The CCR4 damage response network has a number of genetic and/or physical interactions with characterized repair genes (including RAD9, RAD52, RAD6, RAD27, and MUS81) (Fig. 1). Furthermore, members of the PAF complex (HPR1) as well as RLR1 play a role in transcription elongation and confer a hyperrecombination phenotype when deleted. The repair genes RAD9, RAD52, RAD6, RAD27, and MUS81 participate in another interactive damage response network that includes a large number of the IR resistance genes detected in our screening and elsewhere (Fig. 1). In total, 68 genes form an overlapping interactive network that includes previously characterized repair genes and those from our combined collection of newly identified radiation resistance genes (Fig. 1).
We have also found from our combined studies IR resistance genes that belong to smaller groups within which the genes and/or protein products interact genetically or physically. Six interacting genes within the nuclear pore complex (NUP84, NUP120, NUP133, NUP170, NUP188, and ASM4) are sensitive to IR following deletion (12) (see Table S1 in the supplemental material). Another group of six IR toleration genes (PDR13, ZUO1, SRO9, TIF4631, SCP160, and BFR1) can be grouped through genetic and/or physical interactions but share no apparent common function. A group of interacting IR resistance genes (RVS161, RVS167, SAC6, and SRV2) have been implicated in actin-related, cytoskeletal functions. Recently these actin-related genes have been found to interact with the repair protein Mus81 through Rvs167 (Fig. 1). Three groups (a group consisting of NAT1, NAT3, and ARD1, a group consisting of BUD32, DIA4, and YML036W, and a group consisting of RAD6, YPL055C [LGE1], and BRE1) containing three IR resistance genes each were also found to interact physically or genetically. Finally, three pairs of IR resistance genes (pair CIS3 and BUR2, pair BEM1 and AKR1, and pair RAD1 and RAD10) were also found to interact genetically and/or physically. Thus, 47% (94/200) of the IRS gene deletions show genetic or physical interactions as part of a large damage response network or within smaller interactive groups.
Members of the CCR4 damage response network have overlapping functions in cell size homeostasis and zymocin resistance.
Recently, genome-wide screenings have identified gene deletions that are required to maintain cell size homeostasis (39, 73). Surprisingly, a large number (80/200 = 40%) of the gene deletions that have been identified as IRS from our combined studies have also been characterized as having abnormally small or large cell volumes compared to the results seen with WT cells (Table 2). Many of these genes are members of the CNOT or PAF complexes and are thought to modulate cell size by altering expression of G1 cyclins required to progress from G1 to S phase of the cell cycle. Of the genes that interact with CCR4 (see Fig. 1, upper panel), deletion of 16 (POP2, DBF2, NOT4, PAF1, HPR1, SRB5, RLR1, ANC1, RPB9, SPT10, HFI1, PAT1, TPS1, HOF1, YDJ1, and BCK1) (in addition to CCR4) has been shown to cause altered cell size homeostasis. With the exception of BCK1 and TPS1, all of these gene deletions result in cells that are larger than WT cells. Since the cln3 mutant strain also produces large cells, many of these gene deletions appear to affect the G1- to S-phase transition by delaying CDC28-dependent Start function.
TABLE 2.
IR-sensitive gene deletions that show overlapping defects in G1-regulated responses (including maintenance of cell size homeostasis and sensitivity to zymocin)
| Cell size homeostasis defecta | Sensitivity to zymocinb | IR-sensitive gene deletionc |
|---|---|---|
| Large | Yes (n = 39) | ADK1, AKR1, ANC1, APN1, ASF1, BEM1, BFR1, BUR2, CCR4, CDC40, CLC1, DEG1, DHH1, EAP1, EST1, HFI1, HPR1, HTL1, MMS22, NOT3, NOT4, NOT5, PAT1, POP2, RAD50, REF2, RPB9, RSC1, RSC2, RTF1, RVS161, SCP160, SRB5, VID21, VID31, YBL006C, YCL016C, YLR322W, YPL055C |
| Large | No (n = 2) | FUN12, YML014Wd |
| Large | NDe (n = 18) | ARP5, ARP8, BDF1, BRE1, BUD32, CDC73, DBF2, FAB1, HOF1, PAF1, PLC1, RLR1, SLX8, SPT10, SRV2, YDJ1, YDR532C, YLR358C |
| Small | Yes (n = 10) | BCK1, DIA4, MEC3, MRPL31, RSA1, SFP1, TOM37, ZUO1, YGR165W, YJL188C |
| Small | No (n = 2) | LOC1, MRT4 |
| Small | ND (n = 10) | ATP4, GLO3, MAP1, MDM10, RIM1, RPL34B, SCO1, TPS1, YDR417C, YGL218W |
Yeast deletion strains that fail to maintain cell size homeostasis (i.e., cells are either larger or smaller than wild-type cells) as described by Jorgensen et al. (39) and Zhang et al. (73).
Enhanced sensitivity of diploid deletion strains to zymocin was determined using the zymocin eclipse assay or dilution plating to zymocin-containing plates as previously described by Kitamoto et al. (40). Sensitivity of the dhh1 Δ to zymocin was previously determined by Westmoreland et al. (72). Haploid deletion strains previously described by Kitamoto et al. (40) as hypersensitive to zymocin are highlighted in boldface characters.
IR-sensitive diploid deletion strains identified in this study and by Bennett et al. (8). The cdc73Δ strain was IR sensitive only at a high dose (120 krads).
The yml014wΔ strain showed enhanced resistance to zymocin (i.e., enhanced growth rate and survival) compared to the WT.
ND, not yet determined.
Many of the IRS strains that interact with Ccr4 and are defective for RNA Pol II transcription (strains CCR4, POP2, NOT3, NOT4, NOT5, RTF1, SRB5, SPT20, ADA2, GCN5, and DHH1) are also hypersensitive to the killer toxin zymocin that is produced by the yeast K. lactis. This toxin causes a prolonged G1 arrest and lethality in haploid S. cerevisiae. The overlap between IRS deletion strains that are also sensitive to zymocin and show defects in cell size control suggests that these mutant phenotypes all share a common underlying molecular defect. Furthermore, the overlap of mutants sensitive to both zymocin and IR suggests that the presence of zymocin might induce DNA DSB damage. Since abnormal cell cycle regulation at the G1/S phase boundary has been observed for zymocin-hypersensitive cells and for cells with altered cell size homeostasis, this suggests that IRS mutants that share these phenotypes might also have abnormal regulation at the G1/S boundary in response to IR. Interestingly, deletions of four genes (MEC3, SFP1, BCK1, and MRT4) that have been previously associated with defective damage checkpoint arrest produced cells that were abnormally small compared to WT cells (Table 2). Therefore, IRS deletions that overlap with those that fail to inhibit G1- to S-phase transition in response to growth signals may represent a subset of genes required for DNA damage-dependent checkpoint functions. Taken together, these results suggest that many of the newly identified IRS gene deletions might exhibit defects in cell cycle transition at the G1/S boundary. Since individual deletions of at least seven members of the CNOT complex render cells IRS (and since CCR4 appears to be the hub of an interactive damage response network), we investigated in detail the mechanistic role the CNOT transcriptional complex plays in radiation resistance in diploid yeast cells.
Two CCR4-dependent transcription complexes are required for toleration of radiation in diploid cells. In our previous IR screening, we determined that two core members (CCR4 and DHH1) of the CNOT complex were required for radiation resistance in diploid yeast (8). However, several members of the CNOT complex were not present in our first screening (which included only 3,670 of the 4,746 nonessential genes). Screening the remaining genes identified another two members (NOT4 and NOT5) of the CNOT complex that were IRS. In our previous screening, we also found enhanced IR sensitivity for the isogenic diploid hpr1Δ and rtf1Δ strains. Both these genes are members of the PAF transcription complex, which is distinct from the CNOT complex even though both complexes contain Ccr4 (17, 19, 54). Deletion of HPR1 has been shown to cause hyperrecombination but does not result in radiation sensitivity in haploid cells (2, 3). These results suggest that the two CCR4-dependent transcriptional complexes (CNOT and PAF) are required for IR resistance in diploid yeast.
