Abstract
Fungi have an absolute requirement for K+, but K+ may be partially replaced by Na+. Na+ uptake in Ustilago maydis and Pichia sorbitophila was found to exhibit a fast rate, low Km, and apparent independence of the membrane potential. Searches of sequences with similarity to P-type ATPases in databases allowed us to identify three genes in these species, Umacu1, Umacu2, and PsACU1, that could encode P-type ATPases of a novel type. Deletion of the acu1 and acu2 genes proved that they encoded the transporters that mediated the high-affinity Na+ uptake of U. maydis. Heterologous expressions of the Umacu2 gene in K+ transport mutants of Saccharomyces cerevisiae and transport studies in the single and double Δacu1 and Δacu2 mutants of U. maydis revealed that the acu1 and acu2 genes encode transporters that mediated high-affinity K+ uptake in addition to Na+ uptake. Other fungi also have genes or pseudogenes whose translated sequences show high similarity to the ACU proteins of U. maydis and P. sorbitophila. In the phylogenetic tree of P-type ATPases all the identified ACU ATPases define a new cluster, which shows the lowest divergence with type IIC, animal Na+,K+-ATPases. The fungal high-affinity Na+ uptake mediated by ACU ATPases is functionally identical to the uptake that is mediated by some plant HKT transporters.
Potassium and Na+ were present in the sea, where the early evolution of life took place, but only K+ was selected to maintain the electrical and osmotic equilibria of the cells. Constricted by the K+ environment, further evolution of the cellular physiology added to the original requirements; the K+ dependence on many physiologic processes that adapted to the cation was present. This K+ dependence of living cells was not a restriction for life in the sea, where K+ was abundant, but involved a nutritional problem for the start of terrestrial life on the rocks emerging from the sea, probably in the Precambrian (27). Therefore, before plants and fungi could colonize terrestrial environments (12, 26) they had to sustain a complex process of physiological adaptations to a new medium in which K+ and Na+ concentrations were very low.
A crucial role in this terrestrial adaptation was played by high-affinity K+ transporters, whose existence has been well established for several plant and fungal species (44); but very little is known of other cooperative processes that may alleviate these strict K+ requirements. Among these, an emerging notion is that Na+ can be used as a substitute for K+. It has been known for a long time that Na+ enhances the growth of fungi (17) and plants (14, 28, 49) under conditions of K+ deficiency, but this effect had been considered accidental or under a weak physiological regulation. In contrast with this view, the characterization of high-affinity Na+ uptake in several plants and the fact that it takes place only when K+ has been exhausted (21) strongly suggest that Na+ plays a physiologically programmed role in plants given an insufficient supply of K+. Although this conclusion is simple, the process and regulation of high-affinity Na+ uptake may be complex and difficult to dissociate from K+ uptake, because the plant transporters that mediate this Na+ uptake (21) belong to a large family of K+ transporters (44). Notably, at least one of them, HKT1, from wheat, has been shown to function as an Na+-K+ symporter (47).
The notion of a physiologically programmed use of Na+ as a substitute for K+ and the mechanistic links between K+ and Na+ uptake raise the question of whether this may be a process that is not exclusive to plants but rather a general one that exists in nonanimal eukaryotic cells adapted to terrestrial environments. If this were the case, it should exist in fungi because of the parallel life of fungi and plants as soil inhabitants and their concurrent colonization of terrestrial environments. We investigated this possibility and found that high-affinity Na+ uptake does occur in fungi and is mediated by transporters whose sequences define a novel phylogenetic cluster of P-type ATPases that is close to that of animal Na+,K+- and H+,K+-ATPases. We show that these ATPases mediate high-affinity K+ uptake primarily and Na+ uptake secondarily, when K+ is at very low concentrations.
MATERIALS AND METHODS
Strains and growth conditions.
Ustilago maydis strain FB1 (a1b1) (7) and Pichia sorbitophila strain CBS7064 (6) were used throughout this study. The Escherichia coli strain DH5α was routinely used for plasmidic DNA propagation. The Saccharomyces cerevisiae strains used were W303.1A (Mat a ade2 ura3 trp1 leu2 his3) and its derivatives WΔ3 (Mat a ade2 ura3 trp1 trk1Δ::LEU2 trk2Δ::HIS3), which is deficient in the endogenous K+ uptake systems TRK1 and TRK2 (25), and MA5 (Mat a ade2 ura3 trp1 trk1Δ::LEU2 trk2Δ::HIS3 ena1-4Δ::HIS3 nha1Δ::LEU2), a strain obtained for this work by crossing the aforementioned WΔ3 with B31 (Mat a ade2 ura3 trp1 ena1-4Δ::HIS3 nha1Δ::LEU2) (6), which is deficient in the Na+ efflux systems ENA1-4 and NHA1. Fungal strains were normally grown either in the complex medium YPD (1% yeast extract, 2% peptone, 2% glucose) or in the minimal SD medium (48), supplemented with 50 mM K+ when used for the K+ uptake mutants. Growth at low K+ and Na+ concentrations occurred on arginine phosphate (AP) medium (46) supplemented with the indicated K+ and Na+ concentrations (the basal medium contains 5 μM K+ and 20 μM Na+).
Recombinant DNA techniques.
Manipulation of nucleic acids was performed by standard protocols or, when appropriate, according to the manufacturer's instructions. PCRs were performed with a Perkin-Elmer thermocycler by using the Expand-High-Fidelity PCR system (Roche Molecular Biochemicals). Some of the PCR fragments were first cloned into the PCR2.1-TOPO vector by using the TOPO TA cloning kit (Invitrogen). For yeast expressions, the genes were cloned into vector pYPGE15 (16). DNA sequencing was performed with an automated ABI PRISM 377 DNA sequencer (Perkin-Elmer). DNA and total RNA were prepared by using the DNeasy and RNeasy plant kits (Qiagen), respectively. PCR amplifications of mRNA fragments were carried out with double-stranded cDNA synthesized from total RNA by using the cDNA Synthesis System kit (Roche). The full-length ACU1 cDNA of P. sorbitophila was obtained by using the 5′/3′RACE Kit (Roche) as described earlier for other fungal ATPases (9). The full-length Umacu1 and Umacu2 cDNAs were obtained by reverse transcription-PCR from RNA extracted from K+-starved cells of U. maydis, by using specific primers whose sequences contained the predicted START and STOP codons. The three genes Umacu1, Umacu2, and PsACU1 were amplified from genomic DNA by PCR with the same primers as for the cDNAs. The pseudogene of Candida albicans was amplified from genomic DNA of strain SC5314 (37). The substitution of the AAG codon for the stop TAG codon at position 1066 was carried out by PCR in two steps by using primers that included one mutated nucleotide. First, we amplified a 1,110-bp DNA fragment by using a sense-specific primer upstream of the start codon NaK1 (5′-TTCAAGAAATCAACAAATAACATC-3′) and the antisense mutator primer NaKMUT-R (5′-ATCTAATAATTTCCTTTTGAAGTG-3′) (the changed nucleotide is underlined). We then amplified a 2,210-bp DNA fragment by using a sense mutator primer NaKMUT (5′-CCACTTCAAAAGGAAATTATTAGA-3′) (the changed nucleotide is underlined) and the antisense specific primer downstream of the stop codon NaK2 (5′-CTATAACTTCTTCCCCACCAATGA-3′). In a second step, a PCR was carried out by using the two DNA fragments obtained above as templates and primers NaK1 and NaK2, which amplify the complete gene.
Real-time PCR assays.
