Abstract
The genome of feline calicivirus (FCV) is an ∼7.7-kb single-stranded positive-sense RNA molecule that is polyadenylated at its 3′ end and covalently linked to a VPg protein (calculated mass, 12.6 kDa) at its 5′ end. We performed a mutational analysis of the VPg protein in order to identify amino acids potentially involved in linkage to the genome and replication. The tyrosine residues at positions 12, 24, 76, and 104 were changed to alanines by mutagenesis of an infectious FCV cDNA clone. Viruses were recovered when Tyr-12, Tyr-76, or Tyr-104 of the VPg protein was changed to alanine, but virus was not recovered when Tyr-24 was changed to alanine. Growth properties of the recovered viruses were similar to those of the parental virus. We examined whether the amino acids serine, threonine, and phenylalanine could substitute for the tyrosine at position 24, but these mutations were lethal as well. A tyrosine at this relative position is conserved among all calicivirus VPg proteins examined thus far, suggesting that the VPg protein of caliciviruses, like those of picornaviruses and potyviruses, utilizes tyrosine in the formation of a covalent bond with RNA.
Feline calicivirus (FCV), a member of the genus Vesivirus in the family Caliciviridae, is one of the major etiologic agents of respiratory illness in cats (9, 14). The genome of FCV is an ∼7.7-kb single-stranded positive-sense RNA molecule that is covalently linked to a protein designated VPg (virion protein, linked to genome) at its 5′ end and polyadenylated at its 3′ end (4, 12). The FCV genome is organized into three major open reading frames (ORFs). ORF1 encodes a 200-kDa polyprotein that is processed by the virus-encoded 3C-like cysteine proteinase into p5.6, p32, p39 (NTPase), p30, p13 (VPg), and p76 (Pro-Pol) (28, 32). ORF2 encodes a 73-kDa capsid precursor (preVP1) that is also cleaved by the cysteine proteinase to yield the 14-kDa capsid leader and the 60-kDa major mature capsid protein VP1 (5, 31). ORF3 encodes a 12-kDa basic protein (VP2) of unknown function (11).
Early after FCV infection, the genomic RNA interacts with the cellular machinery to initiate translation. The VPg protein may play an important role in this early event, because treatment of genomic calicivirus RNA with proteinase K abolishes its infectivity and, in addition, decreases its translation efficiency in vitro (2, 12). The infectivity of RNA transcribed from a full-length cDNA clone of FCV is dependent on its synthesis in the presence of a cap analog, which suggests that the cap might substitute for the role of VPg in the initiation of infection (27). Of interest, sequence similarities between the calicivirus VPg protein and the eukaryotic translation initiation factor eIF1A have been described (32) and recent studies have reported an interaction between the norovirus VPg protein and eIF3 (6). Although the calicivirus VPg protein has been implicated in translation, its function is not clear. The recent observation that the VPg protein of rabbit hemorrhagic disease virus (RHDV) was uridylylated in vitro by recombinant RHDV polymerase was the first evidence that the calicivirus VPg protein may also function directly in RNA replication (20).
