Abstract
The herpes simplex virus type 1(JMP) [HSV-1(JMP)] mutant was selected for its ability to grow and form plaques in receptor-negative J cells. It enters J cells through a novel gD-dependent pathway, independent of all known HSV receptors, nectin1, nectin2, and HveA. Evidence that the pathway is dependent on a nectin3 binding site on HSV-1(JMP) and requires three mutations in gD rests on the following. We derived monoclonal antibodies to nectin3 and show that J cells express nectin3. HSV-1(JMP) entry and cell-to-cell spread were inhibited by soluble nectin3-Fc, demonstrating that virions carry a binding site for nectin3. The site is either directly involved in HSV-1(JMP) entry, or nectin3 binding to its site affects the gD domains involved in entry (entry site). HSV-1(JMP) entry and cell-to-cell spread in J cells were also inhibited by soluble nectin1-Fc, showing that the nectin1 binding site on gDJMP overlaps with the entry site or that nectin1 binding to gD affects the entry site. gDJMP carries three mutations, S140N, R340H, and Q344R. The latter two lie in the C tail and are present in the parental HSV-1(MP). HSV-1 strain R5000 carrying the S140N substitution was not infectious in J cells, indicating that this substitution was not sufficient. We constructed two recombinants, one carrying the three substitutions and the other carrying the two C-tail substitutions. Only the first recombinant infected J cells with an efficiency similar to that of HSV-1(JMP), indicating that the three mutations are required for the novel entry pathway. The results highlight plasticity in gD which accounts for changes in receptor usage.
Herpes simplex virus (HSV) enters cells through a gD-dependent pathway (37, 49). gD is the receptor binding glycoprotein and interacts with one of three receptors that belong to unrelated families (9, 59). The three receptors are nectin1 (previously herpesvirus entry mediator C [HveC]) (13, 15, 29, 40), HveA (also named HVEM) (47), and modified heparan sulfate (58). nectin1 belongs to the immunoglobulin (Ig) superfamily, whereas HveA belongs to the tumor necrosis factor receptor family. nectin1 is expressed ubiquitously in human tissues and is present at high levels in tissues targeted by HSV (15, 29), including sensory neurons (54). By contrast, the distribution of HveA appears to be more restricted (47). The physical interaction of gD with its receptors has been studied in detail. The crystal structure of the gD-HveA complex has been resolved (10, 11); the HveA binding site on gD lies in the first 32 residues (11, 17). The nectin1 binding site on gD appears to be discontinuous and more widespread (64). In turn, the gD binding site on nectin1, which is the site involved in HSV entry (herein defined as the HSV entry site), maps to the C-C′-C" ridge of the most N-terminal V-type domain (12, 35). Critical residues lie between residues 69 and 75 and residues 77 and 85 and at residues 34 and 243 (41, 44, 46). The interaction of gD with nectin1 or HveA is a requirement also for cell-to-cell spread of virus (14) and for mediation of cell-cell fusion (6, 50, 61). Similar to virus entry, the latter activities require the participation of three additional fusogenic glycoproteins, gB, gH, and gL (5, 7, 25, 30).
The nectins form a family of intercellular adhesion molecules that includes five members named nectin1 to nectin4 and poliovirus receptor (1, 20, 40, 51-53). Nectins can be expressed as transmembrane or soluble proteins by alternative splicing of their RNAs (34, 38). The isoforms share a sequence formed by three Ig-type domains that constitutes the ectodomain of the transmembrane isoforms. Nectins form homo-cis-dimers on the cell surface and mediate cell-cell adhesion by forming trans-homo- or trans-heterodimers with nectins expressed on adjacent cells (22, 60). Each nectin is specialized with respect to the nectins with which it can form trans-dimers. Thus, nectin1 forms trans-heterodimers with nectin3 and with nectin4 but with a lower affinity; it does not form heterodimers with nectin2. nectin3 binds to nectin2 and poliovirus receptor (22, 53). nectin1 serves as a receptor for wild-type (wt) HSV-1 and HSV-2, pseudorabies virus, and bovine herpesvirus 1 (BHV-1) (15, 29). Specialized mutations at residues 25 and 27 (called unrestricted or rid mutations) confer the ability to interact with nectin2, a homolog of nectin1 that does not interact with wt gD, on HSV-1 gD (36, 39, 63). The interaction is at a lower affinity than that between wt gD and nectin1 yet sufficient to mediate virus entry. The unrestricted gD mutants maintain the abilities to interact with nectin1 and to enter cells through this receptor; therefore, they can use two alternative receptors. The affinity exhibited by different gDs to different nectins may vary from very high (e.g., wt gD1 and nectin1) to very low or undetectable (e.g., wt gD and murine nectin1, BHV-1 gD and human nectin1, and nectin1 mutants and wt gD). Remarkably, nectins may serve as receptors even when their affinity to gD is very low or undetectable (18, 41, 43, 45, 46). Recently, two novel members of the nectin family, nectin3 and nectin4, were identified and characterized with respect to intercellular adhesion properties (22, 52, 53). Neither one was reported to serve as a receptor for HSV or animal alphaherpesviruses.
The objective of this study was to isolate an HSV mutant able to enter cells through a pathway independent of the known receptors, nectin1, nectin2 and HveA. The receptor-negative J cells (15) were exposed to a number of HSV-1 strains, including the wild-type cyto-aggregating HSV-1(F) (21) and two syncytial strains, and passaged in a blind manner. Only cells exposed to HSV-1(MP) yielded a mutant, designated HSV-1(JMP), able to grow and form plaques in J cells. We report the following: (i) HSV-1(JMP) infects J cells through a gD-dependent entry pathway independent of nectin1, HveA, and nectin2; (ii) J cells express nectin3; (iii) the pathway of HSV-1(JMP) entry into J cells was inhibited by soluble nectin3, implying that virions bind nectin3, i.e., they carry a nectin3 binding site, and that this site is either directly involved in entry or lies close to the entry site and affects it; (iv) gD of HSV-1(JMP) carries three mutations, all of which are required for the enhanced entry of HSV-1(JMP) into J cells.
MATERIALS AND METHODS
Cells and viruses.
Cells were grown in Dulbecco's modified Eagle medium supplemented with 5% fetal calf serum. J cells, a derivative of BHK-tk− cells that lack gD receptors, were described previously (15). Viruses were grown in Vero or BHK cells and routinely titrated by plaque assay in Vero cells overlaid with medium containing 0.2% pooled human gamma globulins. Virions were obtained by high-speed centrifugation (1 h, 100,000 × g) of infected-cell medium. HSV-1(MP), HSV-1(F), HSV-1(HFEM), R5000, and R5001 were described previously (21, 32, 56). The gD-minus virus FgDβ was grown in a clone of rabbit skin cells carrying an inducible gD, called R6 cells (37). For construction of COS cells expressing human nectin3, the nectin3 cDNA was subcloned from pFLV3.1 (52) to pCDNA3.1; the transfected COS cells were selected with neomycin G418.
Antibodies.
HD-1 is a monoclonal antibody (MAb) directed against gD with potent neutralizing activity (49). MAb H170 directed against gD, rabbit polyclonal antibody (PAb) against gM, and MAb 6E2 against human herpesvirus 6 (HHV-6) were described previously (3, 24, 48). MAb DL11 that fails to react with gD carrying the S140N substitution was described previously (16). Anti-human IgGs coupled to peroxidase and fluorescein isothiocyanate (FITC)-conjugated anti-mouse antibodies were from Dako and Jackson, respectively. Goat anti-mouse antibodies coupled to phycoerythrin and to peroxidase were from Beckman-Coulter-Immunotech (Marseille, France). Antibodies against the Fc fragment of human IgGs were from Sigma Chemical Co. (Milan, Italy). When indicated, IgGs were affinity purified on columns of proteins A and G (Pierce).
Soluble nectin3 [N3(VCC)-Fc] and other soluble receptors.
