Abstract
In the yeast Saccharomyces cerevisiae, the transcription factor Yap1p is a central determinant of resistance to oxidative stress. Previous work has demonstrated that Yap1p is recruited from the cytoplasm to the nucleus upon exposure to the oxidants diamide and H2O2 in a process that requires the transient covalent linkage of the glutathione peroxidase Gpx3p to Yap1p. Genetic and biochemical analyses indicate that while both oxidants trigger nuclear accumulation of Yap1p, the function and regulation of this transcription factor is different under these two different oxidative stresses. Ybp1p (Yap1p-binding protein) has recently been demonstrated to be required for Yap1p-mediated H2O2 resistance but not diamide resistance. A Ybp1p homologous protein (Ybh1p/Ybp2p) was also detected in the S. cerevisiae genome. Here we compare the actions of these two closely related proteins and provide evidence that while both factors influence H2O2 tolerance, they do so by nonidentical mechanisms. A double mutant strain lacking both YBP1 and YBH1 genes is more sensitive to H2O2 and more defective in activation of Yap1p-dependent gene expression than either single mutant. Ybp1p has a more pronounced effect on these phenotypes than does Ybh1p. Protein-protein interactions between Yap1p and Ybp1p could be detected by either the yeast two-hybrid or coimmunoprecipitation approach while neither technique could demonstrate Yap1p-Ybh1p interactions. Overexpression experiments indicated that high levels of Ybh1p but not Ybp1p could bypass the H2O2 hypersensitivity of a gpx3Δ strain. Together, these data argue that these two homologous proteins act as parallel positive regulators of H2O2 tolerance.
Detoxification of reactive oxygen species (ROS) is an essential ability for viability in an aerobic environment. Organisms from bacteria to humans have multiple systems devoted to prevention of potentially lethal change in the intracellular redox potential that would otherwise occur upon accumulation of ROS (recently reviewed in reference 16). Activation of these systems requires a mechanism to sense inappropriate changes in ROS levels and mount a compensatory response.
The yeast Saccharomyces cerevisiae has been extensively used to analyze the molecular basis of the response of a eukaryotic organism to ROS-based environmental challenges. A central feature of the response to oxidative stress in S. cerevisiae is transcriptional induction of a variety of antioxidant genes that act to lower the toxic levels of ROS and ROS-damaged macromolecules (reviewed in references 10 and 21). One of the key regulators of the transcriptional response to oxidative stress is the basic region leucine zipper-containing transcription factor Yap1p (see references 30 and 35 for reviews).
Yap1p is primarily located in the cytoplasm but is rapidly recruited to the nucleus upon imposition of oxidative stress elicited by either H2O2 or diamide exposure (26). Evidence has been provided that two different cysteine-rich domains (CRD) located in the amino-terminal (n-CRD) and carboxy-terminal (c-CRD) regions of the protein are required for the normal response to oxidative challenge (4, 37). Diamide is believed to form disulfide bonds between closely linked cysteine residues present either in the n-CRD or c-CRD while H2O2 has been shown to induce a disulfide bond between a cysteine residue located in the n-CRD and one in the c-CRD (7, 24). While either oxidant will cause nuclear localization of Yap1p, mutant forms of Yap1p exhibit oxidant-specific behaviors to these two stress agents. For example, a mutant lacking a key cysteine residue in the c-CRD is constitutively located in the nucleus and confers hyperresistance to diamide yet fails to provide normal tolerance to H2O2 (4).
Recent studies have provided evidence for H2O2-specific factors that are required for normal regulation of Yap1p during H2O2-induced oxidative stress. The glutathione peroxidase homologue Gpx3p/Hyr1p (1, 17) has been demonstrated to form a covalent intermediate with Yap1p upon H2O2-induced but not diamide-induced stress (8). This covalent intermediate is first formed between a cysteine residue in the Yap1p c-CRD and one in Gpx3p. Gpx3p is then thought to act essentially as a leaving group to allow formation of an intramolecular disulfide bond between the n- and c-CRDs. This form of Yap1p is referred to as the oxidized form and exhibits a higher mobility on nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) than a Yap1p form lacking this intramolecular disulfide (reduced form) (7).
A second H2O2-specific regulator of Yap1p has been designated Ybp1p (Yap1p-binding protein) (36). The function of this protein is unknown, but in response to H2O2 stress, ybp1Δ mutant strains cannot form the oxidized form of Yap1p, recruit Yap1p to the nucleus, or support normal gene activation and oxidant resistance. Loss of YBP1 has no effect on Yap1p function in response to diamide stress. Two groups have demonstrated that Yap1p interacts with Ybp1p by using global protein-protein interaction strategies (12, 18), and this interaction has been mapped to the C terminus of Yap1p (36). No information is available for the region of Ybp1p involved in Yap1p binding. A homologue of Ybp1p designated Ybp2p is also present in the S. cerevisiae genome (36).
In this work, we evaluate the participation of Ybp2p in Yap1p-mediated processes by comparison to Ybp1p. While Ybp1p and Ybp2p both influence tolerance to H2O2, Ybp1p has a much greater role in this resistance phenotype. Additionally, by several different criteria, Ybp2p does not appear to directly interact with Yap1p, unlike Ybp1p. Our findings are most consistent with Ybp2p acting in a pathway parallel to that of Ybp1p. We propose that Ybp2p be renamed Ybh1p (Ybp1p homologue) to indicate that, while these two factors share strong sequence identity, their actions in the cell are nonidentical.
MATERIALS AND METHODS
Yeast strains and media.
The strains used in this study are listed in Table 1. All strains were derived from either SEY6210 or BY4742. Yeast cells were grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) or synthetic complete (SC) medium at 30°C with shaking. SC medium was prepared as described by the manufacturer (Bio 101) from stocks in which particular amino acids or nucleic acids were deleted. Medium containing either gradient or different concentrations of diamide or hydrogen peroxide was prepared by the addition of the required amounts of drugs after autoclaving the media and just before pouring the plates or immediately prior to the growth experiment. The YBP1 open reading frame (ORF) was disrupted in SEY6210 by using PCR-mediated gene disruption with the KanMX2 module with primers Ybr216cDEL1 and Ybr216cDEL2, yielding KGS1 (ybp1Δ::KanMX2). Primer sequences are available on request. The YBH1 (YBP2/YGL060w) ORF was also disrupted in SEY6210 and KGS1, with primers Ygl060wDEL1 and Ygl060wDEL2, to yield KGS2 (ybh1Δ::natR) and KGS3 (ybp1Δ::KanMX2 ybh1Δ::natR), respectively. The single and double deletion mutants of YBP1 and YBH1 were also made in BY4742, yielding KGS4 (ybp1Δ::KanMX2), KGS5 (ybh1Δ::natR), and KGS6 (ybp1Δ::KanMX2 ybh1Δ::natR). All of the disruptions were confirmed by PCR. The YBP1 disruptions were confirmed by primers 216cF-confirm and 216cR-confirm while disruptions of YBH1 were confirmed by using the primers Ygl060w-FOR and Ygl060w-REV. Strains containing green fluorescent protein (GFP) fusions to YBP1 and YBH1 were generated in BY4742 (YSAR49 and YSAR52) by amplification of pFA6a-GFP (S65T)-TRP1 (28) with primers P28F and P23R for YBP1 tagging and primers P29F and P37R for YBH1 tagging. Strains containing hemagglutinin (HA) tag fusions in BY4742 (YSAR48 and YSAR51) were constructed with the same primers, with pFA6a-3X HA-TRP1 (28) as a template. The resulting 2,001-bp (GFP) or 1,395-bp (3× HA) fragments were gel purified and transformed into SEY6210 or BY4742 Δtrp1 and plated on SC media lacking tryptophan (SC-W). Transformants were screened by PCR to verify insertions.
