Abstract
During an infection, the coordinated orchestration of many factors by the invading organism is required for disease to be initiated and to progress. The elucidation of the processes involved is critical to the development of a clear understanding of host-pathogen interactions. For Candida species, the inactivation of many fungal attributes has been shown to result in attenuation. Here we demonstrate that the Candida glabrata homolog of the Saccharomyces cerevisiae transcription factor gene ACE2 encodes a function that mediates virulence in a novel way. Inactivation of C. glabrata ACE2 does not result in attenuation but, conversely, in a strain that is hypervirulent in a murine model of invasive candidiasis. C. glabrata ace2 null mutants cause systemic infections characterized by fungal escape from the vasculature, tissue penetration, proliferation in vivo, and considerable overstimulation of the proinflammatory arm of the innate immune response. Compared to the case with wild-type fungi, mortality occurs much earlier in mice infected with C. glabrata ace2 cells, and furthermore, 200-fold lower doses are required to induce uniformly fatal infections. These data demonstrate that C. glabrata ACE2 encodes a function that plays a critical role in mediating the host-Candida interaction. It is the first virulence-moderating gene to be described for a Candida species.
The number of invasive life-threatening infections caused by the members of the fungal genus Candida has increased dramatically over the past 20 years (1, 2). Candida species now cause more bloodstream infections than streptococci, Enterococcus faecalis, and all individual gram-negative bacterial species, including Escherichia coli (30). Indeed, candidemia now accounts for 10 to 15% of all bloodstream infections (18), one of the top 15 causes of mortality in the United States, resulting in over 200,000 deaths per year (24). Given the continuing use of broad-spectrum antibiotics and indwelling intravenous catheters, the two major risk factors for the development of candidemia, plus the inexorable rise of at-risk patients, it seems unlikely that this threat will diminish (6). Candida species thus exert a significant clinical and economic impact.
Candida albicans remains the most commonly isolated species, but Candida glabrata now accounts for almost 20% of all cases of systemic candidiasis (6) and 30% of urinary tract infections (3). This increase in the incidence of C. glabrata infections is a major cause for concern, as this species is often more resistant to antifungal agents and results in a higher overall mortality rate than other Candida species (11, 26).
Analyses of Candida virulence facilitate the elucidation of critical aspects of the host-pathogen interaction, which may result in the development of better options for therapeutic and/or diagnostic interventions. The majority of analyses have focused on C. albicans, although we have recently shown that the transcriptional regulator Ste12 plays a role in C. glabrata virulence (5). These investigations have demonstrated that genes which encode functions in many diverse areas of Candida biology, including environmental sensing, transcriptional responsiveness, adhesion, and morphogenesis, are crucial for the initiation and/or progression of candidiasis (14, 21). Like most microbial virulence studies, these experiments have focused on the identification of traits that are essential for virulence. The converse has not been explored, i.e., the identification of genes whose inactivation results in an augmented ability to cause disease. These so-called antivirulence genes have recently been described as crucial determinants of the host-pathogen interaction for several different microbial species (9). We prefer and will use hereafter the term “virulence moderating.” The inactivation of virulence-moderating genes can result in an enhanced capacity of the organism to cause tissue damage, to increase in vivo proliferation, and to require lower lethal doses, resulting in more severe disease and higher mortality rates (16, 29). These virulence-moderating genes represent a new and exciting aspect of microbial virulence. Their identification and subsequent functional characterization in Candida species may have the power to reveal novel insights into host-fungus interactions.
In order to perform a global analysis of C. glabrata virulence that would reveal potential virulence-moderating genes and genes encoding functions essential for virulence, we constructed a 9,600-member signature tag (15) library of insertional mutants (19). A preliminary analysis of this library revealed the presence of a strain (10G6) with an apparent increased ability to persist in vivo in a murine model of candidiasis (5). We hypothesized that this strain may contain a disruption of a C. glabrata virulence-moderating gene. Our objective for this study was to test this hypothesis. Specifically, we sought to determine the site of plasmid insertion in C. glabrata 10G6, to construct independent null and reconstituted mutants of any disrupted gene, and to determine the impact of these mutations on the ability of C. glabrata to cause disease. Our data show that C. glabrata 10G6 has an inactivated allele of the C. glabrata homolog of the Saccharomyces cerevisiae transcriptional activator-encoding gene ACE2. Furthermore, we demonstrate that C. glabrata ace2 mutants are hypervirulent in a murine model of candidiasis. This hypervirulence is independent of the ace2 clumpy growth phenotype, at the time of infection, and is accompanied by a massive overstimulation of the proinflammatory arm of the innate immune response. C. glabrata ACE2 is thus the first virulence-moderating gene to be reported for a Candida species.