The soluble receptors carrying either the entire ectodomain (VCC) or the single N-terminal V domain (V) of nectin1 (N1), nectin2 (N2), nectin3 (N3), nectin4 (N4), and CD28 were named either nectin-Fc or N1(VCC)-Fc, N3(VCC)-Fc, N3(V)-Fc, N4(VCC)-Fc, and N2(V)-Fc, respectively. N4Δ-Fc is an inactive form of soluble nectin4, which carries the first 141 residues joined to the Fc portion. Their construction and production were described previously (13, 39, 53). Briefly, the PCR amplification products were cloned in the COS Fc Link vector (SmithKline Beecham) and transfected in COS cells with FuGENE6, according to the manufacturer's instructions. Soluble proteins were affinity purified from supernatants on Affigel protein A, as detailed elsewhere (13).
Derivation of MAbs to nectin3.
The anti-nectin3 MAbs were obtained after three successive intraperitoneal injections of BALB/c mice with 10 μg of recombinant soluble N3(VCC)-Fc. Ten micrograms of N3(VCC)-Fc was injected intravenously 3 days before the cell fusion assay. Splenocytes were fused with the murine myeloma hypoxanthine phosphoribosyltransferase-negative cell line X63Ag853 using polyethylene glycol 1500. Hybridomas were selected in medium containing hypoxanthine, aminopterine, and thymidine (Sigma Chemical Company) and 10% hybridoma cloning factor (Origen). Five hundred clones were tested for differential reactivity to COS cells and COS cells expressing the human nectin3 cDNA clone. Hybridomas that produced reactive supernatants were then cloned and expanded. MAbs were then assayed for the ability to detect cell surface expression of nectin3 by fluorescence-activated cell sorting (FACS) and for the ability to react with recombinant soluble nectin3-Fc in an enzyme-linked immunosorbent assay (ELISA). The positive clones were designated MAbs N3.82.5 and N3.12.5.
Plating efficiency and plaque size reduction assays.
Cells were infected with the different viruses for 90 min at 37°C. The viral inoculum was removed, the adsorbed virus that had not yet entered cells was inactivated by means of a pH 3 citrate buffer wash (50 mM sodium citrate-4 mM KCl, adjusted to pH 3.0 with HCl), and cells were overlaid with medium containing 0.2% pooled human gamma globulins. Cells were fixed at 48 h postinfection, and plaques, or singly infected cells, were detected by immunostaining with anti-gM PAb (3), followed by anti-rabbit antibodies conjugated to alkaline phosphatase. For plaque size reduction assay, J cells were infected with HSV-1(JMP) virions for 90 min at 37°C, the viral inoculum was removed, cells were overlaid with medium containing soluble receptors at a 2 μM concentration, and fixed 48 h later. Plaques were detected by immunostaining with anti-gM PAb. Digital micrographs were imported into Adobe Photoshop, and the plague areas were determined by means of the Histogram program and expressed in pixels, as detailed elsewhere for syncytia (2).
Infectivity inhibition by MAb HD-1 and soluble receptors.
Aliquots of virions were incubated with MAb HD1 or with different amounts of soluble receptors for 1 h at 37°C, prior to virus absorption to cell monolayers for 90 min at 37°C. The viral inoculum was removed, and cells were overlaid with medium containing 0.2% pooled human gamma globulins supplemented with MAb HD1 or soluble receptors. Plaques or singly infected cells were detected by immunostaining with anti-gM PAb.
Reverse transcription-PCR (RT-PCR).
Total RNA was extracted from J cells by using a RNAeasy extraction kit (Qiagen) and retrotranscribed with avian myeloblastosis virus reverse transcriptase (Roche cDNA synthesis kit). The primers for PCR amplification follow: 5′-GAGGG TACCC AGGCA GTGCT TCGAG-3′ and 5′-GTGAG GCTTT CCTTG AAGCG GTCCA TGTGG-3′ for nectin1; 5′-CACCT CCGCT GCTGC TGCTG CTCTT CCC-3′ and 5′-CAGTT GTAGA GGACT GGGCA TTTCC-3′ for nectin3, and 5′-TGACG GGGTC ACCCA CACTG TGCCC ATCTA-3′ and 5′-AGTCA TAGTC CGCCT AGAAG CATTT GCGGT-3′ for β-actin. For nectin1, PCR was performed as follows: (i) 10 cycles of PCR, with 1 cycle consisting of 30 s at 95°C, 30 s at 55°C, and 1 min at 68°C; and (ii) 35 cycles, with 1 cycle consisting of 30 s at 95°C, 30 s at 65°C, and 1 min at 68°C. For nectin3, 35 cycles of PCR amplification were done, with 1 cycle consisting of 1 min at 94°C, 1 min at 60°C, and 1 min at 68°C. For β-actin, 35 cycles of PCR amplification were performed, with 1 cycle consisting of 1 min at 94°C, 1 min at 56°C, and 1 min at 68°C (15, 62). Samples were routinely subjected to an initial denaturation step of 5 min at 94°C. Plasmids carrying nectin1 (pCF18) or human nectin3 (pFLR3V.1) cDNA served as positive controls for nectin1 or nectin3 amplification, respectively (15, 52).
FACS analysis.
To measure the binding of gD, J cells (106) were trypsinized, fixed with 4% paraformaldehyde for 5 min at room temperature, and permeabilized with 0.1% Triton X-100. Cells were then incubated with 500 ng of gDJMP or wt gD for 1 h at 4°C and then with MAb H170 (1:1,000) and an FITC-conjugated anti-mouse antibody (1:100) for 1 h at 4°C and finally resuspended in phosphate-buffered saline (PBS) containing 3.7% paraformaldehyde. Cells were analyzed in a FACScan flow cytometer. gDJMP and wt gD were purified from lysates of HSV-1(JMP)- or HSV-1(F)-infected BHK cells by affinity chromatography with anti-gD MAb30 (4) immobilized to Affigel, as previously described (39). Protein purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining. In order to measure the binding of MAb N3.82.5 to J cells, live cells were incubated with purified IgGs (10 μg/ml) and then with a 1:50 dilution of goat anti-mouse phycoerythrin antibody (Beckman-Coulter-Immunotech).
ELISA.
A sandwich ELISA was performed to detect the binding of MAbs N3.82.5 and N3.12.5 to nectin3-Fc. The wells (in 96-well trays) were coated with anti-human Fc IgGs (Sigma Chemical Company) at 5 μg/ml. After nonspecific binding sites were blocked with 1% bovine serum albumin (BSA), N3(VCC)-Fc, N3(V)-Fc, or N4Δ-Fc (negative control) was added to the wells at a 10 nM concentration in PBS containing 1% BSA for 1 h at room temperature. The excess soluble receptors were removed, and the bound soluble receptors were reacted with the hybridoma supernatants, followed by goat anti-mouse antibodies conjugated to peroxidase and One Step ABTS (Pierce). To measure the binding of gDJMP to soluble N3(VCC)-Fc, the wells in 96-well trays were coated with purified gDJMP (80 ng/well), nonspecific binding sites were blocked with 3% BSA in PBS; the gD-coated wells were then reacted with soluble receptors N3(VCC)-Fc, N1(VCC)-Fc, N2(VCC)-Fc, N4(VCC)-Fc, and CD28-Fc for 1 h at 37°C. Binding was detected by incubation with anti-human Fc antibodies coupled to peroxidase (1:6,000), followed by o-phenylenediamine (Sigma Chemical Company) at 0.5 mg/ml and reading the optical density at 490 nm.
Construction of HSV-1(JMP) recombinants.