TABLE 1.
Yeast strains used
| Strain | Genotype | Source or reference |
|---|---|---|
| SEY6210 | MATα leu2-3,-112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel− | Scott Emr |
| BY4742 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 | Open Biosystems |
| KGS1 | MATα leu2-3,-112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel−ybp1Δ::KanMX | This study |
| KGS2 | MATα leu2-3,-112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel−ybh1Δ::natR::M | This study |
| KGS3 | MATα leu2-3,-112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel−ybh1Δ::KanMX2 ybh1Δ::natR | This study |
| KGS4 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 ybp1Δ::KanMX2 | This study |
| KGS5 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 ybh1Δ::natR | This study |
| KGS6 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 ybp1Δ::KanMX2 ybh1Δ::natR | This study |
| YSAR48 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 trp1Δ::KanMX2 YBP1-3X-HA::TRP1 | This study |
| YSAR49 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 trp1Δ::KanMX2 YBP1-GFP (S65T)::TRP1 | This study |
| YSAR51 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 trp1Δ::KanMX2 YBH1-3X-HA::TRP1 | This study |
| YSAR52 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 trp1Δ::KanMX2 YBH1-GFP (S65T)::TRP1 | This study |
| SM13 | MATα leu2-3,-112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 Mel−yap1-Δ2::hisG | 37 |
| gpx3Δ | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 gpx3-Δ::KanMX2 | Open Biosystems |
| PJ69-4A | MATatrp1-Δ901 leu2-3,112 ura3-52 his3-Δ200 gal4 Δgal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | 20 |
Plasmids.
Plasmids used in this study are listed in Table 2. The 3.0-kb fragment containing the YBP1 ORF along with its promoter was amplified by using the primers P16F_YBR216c and P17R_YBR216c and cloned in the pCR 2.1-TOPO vector (Invitrogen). The 2.8-kb KpnI-SalI fragment containing the YBP1 gene was excised from the pCR2.1-TOPO clone and transferred to low- and high-copy-number plasmids pRS314 (33) and pRS426 (2) at the KpnI-SalI sites. The ORFs coding for GPX3, YBP1, and YBH1 were cloned under control of the phosphoglycerate kinase (PGK) promoter in plasmid pXTZ134 (41) at the BamHI site. The primers used for amplifying the genes for GPX3, YBP1, and YBH1 were GPX3-For, GPX3-Rev, PGK-216-p-for, PGK-216-Rev, Ygl060w clone-F, and Ygl060w clone-R, respectively. For the two-hybrid assay, full-length ORFs for YBP1, YBH1, and GPX3 were cloned in the plasmid pGBD-C1 (20). Three different DNA fragments of YBP1 gene, from bp 1 to 560, 558 to 1250, and 1250 to 2025, were also cloned in the EcoRI-BamHI sites of plasmid pGBD-C1. The full-length YAP1 ORF was cloned in plasmid pGAD-C1 as pJT20.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant features | Source or reference |
|---|---|---|
| pKGE1 | 2μm URA3 YBP1 | This study |
| pKGE2 | PGK-HA-YBP1 | This study |
| pKGE3 | PGK-HA-YBH1 | This study |
| pKGE4 | PGK-HA-GPX3 | This study |
| pKGE5 | GBD-YBP1 | This study |
| pKGE6 | GBD-YBH1 | This study |
| pKGE7 | GBD-GPX3 | This study |
| pKGE8 | GBD-YBP1 (bp 1-560) | This study |
| pKGE9 | GBD-YBP1 (bp 558-1250) | This study |
| pKGE10 | GBD-YBP1 (bp 1250-2025) | This study |
| pJT20 | GAD-YAP1 | This study |
| pAW 14 | URA3 TRX2-lacZ | 38 |
| pNT13 | URA3 GFP-YAP1 | 4 |
β-Galactosidase reporter assay.
The strains carrying single and double deletions of YBP1 and YBH1 were transformed with low-copy-number plasmids containing either TRX2-lacZ (4) or GSH1-lacZ (38) gene fusions. Transformants were grown to log phase in selective medium with or without hydrogen peroxide and diamide. An equal number of cells from each strain were harvested, washed, and assayed for β-galactosidase activity as described previously (14). All assays were carried out in triplicate, and activity was expressed as β-galactosidase units per OD600 (optical density at 600 nm) unit of cells.
Plate growth assays.
To examine the growth on plates, overnight grown cells were subcultured in the same medium and were grown to an OD600 of ∼1.0. An equal number of cells were then plated as spots on gradient plates of hydrogen peroxide (up to 6 mM) and diamide (up to 2 mM). For complementation assays with plasmids, transformants were grown overnight in selective medium, then diluted in the same medium, and grown to an OD600 of ∼1.0. Transformants were then plated on selective medium plates containing a gradient of hydrogen peroxide.
Fluorescence microscopy.
Log-phase cells expressing GFP-tagged Yap1p were examined for localization of Yap1p in different deletion backgrounds, with and without induction with H2O2 at different time intervals. Cells were grown to an OD600 between 0.7 and 1.0 in SC-Ura and induced for 0, 5, 20, and 45 min with 1 mM H2O2. The strains carrying Ybp1p-GFP or Ybh1p-GFP were grown in a similar manner, but only the fluorescence profiles seen in nonstressed cells are shown here, as the presence of H2O2 did not change the localization of either fusion protein (data not shown). Five microliters of concentrated culture was placed on a glass slide, overlaid with a coverslip, and examined under the 100× oil objective of an Olympus BX60 microscope. Nuclei were visualized by staining with 4′,6′-diamidino-2-phenylindole (DAPI). Pictures were captured on a Hamamatsu digital camera.
Two-hybrid assays.
Two-hybrid assays were performed to test for interactions between Ybp1p, Ybh1p, Gpx3p, and Yap1p. Full-length and different deletion constructs of Ybp1p, as well as full-length Ybh1p and Gpx3p, were tested for their interaction with Yap1p in the presence and absence of hydrogen peroxide. The two-hybrid studies were done in strain PJ69-4A (20), and interactions were determined on the basis of the ability of cells to grow in the absence of histidine and adenine. The strain PJ69-4A was transformed with pYAP1-GAD along with different constructs in pGBD-C1. The transformants were selected first on SC-Leu-Ura medium to ensure that both the GAD and GBD plasmids were present. These transformants were then streaked on SC-Leu-Ura-His plus 10 mM 3-aminotriazole and on SC-Leu-Ura-Ade medium with or without hydrogen peroxide.
Western analysis.