The results of real-time PCR assays reported in Table 1 were obtained for cells that were grown either in YPD or in AP minimal medium with 3 mM KCl. The latter were also exposed to the same medium with the following modifications (growth times are in parentheses): addition of 10 mM tartaric acid and pH brought to 3.5 with arginine (2 h); addition of 0.5 mM Ca2+ and pH brought to 8.0 with arginine (2 h); addition of 100 mM NaCl (2 h); without K+ (4 h); and without arginine and pH brought to 6.5 with KOH (6 h). The real-time PCR assays were performed as described previously (21), except that the standard DNA solutions corresponded to the genes studied in this report, acu1, acu2, and actin genes of U. maydis. These genes do not have introns, and the primers could not be designed to span over two gene exons. Therefore, the RNA preparations were treated with RNase-free DNase I (Roche) (40 U in 100 μl) for 1 h at 37°C. After the treatment, the RNA was purified following the method described in the RNeasy plant kit (Qiagen). The means and standard deviations of the ratios of the transporter to the actin cDNA contents were estimated as described in reference 34. Statistical analysis of the data reported in Table 1 proved that two values are significantly different if the largest is double or more than double the smallest. PCR primers were designed to amplify the following fragments of the Umacu1 and Umacu2 cDNAs (the numbering follows that of the databases): Umacu1, 2715 to 2892; Umacu2, 3208 to 3343; for the actin cDNA the primer sequences were 5′-GTGCCCATCTACGAAGGTTACT-3′ for UmACT1-2 and 5′-CGGCAGTGGTGGTGAAGGGGTAG-3′ for UmACT1-1R.
TABLE 1.
Effects of growth conditions on Umacu1 and Umacu2 transcripts
| Gene | Ratio (10−3) of transporter transcript abundance to actin transcript abundance for cells grown in:
|
||||||
|---|---|---|---|---|---|---|---|
| YPD | AP control medium | AP medium with indicated modificationa
|
|||||
| 100 mM Na+ | pH 3.5 | pH 8.0 | K+ starvation | N starvation | |||
| Umacu1 | <0.01 | 2.3 | 0.08 | 1.5 | 0.07 | 150 | 0.12 |
| Umacu2 | 0.17 | 8.1 | 0.31 | 6.7 | 0.23 | 530 | 0.93 |
Cells were grown in AP medium with 3 mM K+ and transferred to the indicated modified medium for 2 to 6 h as described in the text.
Disruption of acu1 and acu2 genes.
To obtain the Δacu1 mutant, we constructed a disruption plasmid by ligation of two DNA fragments flanking the acu1 open reading frame (ORF) into pNEBNat(+), a U. maydis integration vector containing a nourseothricin resistance cassette (35). The 5′ fragment spans from nucleotide −727 to nucleotide +153 (with the adenine of the ATG start codon considered nucleotide +1) and was obtained by PCR by using the primers Snb1b12UATG (5′-CGTACGTATCTCTCTCTGACTTGGCCGGTT-3′) and Nb1b12LATG (5′-CGATGCATTGTGGGAGCGGGTGGTCTGAGT-3′), which include, respectively, SnaBI and NsiI restriction sites for the appropriate cloning into the vector. The 3′ fragment spans from nucleotide +3276 to nucleotide +4172 and was obtained by PCR amplification using the primers Pb1b12USTOP (5′-CGCTGCAGACCGAATGATGCGACTGTAACT-3′) and Snb1b12LSTOP (5′-CGTACGTACATCAGCAGCAAAACCCAGTGT-3′), which include, respectively, PstI and SnaBI sites for the appropriate cloning into the vector. The resulting plasmid was linearized with XhoI and StuI and transformed into the FB1 U. maydis wild-type strain. Transformants were selected in YPD medium in the presence of nourseothricin (Hans-Knöll-Institute, Jena, Germany) (150 μg ml−1) and were screened for the loss of the wild-type copy of the gene by PCR analysis. The Δacu2 mutant was obtained by transforming a DNA fragment constructed with the acu2 gene border sequences flanking the hygromycin B resistance cassette into the FB1 U. maydis strain. The DNA fragment was constructed in two steps by PCR. In the first step, we amplified the two 5′ and 3′ flanking fragments of acu2, to which we added extension sequences that were identical to either the 5′ or the 3′ end of the hygromycin resistance cassette; and in the second step, we carried out a PCR protocol with three DNA templates, i.e., the two flanking fragments of acu2 described above and the hygromycin cassette obtained by NotI digestion of the U. maydis integration vector pSMUT (13). The 5′ fragment of acu2 spans from nucleotide +76 to nucleotide +661 (with the adenine of the ATG considered nucleotide +1) and was obtained by PCR by using the primers ATG10b23U (5′-GCCCCTACCAACAACGCCTCAAAG-3′) and ATG10b23LHyg (5′-CGATTCACGTTTTGTAGCACACTGACGACCACGACCAACAGCAC-3′) (the underlined nucleotides correspond to the sequence of the hygromycin B resistance cassette). The 3′ fragment of acu2 spans from nucleotide +2615 to nucleotide +3344 and was obtained by PCR by using the primers STOP10b23UHyg (5′-CCTTGCAGCACATCCCCCTTTCACGCTTTGCTCTACGGACGACT-3′), (the underlined nucleotides correspond to the sequence of the hygromycin B resistance cassette) and STOP10b23L (5′-AGCTTTCTCGTCTCCTCCAACA-3′). For the second PCR protocol, we used the ATG10b23U and STOP10b23L primers. Transformants were selected in YPD medium supplemented with hygromycin B (Sigma-Aldrich) (50 μg ml−1) and were screened for loss of the wild-type copy by PCR analysis. The double mutant Δacu1 Δacu2 was constructed by transforming Δacu1 with the linearized DNA construct used for the disruption of Δacu2 and by selection in YPD medium supplemented with hygromycin B.
Cation uptake experiments.
Cation uptake experiments were all performed with K+-starved cells. First, cells were grown in AP medium supplemented with 3 mM K+, washed in water, and then inoculated in K+- and Na+-free AP medium for 5 h. These starved cells were spun down and suspended in 10 mM morpholineethanesulfonic acid (MES) brought to pH 6.0 with ultrapure Ca(OH)2 and supplemented with 2% glucose (testing buffer). K+-starved cells with a high Na+ content for measuring K+ and Na+ exchange were prepared in the same way as regular K+-starved cells, except for an Na+ loading period of 30 min in 100 mM Na+ AP medium before the starvation step. The uptake tests were started by the addition of the selected cation, and then, at intervals, samples of the suspended cells were either centrifuged or filtered to determine either the time course of the depletion of the cation in the testing buffer or the increase of the cation content inside the cells. In both cases, the cation contents were determined by atomic emission spectrophotometry. Both methods, as well as the statistical analyses of the results, have been described previously (3, 24, 46). For each experiment, four or five independent repetitions were carried out.
Protein alignments and phylogenetic tree generations.
Protein sequence alignments and phylogenetic trees were obtained by using the Clustal X program (53).
Database searches and phylogenetic tree generations.
BLAST searches were carried out in the following databases: http://www.ncbi.nlm.nih.gov/ for S. cerevisiae and Schizosaccharomyces pombe; http://pedant.gsf.de for Neurospora crassa; Bayer CropScience AG, Monheim, Germany, for U. maydis; http://wwwgenome.wi.mit.edu/annotation/fungi/magnaporthe/ for Magnaporthe grisea; http://genolist.pasteur.fr for C. albicans; http://www.tigr.org/tdb/e2k1/afu1/ for Aspergillus fumigatus; and http://cogeme.ex.ac.uk for Mycosphaerella graminicola and Fusarium sporotrichioides. The genomic sequence of U. maydis has been published at http://www-genome.wi.mit.edu/annotation/fungi/ustilago_maydis/ recently.