The gD open reading frame was PCR amplified from HSV-1(JMP) or HSV-1(MP) DNA with primers gDBamHIforw (5′-CGGCC CCCAA TAAGG ATCCC GGTAG CCCGG CCGTG TGAC-3′) and gDSalIrev (5′-TTATA TGGAG TTAAG GTGTC GACCC AACCC CGCAG ACC-3′) carrying the appropriate restriction site. The amplified open reading frames were digested with BamHI and SalI restriction endonucleases and cloned in a recombination vector called pBluescript-gD-up-and-down. The plasmid carries about 1,500 bp of gD upstream and downstream sequences, namely, fragments from positions 136768 to 138189 and positions 139632 to 141146, respectively, to allow recombination in the viral genome. The pgDJMP and pgDMP plasmids generated were sequenced. BHK cells were cotransfected with FgDβ DNA and pgDJMP or pgDMP plasmids, generating the R-gDJMP and R-gDMP recombinants. For viral DNA preparation, infected cells were lysed in a solution containing 0.1% Nonidet P-40, 0.2% sodium dodecyl sulfate, 5 mM EDTA, and 50 mM β-mercaptoethanol. After two phenol-chloroform extractions, the DNA was precipitated overnight, digested with RNase A, and loaded onto a 5 to 20% (wt/vol) potassium acetate gradient, run at 280,000 × g for 3.5 h at 20°C. The high-molecular-weight fraction was precipitated by adding ethanol. Increasing amounts of the viral DNA were transfected in mammalian cells to determine the concentration that gave the optimal number of plaques.
Digital microscopy.
Digital micrographs were taken in an Axiophot Zeiss microscope equipped with a DC-120 Kodak digital camera and imported in Adobe Photoshop. For plaque size quantification, digital micrographs of infected-cell monolayers in 24-well coverslips were taken at low magnification (×1.25). The areas corresponding to immunostained plaques were quantified by means of the Histogram program of Adobe Photoshop.
RESULTS
Isolation of HSV-1(JMP).
J cells, which are highly resistant to HSV infection due to the absence of receptors (15), were exposed to the following HSV-1 strains: F, MP, HFEM, R5000, and R5001 (21, 32, 55). Viral stocks made of extracellular virions were used at an input multiplicity of 30 to 100 PFU/cell, as titrated in Vero cells. Since there was no sign of infection after 2 or 3 days, cells were trypsinized (1:8) in an blind manner for a number of passages. J cells exposed to HSV-1(MP) were the only cells showing a cytopathic effect at passage 4 to 5. The virus was named HSV-1(JMP). The isolation of an HSV-1(MP) mutant able to grow in J cells was repeated in an independent experiment. None of the other strains, including syncytial strain HFEM, yielded a mutant capable of growing in J cells.
HSV-1(JMP) forms plaques and replicates in J cells.
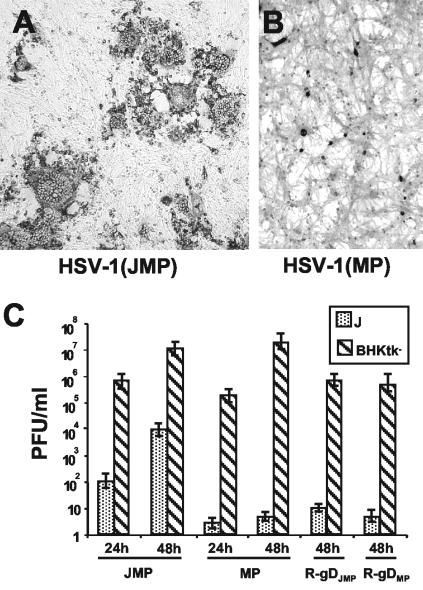
Figure 1 and Table 1 compare the abilities of HSV-1(JMP) and its parent, HSV-1(MP), to form plaques and to replicate in J cells and in BHK-tk− cells. HSV-1(JMP) formed medium-sized syncytial plaques in J cells (Fig. 1A), whereas HSV-1(MP) produced singly infected cells (Fig. 1B). The ability of HSV-1(JMP) to form plaques in J cells relative to BHK-tk− cells (expressed as J/BHK-tk− ratio) was about 10-fold higher than that of the parental HSV-1(MP) (Table 1). Figure 1C compares the ability of HSV-1(JMP) and its parent, HSV-1(MP), to replicate in J and BHK-tk− cells. J and BHK-tk− cells were exposed to 10 and 0.01 PFU/cell, according to the titer determined in Vero cells, respectively. Cells were frozen 24 and 48 h later, and progeny virus was titrated in Vero cells. Both viruses replicated in BHK-tk− cells at similar yields. HSV-1(JMP) replicated in J cells, whereas the parental HSV-1(MP) did not replicate at all. Cumulatively, these data indicate that HSV-1(JMP) could infect, replicate, and spread in J cells, although at a lower efficiency than in BHK-tk− cells. The parental HSV-1(MP) infected J cells with a 10-fold-lower efficiency than HSV-1(JMP), but it failed to replicate and to form plaques. HSV-1(JMP) showed the same tropism as HSV-1(MP) toward a variety of cells (BHK, Vero, and 143-tk−) (data not shown), which suggests that alternate receptors are used in different cell lines.
FIG. 1.
(A and B) Digital micrographs of plaques or singly infected cells in J cells infected with HSV-1(JMP) (A) or HSV-1(MP) (B). Infection was revealed by immunostaining with PAb directed against gM (1:1,500). The micrographs in panels A and B are at the same magnification. (C) Growth of HSV-1(JMP) and HSV-1(MP) and recombinants R-gDJMP and R-gDMP in J cells and BHK-tk− cells. J cells were infected with 10 PFU/cell, whereas BHK-tk− cells were infected with 0.01 PFU/cell, according to the titer determined in Vero cells. Viruses were absorbed to cells for 90 min at 37°C; excess virus was removed, and the remaining infectivity was inactivated with a pH 3 citrate buffer wash. Cells were frozen at 24 and 48 h after infection. The titer of progeny virus was determined by plaque assay in Vero cells.
TABLE 1.
Plating efficiency of JMP and MP viruses and of R-gDJMP and R-gDMP recombinants
| Virus | Plating efficiency on:
|
J/BHK-tk− ratioa | |
|---|---|---|---|
| J cellsb | BHK-tk− cells | ||
| HSV-1(JMP) | 9 × 103 (SP) | 3 × 107 | 3 × 10−4 |
| HSV-1(MP) | 2 × 104 (SC) | 1 × 109 | 2 × 10−5 |
| R-gDJMP | 4 × 104 (SC) | 1 × 108 | 4 × 10−4 |
| R-gDMP | 2 × 104 (SC) | 1 × 109 | 2 × 10−5 |
Plating efficiency on J cells/plating efficiency on BHK-tk− cells.
SP, small plaques; SC, single cells.
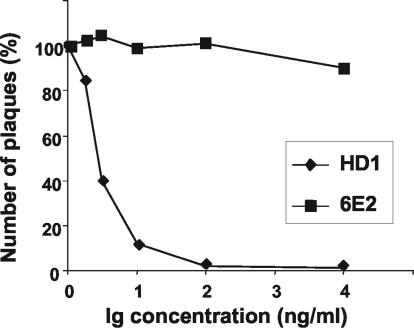
HSV-1(JMP) infectivity is blocked by anti-gD HD1 MAb.
To ascertain whether HSV-1(JMP) entry into J cells was dependent on gD, we measured the effect on plague formation of MAb HD-1, a MAb that blocks wt HSV infectivity (49). Aliquots of HSV-1(JMP) virions were incubated with purified IgGs from MAb HD1 for 1 h at 37°C and then absorbed onto J cells. Plaques were scored 48 h later by immunostaining. As can be seen from Fig. 2, MAb HD-1 blocked the ability of HSV-1(JMP) to infect and form plaques in J cells, whereas an unrelated MAb (directed against HHV-6) (24) had no effect. The results indicate that the pathway of HSV-1(JMP) entry into J cells is gD dependent but nectin1, nectin2, and HveA independent.
FIG. 2.