For Western analysis, cells were grown in 100 ml of YPD or selective medium. Cells were harvested at an OD600 of ∼1.0, washed, and lysed by glass bead lysis in a buffer containing 300 mM sorbitol, 100 mM NaCl, 5 mM MgCl2, 10 mM Tris (pH 7.4), and complete protease inhibitors (Boehringer Mannheim). Cell extracts were then clarified by centrifugation, and the Bradford assay was used to determine the protein concentration. Equal amounts of proteins were resolved by SDS-10% PAGE. The proteins were transferred to a nitrocellulose membrane, blocked with 5% nonfat dry milk in phosphate-buffered saline, and then probed with anti-HA antibodies. Horseradish peroxidase-conjugated secondary antibody and the ECL kit (Pierce) were used to visualize immunoreactive proteins.
Coimmunoprecipitation assay.
Immunoprecipitation assays were completed with lysed spheroplasts. In brief, 100 ml of cells was harvested at an OD600 of 1.0 and resuspended in 10 mM Tris-HCl (pH 8) plus 10 mM dithiothreitol. Cells were incubated at room temperature for 5 min, centrifuged, suspended in spheroplast buffer (1 M sorbitol in 0.5× YPD, 50 mM KPO4) with oxalyticase, and incubated at 30°C for 30 min on a shaker. After chilling on ice, cell suspensions were overlaid on a sucrose cushion (20 mM HEPES, 1.2 M sucrose, 0.02% sodium azide) and pelleted by centrifuging at 4,000 × g for 20 min at 4°C. The resulting spheroplasts (treated and untreated with 1 mM hydrogen peroxide) were suspended in ICB (lysis) buffer (100 mM potassium acetate, 50 mM KCl, 20 mM PIPES, 200 mM sorbitol [pH 6.8]). These lysates were then incubated with anti-Yap1p polyclonal antiserum for 2 h on ice. IgGSorb beads (fixed Staphylococcus aureus cells from the Enzyme Center, Malden, Mass.) were added, and lysates were incubated for an additional 1 h on ice with occasional mixing. The immunoprecipitated proteins were recovered by adding Twirl buffer (8 M urea, 5% SDS, 10% glycerol, and 50 mM Tris [pH 6.8]). These precipitates were then loaded on 10% polyacrylamide gel and probed with anti-HA antibodies (see Fig. 6) or anti-Yap1p antibody to confirm equal precipitation (data not shown). A final control experiment was carried out by probing aliquots of the supernatant fractions from these anti-Yap1p immunoprecipitations to ensure that the 3× HA-tagged proteins were present. Both Ybp1p-3X HA and Ybh1p-3X HA could be seen in these supernatant fractions (data not shown).
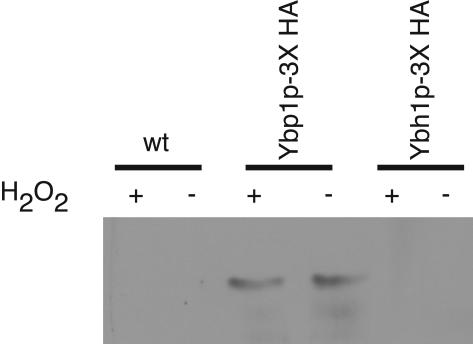
FIG. 6.
Coimmunoprecipitation analysis of Yap1p with Ybp1p and Ybh1p. Whole-cell protein extracts were produced from three different strains: wild-type (wt) cells and wild-type cells expressing 3× HA-tagged forms of Ybp1p (Ybp1p-3X HA) or Ybh1p (Ybh1p-3X HA). Extracts were prepared from cells grown in the presence (+) or absence (−) of H2O2 and then subjected to immunoprecipitation with a rabbit polyclonal antiserum against Yap1p. The resulting immunoprecipitates were then resolved by SDS-PAGE and analyzed by Western blotting with anti-HA antibody.
RESULTS
Differential contributions of YBP1 and YBH1 to Yap1p-dependent phenotypes.
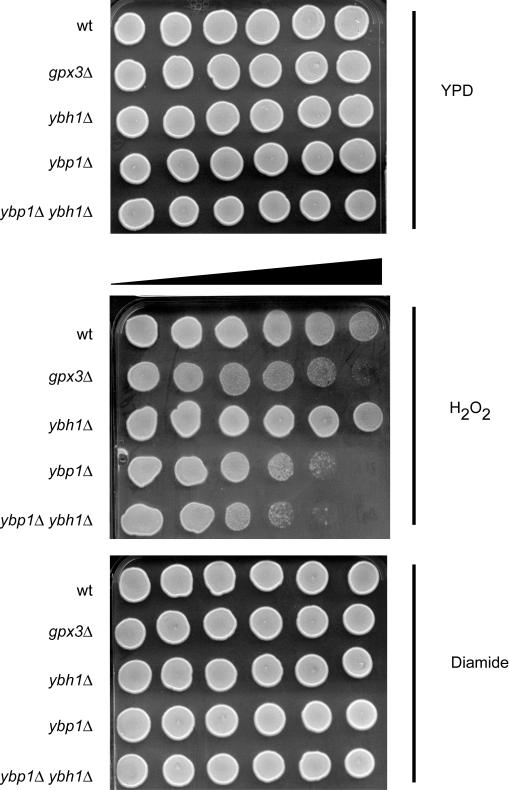
To assess the roles of Ybp1p and Ybh1p in the function of Yap1p, we constructed a set of isogenic strains that contained different dosages of the corresponding genes for these homologous proteins. These strains were then tested for their ability to tolerate oxidative stress imposed by either H2O2 or diamide by use of a gradient plate assay (22). These stress phenotypes were also compared with those of a gpx3Δ mutant strain that has previously been shown to be sensitive to H2O2 (17).
As can be seen in Fig. 1, loss of YBP1 produced an H2O2 hypersensitive phenotype that was more severe than that of the gpx3Δ strain. A strain lacking YBH1 alone did not show any increase in H2O2 sensitivity, but loss of YBH1 from the Δybp1 background further reduced H2O2 tolerance. None of these mutant backgrounds had detectable effects on either diamide resistance or growth in the absence of oxidant.
FIG. 1.
YBH1 is required for normal H2O2 tolerance. An isogenic series of yeast strains were grown to mid-log phase, and a 5-μl spot corresponding to 1,000 cells was placed on rich media (YPD) or rich media containing a gradient of H2O2 or diamide. The increasing concentration of oxidant is denoted by the bar of increasing width. The relevant genetic background of each strain is indicated on the left. The wild-type (wt) strain used is SEY6210.
Yap1p-mediated activation of the γ-glutamylcysteine synthetase-encoding GSH1 gene (32) and the thioredoxin-encoding TRX2 gene (31) has previously been demonstrated to be important in normal oxidative-stress tolerance (13, 25, 34). To examine the influence of Ybp1p and Ybh1p on expression of these two antioxidant loci, lacZ gene fusions to GSH1 and TRX2 were utilized. These fusion genes have previously been shown to accurately reflect transcriptional control of the native genes (4, 38). Low-copy-number plasmids carrying either GSH1- or TRX2-lacZ fusion genes were introduced into the isogenic series of strains containing different dosages of YBP1 and YBH1. Transformants were then grown to mid-log phase and left untreated or subjected to H2O2- or diamide-induced oxidative stress, and expression of β-galactosidase was determined (Fig. 2).