To confirm that the mutations in the CNOT complex were IRS due to single recessive gene deletions and not due to errors in strain construction, we transformed the diploid ccr4Δ and dhh1Δ strains with plasmids containing full-length copies of CCR4 and DHH1. These strains showed WT survival when exposed to a single dose of IR (80 krads; Fig. 2A). In addition, we found that a reconstructed diploid dhh1Δ strain (haploid strains BY4741 and BY4742 [each individually lacking DHH1] were mated) was also IRS (data not shown).
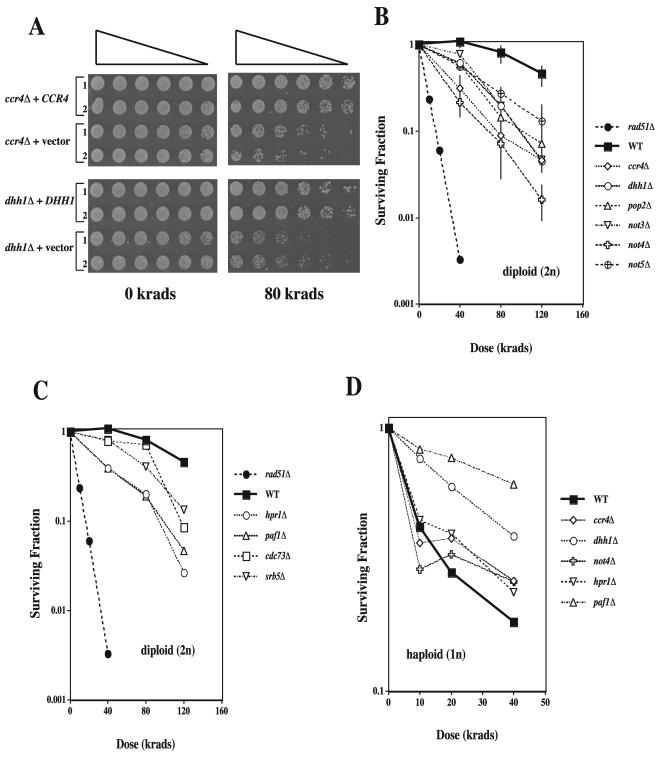
FIG. 2.
Reduced survival of gamma-irradiated diploid stationary cells lacking various members of two CCR4-dependent transcriptional complexes. (A) ccr4Δ and dhh1Δ diploid strains containing the plasmid YEP13-CCR4 or pRS425-DHH1 (49) that express full-length copies of CCR4 and DHH1 or vector alone were grown for 2 days in SC GLU−LEU and serially diluted fivefold. Cells were replica pronged to SC GLU−LEU plates, immediately irradiated with 80 krads of gamma rays, and grown for 3 days at 30°C. Downward-sloping triangles indicate decreasing cell concentration gradients. Complementation of the ccr4Δ and dhh1Δ strains for radiation resistance was similar to that observed for the WT (data not shown). (B) Diploid strains were grown to stationary phase (4 days at 30°C) in liquid YPD such that >95% of cells were in G1 (i.e., unbudded) at the time of irradiation. A dose-dependent decrease in survival of colony-forming ability was seen for diploid strains lacking members of the CNOT complex. Typical error measurements (± 1 standard error) have been shown for WT, ccr4Δ, not4Δ, and not5Δ strains. (C) Diploid cells lacking members of the PAF complex were grown and irradiated as described above. (D) Haploid strains were grownovernight in liquid YPD, diluted 1 to 4 in fresh YPD, and allowed to grow into logarithmic phase with vigorous shaking for 4 h at 30°C. No decrease in survival relative to that of the WT was seen for haploid strains lacking members of either CCR4-dependent transcription complex. The two-component nature of these curves results from the extreme IR sensitivity of haploid unbudded G1-phase cells (see text), while the budded (S and G2 phase) population is radioresistant. The lack of an IRS component in the dhh1Δ and paf1Δ cells was due to a high percentage of radioresistant budded cells in the population at the time of irradiation. All data points represent the averages of three to eight replica experiments.
To determine whether the IR sensitivity of mutants within these two CCR4-dependent complexes was due to altered survival responses and not to slow growth recovery, we compared cell survival of the deletion strains described above to that of the recombination-deficient rad51Δ and WT strains following exposure to various doses of IR (Fig. 2B). On the basis of reported protein interactions of Ccr4 with Dbf2 (45) and Pop2 (32) and Not4 and Not5 with Not3 (44) (as well as the genetic interaction of DHH1 with ELM1) (53), we also used dilution pronging and survival curve analysis to examine dbf2Δ, pop2Δ, not3Δ, and elm1Δ cells for IR sensitivity. We similarly used survival curve analysis to examine strains lacking two core members of the PAF complex (PAF1 and CDC73) for sensitivity to IR. These six deletion strains were not initially identified as IRS, possibly due to insensitivity of the spot-testing screening technique.
As predicted, all CNOT complex mutations (including dbf2Δ, pop2Δ, and not3Δ) demonstrated increased IR sensitivity (Fig. 2B and data not shown). However, the dose-dependent decreases in survival were intermediate between those seen for WT and rad51Δ diploid strains, suggesting they do not play a direct role in recombination. Similar results were found for the srb5Δ, hpr1Δ, and paf1Δ strains (Fig. 2C), but enhanced IR sensitivity was observed only at a high radiation dose (120 krads; Fig. 2C) for the cdc73Δ strain. The survival of these deletion strains was greater than that for the recombination-deficient rad51Δ or rad52Δ strains following IR (Fig. 3A). Therefore, the CNOT and PAF complexes do not appear to play a direct role in recombinational repair (although a minor or indirect role cannot be ruled out without epistasis analysis). The capability of these strains to undergo RAD52-dependent PCR-mediated gene targeting (see below) further suggests they do not directly affect recombination.
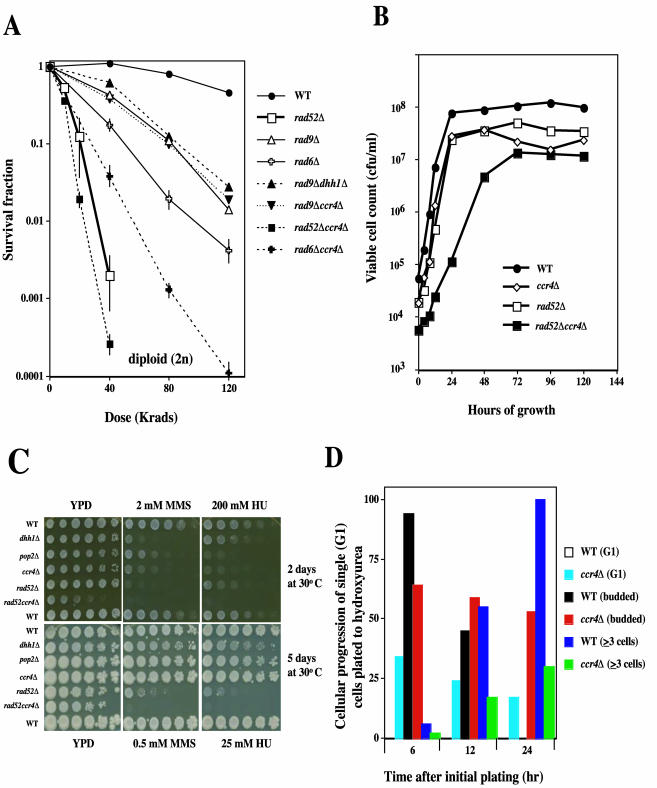
FIG. 3.