Neutralization of HSV-1(JMP) infectivity by MAb HD-1. Aliquots of HSV-1(JMP) virions were incubated with the indicated amounts of purified IgGs from MAb HD-1 or an irrelevant MAb (MAb 6E2 directed against HHV-6) for 1 h at 37°C prior to infection of J cells grown in 24-well dishes. Infected cells were immunostained 48 h later with anti-gM PAb (1:1,500), followed by anti-rabbit antibodies conjugated to alkaline phosphatase (1:3,000). Data are expressed as percentages of the number of plaques obtained in infected cells not exposed to antibodies.
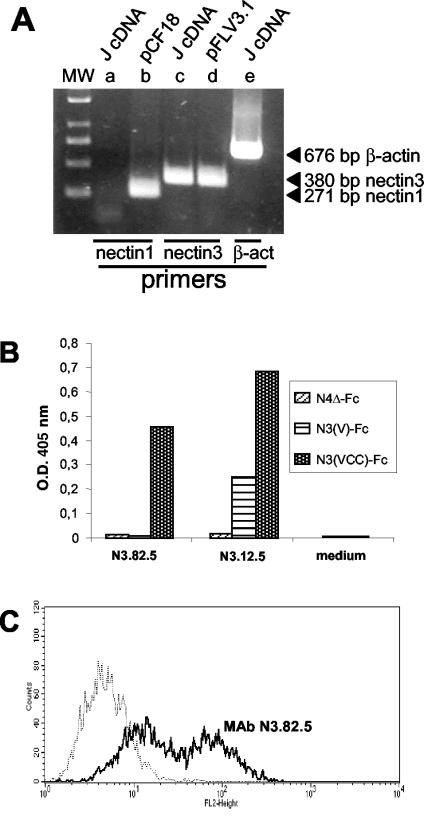
J cells express nectin3 at the cell surface.
J cells lack nectin1, nectin2, and HveA (15). We asked whether they express the nectin3 ortholog of human nectin3. To this end, RT-PCR experiments were performed on RNA extracted from J cells, using primers able to anneal to human and murine nectin3 cDNA. Figure 3A shows that a fragment of the expected size was amplified from the J cell RNA and from the human nectin3 cDNA clone (pFLV3.1). As expected, no fragment was amplified with primers annealing to the nectin1 cDNA clone (pCF18). These results indicate that J cells express a hamster ortholog of human nectin3, which can be amplified with primers designed on human and murine nectin3.
FIG. 3.
(A) RT-PCR amplification of nectin3 from retrotranscribed RNA from J cells (J cDNA). nectin3 cDNA was amplified with primers designed on human and murine nectin3 cDNA. nectin1 primers were designed on the human cDNA. Plasmids carrying human nectin1 (pCF18) or human nectin3 (pFLR3V.1) cDNA served as positive controls for nectin1 and nectin3 amplification, respectively (15, 52). The expected sizes of the amplified fragments are indicated to the right of the gel. β-act, β-actin. (B) Derivation and specificity of MAbs to nectin3. Cell culture media from the indicated hybridomas (N3.12.5 and N3.82.5 to nectin3 and medium from a negative clone [control medium]) were reacted in a sandwich ELISA with soluble nectin3 ectodomain [N3(VCC)-Fc], soluble nectin3 V domain [N3(V)-Fc], or with nectin4Δ-Fc as a negative control. Briefly, 96-well trays were coated with an antibody against the human Fc fragment (Sigma Chemical Co.) at 5 μg/ml. After the nonspecific sites were blocked with PBS containing 1% BSA, N3(VCC)-Fc, nectin3V-Fc, or nectin4Δ-Fc was added at a concentration of 10 nM, and then goat anti-mouse antibodies conjugated to peroxidase and One Step ABTS (Pierce) were added. The optical density (O.D.) at 405 nm was read. MAb N3.82.5 reacted with soluble nectin3 containing the entire ectodomain but failed to react with the soluble nectin3 containing only the V domain; therefore, it was directed against the C domains. (C) Binding of MAb N3.82.5 to unfixed J cells. J cells were reacted with MAb N3.82.5 (thick black line) (10 μg of purified IgG/ml) for 1 h at 37°C, followed by a phycoerythrin-labeled goat anti-mouse antibody (1:50) (Beckman-Coulter-Immunotech). The negative control (thin stippled line) represents reactivity to mouse IgG1 (10 μg/ml), followed by the same secondary phycoerythrin-labeled antibody. Reactivity to MAb N3.82.5 denotes cell surface expression of nectin3.
To assess whether J cells express nectin3 at the protein level and whether nectin3 is a cell surface protein, two MAbs to human nectin3 were derived by immunization of mice with a soluble form of human nectin3 [N3(VCC)-Fc] made of the human nectin3 ectodomain (VCC) fused to the Fc portion of human IgG (52). The two antibodies were named MAb N3.82.5 and MAb N3.12.5. Their specific reactivity to human nectin3 was assessed in a sandwich ELISA, in which two forms of soluble nectin3-Fc carrying the entire ectodomain (VCC) or the N-terminal V domain (V) were immobilized on ELISA plates. As can be seen from Fig. 3B, both MAbs N3.82.5 and N3.12.5 reacted to nectin3. Specifically, MAb N3.82.5 was directed to an epitope contained in the C domains, whereas MAb N3.12.5 was directed to an epitope encompassing the V and C domains. Neither antibody bound N4Δ-Fc, confirming their specificity.
The degree of identity between human and murine nectin3 (57) is 92.9% in the ectodomain, with few substitutions in the V domain. Therefore, it was expected that a MAb directed against human nectin3 might react with the hamster ortholog. MAbs N3.82.5 and N3.12.5 were allowed to react with unfixed J cells, and their reactivity was measured by FACS analysis. Figure 3C shows that J cells reacted with the MAb N3.82.5, confirming the RT-PCR results and demonstrating that hamster nectin3 is expressed on the surfaces of J cells. MAb N3.12.5 did not exhibit any reactivity and may be specific to the human isoform or may not recognize hamster nectin3 by FACS analysis.
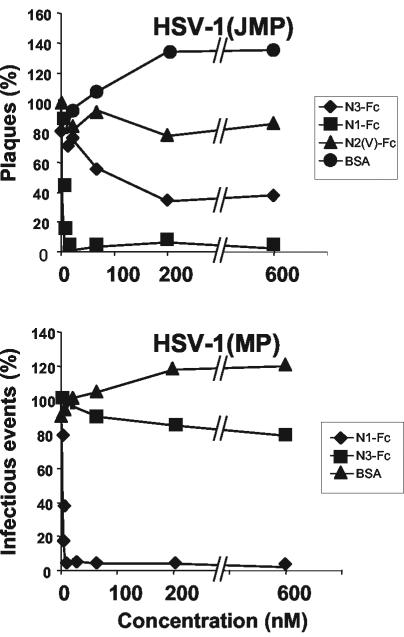
Soluble nectin3 inhibits HSV-1(JMP) infection of J cells.
Having ascertained that J cells express nectin3, we asked whether HSV-1(JMP) infection is mediated by this nectin. The MAbs described above did not prove to be useful to this end, as they were unable to block HSV-1(JMP) infectivity (data not shown). In particular, MAb N3.82.5, which recognized nectin3 at the J cell surface, is directed against the C domains, whereas the gD binding sites on nectin1 or nectin2 map to the V domain (13, 36). The other MAb (N3.12.5) was not reactive to J cells.