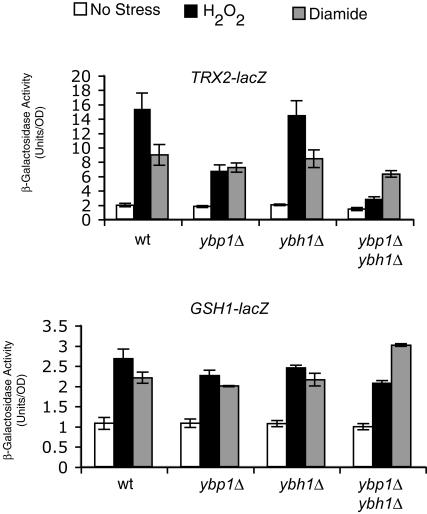
FIG. 2.
Ybp1p and Ybh1p contribute to H2O2-induced expression of TRX2 but not GSH1. The isogenic series of strains described in the legend to Fig. 1 were transformed with low-copy-number vectors carrying either a TRX2-lacZ (top panel) or a GSH1-lacZ (bottom panel) fusion gene. Transformants were grown to mid-log phase, induced by H2O2 or diamide addition or left untreated (no stress), and allowed to grow for ∼1 h. The level of β-galactosidase produced in each transformant was then determined. Relevant genetic backgrounds are indicated as for Fig. 1. wt, wild type.
Expression of TRX2 was strongly reduced in ybp1Δ cells compared to either wild-type or ybh1Δ cells. The decrease in TRX2-lacZ expression was further diminished in the double mutant strain ybp1Δ ybh1Δ. Importantly, diamide-induced TRX2 expression remained constant in these backgrounds. These data indicate that Ybp1p is required for H2O2 but not diamide induction of TRX2 transcription. Ybh1p can also support H2O2-induced expression, but this contribution is masked by the presence of Ybp1p.
Analysis of GSH1-lacZ expression in these same strains produced a surprising result. No significant difference could be detected in either H2O2- or diamide-induced oxidative stress in response to loss of Ybp1p and/or Ybh1p. Although both TRX2 and GSH1 are Yap1p-regulated target genes that are induced by oxidative stress, their dependences on Ybp1p are not equivalent. We believe that the differential requirement of TRX2 and GSH1 for Ybp1p is due to different mechanisms for Yap1p-mediated activation of these genes and consider a possible rationale below (see discussion).
Finally, we examined the ability of these strains to support oxidant-regulated nuclear translocation of Yap1p. Each strain was transformed with a plasmid expressing a GFP-Yap1p fusion protein that was previously shown to faithfully reflect Yap1p subcellular trafficking (4). Transformants were then challenged with H2O2 for various times, and the distribution of GFP-Yap1p was assessed (Fig. 3).
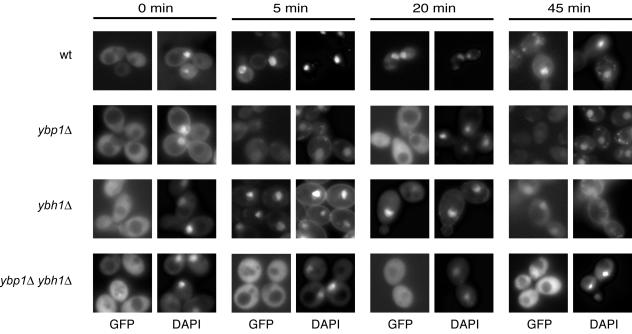
FIG. 3.
Differential effect of ybp1Δ and ybh1Δ on H2O2-induced nuclear recruitment of Yap1p. An isogenic series of strains containing the indicated alleles of YBH1 and YBP1 were transformed with a low-copy-number plasmid expressing a GFP-Yap1p fusion protein under the control of the wild-type (wt) YAP1 promoter (4). Transformants were grown to mid-log phase and challenged with H2O2 for the indicated lengths of time. Cells were then visualized by fluorescence microscopy and filters appropriate for detection of GFP or DAPI fluorescence.
Yap1p rapidly recruits to the nucleus in wild-type cells. Loss of Ybp1p prevented this recruitment while elimination of Ybh1p had no effect on nuclear accumulation of Yap1p. The double ybp1Δ ybh1Δ mutant also showed no ability to deliver Yap1p to the nucleus upon H2O2 exposure. None of these mutant strains exhibited any defect in nuclear localization of Yap1p in response to diamide challenge (data not shown).
These data suggest that the in vivo actions of Ybp1p and Ybh1p are similar, with Ybp1p contributing the major influence to Yap1p function. We next compared the properties of Ybp1 and Ybh1p to directly test whether these proteins worked in similar ways to regulate Yap1p.
Localization and expression of Ybp1p and Ybh1p.
To examine both the subcellular distribution and level of expression of Ybp1p and Ybh1p, we prepared two different epitope-tagged versions of these proteins. Both GFP and 3× HA-tagged forms were generated by integrating appropriate PCR fragments into the genome of an otherwise wild-type strain. We then employed these strains to assess the localization of these proteins as well as to compare their steady-state expression levels (Fig. 4).
FIG. 4.
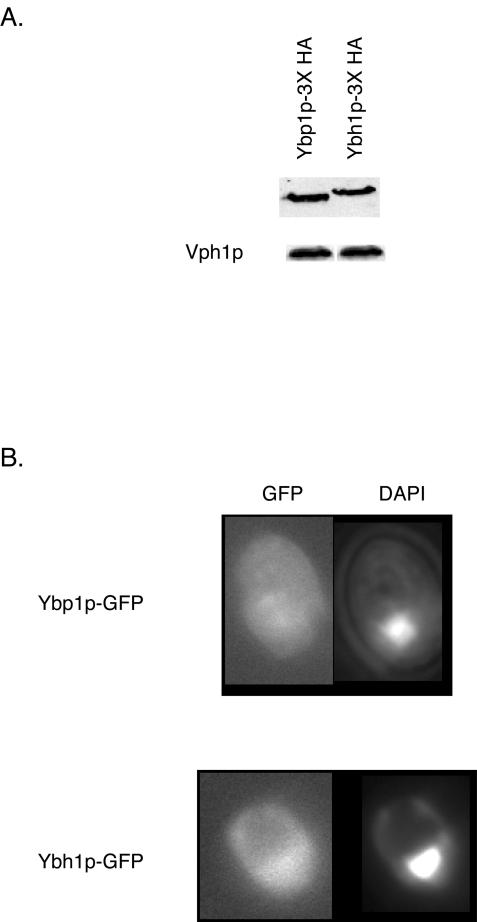
Expression and localization of Ybp1p and Ybh1p. (A) Whole-cell protein extracts were prepared from strains expressing 3× HA-tagged forms of Ybp1p or Ybh1p. Equal amounts of protein were electrophoresed by SDS-PAGE and subjected to Western blot analysis with anti-HA antibody to detect the epitope-tagged proteins. The blot was also probed with antibody directed against the vacuolar ATPase subunit Vph1p to ensure equal protein loading. (B) Strains containing integrated Ybp1p- or Ybh1p-GFP fusion genes were grown to mid-log phase, stained with DAPI, and visualized by fluorescence microscopy.