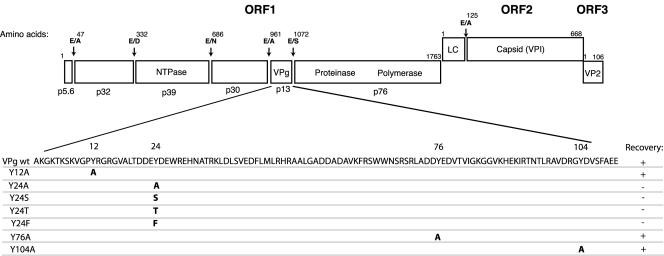
The VPg protein of FCV is 111 amino acids long, maps to amino acids 961 to 1072 of ORF1 (Fig. 1), and has a calculated molecular weight of 12.65 kDa (29, 32). The mature VPg protein is present in FCV-infected cells as an unmodified cleavage product and as a more slowly migrating form (15.5 kDa) that is likely covalently linked to viral RNA (29). Of note, the 15.5-kDa form is also found in virions (29). Cleavage and the release of the mature VPg protein from the ORF1 polyprotein are essential for the growth of the virus (28). The length of the FCV VPg cleavage product (111 amino acids) is similar to those of other caliciviruses, including Pan-1 (in the genus Vesivirus) (7), RHDV (in the genus Lagovirus) (35), and Southampton virus (in the genus Norovirus) (18), which contain 114, 115, and 138 amino acid residues, respectively. Two conserved amino acid motifs, KGK(N/T)K and (D/E)EY(D/E)E, have been identified in comparative sequence alignments of calicivirus VPg proteins (7). The latter motif in the RHDV VPg protein contains the tyrosine residue (Tyr-21) that was involved in in vitro uridylylation by the polymerase and has been proposed as the site where linkage to the viral RNA might occur (20).
FIG. 1.
Summary of mutations in the FCV VPg protein and their effects on recovery of virus. The genome organization and coding assignments of the Urbana strain of FCV (GenBank accession no. L40021) are shown. Dipeptide cleavage sites recognized by the virus-encoded cysteine proteinase are indicated. The four tyrosines (numbered according to the VPg protein amino acid sequence) at positions 12, 24, 76, and 104 were changed to alanine in the infectious cDNA clone pQ14. In addition, the tyrosine at position 24 was changed to serine, threonine, or phenylalanine. For each construct, the ability to recover viable virus is indicated by a plus sign and a lethal mutation is indicated by a minus sign. wt, wild type.
We compared the calicivirus VPg protein to those of other positive-strand RNA viruses with VPg-linked genomes in an effort to identify conserved features. The Caliciviridae, Picornaviridae, Comoviridae, and Potyviridae show evolutionary relatedness in their RNA-dependent RNA polymerase proteins as members of the proposed Supergroup I lineage (15). The picornavirus (1) and potyvirus (22) VPg proteins are linked to RNA via a phosphodiester bond between the β-OH group of tyrosine and the 5′ end of the genome (U for picornaviruses and A predominantly for potyviruses), whereas the comovirus VPg protein is linked to the 5′-terminal U of the genomic RNA by the β-OH group of serine (13). The sizes and putative functions of these VPg proteins vary. The picornavirus and comovirus proteins are relatively smaller (approximately 2 to 6 kDa) than those of the potyviruses and caliciviruses (approximately 13 to 21 kDa). However, a common feature of these VPg proteins is that mutation of the tyrosine or serine involved in the linkage of RNA to the VPg protein is lethal for virus growth and replication (3, 21, 25). The picornavirus VPg protein is uridylylated by the 3D polymerase to form VPg-pU and VPg-pUpU and functions as a primer for RNA synthesis during replication (24, 34). The potyvirus VPg protein has been implicated in translation (17), long-distance movement in plant tissue (26), and possibly replication (8). The function of the comovirus VPg protein is not known, but the comovirus VPg protein is covalently linked to the positive and negative strands of the RNA replicative forms during infection, suggesting that it could be involved in RNA replication (19).