MATERIALS AND METHODS
C. glabrata strains and growth media.
All strains used for this study are listed in Table 1. Fungal cells were routinely cultured in YAPD (2% [wt/vol] peptone, 2% [wt/vol] glucose, 1% [wt/vol] yeast extract, 0.01% [wt/vol] adenine) or SD (0.17% [wt/vol] yeast nitrogen base without amino acids [Difco], 2% [wt/vol] glucose, and an appropriate dropout mix [Clontech, Basingstoke, United Kingdom]) medium at 30 or 37°C. For solid media, 2% (wt/vol) agar was added prior to autoclaving. Other additives were filter sterilized and added at appropriate concentrations to the media after autoclaving.
TABLE 1.
Fungal strains used for this study
| C. glabrata strain | Genotype or description | Source or reference |
|---|---|---|
| ATCC 2001 | Type strain | American Type Culture Collection |
| ΔH1 | his3::URA3 Δura3 | 28 |
| ΔHT6 | his3::URA3 Δura3 Δtrp1 | 28 |
| 10G6 | his3::URA3 Δura3 Δace2-1a | 19 |
| HLS120 | his3::URA3 Δura3 Δtrp1 Δace2::HIS3 | This study |
| HLS122 | his3::URA3 Δura3 Δtrp1 Δace2::HIS3(pCgACT-14 [TRP1]) | This study |
| HLS121 | his3::URA3 Δura3 Δtrp1 Δace2::HIS3(pKH355 [ACE2 TRP1]) | This study |
pTW23 was inserted between nucleotides 452 and 453 of ACE2 in this strain.
Construction and preliminary analysis of a C. glabrata STM library.
To undertake a functional analysis of virulence in C. glabrata, we constructed a 9,600-member STM insertional mutant library (19). Briefly, a pool of signature tags with the overall sequence CTAGGTACCTACAACCTCAAGCTT-(NK)20-AAGCTTGGTTAGAATGGGTACCATG was amplified essentially as described by Hensel et al. (15). The tag pool was digested with KpnI and ligated into pTW23, which contains the C. glabrata HIS3 gene on a pSKII+ backbone (19). We isolated 96 individual plasmids that had strong tag hybridization signals. These tags did not cross hybridize with each other. Each of these plasmids was digested with PstI and transformed into C. glabrata ΔH1 (28) by a modified lithium acetate protocol. The final transformation mix consisted of 10 mM lithium acetate, 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 33.5% (wt/vol) polyethylene glycol 3350, 0.1 mg of bovine serum albumin/ml, and 0.29 mg of salmon sperm DNA/ml. Histidine prototrophs were selected and arrayed into 100 microtiter plates such that each plate contained 96 C. glabrata transformants, with each carrying 1 of the 96 tags.
An initial screening of this library revealed a mutant that had an altered colony morphology and an apparent increased ability to persist in vivo (19). This mutant was designated C. glabrata 10G6.
Characterization of the site of insertion in C. glabrata 10G6.
Plasmid rescue was used to identify the site of pTW23 insertion in C. glabrata 10G6. Genomic DNA was prepared from C. glabrata 10G6, and aliquots of 100 ng were digested to completion with ApaI, NotI, SacI, SmaI, and XhoI. These digested DNA populations were self-ligated and used to transform E. coli XL-10. Ampicillin-resistant colonies were selected, plasmids were recovered, and those that contained flanking DNA were identified. Flanking DNA was sequenced from the backbone M13 forward and reverse primer sites.
Construction of C. glabrata ace2 null and ACE2 reconstituted strains.
For construction of an ACE2 disruption cassette, the entire C. glabrata ACE2 open reading frame, plus its surrounding sequences (positions −1169 to +2368 [the stop codon is at +2112]), was amplified from genomic DNA with primers ACE2-1169 (AGAATTGACCGTTGTCCGTGTAAG) and ACE2+2368 (AATGGGTGAATAAATCCCTCCCTAA). The resultant PCR product was cloned into pGEM-T Easy (Promega, Southampton, United Kingdom) and digested with Eco471I and BamHI to remove 2,055 bp from positions −87 to +1968 with respect to the ACE2 start codon. This plasmid was blunt ended and ligated to C. glabrata HIS3. This disruption cassette was amplified with primers ACE2-1169 and ACE2+2368 and was transformed into C. glabrata ΔHT6 (28). Histidine prototrophs were selected and the ACE2 disruption was confirmed by diagnostic PCR and Southern blot analysis. A representative ace2 strain (HLS120) was selected and transformed with pCg-ACT14 (20) to obtain the fully prototrophic C. glabrata ace2 mutant HLS122.