Comparison of the expression levels of Ybp1p and Ybh1p was carried out with the 3× HA-tagged forms of both proteins. Equal amounts of whole-cell protein extracts were analyzed by Western blotting with an anti-HA antibody. Two important observations were made in this Western blot analysis. First, the steady-state levels of these two proteins are quite similar. Second, although the molecular mass predicted for Ybp1p is 78 kDa and that for Ybh1p is 73 kDa, the mobility of Ybh1p is lower in the SDS-PAGE analysis. By comparison to molecular mass standards, we estimate Ybh1p migrates as an approximately 80-kDa protein. The basis for this altered migration is not understood, but alignment with four other Saccharomyces species suggests the presence of a sequence error in the N-terminal coding sequence of YBH1. If a G residue, found only in the S. cerevisiae sequence at position 939, is deleted, then the N terminus of Ybh1p matches those of the other four species (3, 23). This alteration would increase the predicted molecular mass of Ybh1p to 77 kDa, a better match for its observed migration in SDS-PAGE, but further experiments are required to confirm this idea.
Both Ybp1p- and Ybh1p-GFP fusion proteins were found primarily in the cytoplasm of cells, and this distribution was not detectably influenced by oxidative stress (data not shown). As described by Veal and colleagues (36), the fluorescence produced by the Ybp1p-GFP fusion protein was difficult to detect, a problem not experienced with the Ybh1p-GFP protein. Ybh1p-GFP was easily seen in the cytoplasm of cells and produced readily detectable fluorescence. Our results agree with those recently obtained in a large-scale analysis of GFP fusions to nearly all S. cerevisiae ORFs (15).
These findings provide evidence that the phenotypic differences between ybp1Δ and ybh1Δ strains are not due to differences in expression level of the relevant gene products or detectably different subcellular localization. To explore the basis of the different degree of involvement of Ybp1p and Ybh1p in Yap1p-dependent H2O2 tolerance, we examined the ability of these two proteins to interact with Yap1p.
Carboxy terminus of Ybp1p interacts with Yap1p.
Work from three different groups has provided evidence that Ybp1p binds to Yap1p in several different assays: yeast two hybrid (18), affinity purification (12), and polyhistidine fusion protein precipitation (36). We used the yeast two-hybrid approach to compare the ability of Ybp1p and Ybh1p to bind Yap1p. Full-length clones of the ORFs of YBP1 and YBH1 were fused in-frame with DNA encoding the Gal4p DNA binding domain (GBD) present in the plasmid pGBD-C1 (20). A second plasmid containing the Gal4p transcriptional activation domain (GAD) was used to construct a fusion gene between GAD and full-length YAP1. To localize the region of Ybp1p that interacted with Yap1p, three different subfragments of YBP1 were fused to GBD and tested in this two hybrid assay. We also constructed a GBD-GPX3 fusion gene to determine whether the resulting fusion protein could interact with Yap1p.
These different clones were introduced in pairwise fashion into the Gal4p-dependent reporter strain PJ69-4A (20). This strain contains a Gal4p-responsive HIS3 gene, and we assessed interactions by comparing the ability of different combinations of GBD and GAD fusion plasmids to confer the His+ phenotype on these cells. PJ69-4A also contains Gal4p-regulated ADE2 and lacZ genes that can be used for screening two hybrid interactions. Each interaction was tested on plates with selection for histidine prototrophy in the presence or absence of 1 mM H2O2. We found no difference in the behavior of HIS3 and ADE2 reporters and only report the HIS3 data here (Fig. 5).
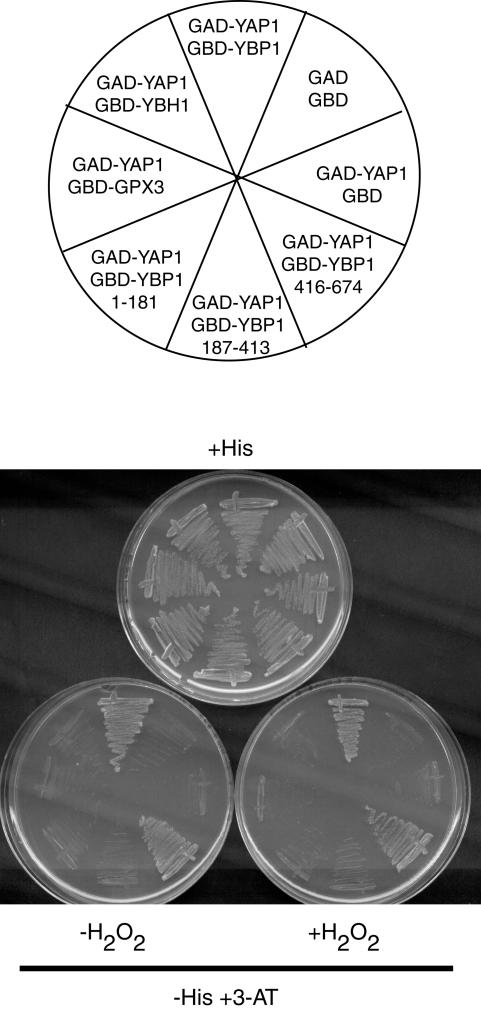
FIG. 5.
Two-hybrid interaction of Ybp1p and Ybh1p with Yap1p. Plasmids expressing fusion proteins with the GBD or GAD of Gal4p were cotransformed into a strain containing a Gal4p-responsive HIS3 gene (20). Full-length fusion proteins were used in every case except for constructs expressing subfragments of the YBP1 coding sequence. The region of Ybp1p expressed is indicated by amino acid residue number. Appropriate transformants were streaked on plates containing histidine (+H) or lacking histidine but containing the His3p inhibitor 3-aminotriazole (+3-AT) in the presence or absence of H2O2.
As seen previously, a clear interaction was seen between Ybp1p and Yap1p. We saw no detectable modification of this interaction in the presence of H2O2. However, full-length Ybh1p failed to support any interaction in this same assay. Similarly, no interaction was seen for Gpx3p. Western blot experiments indicated that the GBD-Ybp1p and -Ybh1p fusion proteins were expressed at similar levels (data not shown).
To further localize the region of Ybp1p involved in binding to Yap1p, GBD-Ybp1p fusion constructs were prepared that expressed amino acids 1 to 181, 187 to 413, and 416 to 674. A positive interaction was seen only for the construct expressing the amino acid 416 to 674 region of Ybp1p and GAD-Yap1p. These data suggest that while either full-length Ybp1p or the C-terminal region of Ybp1p can bind to Yap1p, Ybh1p cannot. However, the two-hybrid assay system requires the use of chimeric protein fusions to the relatively large GBD and GAD that might interfere with the function of the normal gene products. To compare the association of Yap1p with Ybp1p and Ybh1p, we used functional epitope-tagged alleles of these proteins and tested their ability to complex with Yap1p by a coimmunoprecipitation strategy.
Immunological detection of Ybp1p but not Ybh1p in a complex with Yap1p.
To compare the abilities of normally functional Ybp1p and Ybh1p to bind Yap1p, we carried out coimmunoprecipitation analyses with yeast cells. The 3× HA-tagged proteins conferred the same level of H2O2 tolerance as their respective wild-type forms (data not shown). Cells containing these 3× HA-tagged forms of either Ybp1p or Ybh1p were grown to mid-log phase and either left untreated or stressed with H2O2, and protein extracts were prepared. Protein complexes containing Yap1p were recovered by immunoprecipitation with an antibody directed against this transcription factor (37). Immunoprecipitates were then electrophoresed through SDS-PAGE and analyzed by Western blotting with an anti-HA antibody (Fig. 6).