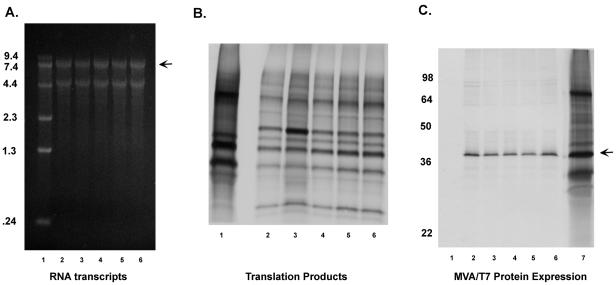
A mutational analysis of the VPg region from the Urbana strain of FCV was initiated in this study to determine whether tyrosine might be involved in the activity of this region. The FCV VPg has four tyrosine residues, at positions 12, 24, 76, and 104, that could potentially link VPg to the viral RNA (Fig. 1). Plasmid constructs in which each tyrosine residue was changed to alanine in the infectious FCV cDNA clone pQ14 were made. Point mutations were introduced by using the QuikChange site-directed mutagenesis kit from Stratagene; the forward-sense primers used for the mutagenesis are shown in Table 1. The mutagenized plasmids were transformed into Escherichia coli, and the entire FCV genome-specific insert of each selected plasmid was sequenced to verify the mutagenesis procedure. The plasmid DNA (containing the FCV genome under the control of the T7 RNA polymerase promoter) was transfected into Crandell Rees feline kidney (CRFK) cells infected with MVA/T7 (30). The recovery of FCV was monitored by passage of the cell culture medium collected at 24 h onto a fresh CRFK monolayer. Cytopathic effects characteristic of FCV were observed in passages derived from constructs in which the Tyr-12, Tyr-76, and Tyr-104 residues of the VPg protein were replaced by alanine (data not shown). After four passages, the recovered viruses were analyzed by reverse transcription-PCR and sequence analysis to verify the presence of the engineered mutation in the RNA genome. Passage of the cell culture material derived from transfection of the construct Y24A did not yield a visible cytopathic effect in a fresh monolayer, and the results of an immunofluorescence assay to monitor capsid expression were negative for both the original transfection and subsequent passages (data not shown). We then examined whether other amino acids with an available hydroxyl group (serine or threonine) or a similar structure (phenylalanine) could substitute for tyrosine residue 24, but these mutations were lethal as well, and no capsid expression was detected by immunofluorescence in the original transfection or in subsequent passages (data not shown). In order to verify that the failure to recover virus was not due to defective RNA or protein synthesis from the mutagenized plasmids, capped RNA transcripts were produced in vitro from the four cDNA clones containing mutations at residue 24 and compared to those derived from the parental pQ14 plasmid (Fig. 2A). The migration of the full-length RNA (Fig. 2A) was similar, as was that of the approximately 5 kb smaller transcript that has been associated with transcription of the FCV cloned genome (10, 33). The capped RNA transcripts were then translated in vitro in the Flexi rabbit reticulocyte system (Promega), and comparison of the synthesized proteins with those derived from wild-type pQ14 showed an overall similarity (Fig. 2B). The RNA transcripts shown in Fig. 2A were transfected into CRFK cells with Lipofectamine Plus (Invitrogen), and progeny virus was recovered only from the wild-type (pQ14-derived) RNA (data not shown). In order to verify that failure to recover virus with the MVA/T7 system was not due to aberrant transcription or protein synthesis in cells, the mutagenized plasmids and wild-type pQ14 were transfected into MVA/T7-infected cells and proteins were radiolabeled with [35S]methionine (>1,000 μCi/mmol; Amersham) for 3 h. Cell lysates were prepared and incubated with p39 (NTPase)- or FCV capsid-specific antibodies for analysis of the immunoprecipitated products by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography as described previously (28, 31). Similar levels of mature p39 protein were observed in both the wild-type pQ14 and the mutagenized plasmid transfections (Fig. 2C), indicating that the levels of RNA transcription and nonstructural protein synthesis in the MVA-T7 system were not affected by the engineered mutations. Consistent with the immunofluorescence experiments described above, viral capsid protein was not detected by immunoprecipitation following transfection of the mutagenized plasmids (data not shown). This experiment confirmed that mature nonstructural proteins were synthesized and present in the transfected cells but that structural protein synthesis did not occur. We failed to find evidence for FCV-specific negative-strand RNA synthesis in cells transfected with either mutant capped transcript RNAs or plasmid DNA in the MVA/T7 expression system when RNA purified from the cells was examined by Northern blot analysis as described previously (10) (data not shown).
TABLE 1.