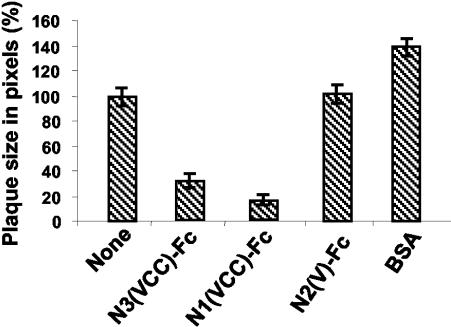
Next, we determined whether HSV-1(JMP) infection was inhibited by soluble nectin3. HSV-1(JMP) virions were preincubated with increasing concentrations of one of two soluble receptors or with BSA prior to infection of J cells. The soluble receptors were N3(VCC)-Fc and N1(VCC)-Fc, carrying the ectodomain (VCC) of nectin3 and nectin1, respectively, and the unrelated construct N2(V)-Fc, carrying the V domain of nectin2. The preincubated virions were laid onto J cells. Infectious events (plagues or singly infected cells) were immunostained with PAb directed against gM 48 h later, and their numbers were counted. Figure 4 shows that N3(VCC)-Fc reduced the number of HSV-1(JMP) plaques in a dose-dependent fashion. About 70% inhibition was achieved at 200 to 600 nM N3(VCC)-Fc. Higher concentrations could not be tested because of extreme difficulties in producing sufficient quantities of N3(VCC)-Fc. Plaque formation was also inhibited by preincubation of virions with the soluble nectin1, N1(VCC)-Fc, consistent with the finding that HSV-1(JMP) maintained the ability to infect a variety of cells, where nectin1 or its ortholog serves as a receptor. Inhibition by N1(VCC)-Fc was more efficient than that by N3(VCC)-Fc, a finding consistent with the possibility that the affinity of gDJMP to nectin3 is lower than that to nectin1. As far as HSV-1(MP) was concerned, infectivity was slightly inhibited by N3(VCC)-Fc and strongly inhibited by N1(VCC)-Fc. The results have four implications. First, the inhibition of HSV-1(JMP) infectivity by soluble nectin3 implies that HSV-1(JMP) carries a binding site for nectin3. Second, this site coincides fully or partly with the virion region involved in entry (the entry site), or nectin3 binding to its site on virions modifies the entry site. Third, the inhibition by soluble nectin1 implies that gDJMP has not lost the ability to interact with nectin1 and that the nectin1 binding site on gDJMP at least partly overlaps the entry site or affects its activity. Fourth, the higher inhibition exerted by soluble nectin1 than by soluble nectin3 argues that the affinity of gDJMP to nectin1 is higher than that to nectin3.
FIG. 4.
Infectivity inhibition by soluble receptors. Aliquots of HSV-1(JMP) virions (100 PFU/well) were incubated with the indicated nanomolar concentrations of soluble N3(VCC)-Fc (N3-Fc), N1(VCC)-Fc (N1-Fc), the V domain of nectin2-Fc [N2(V)-Fc], or BSA for 1 h at 37°C prior to infection of J cells (grown in 24-well dishes). Infectious events (plaques or singly infected cells) were detected by immunostaining with PAb directed against gM (1:1,500), followed by anti-rabbit antibodies conjugated to alkaline phosphatase (1:3,000). Data are expressed as a percentage of the number of plaques or of infectious events, relative to the values obtained in infected cells not exposed to soluble receptors. The values are averages from three replicate samples.
Soluble nectin3-Fc inhibits HSV-1(JMP) cell-to-cell spread.
To assess whether nectin3 can also inhibit the cell-to-cell spread of HSV-1(JMP), J cells were first infected with HSV-1(JMP) and subsequently exposed to soluble N3(VCC)-Fc. Images of the immunostained plaques were acquired by digital microscopy, and the plaque areas were determined as detailed in Materials and Methods. Figure 5 shows that N3(VCC)-Fc reduced plaque size. N1(VCC)-Fc was even more effective in reducing plaque size. Inhibition by nectin3-Fc and by nectin1-Fc has the same implications discussed above for virus entry, i.e., the process of virus spread is gD dependent, and the nectin3 and nectin1 binding sites on virions are either directly involved in cell-to-cell spread or close to the site involved in cell-to-cell spread.
FIG. 5.
Plaque size reduction by soluble receptors. Monolayers of J cells, grown in 24-well dishes, were infected with HSV-1(JMP) (100 PFU/well). At the end of virus adsorption, cells were overlaid with medium containing a 2 μM concentration of the indicated soluble receptor [N3(VCC)-Fc, N1/(VCC)-Fc, or N2(V)-Fc] or BSA. Plaques were scored 48 h later by immunostaining with PAb directed against gM. Digital micrographs were imported into Adobe Photoshop for plaque size determination. The average value (expressed in pixels) obtained in cells not exposed to any soluble receptor is 100%. Means ± standard errors (error bars) from three samples are shown.
Binding of gDJMP to nectin3.
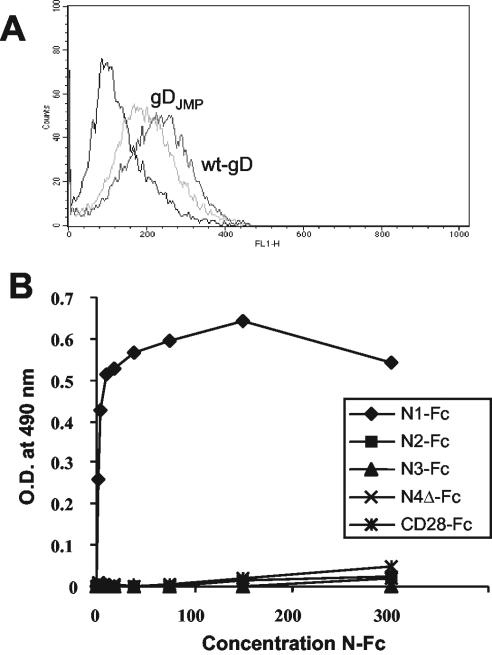
Two series of experiments were performed to detect whether gDJMP binds to J cells or to soluble nectin3. In the first series of experiments, J cells were allowed to react with gDJMP or wt gD, both affinity purified from lysates of infected cells. Binding was detected by FACS analysis with anti-gD H170 MAb (48). Figure 6A shows that the binding of gDJMP or wt gD to J cells was low and, more importantly, that gDJMP and wt gD could not be differentiated, suggesting that the observed binding was nonspecific. In the second series of experiments, gDJMP and wt gD were immobilized onto ELISA plates and reacted with increasing amounts of N3(VCC)-Fc or N1(VCC)-Fc as a positive control. N2(VCC)-Fc, nectin4Δ-Fc, and CD28-Fc served as negative controls. Binding was detected with anti-human antibodies linked to peroxidase. As can be seen from Fig. 6B, the only molecule that interacted with gDJMP at a detectable level in a dose-dependent fashion was N1(VCC)-Fc. The results indicate that binding of gDJMP to nectin3 or to J cells was almost undetectable or not significant in the assays used.
FIG. 6.
(A) FACS analysis of the binding of gDJMP or wt gD to J cells. gDJMP and wt gD (500 ng) [affinity purified from lysates of HSV-1(JMP)-infected BHK cells] were reacted with J cells for 1 h at 4°C. Binding was revealed by incubation with MAb H170 (1:1,000) directed against the N terminus of gD, followed by incubation with an FITC-conjugated anti-mouse antibody (1:100). The negative control (leftmost line) represents reactivity to MAb H170 and an FITC-conjugated anti-mouse antibody. (B) ELISA binding of soluble N1(VCC)-Fc (N1-Fc), N2(VCC)-Fc (N2-Fc), N3(VCC)-Fc (N3-Fc), nectin4Δ-Fc (N4-Fc), and CD28-Fc to gDJMP. Ninety-six wells were coated with gDJMP affinity purified from HSV-1(JMP)-infected BHK cells (80 ng/well) and were reacted with the indicated soluble receptors. Binding was revealed by adding anti-human Fc antibodies coupled to peroxidase (1:6,000) and then o-phenylenediamine and reading the optical density (O.D.) at 490 nm.
HSV-1(JMP) carries three mutations in the gD gene.