CCR4 is a member of the RAD9 checkpoint pathway and exhibits defects in cell cycle progression in S phase. (A) CCR4 and RAD9 show the same epistatic relationship with respect to radiation-induced lethality. Diploid cells were grown to stationary phase and gamma irradiated at various doses as described above. Survival of colony-forming ability was determined on YPD following irradiation. No increase in lethality was seen for the rad9Δ dhh1Δ or rad9Δ ccr4Δ strains compared to the results seen with the rad9Δ strain. However, the rad52Δ ccr4Δ and rad6Δ ccr4Δ strains showed dose-dependent decreases in survival that were greater than that observed for the rad52Δ and rad6Δ strains, respectively. The data points represent the averages of three to eight replica experiments. Error bars are ± 1 standard error. (B) The growth rate of the diploid rad52Δ ccr4Δ strain is decreased compared to the results seen with the isogenic rad52Δ or ccr4Δ strains. Strains growing logarithmically in YPD were diluted into fresh YPD with vigorous shaking at 30°C, and the viable cell counts were determined according to the colony-forming ability of samples selected at the indicated times. All data points represent the averages of three determinations. (C) Strains lacking members of the CNOT complex are sensitive to the replication inhibitors MMS and HU. Strains were grown in liquid YPD for 2 days and serially diluted as described for Fig. 2A. Diluted cells were replica pronged to YPD plates containing either the alkylating agent MMS or the ribonucleotide reductase inhibitor HU at the indicated concentrations and allowed to grow for 2 days (high doses of MMS and HU) or 5 days (low doses of MMS and HU) at 30°C. No growth of the rad52Δ ccr4Δ strain was observed on either the high- or low-dose plates after extended incubation (5 days), indicating a hypersensitivity of the double mutant to these S-phase inhibitors. Similar inhibition results were observed for plates containing zymocin (66%) or doxorubicin (50 μg/ml; data not shown). (D) Prolonged S/M arrest of ccr4Δ cells in the presence of HU. WT and ccr4Δ diploid cells were grown to logarithmic phase and plated to YPD or YPD plus 200 mM HU. Individual G1 cells were micromanipulated into a grid pattern, and the time required for the transition from single (G1) cells to budded (S and G2) cells and into microcolonies (≥3 cells) was determined. More than 50% of the ccr4Δ cells remained budded (in G2) at 24 h after plating to the S-phase-specific inhibitor HU. At 6 h, all of the WT cells initially plated, as single cells had progressed to budded cells or formed microcolonies; by 24 h, all (100%) of the WT cells had progressed to form viable microcolonies on HU. For both strains, 85% of G1 cells (in the absence of HU) progressed to microcolonies following 6 h on YPD plates. Cell progression (vertical bars) was calculated as average percentages from four replicate experiments.
Deletion mutants within the CCR4 dependent transcriptional complexes are radiation resistant in G2.
In yeast, unrepaired DSBs are the primary source of lethality caused by IR. Haploid strains are extremely sensitive in the G1 phase of the cell cycle, since DSBs are not able to utilize recombinational repair due to the lack of an available homolog. WT haploid cells that have replicated their DNA prior to segregation (i.e., G2 cells) are IR resistant due to their ability to repair DSBs by recombination using the undamaged sister chromatid as a template. Diploid cells are IR resistant in G1 because of the presence of homologous chromosomes. Among some of the previously identified IRS diploid deletion strains, the isogenic haploid derivatives were more resistant to radiation (8). We therefore examined the IR sensitivity of logarithmically growing MATa haploid strains lacking individual members (ccr4Δ, dhh1Δ, not4Δ, hpr1Δ, and paf1Δ) of the two CCR4-dependent transcription complexes. These haploid strains did not show enhanced IR sensitivity compared to the WT strain (Fig. 2D), thus indicating that these mutants lacking members of the two CCR4-dependent transcription complexes are recombination proficient for DSBs induced in G2. Furthermore, this suggests that a mechanism other than reduced recombination between sister chromatids in G2 is responsible for the IR sensitivity of diploid mutants.
The resistance of G2 haploid cells led us to consider whether IR sensitivity of the CCR4 diploid mutants is primarily due to killing of the G1 population. We therefore compared the radiation sensitivities of logarithmically growing versus stationary cultures of WT and mutant ccr4Δ cells. A threefold increase in the survival fractions was obtained for irradiated logarithmically growing ccr4Δ diploid cells (∼80% budded) compared to the results seen with stationary cells (<10% budded) which were exposed to 80 krads of IR. The survival fractions for ccr4Δ cells relative to those of WT cells were 0.82 versus 0.27 (means of four experiments [80 krads]) for the logarithmically growing and stationary cells, respectively.
To confirm that the G1 cell population of the ccr4Δ strain was radiosensitive compared to the G2 cell population, we used the tubulin inhibitor benomyl to synchronize WT and ccr4Δ cells in G2 (92 and 88% [means of five experiments] large budded cells for WT and ccr4Δ strains, respectively). These synchronized cells were exposed to IR (80 krads), and viability was determined by plating unirradiated and gamma-irradiated synchronized cells to YPD. We similarly determined the relative survival levels of unirradiated and IR (80 krads)-exposed ccr4Δ and WT cells following release from the benomyl cell cycle block (when irradiated, 59 and 45% of the cells were unbudded [i.e., in G1] for the WT and ccr4Δ strains, respectively). The resulting survival fractions for ccr4Δ cells relative to those for the WT cells were 0.81 for the synchronized G2 cells and 0.57 for the released asynchronous population of G1 and G2 cells. If the G2 cells within the asynchronous ccr4Δ cell population are assumed to have the same survival rate as the WT benomyl-treated cells, then the relative survival of the ccr4Δ G1 cell fraction was 0.28. This relative decrease in the level of survival (compared to that of the WT cells) for the ccr4Δ G1 cells released from the benomyl block was very similar to that obtained for cells that were irradiated as stationary G1 cultures (0.27). These results are consistent with the IR sensitivity of the diploid ccr4Δ strain being primarily associated with the G1 population of cells.
Strains lacking CCR4 are recombination proficient.
We previously reported that targeted chromosomal recombination, which is a RAD52-dependent process, was not significantly decreased in the diploid dhh1Δ strain but was enhanced in the hpr1Δ strain (8). To confirm that diploid CCR4 complex mutants were radiation sensitive due to a defect in a process other than RAD52-dependent recombination, we similarly assayed the ccr4Δ, paf1Δ, and cdc73Δ strains for targeted PCR fragment-mediated recombination at the chromosomal his3Δ-1 locus. For the ccr4Δ strain, the efficiency of targeted recombination at the his3Δ-1 locus was comparable to that observed in the WT strain (1.2 ± 0.7 versus 1.0 ± 0.7). Furthermore, both the diploid paf1Δ and the cdc73Δ strains produced targeted recombination efficiencies that were similar to that of the WT strain (0.65 ± 0.17 and 2.6 ± 1.9, respectively). By comparison, the recombination-deficient rad51Δ strain had a significantly reduced targeted-recombination efficiency (0.016 ± 0.005) whereas the hyperrecombination strain hpr1Δ had an enhanced recombination efficiency (24 ± 10) compared to the WT strain (8). These results suggest that the ccr4Δ, paf1Δ, and cdc73Δ strains are recombination proficient for PCR fragment-mediated targeted recombination at his3Δ-1.
Following exposure of recombination-deficient diploid rad52 cells to IR (20 krads), chromosome integrity is lost as measured by pulse-field gel analysis and only partially restored after prolonged liquid holding (resuspension of cells in water) of the damaged cells for 24 to 48 h (52). However, lost chromosome integrity was completely restored in diploid ccr4Δ cells at 4 to 6 h following irradiation at a much higher dose (40 krads) when chromosome integrity was examined by pulsed-field gel analysis (data not shown). These results further indicate that ccr4Δ cells are recombination proficient and are able to repair IR-induced DSB damage.
CCR4 is a member of the RAD9 checkpoint repair epistasis group.
Epistasis analysis can be used to determine whether two IR resistance genes are members of the same genetic pathway. IR resistance genes are within the same epistasis repair group if the IR sensitivity of a strain containing both mutations is no greater than the sensitivity of the more sensitive of the two single gene mutation strains (26). Since a CCR4 mutation was reported to suppress the IR sensitivity of an allele of rad52 (rad52-20) (61), we determined whether CCR4 was a member of the RAD52 radiation repair epistasis group. Although RAD52 is responsible for the majority of DSB repair in yeast, we also examined whether CCR4 was a member of two other epistasis groups (RAD6 and RAD9) which are responsible for most of the remaining IR repair.