DDBJ/EMBL/GenBank accession numbers.
The new sequences reported in this study have been submitted to DDBJ/EMBL/GenBank databases under the following accession numbers: U. maydis acu1, AJ622829; U. maydis acu2, AJ622830; P. sorbitophila ACU1, AJ622831; C. albicans ACU1, AJ622936; M. grisea ACU1, AJ622937; M. grisea ACU2, AJ622938.
RESULTS
High-affinity Na+ uptake.
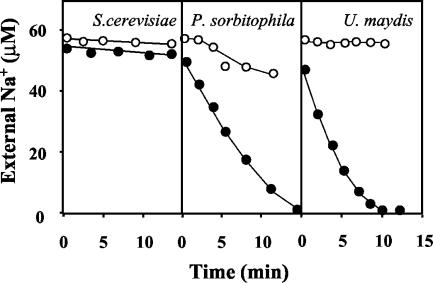
A high-affinity Na+ uptake, equivalent to that described for plants (21), has not been described for fungi. It can be asserted that it does not exist in S. cerevisiae, where alkali cation transport has been extensively investigated; for N. crassa, which is the second most researched fungal model for alkali cation transport studies (44), the low-rate, medium-affinity (Km = 0.1 mM Na+) uptake (36) is not equivalent to that of the plants. Therefore, we investigated Na+ uptake in a collection of fungi and found, after testing very few species, that P. sorbitophila and U. maydis did take up Na+ very rapidly at very low cation concentrations (<10 μM) and that S. cerevisiae did not take it up (Fig. 1). In both P. sorbitophila and U. maydis Na+-influx was strongly inhibited by the presence of micromolar K+ concentrations in a way that was very similar to that described for plants (21). In the same experiments, we found that the two species carried out high-affinity K+ uptake, which, as could be expected, was not inhibited by micromolar Na+ concentrations (not shown).
FIG. 1.
Depletion of external Na+ by P. sorbitophila and U. maydis. K+-starved cells of S. cerevisiae, P. sorbitophila, and U. maydis were suspended in testing buffer, with Na+ or Na+ plus K+ added to the suspension, and the variations of the external concentration of Na+ were recorded at intervals in the absence of K+ (closed circles) or in the presence of 50 μM K+ (open circles). The cell densities (dry weight, in milligrams per milliliter) in the experiments were as follows: S. cerevisiae, 0.7; P. sorbitophila, 0.9; and U. maydis, 0.7.
The Kms of Na+ influxes in P. sorbitophila and U. maydis were very low but difficult to calculate with precision because Na+ influx was influenced by the presence of K+ and by the K+ content of the cells. In experiments with K+-starved cells, the rate of Na+ uptake started to decrease when the Na+ taken up amounted to approximately 50 nmol mg−1. This effect, the rapid initial rate of uptake, and the very low Km reduced the effective sampling time of test experiments to just a few minutes, making the number of effective samples insufficient for a good statistical fitting of the results. However, from the data obtained in experiments using very low Na+ concentrations (<20 μM) we calculated that the Kms of Na+ influx in both U. maydis and P. sorbitophila were below 5 μM Na+.
Although a complete biochemical characterization of Na+ influxes was not tackled, we found that in both U. maydis and P. sorbitophila high-affinity Na+ uptake was not inhibited by Ba2+, which inhibits the corresponding uptake in plants (21). This suggested that a TRK-HKT transporter was not involved. Subsequently, we found that the uncoupler CCCP (carbonyl cyanide m-chlorophenylhydrazone), at 100 μM, inhibited high-affinity Na+ uptake completely in U. maydis, which always respires, but only partially in P. sorbitophila, which can ferment. The latter result suggested that Na+ influx was not a secondary process energized by the membrane potential. Taking into consideration the effects of CCCP in S. cerevisiae (24), it was extremely unlikely that a 100 μM concentration did not completely uncouple the plasma membrane of P. sorbitophila.
Cloning of the genes encoding the high-affinity Na+ transporters.
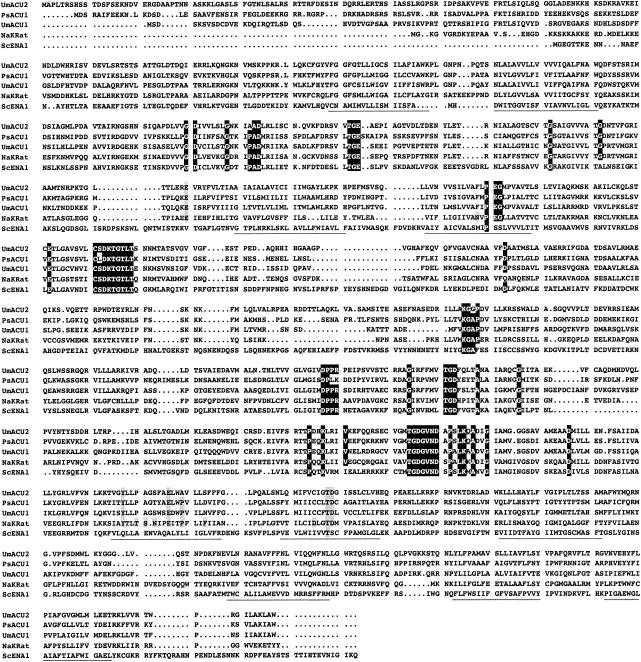
Assuming that the described high-affinity Na+ uptake was not a secondary process and was mediated by P-type ATPases, we carried out BLAST searches in the database GENOLEVURES (http://cbi.labri.u-bordeaux.fr/Genolevures/Genolevures.php3) for P. sorbitophila and in the database of the U. maydis genome sequence (Bayer CropScience). By using as queries the amino acid sequences of the S. cerevisiae and N. crassa ENA ATPases (10), we identified the partial sequence of a gene of P. sorbitophila (accession number, ax0aa002g12cp1) and two complete genes of U. maydis. All of them encoded a novel kind of P-type ATPases, which showed maximum sequence similarity to animal Na+,K+-ATPases. These findings fulfilled minimal conditions of functionality and prompted us to further investigate whether they mediated the Na+ uptake that we were studying. Therefore, we cloned the corresponding genes and cDNAs, i.e., P. sorbitophila ACU1 and U. maydis acu1 and acu2 (mnemonics for alkali cation uptake ATPase). In all three cases, the genes did not have introns and the ORFs comprised 3,218, 3,236, and 3,389 bp starting at the first ATG triplet of the ORFs and could encode polypeptides of 1,073, 1,079, and 1,130 amino acid residues with calculated molecular masses of 122.98, 119.68, and 123.50 kDa, respectively. Sequence analyses of the translated proteins proved that they met all the characteristics of typical P-type ATPases and that they conserved some of the amino acids involved in cation binding in Na+,K+-ATPases (Fig. 2).
FIG. 2.
ACU ATPases are typical P-type ATPases. Alignment of the translated sequences of the U. maydis acu1 and acu2 and P. sorbitophila ACU1 genes with the sequences of one animal Na+,K+-ATPase and one fungal ENA-ATPase. Conserved segments and the 10 putative transmembrane fragments of P-type ATPases (2, 33) are shown in black boxes and underlined, respectively. Amino acids which are essential for the binding of K+ or Na+ and for Na+ or K+ selectivity in Na+,K+-ATPases (30) are shown in grey. Accession numbers: NaKRat, M14511.1; ScENA1, P13587.