Primers used for mutagenesis of VPg in the FCV genome
| Engineered mutation | Primera |
|---|---|
| Tyr-12 to Ala | GTTGGTCCAGCCAGAGGTCGTGG |
| Tyr-24 to Ala | CCTAACTGATGATGAAGCCGATGAATGGAGGGAAC |
| Tyr-76 to Ala | GCTGATGATGCTGAGGACGTC |
| Tyr-104 to Ala | GATCGGGGCGCTGATGTTAGC |
| Tyr-24 to Phe | GATGATGAATTTGATGAATGGAGG |
| Tyr-24 to Ser | GATGATGAATCCGATGAATGGAGG |
| Tyr-24 to Thr | GATGATGAAACCGATGAATGGAGG |
The mutated amino acid codon is underlined, and the engineered nucleotide substitutions are shown in bold type. Only the forward primer sequence of the complementary pair used in the mutagenesis is shown.
FIG. 2.
Analysis of RNA and proteins derived from plasmids containing lethal mutations at VPg residue 24. (A) Capped RNA transcripts were synthesized as previously described (27) from the NotI-linearized plasmid DNA of constructs pQ14 (lane 2), Y24A (lane 3), Y24S (lane 4), Y24T (lane 5), and Y24F (lane 6) and analyzed with a 1% agarose gel (Ambion). The RNA was visualized by ethidium bromide staining. An RNA marker (Invitrogen) was included in lane 1. The arrow indicates the full-length RNA. (B) The capped RNA transcripts were translated in a rabbit reticulocyte lysate in the presence of radiolabeled methionine, and the proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a 10 to 20% polyacrylamide gel. The proteins correspond to RNAs derived from pQ14 (lane 2), Y24A (lane 3), Y24S (lane 4), Y24T (lane 5), and Y24F (lane 6). The previously characterized proteins derived by coupled transcription and translation (TNT) of the FCV ORF1 clone pTMF-1 (32) are shown in lane 1 for comparison. (C) CRFK cells were infected with MVA/T7 and transfected with wild-type pQ14 or mutagenized plasmids. After 5 h, the proteins were radiolabeled with [35S]methionine for 12 h. Cell lysates were prepared and incubated with p39-specific serum, followed by precipitation of antigen-antibody complexes with Sepharose protein A beads. The precipitated proteins were resolved in a 10% Tris-glycine polyacrylamide gel and visualized by autoradiography. Immunoprecipitation of the p39 protein (indicated by an arrow) from MVA/T7-infected cells transfected with plasmids pQ14 (lane 2), Y24A (lane 3), Y24S (lane 4), Y24T (lane 5), and Y24F (lane 6) is shown. Lane 1 contains mock-transfected MVA/T7 cells incubated with p39-specific antibodies, and lane 7 contains radiolabeled TNT products derived from pTMF-1 as shown for panel B.
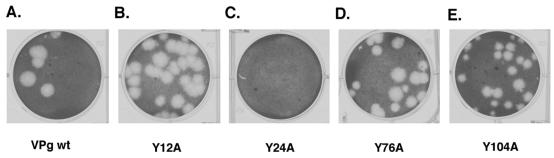
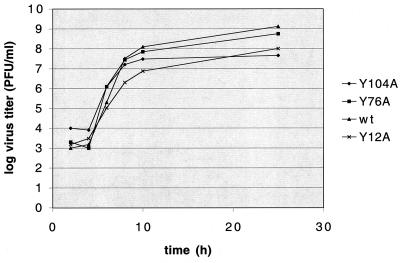
The growth properties of the recovered mutant viruses Y12A, Y76A, and Y104A were compared to those of the wild-type virus. The mutant viruses formed plaques similar in size and morphology to those of the wild-type virus (Fig. 3), although the Y104A plaques (Fig. 3E) appeared to be slightly smaller. The kinetics of virus growth was examined. Confluent CRFK cells (approximately 106 cells) were incubated with each virus at a multiplicity of infection of 0.1 for 1 h at 37°C. The inoculum was removed, cells were washed, and fresh medium was added. Cell culture fluid was harvested 2, 4, 6, 8, 10, and 25 h postinfection, and the titer was determined by plaque assay (23). The mutant viruses showed growth kinetics similar to that of the wild-type virus, with titers at the 25-h end point ranging from 4.5 × 107 (mutant Y104A) to 1.3 × 109 (wild type) (Fig. 4).