IR-induced killing of the diploid rad52Δ ccr4Δ and the rad6Δ ccr4Δ strains was enhanced by an order of magnitude compared to that of the rad52Δ or rad6Δ strain alone (Fig. 3A). However, sensitivity to IR was not enhanced for the rad9Δ ccr4Δ or the rad9Δ dhh1Δ strain compared to that of the rad9Δ strain alone (Fig. 3A), suggesting that the CCR4 and DHH1 genes function in the same pathway as RAD9. Both the diploid ccr4Δ and dhh1Δ survival curves (Fig. 2B) were similar to that expressed by the diploid rad9Δ strain (Fig. 3A). This epistasis analysis therefore indicates that CCR4 is a member of the RAD9 checkpoint pathway for IR-induced cell killing.
Strains lacking CCR4 have an S-phase cell cycle defect following replication stress.
Since the CNOT complex plays a role in cell cycle responses to stress, we examined its impact when a ccr4Δ was combined with a rad52Δ and the double mutant exposed to the replication inhibitor HU. We observed a synergistic decrease in growth rate when CCR4 and RAD52 deletions were combined in the same strain (Fig. 3B); this decrease was not due to loss of mitochondrial function. The generation time of the double mutant was 4.9 h compared to 2.6 and 1.9 h for the rad52Δ and ccr4Δ mutants, respectively (Fig. 3B), and 1.7 h for the WT strain. There was no decreased growth rate for the rad6Δ ccr4Δ or rad9Δ ccr4Δ strain (data not shown).
Diploid strains lacking CCR4 and other members (including DHH1 or POP2) of the CNOT complex are sensitive to HU and MMS compared to WT strains (Fig. 3C). Similar to the diploid strains, haploid strains lacking CCR4 and DHH1 also demonstrated enhanced sensitivity to HU and MMS compared to WT strains (data not shown). Diploid rad52Δ strains are also sensitive to HU (Fig. 3C) because of the inability to repair DSBs produced during HU-induced replication arrest (51). We therefore compared the HU sensitivity of the rad52Δ ccr4Δ double mutant to that of the single rad52Δ and ccr4Δ mutants to determine whether these genes could be placed in the same epistasis group for HU-induced lethality. The rad52Δ ccr4Δ strain was hypersensitive to the killing effects of HU (Fig. 3C). Similar results were observed with MMS, an alkylating agent that is also S phase specific for the induction of DNA damage (Fig. 3C), as well as with doxorubicin, which is a potent topoisomerase inhibitor (data not shown). After extended incubation times at lower doses of HU (25 mM) and MMS (0.5 mM), the enhanced lethality of the slow-growing rad52Δ ccr4Δ strain compared to that of the ccr4Δ and rad52Δ stains is apparent (Fig. 3C). These results indicate that for HU survival, CCR4 is in an epistasis group separate from that defined by the RAD52 repair group. Moreover, CCR4 may be required for cell cycle progression through S phase in the presence of chemical agents that induce replication stress.
When HU blocks DNA replication, WT cells arrest as budded cells until S phase is completed; this is referred to as the S/M checkpoint. To examine whether cell cycle progression of ccr4Δ cells is inhibited by the presence of HU, single G1 cells from logarithmically growing cultures of WT and ccr4Δ strains were examined hourly for cell cycle progression on YPD plates containing 200 mM HU (Fig. 3D). After 6 h on HU, most of the WT and ccr4Δ cells initially plated in G1 progressed into S phase and arrested as budded cells. In the absence of HU, the majority (85%) of G1 cells from these two strains progressed to form microcolonies at 6 h (data not shown). Although all of the WT cells completed S phase and progressed to form viable microcolonies after 24 h of exposure to HU, most (53%) of the ccr4Δ cells remained as large budded cells (Fig. 3D) and most (60%) were lysed (data not shown). Similarly, nearly 65% of the G1 rad52Δ ccr4Δ cells were large budded cells after 6 h; almost all (88%) were lysed after 24 h, however, and none progressed further than the three-cell microcolony stage (data not shown). This severe growth arrest which was followed by cellular lysis was not observed with rad52Δ strains and may account for the enhanced hypersensitivity of the rad52Δ ccr4Δ strain when it is plated to HU (Fig. 3C) compared to the results seen with ccr4Δ or rad52Δ strains alone. Taken together, these results suggest that HU inhibited cell cycle progression in S phase in ccr4Δ cells. Since this effect is enhanced in the absence of RAD52, ccr4Δ strains appear to have a defect in replication or adaptation to the S/M checkpoint which is independent of recombination.
CCR4 and DHH1 are required for cell cycle progression in G1 and G2 following gamma irradiation.
Since CCR4 and the RAD9 checkpoint gene reside within the same epistasis group, we examined cell cycle progression of irradiated ccr4Δ and dhh1Δ cells. In budding yeast, cell cycle arrest following IR damage occurs at G1 as well as G2 stages of the cell cycle and both checkpoints are under the control of the RAD9 gene product (63, 71). However, RAD9 has also been implicated in S-phase checkpoint arrest and is required for the damage-induced transcription of a number of repair genes normally expressed in S phase. Since diploid deletions of CCR4 are IRS in G1 and also show reduced G1 arrest following nitrogen starvation (72), we examined whether the transition from G1 to S phase (i.e., at the G1 checkpoint) was abnormal following IR in diploid strains lacking CCR4 or DHH1 as well as RAD9. A rapid and comparable G1 to S transition was observed for all unirradiated strains (Fig. 4A). As previously reported (31), irradiated rad9Δ cells had a more rapid (1.1 ± 0.5 h earlier on average) G1- to S-phase transition compared to WT cells (Fig. 4A). However, the dhh1Δ and ccr4Δ strains showed prolonged G1 arrest following IR. The transition times from G1 to S phase in these strains were longer (1.4 ± 0.6 and 2.5 ± 1.0 h for dhh1Δ and ccr4Δ, respectively) than that observed in WT (Fig. 4A).
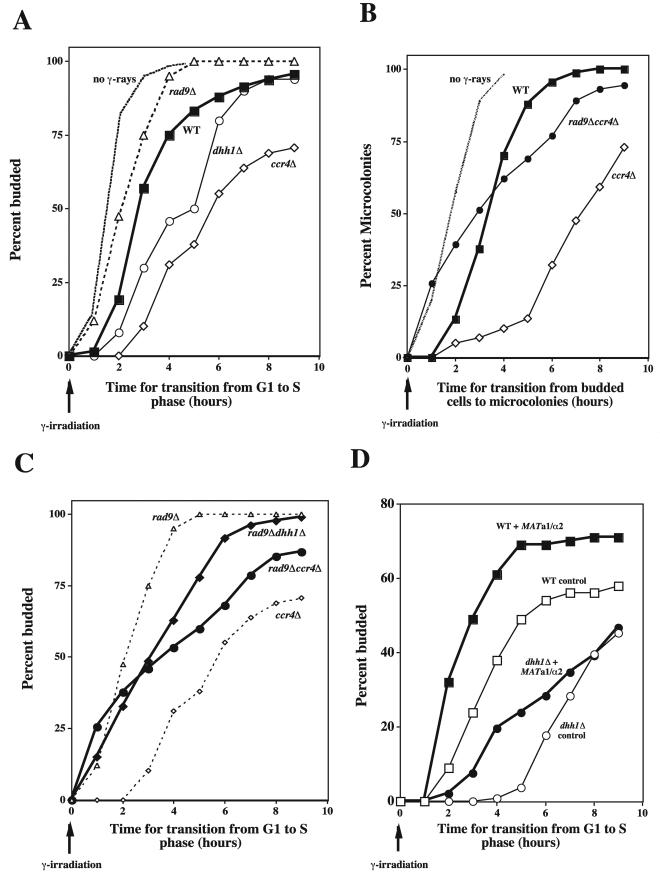
FIG.4.