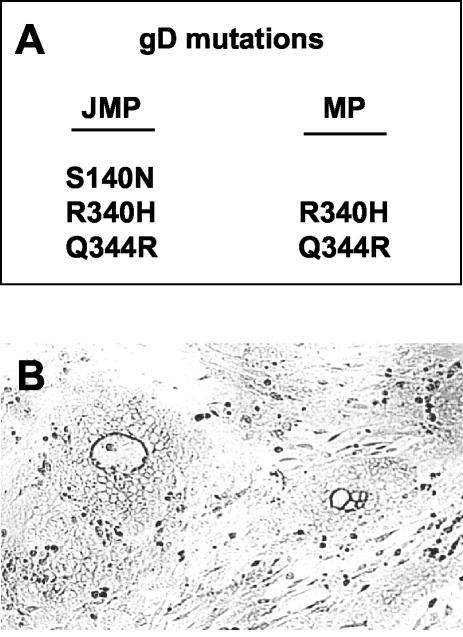
The sequence of the gD gene showed that HSV-1(JMP) carried three mutations: G to A at nucleotide 494, G to A at nucleotide 1094, and AA to GG at nucleotides 1106 and 1107 relative to HSV-1(F). These substitutions predict the S140N, R340H, and Q344R substitutions in mature gD (Fig. 7A). The latter two are located in the cytoplasmic tail of gD and are already present in HSV-1(MP). Previously, the S140N substitution, which creates a new N-glycosylation site, has been detected in two viruses, namely, a mutant resistant to MAb DL11 (16) and R5000, a mutant selected for the ability to grow in a mildly resistant cell clone derived from BHK-tk− cells (56). Staining of HSV-1(JMP) plaques with MAb DL11 confirmed the lack of immunoreactivity to this antibody (Fig. 7B).
FIG. 7.
(A) Amino acid substitutions in gD from HSV-1(JMP) and (MP). (B) Micrograph of HSV-1(JMP) plaques in J cells immunostained with MAb DL11 (1:1,000), followed by anti-mouse antibodies conjugated to alkaline phosphatase (1:3,000).
Role of S140N gD mutation in HSV-1(JMP) infectivity.
To determine the role of the gD substitutions in HSV-1(JMP) infection of J cells, we asked whether the S140N substitution present in HSV-1(JMP) and absent from the parental HSV-1(MP) is responsible for infection of J cells. J cells were exposed to mutant virus R5000, and its recombinant R5001, constructed by transferring the gD gene of R5000 to the backbone of HSV-1(F) (56). Cells were immunostained 18 h later with PAb directed against gM (3). Both viruses were unable to infect J cells (data not shown), ruling out the possibility that the single S140N substitution was responsible for infection of J cells with HSV-1(JMP).
Construction and properties of R-gDJMP and R-gDMP recombinants.
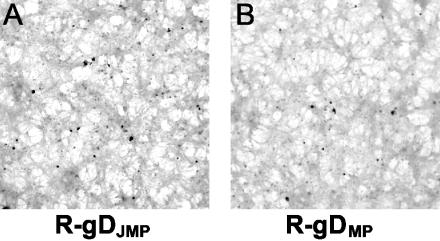
The results with R5000 and R5001 viruses suggested that the S140N substitution did not play a role in HSV-1(JMP) infectivity in J cells or that the S140N substitution necessitates the additional mutations present in gDJMP. To discriminate between these possibilities, recombinants carrying gD genes from HSV-1(JMP) (carrying three substitutions) or from HSV-1(MP) (carrying two substitutions) were constructed by recombination of the respective gD genes into the gD-minus virus FgDβ. The two gD genes were cloned in a plasmid carrying gD gene upstream and downstream sequences to allow recombination. The recombinant viruses were generated by cotransfection of viral and plasmid DNA in BHK cells and designated R-gDJMP and R-gDMP, respectively. Their gD genes were sequenced for accuracy. Cells transfected with FgDβ DNA alone did not yield any virus growing in BHK cells, as expected. The relative plating efficiencies of the recombinants were determined in J and BHK-tk− cells and compared to that of the parental viruses. Either plaques or singly infected cells were scored as infectious events. The results were as follows (Table 1). As mentioned above, the ratio of the plating efficiency in J versus BHK-tk− cells was about 10-fold higher for HSV-1(JMP) than for parental HSV-1(MP). (i) The R-gDJMP recombinant displayed a plating efficiency in J versus BHK-tk− cells very similar to that of its parent HSV-1(JMP). (ii) The R-gDMP recombinant displayed the same plating efficiency in J versus BHK-tk− cells as its parent HSV-1(MP). (iii) R-gDJMP did not form plaques in J cells. As expected, R-gDMP did not form plaques either (Fig. 8). This contrasts with the parental HSV-1(JMP), which formed plaques. The results indicate that the enhanced infectivity of HSV-1(JMP) in J cells relative to the parental HSV-1(MP) can be transferred with the gD gene. However, gD is not sufficient to confer to HSV-1(JMP) the ability to form plaques in J cells.
FIG. 8.
Singly infected cells in monolayers of J cells infected with the R-gDJMP or R-gDMP recombinant, immunostained with PAb directed against gM.
Finally, we measured the ability of the recombinants to replicate in J and BKH-tk− cells. Cells were exposed to the recombinants, and progeny virus produced at 48 h was titrated in Vero cells. It can be seen from Fig. 1 that both recombinants failed to produce progeny virions, i.e., they were defective in replication. This defect may account for the lack of plaque formation observed in Table 1 and Fig. 8.
DISCUSSION
The objective of this study was to isolate an HSV mutant able to enter cells through a pathway independent of the known receptors, nectin1, nectin2, and HveA. J cells, which are highly resistant to HSV infection due to the absence of gD receptors (15), were exposed to a number of HSV-1 strains, including the wild-type cyto-aggregating HSV-1(F) strain (21) and two syncytial strains, and trypsinized in a blind manner. A cytopathic effect developed at passage 4 or 5 only in cells exposed to HSV-1(MP). The mutant able to infect, grow, and form plaques in J cells was designated HSV-1(JMP). The fact that a mutant could be derived only from HSV-1(MP) was not completely surprising inasmuch as this strain carries a number of mutations and is the only HSV strain that was previously reported to infect J cells at a very low level (15). HSV-1(MP) was independently reported to infect CHO cells at very low levels, and CHO cells are very similar to J cells with respect to the absence of HSV receptors and resistance to HSV infection (29).
Compared to its parent, HSV-1(JMP) has acquired the abilities to infect J cells with a ca.10-fold-higher efficiency and to replicate and spread from cell to cell. The HSV-1(JMP) pathway of entry is novel, in that it is gD dependent but independent of all known gD receptors, nectin1, HveA, and nectin2. Evidence that it is dependent on a nectin3 binding site present on HSV virions and requires three mutations in gD rests on the following. We note that mutations that affected postentry step(s) were beyond the purpose of this study.
(i) HSV-1(JMP) entry was gD dependent, as evidenced by the fact that it was inhibited by the neutralizing MAb HD1 directed against gD and by soluble nectin1, a high-affinity ligand for gD. The latter observation is in accordance with the ability of HSV-1(JMP) to infect a number of cells where nectin1 is the receptor and implies that gDJMP has not lost the ability to bind nectin1. The ability of soluble nectin1 to block infection of J cells demonstrates that the nectin1 binding site on gDJMP partly overlaps the gD region involved in entry or that the two sites are distinct, but nectin1 binding to its site on gD influences the entry site.
(ii) nectin3 is a recently discovered member of the nectin family that shares 32.9% identity with nectin1 at the amino acid level in the ectodomain. J cells carry an ortholog of human nectin3, as assessed by RT-PCR amplification using primers designed on the human and murine isoforms We derived two MAbs to human nectin3. One of the antibodies reacted to the hamster ortholog and was able to bind unfixed J cells, providing evidence that J cells express nectin3 on cell surface. It is of note that J cells do not express any of the known HSV gD receptors (15). Their resistance to infection demonstrates that they lack wt gD receptors, even potentially unknown ones.