FIG. 3.
Comparison of the plaque phenotypes of recovered viruses to that of the wild-type (wt) virus. Recovered viruses were assayed for their ability to form plaques in CRFK cells. Serial dilutions of viruses (10−2 to 10−8) were used to infect monolayers of CRFK cells seeded in six-well plates. After 1 h of incubation of the virus inoculum at 37°C, cells were washed and an agarose overlay was added. Cells were then incubated at 37°C for 24 h in a humidified CO2 incubator. The monolayers were fixed with formalin and stained with crystal violet to visualize viral plaques (23).
FIG. 4.
Growth characteristics of mutant viruses in comparison with those of the wild-type (wt) virus. Confluent monolayers of CRFK cells were infected with wild-type or mutant viruses at a multiplicity of infection of 0.1. Cell culture fluid was harvested 2, 4, 6, 8, 10, and 25 h postinfection. Virus titer was determined by end point titration in a plaque assay as described in the legend to Fig. 3.
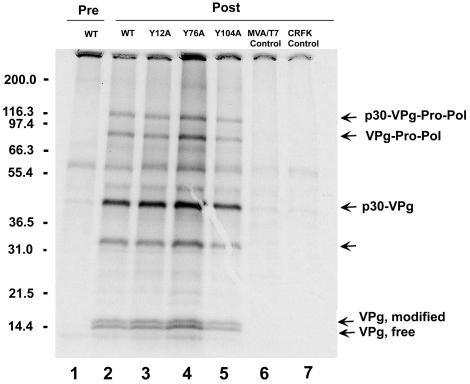
Certain mutations in the VPg proteins of poliovirus and cowpea mosaic virus (a comovirus) were shown to affect proteolytic processing (3, 16). We examined the effects of the engineered FCV VPg mutations on proteolytic processing of the ORF1 polyprotein. Cells infected with the Y12A, Y76A, Y104A, or wild-type virus were analyzed by immunoprecipitation with VPg-specific antibodies. The profiles of the immunoprecipitated proteins from wild-type (Fig. 5, lane 2) and mutant viruses (Fig. 5, lanes 3 to 5) were similar. The two forms of cleaved VPg protein (designated the free and modified forms), as well as its precursors p30-VPg, p30-VPg-Pro, and p30-VPg-Pro-Pol, were observed, indicating that the proteolytic processing of this region was not affected by the nonlethal mutations analyzed in this study. An ∼33-kDa protein which had been observed in a previous study (29) was also precipitated from the virus-infected cells with the VPg-specific antiserum (Fig. 5, lanes 2 to 5). The detection of this 33-kDa protein differs among immunoprecipitation experiments, and the identity of this protein is unclear (data not shown).
FIG. 5.
Effects of mutations on the proteolytic processing of VPg and its precursors. CRFK cells were infected with wild-type (WT) or mutant viruses and radiolabeled with [35S]methionine. Cell lysates were prepared and incubated with VPg-specific serum, followed by precipitation of antigen-antibody complexes and analysis with a 10 to 20% polyacrylamide gel. Lane 1, wild-type virus-infected cell lysate incubated with preimmunization guinea pig serum. Lanes 2 to 6, the VPg-specific postimmunization guinea pig serum was incubated with infected cell lysates prepared from wild-type virus (lane 2), Y12A (lane 3), Y76A (lane 4), Y104A (lane 5), and MVA/T7 (lane 6). Lane 7, mock-infected CRFK cell lysate control incubated with VPg-specific serum. The known precursor proteins containing VPg, as well as free and modified forms of the mature VPg (29), are indicated. The 33-kDa protein is indicated by an unmarked arrow.