To investigate the involvement of the discovered transporters in the nutrition of the fungus under different growth conditions, the transcript abundance of the two genes of U. maydis was tested by real-time PCR (in Table 1 we list the ratios of the content of the cDNA corresponding to each gene to the content of the actin cDNA in the same sample). The rich medium YPD was used as the least stressing growth condition and minimal AP medium with 3 mM K+ as the basal condition over which we imposed high (8.0) or low (3.5) pH, 100 mM Na+, K+ starvation, or nitrogen starvation. The variations of the transcript abundance of the two genes followed similar patterns, although under all conditions the transcripts of acu2 were more abundant than those of acu1. In rich medium, the expression of acu2 was low and acu1 was not expressed. In minimal medium the only condition that clearly triggered strong expressions was K+ (and Na+) starvation. Under all the other conditions the transcript abundances of the ATPases were 10−2 to 10−4 times lower than those of actin, higher at pH 6.5 or 3.5 and lower at 100 mM Na+, pH 8.0, or nitrogen starvation. These results suggest that the UmAcu ATPases have only one function, which is triggered when the cells are suffering K+ starvation.
According to their sequence similarities, the ATPases encoded by the cloned genes could mediate K+ or Na+ uptake. Therefore, in order to study their functions, we inserted the genes in the yeast expression vector pYPGE15 (16) and transformed the constructs into a K+ transport mutant (trk1Δ trk2Δ) (32) of S. cerevisiae. For this mutant the Na+ and K+ uptake rates at low K+ or Na+ concentrations (<2 mM) are undetectable; tests aimed at solving the problem of the inhibition of the Na+ transport by K+ have been described (21). We obtained the transformants for PsACU1 and Umacu2 but not for Umacu1, probably because it was toxic for bacteria. Although many constructs of fungal cDNAs encoding P-type ATPases that are toxic for E. coli can be obtained directly with yeast (9, 10), in the case of Umacu1 all attempts failed.
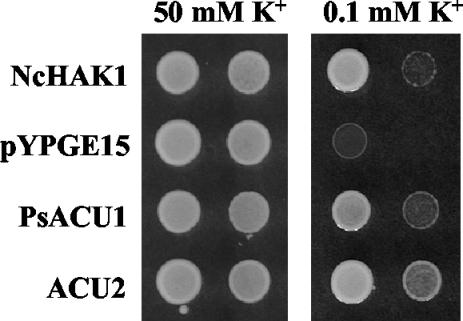
Growth tests with the PsACU1 or Umacu2 yeast transformants revealed that both genes suppressed the defective growth of the trk1Δ trk2Δ mutant at low K+ (the mutant grows poorly at 1 mM K+, and the transformants grew at 0.1 mM K+) in AP medium without Na+ (Fig. 3). These tests are normally carried out at pH 6.5, and at this pH PsACU1 and acu2 showed similar capacities for suppressing the defect of the trk1Δ trk2Δ mutant. At pH 5.0 the trk1Δ trk2Δ mutant is more defective, and the PsACU1 transformant did not grow at 0.3 mM K+, whereas the Umacu2 transformant grew at 0.1 mM K+ as rapidly as mutants expressing the N. crassa HAK1 (high-affinity K+) transporter (25). This suggested that both PsACU1 and Umacu2 encoded K+ uptake transporters and that the transporter encoded by Umacu2 was very active in yeast.
FIG. 3.
PsACU1 and Acu2 mediate K+ uptake, as shown by the suppression of the defective growth of a trk1Δ trk2Δ mutant of S. cerevisiae at low K+. Drops of cell suspensions of two serial dilutions of the mutant transformed with the N. crassa HAK1 cDNA (positive control), the empty pYPGE15 expression vector (negative control), and the PsACU1 and Acu2 genes were inoculated in AP medium supplemented with 50 or 0.1 mM KCl.
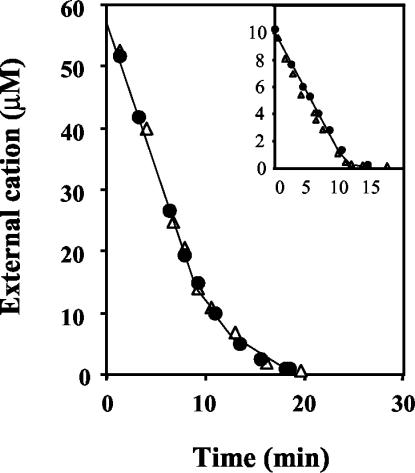
We then carried out uptake experiments in testing buffer. The PsACU1 transformant took up K+ and Na+ but did so too slowly to be studied with our experimental approach. In contrast, in the Umacu2 transformant the uptake was more rapid and we were able to perform uptake kinetic studies that demonstrated that Umacu2 encoded a transporter that mediated K+ or Na+ uptake. The high-affinity uptake kinetics exhibited by this transporter (Kms = 0.5 to 1.5 μM for both cations) and the decrease in rate when modest amounts of cations had been taken up (compare the response to the external cation in the main panel to the one in the inset in Fig. 4) suggested that the product of Umacu2 was involved in the Na+ uptake that we had found in U. maydis. Moreover, Umacu2 encoded a high-affinity uptake system that was similarly effective for Na+ and for K+. Although the uptake test results were less conclusive, it seems likely that PsACU1 encodes an ATPase similar to that encoded by Umacu2.
FIG. 4.
UmAcu2 transports K+ and Na+ in S. cerevisiae. Shown are results of the uptake tests of the trk1Δ trk2Δ mutant of S. cerevisiae transformed with the Umacu2 gene. Convenient amounts of K+ or Na+ were added to suspensions of K+-starved cells in testing buffer, and the variations of the external concentrations of cations were recorded for Na+ (closed circles) and K+ (open triangles). Experiments were started at 60 and 10 μM (inset) to show the regulation of the enzyme by the internal cations and its very low Km. Experiments were performed at the following cell densities (dry weights): main panel, 2.2 mg ml−1 for both the K+ and the Na+ experiments; inset, 0.6 mg ml−1 for both the K+ and the Na+ experiments.
UmAcu2 does not exchange K+ and Na+.
Sequence similarities between the ACU ATPases of U. maydis and P. sorbitophila with animal Na+,K+- and H+,K+-ATPases (see Fig. 6 below) suggested that the mechanism of K+ uptake of ACU ATPases was the exchange of external K+ with internal Na+ or H+. However, K+-Na+ exchanges could be ruled out in the experiments described above (Fig. 4) because K+-starved cells of the trk1 trk2 yeast mutant transformed with Umacu2 took over 100 nmol of K+ mg−1 although the cells had low amounts of Na+ (Umacu2 yeast cells grown with K+ in the absence of added Na+ did not have Na+, but during the starvation process they took up all the Na+ that contaminated the medium; typical acu2 K+-starved cells contained 300 and 30 nmol of K+ and Na+ mg−1, respectively). This suggested that the mechanisms of UmAcu2 could be similar to those of gastric (38) or nongastric (29) H+,K+-ATPases. Because the latter can exchange Na+ for K+ under certain conditions (19, 23, 40), we tested whether Acu2 exchanged Na+ and K+. For this purpose, we transformed the pYPGE15-Umacu2 plasmid into a yeast strain mutant for both K+ uptake and Na+ efflux (trk1Δ trk2Δ ena1-4Δ nha1Δ) and tested K+ uptake and Na+ loss in K+-starved cells that had been previously loaded with Na+, which contained 150 nmol of K+ and Na+ mg−1. In these cells, the rate of K+ uptake was not enhanced by the internal Na+ and Na+ was not lost. However, because it is impossible to prepare a buffer without Na+, the loss of intracellular Na+ had to be measured as the increment of the Na+ content in the testing buffer over the content at the beginning of the experiment. Considering the detection limits under these conditions, the conclusion of several experiments is that in more than 90% of the cycles the enzyme did not exchange Na+ for K+.