For the reconstitution of ACE2 in C. glabrata HLS120, the entire ACE2 open reading frame was released from pGEMT-Easy by SalI-SphI digestion, taking advantage of sites on the plasmid backbone. This was ligated into appropriately digested pCg-ACT14 to give plasmid pKH355. pKH355 was transformed into C. glabrata HLS120, and tryptophan prototrophs were selected. The reconstitution of C. glabrata ACE2 was confirmed by diagnostic PCR and Northern and Southern blot analyses, and a representative ACE2 strain designated HLS121 was selected.
Virulence analysis of C. glabrata ace2 strains.
Virulence analysis was performed essentially as previously described (5). Briefly, groups of up to 12 outbred CD1 mice were immunosuppressed with 200 mg of cyclophosphamide/kg of body weight on day −3 and every fourth day thereafter. Animals were infected with appropriate doses of C. glabrata blastospores in 200 μl of saline (as described in the legends to Fig. 1B and 2B) via tail vein injection.
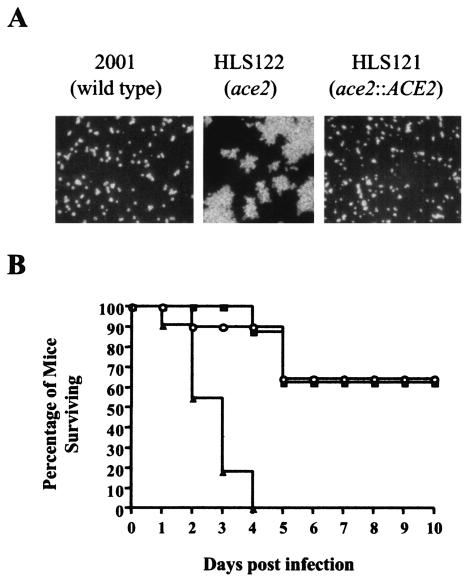
FIG. 1.
Phenotypes of C. glabrata ace2 cells. (A) C. glabrata ace2 cells fail to separate. All strains were cultured for 18 h in liquid SC medium without tryptophan at 37°C, vortexed vigorously for 2 min, and viewed by differential interference microscopy. (B) Clumpy C. glabrata ace2 cells are hypervirulent. Three groups of 12 outbred male CD1 mice were immunosuppressed with cyclophosphamide (200 mg/kg every fourth day), and each mouse was infected with 7 × 107 C. glabrata cells in 200 μl of saline. ▴, C. glabrata ace2 (HLS122); ○, C. glabrata ACE2 (HLS121); ▪, C. glabrata wild type (ATCC 2001). Fungi were not digested with chitinase prior to inoculation.
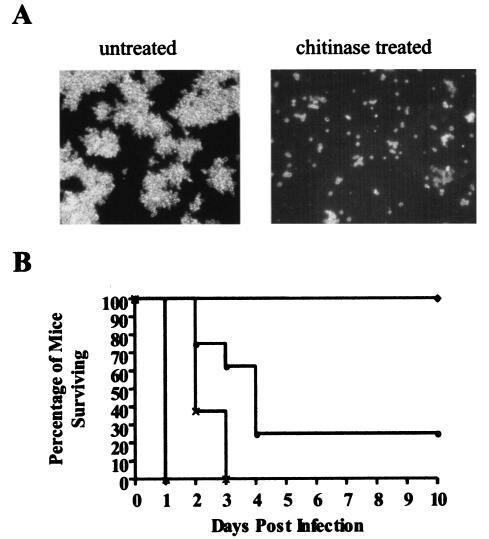
FIG. 2.
Phenotypes of separated C. glabrata ace2 cells. (A) Chitinase treatment of C. glabrata ace2 (HLS122) cells results in breakdown of the large clumps of cells and retrieval of separated yeast cells. Images show C. glabrata HLS122 before and after chitinase digestion, as viewed by differential interference microscopy. (B) Separated C. glabrata ace2 mutants are hypervirulent. Groups of eight outbred male CD1 mice were immunosuppressed with cyclophosphamide (200 mg/kg every fourth day). Mice in each group were infected with 7 × 107 (▴), 1 × 107 (××), or 1 × 106 (•) separated C. glabrata ace2 cells (HLS122) or with 1 × 106 wild-type C. glabrata (ATCC 2001) cells (♦). All fungi were digested with chitinase prior to inoculation.
For the preparation of C. glabrata cells for inoculation, fungi were cultured overnight in 50 ml of YAPD medium or YAPD medium containing 10% (vol/vol) glycerol in 1-liter flasks at 30°C and 180 rpm. Viability was determined for all cultures by trypan blue exclusion and was determined to be >99.9%. C. glabrata ATCC 2001 and HLS121 cells were enumerated with a hemocytometer. For determination of the concentration of C. glabrata HLS122 cells, cultures were vortexed with 0.45-mm-diameter glass beads for 45 s. This resulted in disruption of the large clumps of cells seen for the C. glabrata ace2 mutant (Fig. 1A and 2A) and gave groups of 10 to 20 cells that could be enumerated with a hemocytometer. The original cultures were washed and then diluted in saline to give the desired infectious dose in 200 μl.