(iii) Exposure of HSV-1(JMP) virions to soluble nectin3 inhibited virus entry, and exposure of HSV-1(JMP)-infected J cells to soluble nectin3 inhibited cell-to-cell spread. This property is very similar to the block of HSV-1 infection and cell-to-cell spread by soluble nectin1 (13, 14, 29). The results imply that HSV-1(JMP) virions carry a binding site for nectin3 and that this site is directly involved in virus entry and spread in J cells. Alternatively, the nectin3 binding site lies close to the entry site, such that binding of nectin3 to its site affects the ability of the virus to enter J cells. The results imply that the HSV-1(JMP) pathway of entry is either directly or indirectly dependent on a nectin3 binding site present in virions. Because a cell line simultaneously negative for nectin1, nectin2, HveA, and nectin3 was not available, we could not perform the experiment of transfecting nectin3 cDNA in an otherwise receptor-negative cell and check for acquired infectivity.
(iv) Two lines of evidence favor the view that the nectin3 binding site on HSV-1(JMP) virions lies in gD. First, both nectin1 and nectin2 carry a gD binding site. The extent of homology among the three nectins is on the order of 30% and makes it likely that nectin3 also carries a gD binding site. Second, HSV-1(JMP) infection and spread were inhibited by both soluble nectin3 and soluble nectin1, implying that the binding sites for the two nectins either partly overlap or are located in close proximity so that they influence each other. The inability to detect a physical interaction between gDJMP and nectin3 (Fig. 6) was not surprising and can be accounted for by a low affinity. Recent studies provide examples of interactions between gD and nectin1 below the limits of detection yet sufficient to mediate virus entry. They include murine nectin1 (42, 45), whose binding to HSV-gD was not detected in several assays, BHV-1 gD, whose binding to human nectin1 was also not detected (18), and mutants of nectin1 that bind gD at low affinities (41).
(v) HSV-1(JMP) carries three mutations in gD. S140N was previously detected in R5000 and R5001, a mutant and its recombinant able to replicate in a mildly HSV-resistant cell line (56). This substitution confers resistance to MAb DL11 and creates a novel N-glycosylation site (16). The two other mutations, R340H and Q344R, lie in the cytoplasmic tail of gD and are present in the parental HSV-1(MP), as well as in other strains, e.g., KOS and Patton, but not in association with S140N. We show that the S140N mutation acted in conjunction with mutations in the cytoplasmic tail to confer an increased ability to infect J cells to HSV-1(JMP). Specifically, the R-gDJMP recombinant, carrying all three mutations, infected J cells with an efficiency similar to that of the parental HSV-1(JMP). By contrast, neither the S140N mutation present in R5000 and R5001 nor the R340H and Q344R mutations present in HSV-1(MP) or in its recombinant were sufficient to allow J cell infection with the efficiency exhibited by HSV-1(JMP) (Table 1).
The finding that gDJMP activity was affected by mutations located in the cytoplasmic tail is novel with respect to gD but is not totally surprising. Specifically, Feenstra et al. (23) deleted the cytoplasmic tail of gD and complemented a gD-minus virus with the mutant gD. The deletion caused a 100- to 1,000-fold reduction in infectivity, and the virus formed tiny plaques. Browne et al. (6) replaced the cytoplasmic tail of gD with that of CD8 and, furthermore, generated virions carrying a glycosylphosphatidylinositol-anchored form of gD. Both viruses were infectious; however, the latter showed a decreased rate of entry, and the glycosylphosphatidylinositol-anchored gD was inactive in a cell-cell fusion assay. On the other hand, mutations in the cytoplasmic tail of gB that affect virus entry, the rate of entry, and cell-cell fusion have been known for a long time (19, 26, 27). Furthermore, mutations in the transmembrane domain of gH affect its ability to be expressed at the cell surface and to carry out fusion (31). Altogether the results indicate that the effect of cytoplasmic tail substitutions may be dependent on additional mutations present in the ectodomain and on the type of receptor that the virus employs.
(vi) The approach adopted in this paper was to passage HSV in receptor-negative cells in a blind manner. This approach resulted in the selection of a mutant whose pathway of entry is dependent on nectin3, a cell surface protein in J cells. The implication is that the selected mutations were those that allowed usage of the available receptor. This phenomenon is reminiscent of previous selections that we can reinterpret now, in light of present knowledge on HSV receptors. Thus, the unrestricted mutants U10, U21, rid1, and rid2 were selected for the ability to infect cell lines expressing gD (4, 8, 33). In retrospect, in those cell lines, the gD sequestered its receptor (4, 28), which was most likely nectin1, and rendered the receptor unavailable to HSV (restriction to infection or interference). The mutants able to infect gD-expressing cells were later shown to be capable of using nectin2 through mutations at residue 25 or 27 (39, 63). Similarly, R5000 virus was selected for the ability to infect a mildly resistant cell line derived from BHK-tk− cells; its receptor is unknown (56). Cumulatively, these findings emphasize a mutational plasticity of gD that results in the ability to interact with alternative receptors and accounts for changes in receptor usage.
Acknowledgments
We thank G. Cohen and R. Eisenberg (University of Pennsylvania) for the kind gift of MAb DL11, D. Johnson (Oregon University) for the kind gift of FgDβ, and E. Avitabile (from our laboratory at University of Bologna) for the kind gift of the transfer vector pBluescript carrying gD upstream and downstream sequences. We thank Elisabetta Romagnoli for invaluable help with cell cultures.
This study was supported in part by grants from Cofin-MIUR 2001, 2002, 2003, FIRB autonomous and coordinated projects, INSERM and the Ligue Nationale Français Contre le Cancer, University of Bologna 60%.
REFERENCES
- 1.Aoki, J., S. Koike, I. Ise, Y. Sato-Yoshida, and A. Nomoto. 1994. Amino acid residues on human poliovirus receptor involved in interaction with poliovirus. J. Biol. Chem. 269:8431-8438. [PubMed] [Google Scholar]
- 2.Avitabile, E., G. Lombardi, and G. Campadelli-Fiume. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J. Virol. 77:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1993. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J. Virol. 67:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandimarti, R., T. Huang, B. Roizman, and G. Campadelli-Fiume. 1994. Mapping of herpes simplex virus 1 genes with mutations which overcome host restrictions to infection. Proc. Natl. Acad. Sci. USA 91:5406-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419-1422. [DOI] [PubMed] [Google Scholar]
- 6.Browne, H., B. Bruun, A. Whiteley, and T. Minson. 2003. Analysis of the role of the membrane-spanning and cytoplasmic tail domains of herpes simplex virus type 1 glycoprotein D in membrane fusion. J. Gen. Virol. 84:1085-1089. [DOI] [PubMed] [Google Scholar]
- 7.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. (Erratum, 62:4438.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campadelli-Fiume, G., M. Arsenakis, F. Farabegoli, and B. Roizman. 1988. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J. Virol. 62:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 10.Carfi, A., H. Gong, H. Lou, S. H. Willis, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2002. Crystallization and preliminary diffraction studies of the ectodomain of the envelope glycoprotein D from herpes simplex virus 1 alone and in complex with the ectodomain of the human receptor HveA. Acta Crystallogr. Sect. D 58:836-838. [DOI] [PubMed] [Google Scholar]