Deletion of CCR4 or DHH1 results in a prolonged, RAD9-dependent G1- to S-phase cell cycle transition following IR damage. (A) The time required for cell cycle progression of individual G1 cells into budded (S-phase) cells was determined for the diploid WT, dhh1Δ and ccr4Δ mutants, and the G1 checkpoint mutant rad9Δ strains following exposure to 8 krads of IR. Cells were grown to logarithmic phase, plated to YPD, and gamma irradiated. The rad9Δ cells showed a more rapid transition from single (G1-phase) to budded (S-phase) cells than WT cells. Compared to the results seen with the rad9Δ strain, the average increase in time required for the WT cells to transit from G1 to S was 1.1 ± 0.5 h following IR. Compared to the results seen with the WT strain, both the dhh1Δ and ccr4Δ strains required more time (1.3 ± 0.6 and 2.5 ± 1.0 h, respectively) to transit from G1 to S following IR damage. No difference was seen in the onset of cell cycle progression for unirradiated cells (no gamma rays; averaged pooled data for all strains examined). (B) The time required for cell cycle progression of diploid budded cells (S and G2) into microcolonies following 8 krads of IR. The dhh1Δ and rad9Δ dhh1Δ cells showed delays similar to those seen with the ccr4Δ and rad9Δ ccr4Δ cells; the results obtained with those cells have been omitted for clarity. No difference was seen in the onset of cell cycle progression for unirradiated cells (no gamma rays; averaged pooled data for all strains examined). (C) The time required for cell cycle progression of individual G1 cells into budded (S-phase) cells was determined for the diploid mutant rad9Δ dhh1Δ and rad9Δ ccr4Δ strains following exposure to 8 krads of IR. Data for the rad9Δ and ccr4Δ strains have been included for comparison. Cell cycle progression results for individual G1 cells of the diploid deletion mutant dhh1 and ccr4 strains were compared to the results seen with the WT and G1 checkpoint deletion mutant rad9 strains following exposure to 8 krads of IR. The time required to transit from G1 (unbudded) to S (budded) phase for the rad9Δ dhh1Δ and rad9Δ ccr4Δ strains was significantly shorter than that seen with the dhh1Δ or ccr4Δ single-mutant strains. The progress of the unirradiated cells was identical to that shown in panel A. (D) Effects of MAT expression on G1- to S-phase cell cycle transition following IR damage in haploid cells. WT and dhh1Δ haploid MATa cells were transformed with either pRS315 (control) or pCB115 (MATa1α2). The plasmid pCB115 expresses the MATa1 and MATα2 genes which confer a nonmating, pseudodiploid phenotype on haploid cells containing the plasmid. The time required for G1- to S-phase transition was determined following IR as described above, with the exception that plasmid-containing cells were grown in SC GLU−LEU to maintain the plasmid and subsequently plated to SC GLU-LEU following irradiation.
Similarly, in the dhh1Δ and ccr4Δ strains, we found a prolonged delay in cell cycle progression among the irradiated budded (S plus G2) cell populations following IR compared to the results seen with WT cell populations (Fig. 4B). For all strains examined, the transition of unirradiated budded cells was more rapid than that observed for irradiated cells. Therefore, dhh1Δ and ccr4Δ cells exhibit prolonged cell cycle delay at two morphological checkpoint “landmarks” following IR exposure.
Prolonged damage-induced cell cycle arrest in ccr4Δ and dhh1Δ strains is RAD9 dependent.
Prolonged cell cycle delays at G1 and S phase may result from the persistence of unrepaired DSB damage due to a repair defect; alternatively, cells may be defective in reentering the cell cycle following DNA repair. This latter process has been termed checkpoint adaptation and has been shown to be under genetic control (65). Defects in genes controlling checkpoint adaptation result in prolonged arrest and reduced survival following DNA damage. Therefore, CCR4 and DHH1 could also be required for adaptation to RAD9-dependent checkpoints at G1/S or in S phase following DNA damage. Among the components of the DNA damage checkpoint pathway, RAD9 has been proposed to perform a damage sensor function early in the pathway (48). In rad9Δ cells, unrepaired DSBs have no effect on the rapid onset of cell cycle progression. Therefore, if CCR4 and DHH1 were strictly repair genes their absence would not affect the rapid progression of a rad9Δ strain following damage. To identify whether CCR4 and DHH1 behave like repair- or damage-specific checkpoint adaptation genes, we determined transition times for G1- to S-phase cell cycle progression for rad9Δ ccr4Δ and rad9Δ dhh1Δ diploid strains following IR (Fig. 4C). The double-deletion strains did not show the prolonged G1 arrest that was characteristic of the ccr4Δ and dhh1Δ strains or the rapid G1 to S transition characteristic of rad9Δ single-mutant strains following IR (Fig. 4A and C). Instead, the double-mutant cells transit through the G1 checkpoint earlier (1.3 ± 0.3 and 2.3 ± 0.1 h earlier for the respective double-mutant strains) than the dhh1Δ or ccr4Δ cells. Compared to the rad9Δ strain, the double mutants showed a delayed progression in ∼60% of the G1 cell population (Fig. 4C). This suggests that CCR4 and DHH1 are required in part for G1 checkpoint adaptation in a pathway that requires the damage-sensing function of RAD9. However, ∼40% of the G1 cells in the double mutants progressed as rapidly as the cells of the rad9Δ strain (Fig. 4C), suggesting the presence of a possible second adaptation pathway similar to that seen for cells arrested at the G2 damage checkpoint at which G2 arrest requires two parallel pathways (29). Alternatively, there may also be a minor RAD52-independent repair pathway in which CCR4 contributes to the repair of DSBs.
A more rapid transition of irradiated G2 (budded cells) into microcolonies was also observed among the rad9Δ ccr4Δ and rad9Δ dhh1Δ diploid strains compared to the results seen with the ccr4Δ or dhh1Δ strains (Fig. 4B). Therefore, deletion of RAD9 can greatly suppress the prolonged IR-induced cell cycle delays observed at both G1/S and G2/M for the ccr4Δ strain.
Checkpoint adaptation at G1/S is not dependent on MAT.
Slow adaptation to the DSB-induced checkpoint at G2/M is dependent in part on expression of the mating-type (MAT) transcriptional regulators MATa1 and MATα2 that confer a diploid phenotype (10). We, therefore, examined WT and dhh1Δ haploid cells that were transformed with a plasmid (pCB115) that expresses both MATa1 and MATα2 to determine whether the prolonged G1 arrest in diploid dhh1Δ cells is MAT dependent (Fig. 4D). Similar to the results seen with the diploid strains, a prolonged damage-induced G1 arrest was found in the dhh1Δ haploid strain compared to that seen with the WT. The length of the G1 delay was increased for both the WT and the dhh1Δ haploid strains compared to the results seen with their isogenic diploid counterparts (Fig. 4A). For example, the time required for 50% of the diploid and haploid WT cells to exit G1 was 3 and 5 h, respectively. The corresponding G1 delays for the dhh1Δ diploid and haploid strains were 5 and >9 h, respectively. Following coexpression of the MATa1 and MATα2 transcriptional regulators which are normally jointly expressed in diploids, but not in haploids, the time required for cell cycle progression from G1 to S decreased for both the WT and dhh1Δ haploid stains. For these strains the time of G1 to S transition occurred earlier when the MAT transcriptional regulators were present (1.9 + 0.8 and 1.8 + 0.7 for the WT and dhh1Δ strain, respectively). Thus, there is a “diploid” effect for adaptation to damage in both the WT and the dhh1 mutants that results in a decreased time for G1- to S-phase transition. This comparison suggests that the DHH1-controlled adaptation in diploids is not dependent on MAT expression.
MAT heterozygosity has also been shown to suppress the IR sensitivity of the rad52-20 allele (61) and rad55 deletion mutants (46). Since the radiosensitivity of ccr4Δ and dhh1Δ has been observed with diploids but not with haploids, the diploid IR sensitivity could similarly be MAT-dependent. We therefore examined the relative survival rates of haploid ccr4Δ, dhh1Δ, and WT (MATa) strains expressing MATa1 and MATα2 transcriptional regulators (transformed with pCB115) versus the results seen with identical control strains (transformed with vector alone) following a single acute IR dose (80 krads). No difference in survival rates was seen between any of the haploid control strains or those expressing MATa1 and MATα2 (data not shown). Therefore, MAT expression is not responsible for the IR sensitivity of diploid ccr4Δ or dhh1Δ strains.