For the isolation of separated ace2 mutant cells, overnight cultures of C. glabrata HLS122 were suspended in 200 mM potassium phosphate (pH 6.0)-2 mM CaCl2 containing 12 U of chitinase (Sigma, Poole, United Kingdom)/ml and were incubated at 37°C and 200 rpm until >90% of cells were single, in pairs, or in small groups. Cells were extensively washed and then were counted and diluted to appropriate concentrations in saline prior to inoculation. C. glabrata ATCC 2001 and HLS121 were treated in the same way.
Analysis of immune modulators.
Groups of four neutropenic outbred CD1 mice were infected with 7 × 107 chitinase-treated C. glabrata wild-type (ATCC 2001) or ace2 (HLS122) cells in 200 μl of saline. Four mice were also injected with saline only. All mice were sacrificed at 18 h postinfection and their blood was collected. Sera were separated from the blood and were stored at −20°C until assayed. Interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) levels were determined by use of a CBA mouse inflammation array (BD Biosciences) by the Cell Analysis Facility, Centre for Molecular Microbiology, Imperial College London.
Nucleotide sequence accession number.
ACE2 sequence data have been submitted to the EMBL database under accession number AJ630371.
RESULTS
C. glabrata 10G6 has an inactivated ACE2 allele.
A total of 327 nucleotides flanking the site of pTW23 insertion were identified in C. glabrata 10G6 by sequencing of plasmids rescued from this strain. A 258-bp fragment of this sequence was amplified from C. glabrata ATCC 2001 DNA with the primers GAGACTTGAATATGAATGCCG and TGTTTCAGAAGAACTTGGC. The resultant PCR product was used to screen a C. glabrata genomic library constructed in YEp24 (a kind gift from Dominique Sanglard, Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland). Three clones containing 5.4-kb inserts were obtained, and one was sequenced on both strands at the automated DNA sequencing facility at Imperial College London. This clone contained an uninterrupted 2,112-bp open reading frame, and a comparison with the sequence carried by plasmids rescued from C. glabrata 10G6 indicated that pTW23 had inserted between nucleotides 452 and 453 of the open reading frame in this strain.
A hypothetical translation indicated that the disrupted gene encodes a protein of 704 amino acids containing a zinc finger DNA binding domain (amino acids 384 to 602) that has significant amino acid identities, of ∼39% and ∼25%, respectively, with the S. cerevisiae cell cycle-regulated transcription factors Ace2 and Swi5 (4). We designated this gene C. glabrata ACE2, as it encodes the Ace2-specific motif SGTAIF, and in separate experiments we have identified a C. glabrata ortholog of SWI5 (G. Butler, unpublished observations).
C. glabrata ace2 mutants are hypervirulent.
To determine if inactivation of ACE2 was responsible for the apparent increased ability of C. glabrata 10G6 to persist in vivo, we infected groups of 12 neutropenic CD1 mice with equivalent doses (7 × 107) of ace2 null (HLS122), reconstituted ACE2 (HLS121), and wild-type (ATCC 2001) strains. For these experiments, the strains were not digested with chitinase prior to inoculation. Infection with C. glabrata HLS122 cells resulted in 100% mortality within 4 days, whereas the inoculation of equivalent doses of C. glabrata HLS121 or ATCC 2001 resulted in only 40% mortality at 10 days postinfection (Fig. 1B). These data demonstrate that C. glabrata ace2 mutants have an increased virulence phenotype compared to wild-type and reconstituted ACE2 cells (P < 0.05 by Kaplan-Meier log rank analysis).