Ybp1p-3X HA was found in immunoprecipitates from both stressed and nonstressed cells. However, we were unable to detect association of Ybh1p-3X HA with Yap1p under either condition. Yap1p was present at equivalent levels in all immunoprecipitates, and the HA-tagged proteins could be detected in supernatants from cells expressing these fusion proteins (data not shown). This is consistent with the failure to observe a Ybh1p-Yap1p interaction in the two-hybrid assay and supports the view that Ybh1p does not associate with Yap1p. We next compared the functional properties of Ybp1p and Ybh1p to assess their ability to act in Yap1p-dependent processes in vivo.
Ybh1p but not Ybp1p can bypass a gpx3Δ mutation.
Proper nuclear localization of Yap1p in response to H2O2 stress requires the transient covalent linkage of Gpx3p to this transcription factor (8). Mutant strains lacking Gpx3p are H2O2 hypersensitive and fail to recruit Yap1p to the nucleus in the presence of this oxidant. To determine whether Ybp1p and Ybh1p function in the same H2O2 resistance pathway, we compared the ability of these two proteins to suppress the H2O2 hypersensitivity of a gpx3Δ mutant strain. Plasmids expressing Ybp1p or Ybh1p from the strong, constitutive PGK promoter were introduced into cells lacking both the GPX3 gene and transformants evaluated for their degree of H2O2 tolerance. A PGK-GPX3 fusion and a 2μm plasmid carrying the wild-type YAP1 gene were also tested for their behavior in this genetic background along with a control vector plasmid (Fig. 7).
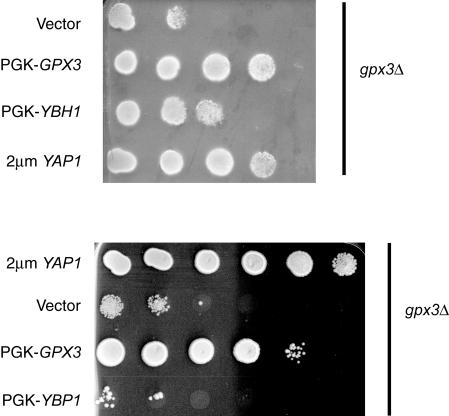
FIG. 7.
Overexpression of Ybh1p but not Ybp1p can suppress the H2O2 sensitivity of a gpx3Δ strain. A strain lacking the GPX3 gene (gpx3Δ) was transformed with plasmids expressing different antioxidant genes. A vector plasmid corresponding to pRS315 containing the PGK promoter (vector) was used to drive the expression of full-length GPX3 (PGK-GPX3), YBP1 (PGK-YBP1), or YBH1 (PGK-YBH1). A high-copy-number plasmid containing the YAP1 gene (2μm YAP1, YEp351-YAP1) (39) was also used to produce high levels of Yap1p. Transformants were analyzed by spot test analysis on H2O2 gradient plates as described previously (4).
Production of Ybh1p from the PGK promoter led to partial suppression of the H2O2 hypersensitivity of the gpx3Δ strain while an analogous clone expressing Ybp1p did not. Expression levels of these proteins were comparable under all conditions as assessed by Western blotting (data not shown). Overproduction of Yap1p was able to elevate H2O2 resistance even in the absence of Gpx3p. As expected, the PGK-GPX3 fusion plasmid restored H2O2 tolerance to the gpx3Δ background. These data argue that, while Ybh1p can influence H2O2 tolerance, it does so in a manner distinct from that of Ybp1p.
Finally, we examined the ability of PGK-driven forms of YBP1 and YBH1 to restore Yap1p nuclear localization to ybp1Δ ybh1Δ cells. As seen in Fig. 3 above, this mutant strain fails to exhibit detectable Yap1p nuclear accumulation after a 10-min challenge with H2O2. PGK-YBP1 and PGK-YBH1 plasmids were introduced into this strain, along with the GFP-Yap1p reporter plasmid and transformants treated with H2O2 for 10 min. Yap1p localization was followed by GFP fluorescence and compared to DAPI-stained DNA (Fig. 8).
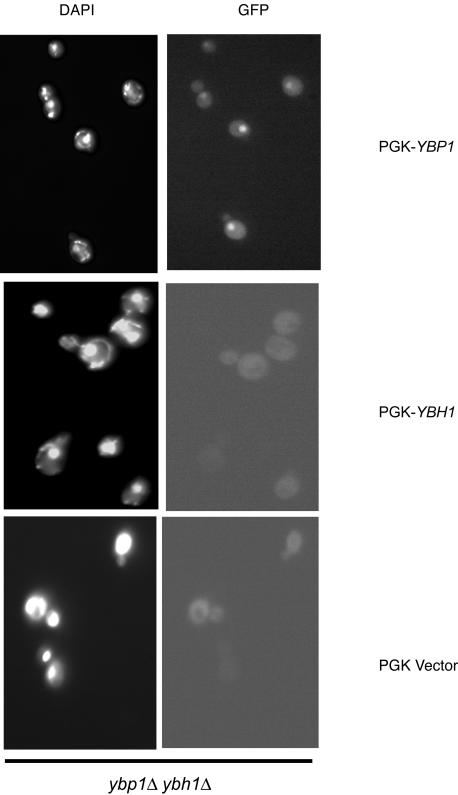
FIG. 8.
Expression of Ybp1p but not Ybh1p can restore H2O2-induced nuclear accumulation of Yap1p. A mutant strain lacking both YBP1 and YBH1 was transformed with a low-copy-number vector containing the PGK promoter driving expression of Ybp1p, Ybh1p, or no additional yeast sequence along with a GFP-Yap1p-containing reporter construct. Transformants were grown to mid-log phase, treated with H2O2 for 20 min, and visualized by fluorescence microscopy.
PGK-directed expression of Ybh1p was unable to restore Yap1p nuclear accumulation in the presence of H2O2 while the PGK-YBP1 fusion plasmid produced normal Yap1p nuclear localization. These functional assays support the interaction results and further indicate that Ybh1p acts in a fashion distinct from that of Ybp1p.
Ybp1p is not required for constitutive nuclear localization of a mutant form of Yap1p.