DISCUSSION
The recent availability of the complete isogenic haploid and diploid yeast deletion strain collections has facilitated the rapid genome-wide identification of new genetic determinants required for yeast to survive exposure to a variety of physical or chemical environmental agents. An inherent feature of these functional genomic screenings is that they often lead to the listing of a large number of new yeast genes required to tolerate a specific inhibitory agent. Often these lists contain a bewildering array of seemingly unrelated genes that define many distinct functional groupings. We therefore used a reductionist approach to identify from our gene list the largest group of IR resistance genes that share a common underlying mechanistic function. Using recent advances in genome-wide determinations of proteomic and genomic interactions (30, 33, 37, 67, 69), we could place many of our newly identified IR resistance genes in a large interactive network (Fig. 1).
This approach has successfully shown that the CCR4 radiation response network is required for survival following IR damage. At least 13 interactions between CCR4 and other radiation resistance genes are present in this network. The CCR4 network also interconnects to another established repair network (67) through at least five well-characterized recombination and repair genes (including RAD9), as described in this study (Fig. 1). Separate experimental screenings have shown that many of the IRS gene deletions within the CCR4 damage response network are also required for maintaining cell size homeostasis and/or zymocin resistance (39, 40, 73). Furthermore, the CNOT mutants ccr4Δ and dhh1Δ (72), as well as pop2Δ, not4Δ, and not5Δ (data not shown), all demonstrated reduced viability following 4 to 5 days of nitrogen starvation. Since these are all G1/S-regulated responses, we propose that the radiation sensitivity of mutants in the CCR4 network also results from defects in cell cycle progression at G1/S. By examining two central members of the CCR4 damage response network, CCR4 and DHH1, we found that they are indeed required for cell cycle progression following RAD9-dependent checkpoint arrest, which is consistent with the apparent G1 sensitivity of the diploid mutants. Furthermore, ccr4Δ strains are also sensitive to the S-phase-specific replication inhibitor HU and show a prolonged arrest at the S/M checkpoint following exposure to HU. This indicates that ccr4Δ strains have cell cycle progression defects in both G1 and S phase following DNA damage.
CCR4-mediates IR resistance in the G1 phase of the cell cycle.
Ccr4 is a highly conserved protein that has multiple roles in the control of mRNA metabolism (including transcription initiation, mRNA elongation, and degradation) (21, 22, 68). It is a core component of two distinct transcriptional complexes that affect diverse processes in yeast. One complex (CNOT) is a global regulator of gene expression that can have both positive and negative effects on RNA Pol II-mediated transcription and is required for the G1 arrest following nitrogen starvation (72) as well as hypersensitivity to zymocin (40). In ccr4Δ diploid strains there is reduced sensitivity to IR when they are irradiated as benomyl-arrested cultures containing a high percentage of G2 cells compared to IRS stationary G1 cultures, further supporting the importance of CCR4 in dealing with damage in the diploid G1 phase. IRS members of the CCR4 network were previously undetected, because all prior radiation screenings utilized haploids in which WT G1 cells are IRS due to the lack of recombinational repair. Therefore, screening of the diploid strain collection has facilitated the detection of a new set of IRS mutants enriched for checkpoint or repair defects specific to the G1 and S phases of the cell cycle.
Similar to the results of this study, IR-induced loss in survival has been observed in diploid but not haploid strains lacking the DNA helicases SGS1 or HPR5 (SRS2) (28). In a separate screening using the same diploid deletion collection, moreover, six deletion strains with identical phenotypes (i.e., IR sensitivity in diploid but not haploid strains) were identified (27). These include five IRS deletion strains (SHE1, ARP8, RSC1, YDR014W, and YNR068C) identified in this study or previously (8). For three of these mutants (ydr014WΔ, she1Δ, and arp8Δ) plasmid expression of the deleted gene restored radiation resistance, indicating that the genomic mutation was indeed responsible for radiation sensitivity (27). Furthermore, deletion of YDR014W was found in both screenings to result in IRS as a diploid. This gene was renamed RAD61, because both diploids and haploids showed enhanced IR sensitivity compared to the WT (27). These results suggest that diploid screenings might be useful for the discovery of new mutants that function specifically in the G1 or early S phases of the cell cycle.
Rapid reentry into the cell cycle following RAD9-dependent checkpoint arrest requires CCR4 and DHH1.
We have shown a prolonged delay in cell cycle transition from G1 to S as well as delay at the G2/M phase of the cell cycle for ccr4Δ and dhh1Δ strains following IR or HU. Furthermore, both the rad9 and ccr4 deletion mutants are within the same epistasis group, as determined for IR-induced cell lethality. Moreover, the prolonged cell cycle arrest of these mutants is RAD9 dependent, further indicating these genes have checkpoint-associated functions within the RAD9 pathway. IR sensitivity resulting from prolonged radiation or DSB-induced arrest at the G2/M checkpoint has been described for numerous mutated genes (including CDC5, YKU70, RDH54, BCK1, MRT4, RAI1, SIR2, SIR3, and SIR4) (9). These genes have been described as checkpoint adaptation genes, since they are required for reentry into the cell cycle following checkpoint arrest. Both CCR4 mutations (this study) and MAT heterozygosity (10) delay reentry into the cell cycle following G2/M checkpoint arrest as well as suppressing the radiation sensitivity of the recombination-proficient rad52-20 allele (61). Therefore, IR sensitivity of this unusual rad52-20 allele may be associated with a defect in checkpoint arrest (at G2/M) which can be independently suppressed by the prolonged, damage-induced G2 arrest mediated by ccr4 or MAT.
Transcriptional regulation of the G1/S damage checkpoint.
Damage-induced checkpoint functions in yeast at G1/S are poorly understood. WT yeast undergo a significant damage-induced G1 arrest, and several G1/S checkpoint genes (RAD9, RAD17, RAD24, and MEC1) have been described and characterized (although the precise molecular mechanism of their action is unknown) (31, 55, 63). However, Rad9 is hyperphosphorylated following damage which is dependent on MEC1, RAD17, RAD24, MEC3, and DDC1, suggesting involvement of the products of these genes in checkpoint signal transduction (70). RAD9 is also required for the damage-mediated transcriptional activation of a number of repair or checkpoint genes (including RAD6, RAD18, RAD51, RAD54, RAD53, DUN1, and others) (1). Since CCR4 and RAD9 share the same epistasis group, it is possible that transcription functions of CCR4 are also required for this “SOS-like” transcriptional regulation. Since the CNOT genes are also involved in nitrogen starvation-induced G1 arrest (72), cell-size regulation (homeostasis), and adaptation to the IR-induced G1 checkpoint, however, it is likely that CCR4 can regulate G1/S cell cycle functions through Cdc28 activation or inhibition. Preexisting functional data for genes that interact within the CCR4 network suggest a molecular model in which transcriptional and posttranscriptional regulation of G1- or S-phase-specific cyclins could account for the cell cycle responses of these deletions to a variety of environmental perturbations. Environmental agents (including IR) (see below) that cause cellular stress are proposed to activate the protein kinase C-mitogen-activated protein (PKC-MAP) signaling pathway that, in turn, activates Ccr4-dependent transcription complexes. This would transiently down regulate a critical subset of cyclin genes involved in G1- and S-phase transition.
The PKC1 stress response pathway is involved in recombination and checkpoint processes.
Ccr4p is a component of the PAF1-CDC73 transcriptional complex which is a downstream effector of the PKC-MAP kinase pathway (17). Genes in this pathway play an essential role in the maintenance of cell wall integrity in response to external environmental signals (including heat shock, hypoosmotic shock, and treatment with α-factor) and possibly to the K. lactis killer toxin zymocin (40). IR appears to be similarly sensed by the PKC-MAP kinase pathway, since the IRS MAP kinase mutant bck1Δ expressed a prolonged G2 arrest followed by cellular lysis in response to IR (9). Since lethality in irradiated ccr4Δ or dhh1Δ strains could not be rescued by plating to YPD plates containing sorbitol (1 M; data not shown), IR-induced lethality in these strains is not mediated solely by the loss of cell wall integrity.