C. glabrata ace2 hypervirulence phenotype is separable, at the time of infection, from the clumpy growth phenotype.
C. glabrata ace2 mutants do not undergo cell separation after cell division, instead forming large clumps of cells (Fig. 1A and 2A). We were concerned that the increased virulence phenotype of C. glabrata ace2 cells was therefore caused by vascular occlusion initiated at the time of infection. To test this hypothesis, we developed a chitinase digestion protocol that enables the separation of C. glabrata HLS122 cells (Fig. 2A). These separated C. glabrata ace2 yeast cells were compared with chitinase-digested wild-type C. glabrata ATCC 2001 and reconstituted ACE2 HLS121 cells for the ability to cause disease in a neutropenic murine model. These experiments revealed that the increased virulence of separated C. glabrata ace2 cells was even more marked than that of the untreated clumpy HLS122 cells (Fig. 2B). Infection with 7 × 107 chitinase-treated C. glabrata ace2 cells resulted in 100% mortality after 18 h, compared to 4 days with an equivalent dose of untreated clumpy ace2 cells (P < 0.05 by Kaplan-Meier log rank analysis); 100% mortality after 3 days with a dose of 1 × 107 cells; and 70% mortality within 5 days with a dose of 1 × 106 cells (Fig. 2B). Surprisingly, chitinase-treated wild-type cells had an increased virulence phenotype compared to untreated C. glabrata ATCC 2001 cells. However, chitinase-treated separated C. glabrata ace2 cells were still more virulent than similarly treated wild-type or reconstituted ACE2 cells. A dose of 7 × 107 chitinase-treated wild-type or reconstituted ACE2 cells caused 100% mortality after 3 days (survival curves not shown), compared with 18 h for the C. glabrata ace2 strain HLS122 (P < 0.05 by Kaplan-Meier log rank analysis). Similarly, a dose of 106 chitinase-treated wild-type or reconstituted ACE2 cells resulted in no mortality during the study period (Fig. 2B), compared with the 70% mortality for C. glabrata ace2 mutant-infected animals (P < 0.05 by Kaplan-Meier log rank analysis). Infection with 2 × 108 wild-type C. glabrata ATCC 2001 cells is required to cause 100% mortality within 6 days (5). These data demonstrate that C. glabrata ace2 cells can cause a mortality rate similar to that caused by wild-type or reconstituted ACE2 cells at almost 200-fold lower doses. C. glabrata ace2 mutants are therefore hypervirulent, and importantly, these data demonstrate that this enhanced virulence is separable, at the time of infection, from the ace2 clumpy growth phenotype.
C. glabrata ace2 hypervirulence is not due to vascular occlusion.
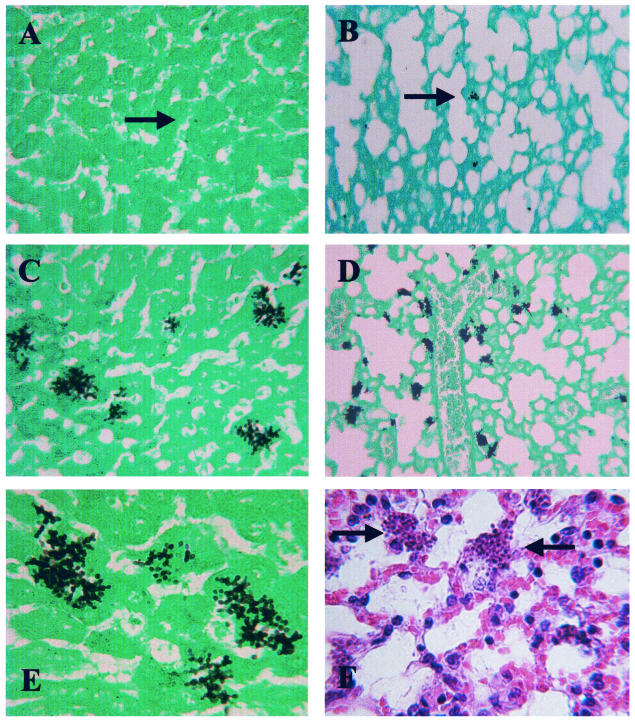
A histological examination of tissues (Fig. 3) revealed that organs from mice infected with separated C. glabrata ace2 cells had much higher tissue burdens than those seen during infections with similarly treated wild-type cells (Fig. 3A to D). In all organs, the tissue parenchyma was invaded and fungi were not restricted to the vasculature (Fig. 3E). Histology also revealed that C. glabrata ace2 cells can proliferate in vivo. Mice were infected with single or small groups of C. glabrata ace2 cells, but microcolonies formed in the tissues (Fig. 3C to E). Interestingly, little or no cellular inflammatory response was seen around sites of either wild-type or C. glabrata ace2 infection (Fig. 3F). These data demonstrate that C. glabrata ace2 cells are not restricted to the bloodstream and therefore that hypervirulence does not result simply from vascular occlusion.
FIG. 3.
Histological appearance of C. glabrata candidiasis. Representative appearances of the liver (A) and lungs (B) of mice infected with wild-type C. glabrata (ATCC 2001) cells demonstrate that these tissues are only sparsely colonized with fungi. Only a few single blastospores or very small microcolonies are visible (arrows). Representative appearances of the liver (C) and lungs (D) of mice infected with C. glabrata ace2 (HLS122) cells demonstrate that these tissues have a substantial fungal burden. Many microcolonies are visible in each field, demonstrating the increased proliferation of C. glabrata ace2 cells in vivo. (E) Representative appearance of C. glabrata ace2 cells in the liver showing invasion of the tissue parenchyma and escape from the vasculature. (F) Representative appearance of C. glabrata ace2 cells in the lungs demonstrating the lack of a cellular inflammatory reaction around C. glabrata ace2 microcolonies (arrows). All tissues were recovered from neutropenic CD1 mice culled after 18 h, except for the tissue shown in panel B, which was from a mouse culled after 4 days. The tissues shown in panels A and B were from mice infected with 2 × 108 C. glabrata ATCC 2001 cells. Tissues shown in panels C to F were from mice infected with 7 × 107 C. glabrata ace2 cells. All cells were treated with chitinase prior to infection. The sections in panels A to E were stained with Grocott and Light Green, and the section in panel F was stained with hematoxylin and eosin.