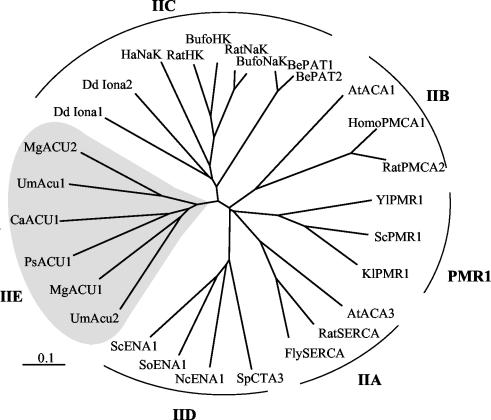
FIG. 6.
Phylogenetic relationships of fungal ACU ATPases with other P-type ATPases. Abbreviations, Um, U. maydis; Mg, M. grisea; Ps, P. sorbitophila; Ca, C. albicans; Dd, D. discoideum; Ha, H. akashiwo; Be, B. emersonii; At, Arabidopsis thaliana; Yl, Yarrowia lipolytica, Sc, S. cerevisiae; Kl, Kluyveromyces lactis; Sp, S. pombe; Nc, N. crassa; So, Schwanniomyces occidentalis. Accession numbers are as follows (in clockwise order): UmACU2, AJ622830; MgACU1, AJ622937; PsACU1, AJ622831; CaACU1, AJ622936; UmACU1, AJ622829; MgACU2, AJ622938; DdIona1, AC116977.2; DdIona2, GC1b09e04.r1 (http://genome.imb-jena.de); HaNaK, AB017481.2; ratHK, HKU94912.1; bufoHK, 2003318A; ratNaK, M14511.1; bufoNaK, S24650; BePAT1, T43025; BePAT2, AF205944.1; AtACA1, L08468.1; homoPMCA1, M95541.1; ratPMCA2, J03754.1; YlPMR1, U75447.1; ScPMR1, P13586; KlPMR1, AJ001018.1; AtACA3, AAF75073; ratSERCA, 1910193A; flySERCA, N62892.1; SpCTA3, P22189; NcENA1, AJ243520; SoENA1, AF030860; and ScENA1, P13587.
Disruption of the Umacu1 and Umacu2 genes.
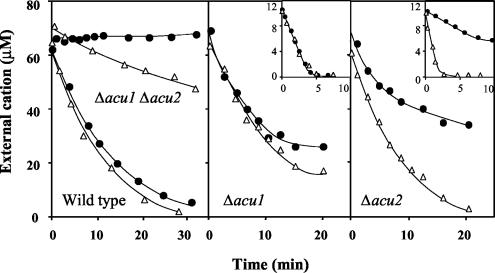
The transport functions of the Acu1 and Acu2 ATPases in U. maydis were then further investigated by undertaking the single and double disruptions of the genes. The hypothesis that the UmAcu1 and UmAcu2 ATPases mediated high-affinity K+ or Na+ uptake systems in U. maydis was confirmed with the Δacu1 Δacu2 double mutant. Here, high-affinity Na+ uptake completely disappeared and K+ uptake was slow in the micromolar range of concentrations (Fig. 5). By sequence analysis we further identified the presence of a trk1 gene in the genome of U. maydis, which suggested that K+ uptake in the Δacu1 Δacu2 double mutant could be mediated by a Trk1 transporter. In a preliminary study we found that the K1/2 of the rate of K+ uptake in the Δacu1 Δacu2 double mutant was approximately 0.3 mM K+.
FIG. 5.
Acu1 and Acu2 transport K+ and Na+ in U. maydis as shown by results for K+ and Na+ uptake in the wild-type strain, Δacu1 and Δacu2 single mutants, and Δacu1 Δacu2 double mutant of U. maydis. In the single mutants the experiments were started at 60 and 10 μM (inset) to show the regulation of the enzyme by the internal cations and its very low Km. Experiments, as described in the legend to Fig. 4 for measurement of Na+ (closed circles) and K+ (open triangles), were performed at the following cell densities (dry weights): wild-type strain, 0.49 mg ml−1 for both the K+ and Na+ experiments; Δacu1 Δacu2, 0.53 mg ml−1 for the K+ experiment and 0.94 mg ml−1 for the Na+ experiment; Δacu1 main panel, 0.58 mg ml−1 for both the K+ and Na+ experiments; Δacu1 inset, 0.30 mg ml−1 for both the K+ and Na+ experiments; Δacu2 main panel, 0.51 mg ml−1 for both the K+ and Na+ experiments; and Δacu2 inset, 0.60 mg ml−1 for both the K+ and Na+ experiments.
In experiments of K+ or Na+ depletion the single mutants Δacu1 and Δacu2 took up K+ or Na+ with high affinity, exhibiting rates that were very similar to those exhibited by the wild-type strain, except for Na+ uptake in the Δacu2 mutant, which was slower (Fig. 5). The existence of K+ uptake in the double mutant did not inconvenience the study of K+ uptake in the single Δacu1 and Δacu2 mutants because the K+ uptake rate in the double-mutant strain was much slower than in the single-mutant strains. Therefore, considering that our strains are completely isogenic, it was evident that high-affinity K+ and Na+ influxes were mediated by the Acu2 ATPase in the Δacu1 strain and by the Acu1 ATPase in the Δacu2 strain. To sum up, K+ uptake in K+-starved cells of U. maydis was dominated by the concurrent functions of Acu1 and Acu2 ATPases, whereas other transport systems, including Trk1, made a minor contribution. In contrast, high-affinity Na+ uptake was dominated only by Acu2 because the double mutant did not take up Na+ and Na+ uptake in the micromolar range of concentrations was much slower in the Δacu2 than in the Δacu1 strain.
The Kms for K+ and Na+ transport mediated by UmAcu2 and K+ uptake mediated by UmAcu1 could not be calculated with precision because, as was described for the wild-type strain, the very low Km and the high uptake rates, which decreased when the cells had taken up 50 nmol of K+ or Na+ mg−1, did not allow us to take enough samples for a good statistical fitting. In any case, the depletion curves of K+ and Na+ when the experiments were started at low concentrations (insets in Fig. 5) and the initial rates of Na+ uptake at constant cation concentrations (analyzing the cells) suggested that the Km values for K+ and Na+ of the UmAcu2 ATPase (Δacu1 strain) were very similar to those found for the yeast mutant expressing this ATPase (Km = 0.5 to 1.5 μM for both cations). The Km for K+ uptake mediated by UmAcu1 (Δacu2 strain) was also 0.5 to 1.5 μM K+, but the kinetics of the Na+ uptake mediated by this ATPase exhibited a much lower affinity (Km = 100 μM Na+, approximately). Consistent with the finding in the wild-type strain, Na+ uptake was inhibited by the presence of micromolar K+ concentrations in both the Δacu1 and the Δacu2 strains (not shown). However, unlike the inhibition of Na+ uptake by K+ in plant HKT transporters, which is the result of a blocking effect of K+ (21), the inhibition of Na+ uptake by K+ in the UmAcu1 and UmAcu2 ATPases might be explained by competitive inhibition of two cations that are both transported.
Experiments similar to those whose results are shown in Fig. 4 but which used Rb+ or Li+ demonstrated that the UmAcu2 ATPase transported Rb+ almost as effectively as it did K+ but did not transport Li+. Ammonium transport tests were not carried out, but we assessed the affinity of the UmAcu1 and UmAcu2 ATPases for NH4+ by measuring K+ uptake in the presence of NH4+. In experiments such as those whose results are shown in the insets of Fig. 5, the presence of 0.1 mM NH4+ did not produce any noticeable effect, and the K+ left in the external medium was still undetectable (<0.05 μM) at the end of the experiments. Even at 1 mM ammonium, the only appreciable effect was that K+ uptake ceased at a concentration of 0.1 μM. These results indicated that ammonium (at a concentration of up to 1 mM) is not transported or is poorly transported by the UmAcu ATPases.