Our data indicate that the conserved tyrosine at position 24 of the FCV VPg protein is essential for FCV replication. This observation is consistent with the recent finding that the analogous tyrosine in the VPg protein of RHDV is the site of in vitro uridylylation by the polymerase (20) and that linkage to the RNA may be mediated by this amino acid. Further studies are in progress to confirm the nature of the biochemical linkage between the FCV VPg protein and RNA and to determine the role of VPg in calicivirus replication.
Acknowledgments
We thank Gaël Belliot (NIAID, NIH) for helpful comments and Albert Z. Kapikian (NIAID, NIH) for support of this project.
REFERENCES
- 1.Ambros, V., and D. Baltimore. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263-5266. [PubMed] [Google Scholar]
- 2.Burroughs, J. N., and F. Brown. 1978. Presence of a covalently linked protein on calicivirus RNA. J. Gen. Virol. 41:443-446. [DOI] [PubMed] [Google Scholar]
- 3.Carette, J. E., A. Kuujawa, K. Guhl, J. Verver, J. Wellink, and A. Van Kammen. 2001. Mutational analysis of the genome-linked protein of cowpea mosaic virus. Virology 290:21-29. [DOI] [PubMed] [Google Scholar]
- 4.Carter, M. J., I. D. Milton, J. Meanger, M. Bennett, R. M. Gaskell, and P. C. Turner. 1992. The complete nucleotide sequence of a feline calicivirus. Virology 190:443-448. [DOI] [PubMed] [Google Scholar]
- 5.Carter, M. J., I. D. Milton, P. C. Turner, J. Meanger, M. Bennett, and R. M. Gaskell. 1992. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch. Virol. 122:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daughenbaugh, K. F., C. S. Fraser, J. W. B. Hershey, and M. E. Hardy. 2003. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 22:2852-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunham, D. M., X. Jiang, T. Berke, A. W. Smith, and D. O. Matson. 1998. Genomic mapping of a calicivirus VPg. Arch. Virol. 143:2421-2430. [DOI] [PubMed] [Google Scholar]
- 8.Fellers, J., J. Wan, Y. Hong, G. B. Collins, and A. G. Hunt. 1998. In vitro interactions between a potyvirus-encoded, genome-linked protein and RNA-dependent RNA polymerase. J. Gen. Virol. 79:2043-2049. [DOI] [PubMed] [Google Scholar]
- 9.Gaskell, R. M. 1985. Induced upper respiratory tract diseases, 1st ed. Blackwell, Oxford, England.