C. glabrata ace2 mutants overstimulate the proinflammatory arm of the innate immune response.
Minimal IL-6, TNF-α, and IFN-γ levels were detected in sera collected after 18 h from neutropenic CD1 mice injected with saline (Table 2). Levels of the classic proinflammatory cytokines IL-6 and TNF-α were 16-fold (P < 0.05; t test) and 38-fold (P < 0.05; t test) higher, respectively, in mice infected with separated C. glabrata HLS122 ace2 mutant cells than in animals infected with chitinase-treated wild-type cells. The level of IFN-γ was 4.15-fold (P < 0.05; t test) higher in mice infected with wild-type C. glabrata than in animals infected with the ace2 mutant HLS122. This analysis suggests that C. glabrata ace2 cells can induce severe sepsis and that this may underpin the hypervirulence phenotype.
TABLE 2.
IL-6, TNF-α, and IFN-γ levels detected in mice at 18 h postinfection with saline or with wild-type or ace2 mutant C. glabrata cells
| Infection | Concn of cytokine (pg/ml)a
|
||
|---|---|---|---|
| IL-6 | TNF-α | IFN-γ | |
| Saline | 1.5 (0.1) | 5.7 (0.5) | UDb |
| C. glabrata ATCC 2001c | 115.4 (54.3)d | 45.2 (3.9)d | 21.1 (12.9) |
| C. glabrata ace2c | 1,857.8 (867)e | 1,723.1 (931)e | 5.1 (2.5)f |
Data are means and standard deviations (in parentheses) of cytokine levels detected in four mice.
UD, undetected.
Cells were treated with chitinase prior to inoculation.
Significantly different from levels in saline-infected mice (P < 0.05; t test).
Significantly different from levels in saline- and wild-type-infected mice (P < 0.05; t test).
Significantly different from levels in wild-type-infected mice (P < 0.05; t test).
DISCUSSION
This study demonstrates that the inactivation of C. glabrata ACE2 results in a strain that is hypervirulent in a murine model of candidiasis. Infection with C. glabrata ace2 cells results in an overwhelming, fatal, systemic disease. The infectious dose required to obtain 100% mortality is 200-fold lower for C. glabrata ace2 mutants than for wild-type cells. However, C. glabrata ace2 mutants do not undergo cell separation after cell division, instead forming large clumps of cells (Fig. 1A and 2A). We were concerned that the increased virulence phenotype of C. glabrata ace2 cells was therefore caused by vascular occlusion initiated at the time of infection. In S. cerevisiae, ACE2 regulates the expression of the chitinase gene CTS1 (8). In the absence of ACE2, CTS1 is not expressed, the chitin ring laid around the site of mother-daughter budding is not degraded after cell division, and daughters do not separate from their mothers (23). We were unable to detect a CTS1 transcript in C. glabrata HLS122 cells by Northern blot analysis (data not shown), so it is likely that the failure of these cells to separate is also caused by a reduction in CTS1 expression. We therefore developed a chitinase digestion protocol that enables the separation of C. glabrata HLS122 cells (Fig. 2A). These separated C. glabrata ace2 yeast cells were compared with similarly treated wild-type C. glabrata ATCC 2001 and reconstituted ACE2 HLS121 cells for the ability to cause disease in a neutropenic murine model. The hypervirulence phenotype of separated C. glabrata ace2 cells was even more marked than that of untreated clumpy HLS122 cells (Fig. 1B and 2B). Separated cells resulted in 100% mortality within 18 h, compared to 5 days when an equivalent dose of clumpy C. glabrata ace2 cells was inoculated. Histological analysis revealed that C. glabrata cells were not restricted to the vasculature but escaped into the tissue parenchyma. We also demonstrated that chitinase-digested wild-type C. glabrata ATCC 2001 cells had an increased virulence phenotype compared with untreated cells. It is therefore possible that the increase in virulence of separated ace2 cells compared to clumpy ace2 cells was at least partially due to the chitinase treatment. This possibility notwithstanding, our observations suggest that the increased virulence phenotype of C. glabrata ace2 mutants is not due simply to vascular occlusion but is a result of a change in an attribute of these cells and/or the host response to them. The elucidation of this attribute and the host changes that may be induced is a priority and has the potential to reveal novel insights into the host-C. glabrata interaction. Taken together, these data support the view that C. glabrata ACE2 encodes a virulence-moderating gene (13), the first to be described for a Candida species.