ACU ATPases possibly exist in many fungi.
In the genome of S. cerevisiae there are no genes encoding ATPases that are similar to the ACU ATPases of P. sorbitophila and U. maydis (18), and we also found that they are absent in the genomes of N. crassa and S. pombe. However, we identified one gene in C. albicans and two genes in M. grisea that suggested that these ATPases may exist in many other fungi. Fragments of putative genes were also found in M. graminicola (COGEME accession number, mga0392f), F. sporotrichioides (COGEME accession number, FS-t2a06fs.fl), and A. fumigatus (gnl|TIGR_5085|contig:4901:a_fumigatus and gnl|TIGR_5085|contig:4951:a_fumigatus). Interestingly, the C. albicans sequence was apparently a pseudogene, because there was a stop codon in position 1066 (with the adenine of the first ATG codon in the ORF considered nucleotide +1). The complete pseudogene comprised 3,243 bp and apparently did not have introns. We cloned the putative gene and sequenced it, finding that the sequence in the database was correct. The mutation of the TAG stop codon to AAG, which encodes a lysine residue, as in PsACU1, was sufficient to restore the activity of the pseudogene. The mutated pseudogene suppressed the defective growth of the trk1Δ trk2Δ mutant at low K+ in a manner almost identical to that described for PsACU1 (Fig. 3).
A phylogenetic study of the ACU ATPases along with other P-type ATPases revealed that ACU ATPases were more related to animal Na+,K+- or H+,K+-ATPases (type IIC [2]) than to Ca2+- or ENA-ATPases (types IIA, IIB, and IID). However, they are rather divergent from type IIC sequences, including those of Heterosigma akashiwo, Blastocladiella emersonii (20), and Dictyostelium discoideum (Fig. 6), and from the prokaryotic sequences recorded elsewhere (11). The divergence, estimated as the number of amino acid changes per 100 positions, between animal Na+,K+-ATPases and fungal ACU ATPases is similar to that existing between plasma membrane and SERCA Ca2+-ATPases (types IIA and IIB [2]), and for that reason we propose naming fungal ACU ATPases type IIE (Fig. 6).
Several high-affinity K+ transporters coexist in fungi.
High-affinity K+ uptake in fungi is mediated by HAK transporters (4, 25), and this implies a certain degree of functional redundancy if ACU ATPases coexist with HAK transporters. This redundancy is less significant if they coexist with TRK K+ transporters, because these transporters have a lower concentrative capacity and, in fact, TRK transporters coexist with HAK transporters (5, 25). Therefore, to update the models of K+ uptake in fungi we investigated the presence of TRK and HAK transporters and ACU ATPases in those fungal species for which the complete genomic sequences are available, i.e., S. cerevisiae, S. pombe, C. albicans, U. maydis, and M. grisea. Table 2 summarizes the results, which revealed that the models of K+ uptake in fungi are very diverse. The only type of K+ transporters that seems to be present in all fungi is TRK, probably because these transporters have a role in controlling the membrane potential (5, 32). The HAK transporters or ACU ATPases may be present or not, either together or independently.
TABLE 2.
Number of TRK, HAK, and ACU genes in the genomes of fungi for which the complete sequences are availablea
| Species | No. of genes in genome
|
||
|---|---|---|---|
| TRK | HAK | ACU | |
| S. cerevisiae | 2 | 0 | 0 |
| S. pombe | 2 | 0 | 0 |
| N. crassa | 2 | 1 | 0 |
| U. maydis | 1 | 0 | 2 |
| M. grisea | 2 | 1 | 2 |
| C. albicans | 1 | 1 | —b |
BLAST searches were carried out with NcTRK1, NcHAK1, and PsACU1 protein sequences against the translated genomic sequences in the databases given in Materials and Methods.
—, this species has a pseudogene, as explained in the text.
DISCUSSION
The existence of active K+ uptake in fungi and plants was established many years ago because, under certain growth conditions, the membrane potential is positive relative to the equilibrium potential of K+ across the plasma membrane (44). When this existence was established, the most persistent notion, which arose from reasoning by analogy with the Na+ pump of animal cells (15), was that a K+-ATPase mediated K+ uptake (31, 39, 51, 52, 55, 56). However, the incomplete biochemical support for the K+-ATPase and the findings that active K+ uptake was mediated by a K+-H+ symporter in N. crassa (45) caused the attention given to active K+ uptake in fungi and plants to be addressed almost exclusively to secondary transporters. Later, the identification of the HKT (47) and HAK (4) transporters, which were able to mediate highly concentrative K+ uptake, led to the complete disappearance of the hypothesis of the K+-ATPase. This was unfounded, in light of the results of the present report. The study of K+ uptake in the Δacu1 and Δacu2 mutants in comparison to the Δacu1 Δacu2 double mutant in U. maydis, as well as in the S. cerevisiae trk1 trk2 mutant expressing UmAcu2p, demonstrates that the Umacu1 and Umacu2 genes encode high-affinity K+ transporters. This conclusion, together with the unmistakable similarity of the translated sequences of all ACU proteins to P-type ATPases (Fig. 2 and 6), allows us to classify the ACU transporters as K+-ATPases. The extraordinary capacity exhibited by the UmAcu1 and UmAcu2 ATPases (as observed in the Δacu2 and Δacu1 mutants, respectively) for depleting the K+ of the external medium and concentrating it in the cells more than 106-fold, their very low Km (possibly as low as 0.5 μM K+), and their almost exclusive expression under conditions of K+ starvation suggest that the function of these ATPases is to take up K+ from media in which the cation is at extremely low concentrations—a function that is similar to that reported for the Kdp-ATPase of E. coli (43).
The old discussion about plant K+-ATPases raises a question about the existence of ACU ATPases in plants. They do not exist in Arabidopsis or rice (8), and we did not find in databases a plant gene sequence that could encode an ACU ATPase. However, these findings cannot be taken as a definitive proof. First, Arabidopsis and rice are not representative of all plant species and ACU ATPases do not exist in all fungi. Second, the absence of expressed sequence tags with ACU sequences in databases can be expected if ACU mRNAs are expressed only in K+-starved plants and consequently, the corresponding cDNAs do not exist in current libraries.
In U. maydis, the Acu2 ATPase mediated a high-rate, high-affinity Na+ uptake and Acu1 was also an Na+ transporter, albeit with lower activity. These findings, the sequence similarities, and the phylogenetic relationships (Fig. 6) strongly suggest that, in general, fungal ACU ATPases have the function of K+ or Na+ uptake described here for the U. maydis ATPases. However, neither the question of whether some ACU ATPases transport K+ or Na+ exclusively nor that of whether Na+ transport may be the primary function in some of them can be answered now. The simplest hypothesis, however, is that both functions take place concurrently, though with different degrees of discrimination between K+ and Na+, and that ACU ATPases do not exchange Na+ and K+. The latter, which was demonstrated for UmAcu2, is also a predictable consequence of the former, to avoid a futile Na+-Na+ exchange. According to this, ACU ATPases would resemble gastric H+,K+-ATPases, in which Na+ acts as a K+ analog (50), and would be different from nongastric H+,K+-ATPases, which may function as Na+,K+-ATPases (19, 23, 40).