The PKC-MAP kinase stress pathway genes similarly play overlapping roles in the signaling of checkpoint arrest and DNA repair, since the MAP kinase mutant bck1 expresses a prolonged IR-induced G2 arrest (9) and PKC1 mutations result in hyperrecombination (35, 47). Moreover, IRS mutations in members (including PAF1) of the PAF1-CDC73 complex have been reported to enhance intrachromosomal recombination (17); in the case of hpr1Δ mutants, this is RAD52 dependent (3). While the interactive roles that genes within the PKC-MAP kinase pathway play in the stress and DNA damage responses are complex and remain to be fully elucidated, similar overlapping roles in stress response and DNA damage-induced checkpoints has been described for mammalian genes such as p38 (57).
CCR4 has a role in the repair of S-phase damage.
The rad52Δ ccr4Δ strain showed a severe synthetic growth defect compared to either the rad52Δ or ccr4Δ strains alone. A slower growth rate for rad52Δ mutants has been attributed to a requirement for recombination functions to repair replication-induced DSBs, especially in mutants partially defective in replication (51). Thus, the greatly decreased growth rate of the rad52Δ ccr4Δ strain may be due to the influence of CCR4-mediated checkpoint adaptation on another replication and/or repair system that acts on spontaneous or replication-associated DSBs occurring in S phase. The fact that rad52Δ ccr4Δ strains are more sensitive to MMS or HU than either rad52Δ or ccr4Δ strains supports the idea that CCR4 regulates a pathway other than recombination that is required for the repair of S-phase damage. An explanation for such an RAD52-independent repair system could be that CCR4 participates in the transcriptional induction of genes required for DNA replication in the presence of S-phase damage. We have shown that ccr4Δ strains are defective in spontaneous and MMS-induced activation of the ribonucleotide reductase (RNR3) promoter (72). It is possible that similar to hrr25 mutant strains (34), ccr4Δ strains are HU sensitive due to a defect in the transcriptional induction of RNR genes in response to ribonucleotide depletion following HU treatment. Furthermore, ccr4 mutants were also shown to reduce transcription of the damage-inducible repair gene RAD51 (61) which is expressed specifically in G1 at Start (50) as well as in response to DNA damage. Not surprisingly, RAD9 is also required for the DNA damage-induced transcriptional activation of RAD51 and RNR3 (1), further indicating that RAD9 and CCR4 share the same damage response pathway.
Other radiation resistance genes.
Many of the newly described IR resistance genes do not appear to be associated with the CCR4 damage response network. This may be due in part to an incomplete functional understanding of the complex interrelationships within the yeast genome as a whole. For example, the radiation resistance gene SCP160 is associated with five other IR resistance genes (see Results) that share no other apparent common function. Recently, Scp160p has been shown to be part of a mRNP complex that binds to a number of specific mRNAs (including that of DHH1) (43). Since the SCP160 deletion can affect abundance and distribution of these mRNAs, the IR sensitivity caused by scp160Δ may be indirect (affecting the expression or cellular distribution of Dhh1p). Similar to the results seen with SCP160, many of the new IR resistance genes may not have direct checkpoint or repair functions (see reference 8 for a review). Instead, their roles may be indirect, affecting metabolism, trafficking, and/or the abundance of critical DNA repair proteins with a variety of basic cellular functions. These include chromatin organization (ARP5 and ARP8), nuclear pore function (ASM4), Golgi and vacuolar function (FAB1 and VPS33), transcription (YAF9, BDF1, and others), cytoskeletal organization (SRV2), mitochondrial function (MDM10 and MDM20), and protein synthesis (NAT1, EAP1, TIF4631, and others).
Many newly identified IRS gene deletions (including those of members of the CCR4 damage response network) are highly conserved in eukaryotes and have been implicated in cancer (see Table S1 in the supplemental material). For example, the proteins Dhh1p (4, 36), Pop2p (23), Yaf9p (64), and Bdf1p (25) are all transcription factors with human orthologs that are putative tumor suppressor genes or translocation-activated fusion oncoproteins implicated in cancers of myeloid or epithelial origin. By virtue of their evolutionarily conserved nature and association with cancer onset and/or progression, these proteins may play a critical and therefore more direct role in the maintenance of genomic integrity following DNA damage in human or yeast cells.
Supplementary Material
Acknowledgments
We thank M. Collart, T. Weinert, K. Lewis, W. Xiao, and R. Schaffrath for the generous gift of yeast plasmids and strains. The technical assistance of A. Kelkar, D. Bailey, J. Westmoreland, J. Sterling, and A. Falae in some of the experiments is greatly appreciated. We also thank M. Kupiec, Y. Jin, and K. Lewis for critical comments during the preparation of the manuscript.
This work was supported in part by a grant from the DOD Breast Cancer Research Program (DAMD17-03-1-0232) to C.B.B.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aboussekhra, A., J. E. Vialard, D. E. Morrison, M. A. de la Torre-Ruiz, L. Cernakova, F. Fabre, and N. F. Lowndes. 1996. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 15:3912-3922. [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera, A., and H. L. Klein. 1989. Genetic and molecular analysis of recombination events in Saccharomyces cerevisiae occurring in the presence of the hyper-recombination mutation hpr1. Genetics 122:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera, A., and H. L. Klein. 1988. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akao, Y., O. Marukawa, H. Morikawa, K. Nakao, M. Kamei, T. Hachiya, and Y. Tsujimoto. 1995. The rck/p54 candidate proto-oncogene product is a 54-kilodalton D-E-A-D box protein differentially expressed in human and mouse tissues. Cancer Res. 55:3444-3449. [PubMed] [Google Scholar]
- 5.Barth, H., and M. Thumm. 2001. A genomic screen identifies AUT8 as a novel gene essential for autophagy in the yeast Saccharomyces cerevisiae. Gene 274:151-156. [DOI] [PubMed] [Google Scholar]
- 6.Begley, T. J., A. S. Rosenbach, T. Ideker, and L. D. Samson. 2002. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol. Cancer Res. 1:103-112. [PubMed] [Google Scholar]
- 7.Bennett, C. B., A. L. Lewis, K. K. Baldwin, and M. A. Resnick. 1993. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. USA 90:5613-5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29:426-434. [DOI] [PubMed] [Google Scholar]
- 9.Bennett, C. B., and M. A. Resnick. 2003. Finding genes that affect signaling and toleration of DNA damage, especially DNA double-strand breaks. Elsevier Science, San Diego, Calif.