The potent activation of pattern recognition receptors that mediate innate and adaptive immune responses can lead to overwhelming septicemia; rising levels of the proinflammatory cytokines IL-6 and TNF-α can reflect their activation (7, 17). S. cerevisiae strains with inactivated SSD1 alleles induce threefold higher levels of TNF-α and IL-6 from peritoneal macrophages than do wild-type cells. This is accompanied by major alterations in the cell wall composition and by increased virulence in DBA/2 mice (29). Similarly, a proteomic analysis of C. glabrata revealed >200 differences between wild-type and ace2 cells (A. J. Brown, B. Dujon, and K. Haynes, unpublished data). These data demonstrate that C. glabrata ACE2 plays a role in the regulation of (among other things) protein folding and degradation and cell wall biogenesis. We therefore speculated that the inactivation of C. glabrata ACE2 may result in the appearance (or exposure) of a fungal component that can potently and inappropriately activate pattern recognition receptors. To investigate this hypothesis, we determined the levels of circulating IL-6, TNF-α, and IFN-γ at 18 h postinfection with chitinase-treated ace2 or wild-type C. glabrata cells (Table 2). Both IL-6 and TNF-α are known to be critical for the host defense against Candida species (22). The acute induction of IL-6 and TNF-α is known to reflect the severity of infection in humans (10, 13). It is also a typical consequence of severe sepsis (23). The lack of IFN-γ induction is also consistent with severe sepsis but suggests lymphocyte apoptosis rather than necrosis (27). Our observations are consistent with the hypothesis that C. glabrata ace2 cells induce severe sepsis. This provides a testable mechanistic explanation for the hypervirulence phenotype of C. glabrata ace2 cells.
Why has C. glabrata ACE2 been retained when its deletion results in such a dramatic increase in virulence and concomitant ability to proliferate in vivo? Ace2 is not an essential protein, but it does play a crucial role in cell cycle progression and is required for cell separation (23, 25). Many fungal mother and daughter cells, including those of the pathogens Aspergillus fumigatus and hyphal C. albicans, do not separate after cell division. However, these species do have efficient methods to ensure successful dispersal. A. fumigatus produces asexual conidia in prodigious numbers, while C. albicans hyphae can give rise to yeast cells that are able to separate from their progenitor (12). The only known method for C. glabrata to effect dispersal is via the separation of mother and daughter cells. This is prevented in ace2 mutants and provides, in addition to the many other advantageous functions of Ace2, a compelling evolutionary rationale for the retention of the ACE2 gene.
Acknowledgments
We thank K. Kitada (Nippon Roche, Kanagawa, Japan) and D. Sanglard (Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland) for supplying materials and G. Janbon (Institut Pasteur, Paris, France) and two anonymous referees for helpful comments. We especially thank F. Odds and D. MacCallum (University of Aberdeen, Aberdeen, Scotland) for confirming our in vivo observations.
This work was supported by the MRC, BBSRC, The Wellcome Trust, and Fungal Research Trust.
REFERENCES
- 1.Abi-Said, D., E. Anaissie, O. Uzun, I. Raad, H. Pinzcowski, and S. Vartivarian. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24:1122-1128. [DOI] [PubMed] [Google Scholar]
- 2.Beck-Sague, C., and W. R. Jarvis. 1993. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980-1990. National Nosocomial Infections Surveillance System. J. Infect. Dis. 167:1247-1251. [DOI] [PubMed] [Google Scholar]
- 3.Berrouane, Y. F., L. A. Herwaldt, and M. A. Pfaller. 1999. Trends in antifungal use and epidemiology of nosocomial yeast infections in a university hospital. J. Clin. Microbiol. 37:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, G., and D. J. Thiele. 1991. ACE2, an activator of yeast metallothionein expression which is homologous to SWI5. Mol. Cell. Biol. 11:476-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calcagno, A., E. Bignell, P. Warn, M. D. Jones, D. W. Denning, F. A. Mühlschlegel, T. R. Rogers, and K. Haynes. 2003. Candida glabrata STE12 is required for wild-type virulence and nitrogen starvation induced filamentation. Mol. Microbiol. 50:1309-1318. [DOI] [PubMed] [Google Scholar]
- 6.Calderone, R. A. 2002. Candida and candidiasis. ASM Press, Washington, D.C.