The expressions of the P. sorbitophila gene and C. albicans mutant gene in S. cerevisiae were only moderately active, but this is not surprising because there are many examples of P-type ATPases that do not even have functional expression in S. cerevisiae (9, 11). Gastric and nongastric H+,K+-ATPases, as well as Na+,K+-ATPases, function with a second β-subunit (22), but it is unlikely that the lack of a β-subunit causes the aforementioned moderate functional expression of PsACU1. The existence of β-subunits should apply to all ACU ATPases, and the remarkable function of UmAcu2 in S. cerevisiae rules out the requirement of a second subunit. Furthermore, we did not find genes encoding proteins similar to the animal β-subunit in the genomes of U. maydis and M. grisea.
In functional terms, the Na+ uptake mediated by ACU ATPases is similar to the high-affinity Na+ uptake of plants, which is mediated by HKT secondary transporters (21). The mechanistic differences between the two transporters demonstrate that high-affinity Na+ uptake developed independently in fungi and in plants and suggest that Na+ uptake probably fulfilled a very important biological function in the evolution of plants and fungi. Such an important function does not apply to all contemporary fungi, because high-affinity Na+ uptake does not exist in some of them (e.g., S. cerevisiae). Therefore, a likely hypothesis is that the importance of Na+ uptake and, hence, the selective pressure that determined the characteristics of the fungal ACU ATPases and plant HKT transporters took place early in the evolution of fungi and plants. This probably occurred when associations of fungi and plants colonized the harsh environment of the emerged land and continental water in the Cambrian era (12) or even earlier (26, 27). In these environments, the mineral nutrients were at very low levels and Na+ was probably used as a substitute for K+. Although K+ is indispensable in fungi and plants, Na+ can substitute for a substantial amount of K+ with very little toxicity. In S. cerevisiae Na+ can substitute for about one-half of the normal K+ content with very little toxicity (17). Because in S. cerevisiae most of the Na+ is confined to the inside of the vacuole (54; K. Venema, personal communication), it can be expected that cells with a big vacuole can sequester in it significant amounts of Na+, thus avoiding its toxic effects in the cytoplasm (1) and reducing notably the K+ requirements. This use of Na+ could have been a selective advantage for millions of years in the oligotrophic soils of Paleozoic ages (41, 42), and it may persist in many contemporary oligotrophic environments in which fungi and plants thrive.
K+ uptake in fungi is especially complex because they thrive in very diverse environments, which implies a great variability in the K+ concentrations to which fungi are exposed. For example, K+ concentrations are very low in many soils but very high in decaying plant materials or in the environment of fungus-plant symbioses (10). Some species meet all these environmental variations, such as plant pathogens that also grow in soil, whereas other species are adapted to more constant environments, such as fermenting yeast. Therefore, the variability of the transport systems among different fungal species (Table 2) is not surprising, although it cannot currently be explained on functional grounds. The absence of HAK transporters and ACU ATPases in S. cerevisiae may be explained by the high K+ content of fruit, an environment in which the organism thrives, but the coexistence of ACU ATPases, which exhibited excellent performances for transporting both K+ and Na+, and HAK transporters, which transport only K+ (44), in M. grisea does not have a simple explanation now.
Acknowledgments
We thank José Pérez-Martín for introducing the Madrid group to the study of U. maydis (the transformations of U. maydis reported here were carried out in his laboratory), Regine Kahmann for the gift of plasmids pNEBHyg(+)dEcoRI and pNEB.Nat(+), Flora Banuett for the gift of strain FB1, and Jesús Pla for the gift of a sample of C. albicans DNA. The technical assistance of Marcel Veldhuizen and Ursula González-Espinosa is acknowledged. We also thank Bayer CropScience for making Ustilago sequences available prior to publication, the Magnaporthe Sequencing Project Ralph Dean, Fungal Genomics Laboratory at North Carolina State University (www.fungalgenomics.ncsu.edu), and the Whitehead Institute/MIT Center for Genomic Research (www-genome.wi.mit.edu) for granting us the use of the M. grisea database (release 2.1-1/9/2003).
This work was supported by the Consejería de Educación y Cultura de la Comunidad de Madrid (Programa de Grupos Estratégicos and postdoctoral fellowship to B.B.) and by the Ministerio de Ciencia y Tecnología (grant no. AGL2001-2716).
REFERENCES
- 1.Apse, M. P., G. S. Aharon, W. A. Snedden, and E. Blumwald. 1999. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256-1258. [DOI] [PubMed] [Google Scholar]
- 2.Axelsen, K. B., and M. G. Palmgren. 1998. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46:84-101. [DOI] [PubMed] [Google Scholar]
- 3.Bañuelos, M. A., B. Garciadeblas, B. Cubero, and A. Rodríguez-Navarro. 2002. Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 130:784-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bañuelos, M. A., R. D. Klein, S. J. Alexander-Bowman, and A. Rodríguez-Navarro. 1995. A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO J. 14:3021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bañuelos, M. A., R. Madrid, and A. Rodríguez-Navarro. 2000. Individual functions of the HAK and TRK potassium transporters of Schwanniomyces occidentalis. Mol. Microbiol. 37:671-679. [DOI] [PubMed] [Google Scholar]
- 6.Bañuelos, M. A., J. Ramos, F. Calero, V. Braun, and S. Potier. 2002. Cation/H+ antiporters mediate potassium and sodium fluxes in Pichia sorbitophila. Cloning of the PsHHA1 and PsNHA2 genes and expression in Saccharomyces cerevisiae. Yeast 19:1365-1372. [DOI] [PubMed] [Google Scholar]
- 7.Banuett, F., and I. Herskowitz. 1989. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baxter, I., J. Tchieu, M. R. Sussman, M. Boutry, M. G. Palmgren, M. Griskov, J. F. Harper, and K. B. Axelsen. 2003. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 132:618-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benito, B., B. Garciadeblas, and A. Rodríguez-Navarro. 2000. Molecular cloning of the calcium and sodium ATPases in Neurospora crassa. Mol. Microbiol. 35:1079-1088. [DOI] [PubMed] [Google Scholar]
- 10.Benito, B., B. Garciadeblás, and A. Rodríguez-Navarro. 2002. Potassium- or sodium-efflux ATPase, a key enzyme in the evolution of fungi. Microbiology 148:933-941. [DOI] [PubMed] [Google Scholar]
- 11.Benito, B., and A. Rodríguez-Navarro. 2003. Molecular cloning and characterization of a sodium-pump ATPase of the moss Physcomitrella patens. Plant J. 36:382-389. [DOI] [PubMed] [Google Scholar]
- 12.Blackwell, M. 2000. Terrestrial life—fungal from the start? Science 289:1884-1885. [DOI] [PubMed] [Google Scholar]
- 13.Bölker, M., H. U. Böhnert, K. H. Brown, J. Görl, and R. Kahmann. 1995. Tagging pathogenicity genes in Ustilago maydis by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 248:547-552. [DOI] [PubMed] [Google Scholar]
- 14.Box, S., and D. P. Schachtman. 2000. The effect of low concentration of sodium on potassium uptake and growth of wheat. Aust. J. Plant Physiol. 27:175-182. [Google Scholar]