- 10.Bennett, C. B., J. R. Snipe, J. W. Westmoreland, and M. A. Resnick. 2001. SIR functions are required for the toleration of an unrepaired double-strand break in a dispensable yeast chromosome. Mol. Cell. Biol. 21:5359-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett, C. B., T. J. Westmoreland, J. R. Snipe, and M. A. Resnick. 1996. A double-strand break within a yeast artificial chromosome (YAC) containing human DNA can result in YAC loss, deletion or cell lethality. Mol. Cell. Biol. 16:4414-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birrell, G. W., J. A. Brown, H. I. Wu, G. Giaever, A. M. Chu, R. W. Davis, and J. M. Brown. 2002. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. USA 99:8778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birrell, G. W., G. Giaever, A. M. Chu, R. W. Davis, and J. M. Brown. 2001. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. USA 98:12608-12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonangelino, C. J., E. M. Chavez, and J. S. Bonifacino. 2002. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2486-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunborg, G., M. A. Resnick, and D. H. Williamson. 1980. Cell-cycle-specific repair of DNA double strand breaks in Saccharomyces cerevisiae. Radiat. Res. 82:547-558. [PubMed] [Google Scholar]
- 16.Chan, T. F., J. Carvalho, L. Riles, and X. F. Zheng. 2000. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc. Natl. Acad. Sci. USA 97:13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang, M., D. French-Cornay, H.-Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, C., and R. D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81-85. [DOI] [PubMed] [Google Scholar]
- 19.Costa, P. J., and K. M. Arndt. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dasika, G. K., S. C. Lin, S. Zhao, P. Sung, A. Tomkinson, and E. Y. Lee. 1999. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene 18:7883-7899. [DOI] [PubMed] [Google Scholar]
- 21.Denis, C. L., Y. C. Chiang, Y. Cui, and J. Chen. 2001. Genetic evidence supports a role for the yeast CCR4-NOT complex in transcriptional elongation. Genetics 158:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denis, C. L., and T. Malvar. 1990. The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics 124:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fidler, C., J. S. Wainscoat, and J. Boultwood. 1999. The human POP2 gene: identification, sequencing, and mapping to the critical region of the 5q− syndrome. Genomics 56:134-136. [DOI] [PubMed] [Google Scholar]
- 24.Foiani, M., A. Pellicioli, M. Lopes, C. Lucca, M. Ferrari, G. Liberi, M. Muzi Falconi, and P. Plevani. 2000. DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat. Res. 451:187-196. [DOI] [PubMed] [Google Scholar]
- 25.French, C. A., I. Miyoshi, J. C. Aster, I. Kubonishi, T. G. Kroll, P. Dal Cin, S. O. Vargas, A. R. Perez-Atayde, and J. A. Fletcher. 2001. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am. J. Pathol. 159:1987-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Game, J. C. 2000. The Saccharomyces repair genes at the end of the century. Mutat. Res. 451:277-293. [DOI] [PubMed] [Google Scholar]
- 27.Game, J. C., G. W. Birrell, J. A. Brown, T. Shibata, C. Baccari, A. M. Chu, M. S. Williamson, and J. M. Brown. 2003. Use of a genome-wide approach to identify new genes that control resistance of Saccharomyces cerevisiae to ionizing radiation. Radiat. Res. 160:14-24. [DOI] [PubMed] [Google Scholar]
- 28.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 29.Gardner, R., C. W. Putnam, and T. Weinert. 1999. RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 18:3173-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 31.Gerald, J. N., J. M. Benjamin, and S. J. Kron. 2002. Robust G1 checkpoint arrest in budding yeast: dependence on DNA damage signaling and repair. J. Cell Sci. 115:1749-1757. [DOI] [PubMed] [Google Scholar]
- 32.Hata, H., H. Mitsui, H. Liu, Y. Bai, C. L. Denis, Y. Shimizu, and A. Sakai. 1998. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 34.Ho, Y., S. Mason, R. Kobayashi, M. Hoekstra, and B. Andrews. 1997. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, K. N., and L. S. Symington. 1995. Suppressors of a Saccharomyces cerevisiae pkc1 mutation identify alleles of the phosphatase gene PTC1 and of a novel gene encoding a putative basic leucine zipper protein. Genetics 141:1275-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikeda, T., K. Ikeda, K. Sasaki, K. Kawakami, and J. Takahara. 1999. The inv(11) (p15q22) chromosome translocation of therapy-related myelodysplasia with NUP98-DDX10 and DDX10-NUP98 fusion transcripts. Int. J. Hematol. 69:160-164. [PubMed] [Google Scholar]
- 37.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacob, S., and F. Praz. 2002. DNA mismatch repair defects: role in colorectal carcinogenesis. Biochimie 84:27-47. [DOI] [PubMed] [Google Scholar]
- 39.Jorgensen, P., J. L. Nishikawa, B. J. Breitkreutz, and M. Tyers. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297:395-400. [DOI] [PubMed] [Google Scholar]
- 40.Kitamoto, H. K., D. Jablonowski, J. Nagase, and R. Schaffrath. 2002. Defects in yeast RNA polymerase II transcription elicit hypersensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol. Genet. Genomics 268:49-55. [DOI] [PubMed] [Google Scholar]
- 41.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 42.Lewis, L. K., and M. A. Resnick. 2000. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res. 451:71-89. [DOI] [PubMed] [Google Scholar]
- 43.Li, A. M., A. Watson, and J. L. Fridovich-Keil. 2003. Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res. 31:1830-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, H. Y., V. Badarinarayana, D. C. Audino, J. Rappsilber, M. Mann, and C. L. Denis. 1998. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17:1096-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, H. Y., J. H. Toyn, Y. C. Chiang, M. P. Draper, L. H. Johnston, and C. L. Denis. 1997. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 16:5289-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovett, S. T., and R. K. Mortimer. 1987. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics 116:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo, H. R., A. Saiardi, H. Yu, E. Nagata, K. Ye, and S. H. Snyder. 2002. Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase c1 mutant yeast. Biochemistry 41:2509-2515. [DOI] [PubMed] [Google Scholar]
- 48.Lydall, D., and T. Weinert. 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270:1488-1491. [DOI] [PubMed] [Google Scholar]
- 49.Maillet, L., and M. A. Collart. 2002. Interaction between Not1p, a component of the Ccr4-not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J. Biol. Chem. 277:2835-2842. [DOI] [PubMed] [Google Scholar]
- 50.Mendenhall, M. D., and A. E. Hodge. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1191-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merrill, B. J., and C. Holm. 1999. A requirement for recombinational repair in Saccharomyces cerevisiae is caused by DNA replication defects of mec1 mutants. Genetics 153:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore, C. W., J. McKoy, M. Dardalhon, D. Davermann, M. Martinez, and D. Averbeck. 2000. DNA damage-inducible and RAD52-independent repair of DNA double-strand breaks in Saccharomyces cerevisiae. Genetics 154:1085-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moriya, H., and K. Isono. 1999. Analysis of genetic interactions between DHH1, SSD1 and ELM1 indicates their involvement in cellular morphology determination in Saccharomyces cerevisiae. Yeast 15:481-496. [DOI] [PubMed] [Google Scholar]
- 54.Mueller, C. L., and J. A. Jaehning. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22:1971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neecke, H., G. Lucchini, and M. P. Longhese. 1999. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 18:4485-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ni, L., and M. Snyder. 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12:2147-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearce, A. K., and T. C. Humphrey. 2001. Integrating stress-response and cell-cycle checkpoint pathways. Trends Cell Biol. 11:426-433. [DOI] [PubMed] [Google Scholar]
- 58.Resnick, M. A., and B. S. Cox. 2000. Yeast as an honorary mammal. Mutat. Res. 451:1-11. [DOI] [PubMed] [Google Scholar]
- 59.Resnick, M. A., and P. Martin. 1976. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol. Gen. Genet. 143:119-129. [DOI] [PubMed] [Google Scholar]
- 60.Ruby, S. W., and J. W. Szostak. 1985. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol. Cell. Biol. 5:75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schild, D. 1995. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics 140:115-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Setlow, R. B. 2001. Human cancer: etiologic agents/dose responses/DNA repair/cellular and animal models. Mutat. Res. 477:1-6. [DOI] [PubMed] [Google Scholar]
- 63.Siede, W., A. S. Friedberg, and E. C. Friedberg. 1993. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:7985-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Super, H. J., J. Martinez-Climent, and J. D. Rowley. 1995. Molecular analysis of the Mono Mac 6 cell line: detection of an MLL-AF9 fusion transcript. Blood 85:855-856. [PubMed] [Google Scholar]
- 65.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 66.Tokunaga, M., J. E. Norman, Jr., M. Asano, S. Tokuoka, H. Ezaki, I. Nishimori, and Y. Tsuji. 1979. Malignant breast tumors among atomic bomb survivors, Hiroshima and Nagasaki, 1950-74. J. Natl. Cancer Inst. 62:1347-1359. [PubMed] [Google Scholar]
- 67.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 68.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 69.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 70.Vialard, J. E., C. S. Gilbert, C. M. Green, and N. F. Lowndes. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 72.Westmoreland, T. J., J. A. Olson, W. Y. Saito, G. Huper, J. R. Marks, and C. B. Bennett. 2003. DHH1 regulates the G1/S checkpoint following DNA damage or BRCA1 expression in yeast. J. Surg. Res. 113:62-73. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, J., C. Schneider, L. Ottmers, R. Rodriguez, A. Day, J. Markwardt, and B. L. Schneider. 2002. Genomic scale mutant hunt identifies cell size homeostasis genes in S. cerevisiae. Curr. Biol. 12:1992-2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.