- 7.Cohen, J. 2002. The immunopathogenesis of sepsis. Nature 420:885-891. [DOI] [PubMed] [Google Scholar]
- 8.Doolin, M. T., A. L. Johnson, L. H. Johnston, and G. Butler. 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40:422-432. [DOI] [PubMed] [Google Scholar]
- 9.Foreman-Wykert, A. K., and J. F. Miller. 2003. Hypervirulence and pathogen fitness. Trends Microbiol. 11:105-108. [DOI] [PubMed] [Google Scholar]
- 10.Girardin, E., G. E. Grau, J. M. Dayer, P. Roux-Lombard, and P. H. Lambert. 1988. Tumor necrosis factor and interleukin-1 in the serum of children with severe infectious purpura. N. Engl. J. Med. 319:397-400. [DOI] [PubMed] [Google Scholar]
- 11.Gleason, T. G., A. K. May, D. Caparelli, B. M. Farr, and R. G. Sawyer. 1997. Emerging evidence of selection of fluconazole-tolerant fungi in surgical intensive care units. Arch. Surg. 132:1197-1201. [DOI] [PubMed] [Google Scholar]
- 12.Gow, N. A., A. J. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 13.Hatherill, M., S. M. Tibby, C. Turner, N. Ratnavel, and I. A. Murdoch. 2000. Procalcitonin and cytokine levels: relationship to organ failure and mortality in pediatric septic shock. Crit. Care Med. 28:2591-2594. [DOI] [PubMed] [Google Scholar]
- 14.Haynes, K. 2001. Virulence in Candida species. Trends Microbiol. 9:591-596. [DOI] [PubMed] [Google Scholar]
- 15.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 16.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42:453-467. [DOI] [PubMed] [Google Scholar]
- 17.Janeway, C. A. J., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 18.Jarvis, W. R. 1995. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin. Infect. Dis. 20:1526-1530. [DOI] [PubMed] [Google Scholar]
- 19.Kamran, M. F. 2003. The identification and isolation of putative virulence determinants of Candida glabrata. Ph.D. thesis. University of London, London, United Kingdom.
- 20.Kitada, K., E. Yamaguchi, and M. Arisawa. 1996. Isolation of a Candida glabrata centromere and its use in construction of plasmid vectors. Gene 175:105-108. [DOI] [PubMed] [Google Scholar]
- 21.Navarro-Garcia, F., M. Sanchez, C. Nombela, and J. Pla. 2001. Virulence genes in the pathogenic yeast Candida albicans. FEMS Microbiol. Rev. 25:245-268. [DOI] [PubMed] [Google Scholar]
- 22.Netea, M. G., C. A. Van Der Graaf, A. G. Vonk, I. Verschueren, J. W. Van Der Meer, and B. J. Kullberg. 2002. The role of Toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J. Infect. Dis. 185:1483-1489. [DOI] [PubMed] [Google Scholar]
- 23.O'Conallain, C., M. T. Doolin, C. Taggart, F. Thornton, and G. Butler. 1999. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pittet, D., N. Li, R. F. Woolson, and R. P. Wenzel. 1997. Microbiological factors influencing the outcome of nosocomial bloodstream infections: a 6-year validated, population-based model. Clin. Infect. Dis. 24:1068-1078. [DOI] [PubMed]
- 25.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 26.Viscoli, C., C. Girmenia, A. Marinus, L. Collette, P. Martino, B. Vandercam, C. Doyen, B. Lebeau, D. Spence, V. Krcmery, B. De Pauw, and F. Meunier. 1999. Candidemia in cancer patients: a prospective, multicenter surveillance study by the Invasive Fungal Infection Group (IFIG) of the European Organization for Research and Treatment of Cancer (EORTC). Clin. Infect. Dis. 28:1071-1079. [DOI] [PubMed] [Google Scholar]
- 27.Voll, R. E., M. Hermann, E. A. Roth, C. Stach, J. R. Kalden, and I. Girkontaite. 1997. Immunosuppressive effects of apoptotic cells. Nature 390:350-351. [DOI] [PubMed] [Google Scholar]
- 28.Weig, M., K. Haynes, T. R. Rogers, O. Kurzai, M. Frosch, and F. A. Mühlschlegel. 2001. A GAS-like gene family in the pathogenic fungus Candida glabrata. Microbiology 147:2007-2019. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler, R. T., M. Kupiec, P. Magnelli, C. Abeijon, and G. R. Fink. 2003. A Saccharomyces cerevisiae mutant with increased virulence. Proc. Natl. Acad. Sci. USA 100:2766-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wisplinghoff, H., H. Seifert, R. P. Wenzel, and M. B. Edmond. 2003. Current trends in the epidemiology of nosocomial bloodstream infections in patients with hematological malignancies and solid neoplasms in hospitals in the United States. Clin. Infect. Dis. 36:1103-1110. [DOI] [PubMed] [Google Scholar]