- 15.Briskin, D. P. 1990. The plasma membrane H+-ATPase of higher plant cells: biochemistry and transport function. Biochim. Biophys. Acta 1019:95-109. [Google Scholar]
- 16.Brunelli, J. P., and M. L. Pall. 1993. A series of yeast/Escherichia coli λ expression vectors designed for directional cloning of cDNAs and cre/lox-mediated plasmid excision. Yeast 9:1309-1318. [DOI] [PubMed] [Google Scholar]
- 17.Camacho, M., J. Ramos, and A. Rodríguez-Navarro. 1981. Potassium requirements of Saccharomyces cerevisiae. Curr. Microbiol. 6:295-299. [Google Scholar]
- 18.Catty, P., A. de Kerchove d'Exaerde, and A. Goffeau. 1997. The complete inventory of the yeast Saccharomyces cerevisiae P-type transport ATPases. FEBS Lett. 409:325-332. [DOI] [PubMed] [Google Scholar]
- 19.Cougnon, M., P. Bouyer, G. Planelles, and F. Jaisser. 1998. Does the colonic H,K-ATPase also act as an Na,K-ATPase? Proc. Natl. Acad. Sci. USA 95:6516-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fietto, L. G., L. Pugliese, and S. L. Gomes. 2002. Characterization and expression of two genes encoding isoforms of a putative Na, K-ATPase in the chytridiomycete Blastocladiella emersonii. Biochim. Biophys. Acta 1576:59-69. [DOI] [PubMed] [Google Scholar]
- 21.Garciadeblás, B., M. E. Senn, M. A. Bañuelos, and A. Rodríguez-Navarro. 2003. Sodium transport and HKT transporters: the rice model. Plant J. 34:788-801. [DOI] [PubMed] [Google Scholar]
- 22.Geering, K. 2001. The functional role of beta subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 33:425-438. [DOI] [PubMed] [Google Scholar]
- 23.Grishin, A. V., and M. J. Caplan. 1998. ATP1AL1, a member of the non-gastric H,K-ATPase family, functions as a sodium pump. J. Biol. Chem. 273:27772-27778. [DOI] [PubMed] [Google Scholar]
- 24.Haro, R., and A. Rodríguez-Navarro. 2002. Molecular analysis of the mechanism of potassium uptake through the TRK1 transporter of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1564:114-122. [DOI] [PubMed] [Google Scholar]
- 25.Haro, R., L. Sainz, F. Rubio, and A. Rodríguez-Navarro. 1999. Cloning of two genes encoding potassium transporters in Neurospora crassa and expression of the corresponding cDNAs in Saccharomyces cerevisiae. Mol. Microbiol. 31:511-520. [DOI] [PubMed] [Google Scholar]
- 26.Heckman, D. S., D. M. Geiser, B. R. Eidell, R. L. Stauffer, N. L. Kardos, and S. B. Hedges. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293:1129-1133. [DOI] [PubMed] [Google Scholar]
- 27.Horodyski, R. J., and L. P. Knauth. 1994. Life on land in the Precambrian. Science 263:494-498. [DOI] [PubMed] [Google Scholar]
- 28.Hylton, I. O., A. Ulrich, and D. R. Cornelius. 1967. Potassium and sodium interrelations in growth and mineral content of Italian ryegrass. Agronomy J. 59:311-314. [Google Scholar]
- 29.Jaisser, F., and A. T. Beggah. 1999. The non-gastric H+-K+-ATPases: molecular and functional properties. Am. J. Physiol. 276:F812-824. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen, P. L., and P. A. Pedersen. 2001. Structure-function relationships of Na+, K+, ATP, or Mg2+ binding and energy transduction in Na,K-ATPase. Biochim. Biophys. Acta 1505:57-74. [DOI] [PubMed] [Google Scholar]
- 31.Leonard, R. T., and C. W. Hotchkiss. 1976. Cation-stimulated adenosine triphosphatase activity and cation transport in corn roots. Plant Physiol. 58:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madrid, R., M. J. Gómez, J. Ramos, and A. Rodríguez-Navarro. 1998. Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 273:14838-14844. [DOI] [PubMed] [Google Scholar]
- 33.Møller, J. V., B. Juul, and M. le Maire. 1996. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta 1286:1-51. [DOI] [PubMed] [Google Scholar]
- 34.Mood, A. M., F. A. Graybill, and D. C. Boes. 1974. Introduction to the theory of statistics, 3rd ed. McGraw-Hill Book Co., London, United Kingdom.
- 35.Müller, P., C. Aichinger, N. Feldbrugge, and R. Kahmann. 1999. The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34:1007-1017. [DOI] [PubMed] [Google Scholar]
- 36.Ortega, M. D., and A. Rodríguez-Navarro. 1986. Sodium ion transport in Neurospora crassa. Physiol. Plant 66:705-711. [Google Scholar]
- 37.Ponzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabon, E. C., and M. A. Reuben. 1990. The mechanism and structure of the gastric H,K-ATPase. Annu. Rev. Physiol. 52:321-344. [DOI] [PubMed] [Google Scholar]
- 39.Rahat, M., and L. Reinhold. 1983. Rb+ uptake by isolated pea mesophyll protoplasts in light and darkness. Physiol. Plant 59:83-90. [Google Scholar]
- 40.Rajendran, V. M., P. Sangan, J. Geibel, and H. J. Binder. 2000. Ouabain-sensitive H,K-ATPase functions as Na,K-ATPase in apical membranes of rat distal colon. J. Biol. Chem. 275:13035-13040. [DOI] [PubMed] [Google Scholar]
- 41.Retallack, G. J. 1997. Early forest soils and their role in Devonian global change. Science 276:483-585. [DOI] [PubMed] [Google Scholar]
- 42.Retallack, G. J., and J. Germán-Heins. 1994. Evidence from paleosols for the geological antiquity of rain forest. Science 265:499-502. [DOI] [PubMed] [Google Scholar]
- 43.Rhoads, D. B., F. B. Waters, and W. Epstein. 1976. Cation transport in Escherichia coli. VIII. Potassium transport mutants. J. Gen. Physiol. 67:325-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodríguez-Navarro, A. 2000. Potassium transport in fungi and plants. Biochim. Biophys. Acta 1469:1-30. [DOI] [PubMed] [Google Scholar]
- 45.Rodríguez-Navarro, A., M. R. Blatt, and C. L. Slayman. 1986. A potassium-proton symport in Neurospora crassa. J. Gen. Physiol. 87:649-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodríguez-Navarro, A., and J. Ramos. 1984. Dual system for potassium transport in Saccharomyces cerevisiae. J. Bacteriol. 159:940-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubio, F., W. Gassmann, and J. I. Schroeder. 1995. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270:1660-1663. [DOI] [PubMed] [Google Scholar]
- 48.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 49.Subbarao, G. V., R. M. Wheeler, G. W. Stutte, and L. H. Levine. 1999. How far can sodium substitute for potassium in red beet? J. Plant Nutr. 22:1745-1761. [DOI] [PubMed] [Google Scholar]
- 50.Swarts, H. G. P., C. H. W. Klaassen, F. M. Schuurmans Stekhoven, and J. J. De Pont. 1995. Sodium acts as a potassium analog on gastric H,K-ATPase. J. Biol. Chem. 270:7890-7895. [DOI] [PubMed] [Google Scholar]
- 51.Sze, H. 1980. Nigericin-stimulated ATPase activity in microsomal vesicles of tobacco cells. Proc. Natl. Acad. Sci. USA 77:5904-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sze, H., and K. A. Churchill. 1981. Mg2+/KCl-ATPase of plant plasma membranes is an electrogenic pump. Proc. Natl. Acad. Sci. USA 78:5578-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J. D. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venema, K., A. Belver, M. C. Marín-Manzano, M. P. Rodríguez-Rosales, and J. P. Donaire. 2003. A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. J. Biol. Chem. 278:22453-22459. [DOI] [PubMed] [Google Scholar]
- 55.Villalobo, A. 1984. Energy dependent H+ and K+ translocation by reconstituted yeast plasma membrane ATPase. Can. J. Biochem. Cell Biol. 62:865-877. [Google Scholar]
- 56.Villalobo, A. 1982. Potassium transport coupled to ATP hydrolysis in reconstituted proteoliposomes of yeast plasma membrane ATPase. J. Biol. Chem. 257:1824-1828. [PubMed] [Google Scholar]