- 10.Green, K. Y., A. Mory, M. H. Fogg, A. Weisberg, G. Belliot, M. Wagner, T. Mitra, E. Ehrenfeld, C. E. Cameron, and S. V. Sosnovtsev. 2002. Isolation of enzymatically active replication complexes from feline calicivirus-infected cells. J. Virol. 76:8582-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbert, T. P., I. Brierley, and T. D. Brown. 1996. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J. Gen. Virol. 77:123-127. [DOI] [PubMed] [Google Scholar]
- 12.Herbert, T. P., I. Brierley, and T. D. Brown. 1997. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J. Gen. Virol. 78:1033-1040. [DOI] [PubMed] [Google Scholar]
- 13.Jaegle, M., J. Wellink, and R. Goldbach. 1987. The genome-linked protein of cowpea mosaic virus is bound to the 5′ terminus of virus RNA by a phosphodiester linkage to serine. J. Gen. Virol. 68:627-632. [Google Scholar]
- 14.Kahn, D. E., and E. A. Hoover. 1976. Infectious respiratory diseases of cats. Vet. Clin. N. Am. 6:399-413. [DOI] [PubMed] [Google Scholar]
- 15.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. (Erratum, 28:546.) [DOI] [PubMed] [Google Scholar]
- 16.Kuhn, R. J., H. Tada, M. F. Ypma-Wong, B. L. Semler, and E. Wimmer. 1988. Mutational analysis of the genome-linked protein VPg of poliovirus. J. Virol. 62:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lellis, A. D., K. D. Kasschau, S. A. Whitham, and J. C. Carrington. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12:1046-1051. [DOI] [PubMed] [Google Scholar]
- 18.Liu, B. L., G. J. Viljoen, I. N. Clarke, and P. R. Lambden. 1999. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 80:291-296. [DOI] [PubMed] [Google Scholar]
- 19.Lomonossoff, G., M. Shanks, and D. Evans. 1985. The structure of cowpea mosaic virus replicative form RNA. Virology 144:351-362. [DOI] [PubMed] [Google Scholar]
- 20.Machin, A., J. M. Martin Alonso, and F. Parra. 2001. Identification of the amino acid residue involved in rabbit hemorrhagic disease virus VPg uridylylation. J. Biol. Chem. 276:27787-27792. [DOI] [PubMed] [Google Scholar]
- 21.Murphy, J. F., P. G. Klein, A. G. Hunt, and J. G. Shaw. 1996. Replacement of the tyrosine residue that links a potyviral VPg to the viral RNA is lethal. Virology 220:535-538. [DOI] [PubMed] [Google Scholar]
- 22.Murphy, J. F., W. Rychlik, R. E. Rhoads, A. G. Hunt, and J. G. Shaw. 1991. A tyrosine residue in the small nuclear inclusion protein of tobacco vein mottling virus links the VPg to the viral RNA. J. Virol. 65:511-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ormerod, E., and O. Jarrett. 1978. A classification of feline calicivirus isolates based on plaque morphology. J. Gen. Virol. 39:537-540. [DOI] [PubMed] [Google Scholar]
- 24.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 25.Reuer, Q., R. J. Kuhn, and E. Wimmer. 1990. Characterization of poliovirus clones containing lethal and nonlethal mutations in the genome-linked protein VPg. J. Virol. 64:2967-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaad, M. C., A. D. Lellis, and J. C. Carrington. 1997. VPg of tobacco etch potyvirus is a host genotype-specific determinant for long-distance movement. J. Virol. 71:8624-8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VPg for infectivity. Virology 210:383-390. [DOI] [PubMed] [Google Scholar]
- 28.Sosnovtsev, S. V., M. Garfield, and K. Y. Green. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 76:7060-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sosnovtsev, S. V., and K. Y. Green. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277:193-203. [DOI] [PubMed] [Google Scholar]
- 30.Sosnovtsev, S. V., S. Sosnovtseva, and K. Y. Green. 1996. Recovery of feline calicivirus from plasmid DNA containing a full-length copy of the genome, p. 125-130. In D. Chasey, R. M. Gaskell, and I. N. Clarke (ed.), Proceedings of the First International Symposium on Caliciviruses, Reading, U.K. European Society for Veterinary Virology and Central Veterinary Laboratory, Weybridge, United Kingdom.
- 31.Sosnovtsev, S. V., S. A. Sosnovtseva, and K. Y. Green. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 72:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sosnovtseva, S. A., S. V. Sosnovtsev, and K. Y. Green. 1999. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 73:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thumfart, J. O., and G. Meyers. 2002. Feline calicivirus: recovery of wild-type and recombinant viruses after transfection of cRNA or cDNA constructs. J. Virol. 76:6398-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 35.Wirblich, C., M. Sibilia, M. B. Boniotti, C. Rossi, H. J. Thiel, and G. Meyers. 1995. 3C-like protease of rabbit hemorrhagic disease virus: identification of cleavage sites in the ORF1 polyprotein and analysis of cleavage specificity. J. Virol. 69:7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]