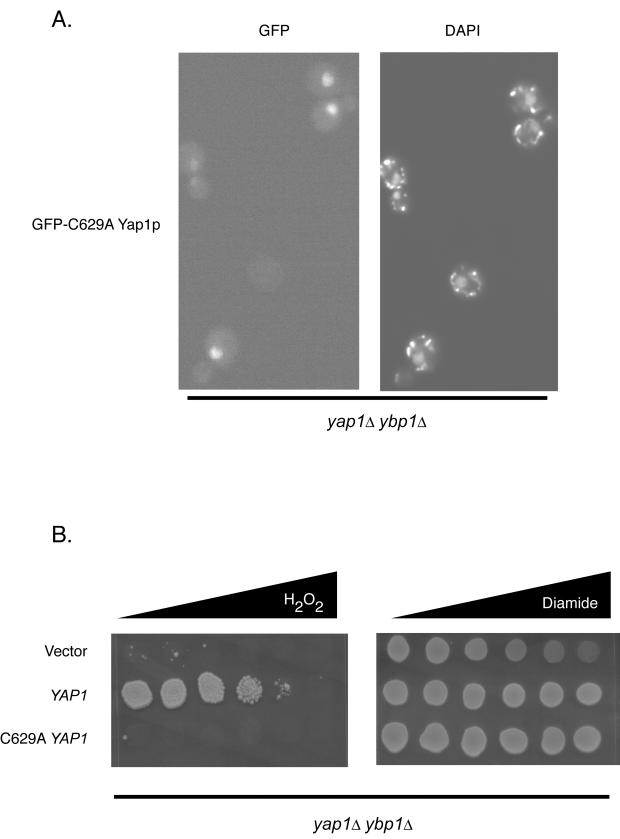
The data presented here and previously (36) indicate that Ybp1p function is required for nuclear localization of Yap1p in the presence of H2O2. To probe the mechanism of action of Ybp1p, we tested the effect of a ybp1Δ mutation on the localization of a mutant form of Yap1p to the nucleus. The C629A allele of Yap1p replaces the cysteine residue at position 629 in the c-CRD with an alanine. In YBP1 cells, C629A Yap1p is localized to the nucleus under H2O2- or diamide-stressed conditions as well as in the absence of oxidative stress (4). Importantly, C629A Yap1p was found to produce hyperresistance to diamide but was hypersensitive to H2O2. We introduced a low-copy-number plasmid expressing Yap1p or C629A Yap1p into a yap1Δ ybp1Δ double mutant strain and tested the H2O2 and diamide tolerance of the resulting transformants. The nuclear localization of C629A Yap1p was also assessed with a GFP fusion to this mutant transcription factor (Fig. 9).
FIG. 9.
Constitutive nuclear localization of a mutant form of Yap1p is independent of Ybp1p function. (A) GFP-C629A Yap1p expressed from a centromeric plasmid was introduced into a yap1Δ ybp1Δ mutant strain. Transformants were grown to mid-log phase, stained with DAPI, and visualized by fluorescence microscopy. (B) Low-copy-number plasmids containing either the wild-type or C629A YAP1 alleles were introduced into a yap1Δ ybp1Δ mutant strain along with an empty vector plasmid (pRS315). Transformants were grown to mid-log phase and analyzed for H2O2 and diamide tolerance as described above by a gradient plate assay.
GFP-C629A Yap1p was found in the nucleus of yap1Δ ybp1Δ cells under nonstressed conditions, as was seen previously in a yap1Δ strain (4). The constitutive nuclear localization of C629A Yap1p is independent of Ybp1p function and was not influenced by the presence of H2O2 (data not shown). Additionally, the oxidant sensitivity of a ybp1Δ strain could not be suppressed by the introduction of a constitutively nuclear C629A Yap1p. Together, these data argue that while regulation of Yap1p nuclear recruitment upon H2O2 stress is an important task of Ybp1p, this localization defect alone is inadequate to explain the H2O2 sensitivity of the ybp1Δ strain.
DISCUSSION
A striking feature of the oxidative stress regulation of Yap1p is the differential behavior of mutant forms of this factor in response to different oxidants. Typically Yap1p mutants that exhibit constitutive nuclear localization confer hyperresistance to diamide-induced oxidative stress (26, 37, 40) but are hypersensitive to the presence of H2O2 in the medium (4, 7, 37). The recent identification of Gpx3p (8) and Ybp1p (reference 36 and this work) as H2O2-specific regulators of Yap1p function provide evidence for the additional complexity of the action of Yap1p during H2O2 stress compared to diamide stress.
These studies also provide the first functional assessment of the action of the Ybp1p homologue Ybh1p. Based on the strong sequence conservation between Ybp1p and Ybh1p, we anticipated that these factors might work in very similar manners. However, the data reported here present several lines of evidence that significant differences exist between the in vivo roles of these two related proteins. First, no evidence could be obtained that Ybh1p actually interacted with Yap1p in the cell. While both two-hybrid data and coimmunoprecipitation approaches were consistent with the presence of the Yap1p-Ybp1p interaction, neither could detect interaction between Yap1p and Ybh1p. Second, PGK-driven expression of Ybp1p but not Ybh1p was able to restore H2O2-induced nuclear accumulation of Yap1p in cells lacking both YBP1 and YBH1. Finally, the H2O2 hypersensitivity of a Δgpx3 strain could be suppressed by overproduction of Ybh1p but not Ybp1p.
These data are most consistent with the idea that Ybh1p acts in a pathway parallel to that of Ybp1p. One attractive model is suggested by the presence of a number of Yap1p homologues in S. cerevisiae (11). At least 7 proteins sharing significant sequence similarity with Yap1p are encoded by the S. cerevisiae genome. Several of these have been demonstrated to have DNA binding and transcriptional regulatory properties similar to Yap1p (11). Ybh1p may exert its normal activity through control of the function of one of these other Yap1p homologues. It is interesting that Ybh1p has previously been implicated in resistance to osmotic stress (6), as have the Yap1p homologues Yap4p/Cin5p and Yap6p (29). An immediate experimental goal is to identify the protein(s) downstream from Ybh1p in its pathway of action.
While the data reported here are consistent with Ybh1p regulating a transcription factor that in turn can carry out some of the functions normally executed by Yap1p, we cannot eliminate the possibility that Ybh1p may act in a Yap1p regulatory pathway that does not involve Gpx3p. Perhaps Ybh1p acts with a Gpx3p-like protein to properly modify Yap1p in response to H2O2. We do not favor this explanation, since we were unable to detect any interaction between Yap1p and Ybh1p, but it remains a possibility.
The differential contribution of Ybp1p and Ybh1p to H2O2 stress induction of TRX2 and GSH1 was surprising. Since Yap1p does not concentrate in the nucleus in ybp1Δ cells, we anticipated that H2O2 induction of TRX2 and GSH1 would be inhibited. The lack of sensitivity of GSH1 expression to YBP1 was unexpected. This differential requirement for Ybp1p to enable H2O2-mediated activation of TRX2 versus GSH1 is reminiscent of the differential effect of constitutively nuclear forms of Yap1p that fail to increase TRX2 expression but dramatically elevate artificial Yap1p-responsive reporter genes (4). We have found that GSH1 expression is also constitutively high in these permanently nuclear mutant forms of Yap1p (unpublished data). Our interpretation of these data is that the mechanism of transcriptional activation by Yap1p is different at TRX2 and GSH1. Importantly, the difference in gene activation by these mutants cannot be explained in terms of nuclear localization.
The persistence of H2O2-inducible expression of GSH1 in ybp1Δ cells could be explained by at least two different possibilities. Since basically all Yap1p localization data come from the use of GFP reporter constructs (both here and elsewhere) (4, 7, 19, 26, 36, 40), resolution of the extent of Yap1p localization is limited by the use of fluorescence microscopy. A low level of nuclear GFP-Yap1p might be masked by the large cytoplasmic GFP signal. With this caveat in mind, it is possible that a low level of Yap1p does accumulate in the nucleus in response to H2O2 challenge. This low level is sufficient for GSH1 activation but inadequate to induce TRX2. Alternatively, it has been suggested by others (9) that H2O2 induction of GSH1 expression does not require Yap1p per se, but rather, Yap1p participates in an indirect role. Further experiments are required to discriminate between these and other models, but clearly, H2O2 regulation of Yap1p action is a complex process involving multiple proteins.
The response of Yap1p to H2O2 challenge is quite different from control of Yap1p by diamide or diethylmaleate, two oxidants that seem likely to influence Yap1p activity exclusively at the level of regulation of nuclear localization. Exposure of cells to diamide is thought to lead to localized oxidation of cysteine residues in Yap1p and prevent the factor from binding to the nuclear exportin Crm1p (27, 40). This causes the protein to accumulate in the nucleus and leads to increased Yap1p target gene expression. Consistent with the notion that the influence of diamide can be considered to be purely at the level of nuclear localization, mutants that are constitutively localized to the nucleus are also invariably hyperresistant to diamide (4, 26, 37).
Ybp1p has previously been demonstrated to be required for the appearance of a disulfide bond linking the n-CRD with the c-CRD (7). It is possible that the formation of this disulfide bond has two different roles in terms of Yap1p regulation. Formation of this bond may inhibit interaction with Crm1p and provide a conformation of Yap1p that is appropriate for the activation of genes like TRX2 that must be induced to support normal tolerance to H2O2. This dual contribution to H2O2 regulation of Yap1p function is entirely consistent with the observed phenotypes of mutants like C629A Yap1p. We speculate that a Ybp1p-requiring folding step is required for Yap1p to assume a particular conformation appropriate for activation of genes like TRX2 but is not necessary for induction of genes like ATR1 (5). ATR1 is a Yap1p-regulated gene but has no detectable role in H2O2 tolerance. The molecular basis of this oxidant- and gene-specific transcriptional activation by Yap1p is currently under investigation.
Acknowledgments
This work was supported by NIH GM57007.
We thank Patricia Bilodeau and Rob Piper for help with immunological methods and valuable discussions.
REFERENCES
- 1.Avery, A. M., and S. V. Avery. 2001. Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J. Biol. Chem. 276:33730-33735. [DOI] [PubMed] [Google Scholar]
- 2.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 3.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 4.Coleman, S. T., E. A. Epping, S. M. Steggerda, and W. S. Moye-Rowley. 1999. Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 19:8302-8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman, S. T., E. Tseng, and W. S. Moye-Rowley. 1997. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J. Biol. Chem. 272:23224-23230. [DOI] [PubMed] [Google Scholar]
- 6.de Jesus Ferreira, M. C., X. Bao, V. Laize, and S. Hohmann. 2001. Transposon mutagenesis reveals novel loci affecting tolerance to salt stress and growth at low temperature. Curr. Genet. 40:27-39. [DOI] [PubMed] [Google Scholar]
- 7.Delaunay, A., A. D. Isnard, and M. B. Toledano. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delaunay, A., D. Pflieger, M. B. Barrault, J. Vinh, and M. B. Toledano. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471-481. [DOI] [PubMed] [Google Scholar]
- 9.Dormer, U. H., J. Westwater, D. W. Stephen, and D. J. Jamieson. 2002. Oxidant regulation of the Saccharomyces cerevisiae GSH1 gene. Biochim. Biophys. Acta 1576:23-29. [DOI] [PubMed] [Google Scholar]
- 10.Estruch, F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24:469-486. [DOI] [PubMed] [Google Scholar]
- 11.Fernandes, L., C. Rorigues-Pousada, and K. Struhl. 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17:6982-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 13.Grant, C. M., F. H. MacIver, and I. W. Dawes. 1996. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 29:511-515. [DOI] [PubMed] [Google Scholar]
- 14.Guarente, L. 1983. Yeast promoter and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101:181-191. [DOI] [PubMed] [Google Scholar]
- 15.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 16.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, Y., T. Matsuda, K. Sugiyama, S. Iszawa, and A. Kimura. 1999. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 274:27002-27009. [DOI] [PubMed] [Google Scholar]
- 18.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izawa, S., K. Maeda, K. Sugiyama, J. Mano, Y. Inoue, and A. Kimura. 1999. Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 274:28459-28465. [DOI] [PubMed] [Google Scholar]
- 20.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 22.Katzmann, D. J., E. A. Epping, and W. S. Moye-Rowley. 1999. Mutational disruption of plasma membrane trafficking of Saccharmyces cerevisiae Yor1p, a homologue of mammalian multidrug resistance protein. Mol. Cell. Biol. 19:2998-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 24.Kuge, S., M. Arita, A. Murayama, K. Maeta, S. Izawa, Y. Inoue, and A. Nomoto. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21:6139-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuge, S., and N. Jones. 1994. YAP1-dependent activation of TRX2 is essential for the response of S. cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuge, S., N. Jones, and A. Nomoto. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuge, S., T. Toda, N. Iizuka, and A. Nomoto. 1998. Crm1 (Xpo1) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells 3:521-532. [DOI] [PubMed] [Google Scholar]
- 28.Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 29.Mendizabal, I., G. Rios, J. M. Mulet, R. Serrano, and I. F. de Larrinoa. 1998. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 425:323-328. [DOI] [PubMed] [Google Scholar]
- 30.Moye-Rowley, W. S. 2003. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot. Cell. 2:381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller, E. G. D. 1991. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J. Biol. Chem. 266:9194-9202. [PubMed] [Google Scholar]
- 32.Ohtake, Y., and S. Yabuuchi. 1991. Molecular cloning of the gamma-glutamylcysteine synthetase gene of Saccharomyces cerevisiae. Yeast 7:953-961. [DOI] [PubMed] [Google Scholar]
- 33.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephen, D. W. S., and D. J. Jamieson. 1997. Amino acid-dependent regulation of Saccharomyces cerevisiae GSH1 gene by hydrogen peroxide. Mol. Microbiol. 23:203-210. [DOI] [PubMed] [Google Scholar]
- 35.Toone, W. M., B. A. Morgan, and N. Jones. 2001. Redox control of AP-1-like factors in yeast and beyond. Oncogene 20:2336-2346. [DOI] [PubMed] [Google Scholar]
- 36.Veal, E. A., S. J. Ross, P. Malakasi, E. Peacock, and B. A. Morgan. 2003. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J. Biol. Chem. 278:30896-30904. [DOI] [PubMed] [Google Scholar]
- 37.Wemmie, J. A., S. M. Steggerda, and W. S. Moye-Rowley. 1997. The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J. Biol. Chem. 272:7908-7914. [DOI] [PubMed] [Google Scholar]
- 38.Wu, A., and W. S. Moye-Rowley. 1994. GSH1, which encodes γ-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 14:5832-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, A., J. A. Wemmie, N. P. Edgington, M. Goebl, J. L. Guevara, and W. S. Moye-Rowley. 1993. Yeast bZIP proteins mediate pleiotropic drug and metal resistance. J. Biol. Chem. 268:18850-18858. [PubMed] [Google Scholar]
- 40.Yan, C., L. H. Lee, and L. I. Davis. 1998. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17:7416-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, X., and W. S. Moye-Rowley. 2001. Saccharomyces cerevisiae multidrug resistance gene expression inversely correlates with the status of the Fo component of the mitochondrial ATPase. J. Biol. Chem. 276:47844-47852. [DOI] [PubMed] [Google Scholar]