- 11.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 12.Cocchi, F., M. Lopez, P. Dubreuil, G. Campadelli-Fiume, and L. Menotti. 2001. Chimeric nectin1-poliovirus receptor molecules identify a nectin1 region functional in herpes simplex virus entry. J. Virol. 75:7987-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocchi, F., M. Lopez, L. Menotti, M. Aoubala, P. Dubreuil, and G. Campadelli-Fiume. 1998. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc. Natl. Acad. Sci. USA 95:15700-15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocchi, F., L. Menotti, P. Dubreuil, M. Lopez, and G. Campadelli-Fiume. 2000. Cell-to-cell spread of wild-type herpes simplex virus type 1, but not of syncytial strains, is mediated by the immunoglobulin-like receptors that mediate virion entry, nectin1 (HveC/HIgR/PRR1) and nectin2 (PRR2). J. Virol. 74:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen, G., W. Wilcox, D. Sodora, D. Long, J. Levin, and R. Eisenberg. 1988. Expression of herpes simplex virus type 1 glycoprotein D deletion mutants in mammalian cells. J. Virol. 62:1932-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connolly, S. A., J. J. Whitbeck, A. H. Rux, C. Krummenacher, S. van Drunen Littel-van den Hurk, G. H. Cohen, and R. J. Eisenberg. 2001. Glycoprotein D homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC (nectin-1) with different affinities. Virology 280:7-18. [DOI] [PubMed] [Google Scholar]
- 19.De Luca, N., D. J. Bzik, V. C. Bond, S. Person, and W. Snipes. 1982. Nucleotide sequences of herpes simplex virus type 1 (HSV-1) affecting virus entry, cell fusion, and production of glycoprotein gb (VP7). Virology 122:411-423. [DOI] [PubMed] [Google Scholar]
- 20.Eberlé, F., P. Dubreuil, M. G. Mattei, E. Devilard, and M. Lopez. 1995. The human PRR2 gene, related to the human poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene 159:267-272. [DOI] [PubMed] [Google Scholar]
- 21.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 22.Fabre, S., N. Reymond, F. Cocchi, L. Menotti, P. Dubreuil, G. Campadelli-Fiume, and M. Lopez. 2002. Prominent role of the Ig-like V domain in trans-interactions of nectins: nectin3 and nectin4 bind to the C-C′-C" β-strands of the nectin1 V domain. J. Biol. Chem. 277:27006-27013. [DOI] [PubMed] [Google Scholar]
- 23.Feenstra, V., M. Hodaie, and D. Johnson. 1990. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J. Virol. 64:2096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foa-Tomasi, L., E. Avitabile, and G. Campadelli-Fiume. 1995. Selection of a monoclonal antibody specific for variant B human herpesvirus 6-infected mononuclear cells. J. Virol. Methods 51:289-296. [DOI] [PubMed] [Google Scholar]
- 25.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster, T., J. Melancon, and K. Kousoulas. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18-29. [DOI] [PubMed] [Google Scholar]
- 27.Gage, P. J., M. Levine, and J. C. Glorioso. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J. Virol. 67:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geraghty, R. J., C. R. Jogger, and P. G. Spear. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147-158. [DOI] [PubMed] [Google Scholar]
- 29.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 30.Gompels, U., and A. Minson. 1986. The properties and sequence of glycoprotein H of herpes simplex virus type 1. Virology 153:230-247. [DOI] [PubMed] [Google Scholar]
- 31.Harman, A., H. Browne, and T. Minson. 2002. The transmembrane domain and cytoplasmic tail of herpes simplex virus type 1 glycoprotein H play a role in membrane fusion. J. Virol. 76:10708-10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoggan, M. D., and B. Roizman. 1959. The isolation and properties of a variant of herpes simplex producing multinucleated giant cells in monolayer cultures in the presence of antibody. Am. J. Hyg. 70:208-219. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, R. M., and P. G. Spear. 1989. Herpes simplex virus glycoprotein D mediates interference with herpes simplex virus infection. J. Virol. 63:819-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koike, S., H. Horie, I. Ise, A. Okitsu, M. Yoshida, N. Iizuka, K. Takeuchi, T. Takegami, and A. Nomoto. 1990. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 9:3217-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krummenacher, C., I. Baribaud, M. Ponce De Leon, J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2000. Localization of a binding site for herpes simplex virus glycoprotein D on herpesvirus entry mediator C by using antireceptor monoclonal antibodies. J. Virol. 74:10863-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krummenacher, C., A. H. Rux, J. C. Whitbeck, M. Ponce-de-Leon, H. Lou, I. Baribaud, W. Hou, C. Zou, R. J. Geraghty, P. G. Spear, R. J. Eisenberg, and G. H. Cohen. 1999. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J. Virol. 73:8127-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez, M., F. Cocchi, E. Avitabile, A. Leclerc, J. Adelaide, G. Campadelli-Fiume, and P. Dubreuil. 2001. Novel, soluble isoform of the herpes simplex virus (HSV) receptor nectin1 (or PRR1-HIgR-HveC) modulates positively and negatively susceptibility to HSV infection. J. Virol. 75:5684-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez, M., F. Cocchi, L. Menotti, E. Avitabile, P. Dubreuil, and G. Campadelli-Fiume. 2000. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 74:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez, M., F. Eberlé, M. G. Mattei, J. Gabert, F. Birg, F. Bardin, C. Maroc, and P. Dubreuil. 1995. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene 155:261-265. [DOI] [PubMed] [Google Scholar]
- 41.Martinez, W. M., and P. G. Spear. 2002. Amino acid substitutions in the V domain of nectin-1 (HveC) that impair entry activity for herpes simplex virus types 1 and 2 but not for pseudorabies virus or bovine herpesvirus 1. J. Virol. 76:7255-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menotti, L., E. Avitabile, P. Dubreuil, M. Lopez, and G. Campadelli-Fiume. 2001. Comparison of murine and human nectin1 binding to herpes simplex virus glycoprotein D (gD) reveals a weak interaction of murine nectin1 to gD and a gD-dependent pathway of entry. Virology 282:256-266. [DOI] [PubMed] [Google Scholar]
- 43.Menotti, L., R. Casadio, C. Bertucci, M. Lopez, and G. Campadelli-Fiume. 2002. Substitution in the murine nectin1 receptor of a single conserved amino acid at a position distal from herpes simplex virus gD binding site confers high-affinity binding to gD. J. Virol. 76:5463-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menotti, L., F. Cocchi, and G. Campadelli-Fiume. 2002. Critical residues in the CC′ ridge of the human nectin1 receptor V domain enable herpes simplex virus entry into the cell and act synergistically with the downstream region. Virology 301:6-12. [DOI] [PubMed] [Google Scholar]
- 45.Menotti, L., M. Lopez, E. Avitabile, A. Stefan, F. Cocchi, J. Adelaide, E. Lecocq, P. Dubreuil, and G. Campadelli-Fiume. 2000. The murine homolog of human Nectin1δ serves as a species nonspecific mediator for entry of human and animal αherpesviruses in a pathway independent of a detectable binding to gD. Proc. Natl. Acad. Sci. USA 97:4867-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milne, R. S., S. L. Hanna, A. H. Rux, S. H. Willis, G. H. Cohen, and R. J. Eisenberg. 2003. Function of herpes simplex virus type 1 gD mutants with different receptor-binding affinities in virus entry and fusion. J. Virol. 77:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 48.Pereira, L., D. V. Dondero, D. Gallo, V. Devlin, and J. D. Woodie. 1982. Serological analysis of herpes simplex virus types 1 and 2 with monoclonal antibodies. Infect. Immun. 35:363-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pereira, L., T. Klassen, and J. R. Baringer. 1980. Type-common and type-specific monoclonal antibody to herpes simplex virus type 1. Infect. Immun. 29:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 51.Racaniello, V. R. 1996. Early events in poliovirus infection: virus-receptor interactions. Proc. Natl. Acad. Sci. USA 93:11378-11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reymond, N., J. Borg, E. Lecocq, J. Adelaide, G. Campadelli-Fiume, P. Dubreuil, and M. Lopez. 2000. Human nectin3/PRR3: a novel member of the PVR/PRR/nectin family that interacts with afadin. Gene 255:347-355. [DOI] [PubMed] [Google Scholar]
- 53.Reymond, N., S. Fabre, E. Lecocq, J. Adelaide, P. Dubreuil, and M. Lopez. 2001. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J. Biol. Chem. 276:43205-43215. [DOI] [PubMed] [Google Scholar]
- 54.Richart, S., S. Simpson, C. Krummenacher, J. Whitbeck, L. Pizer, G. Cohen, R. Eisenberg, and C. Wilcox. 2003. Entry of herpes simplex virus type 1 into primary sensory neurons in vitro is mediated by nectin-1/HveC. J. Virol. 77:3307-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roller, R. J., and D. Rauch. 1998. Herpesvirus entry mediator HVEM mediates cell-cell spread in BHK(TK−) cell clones. J. Virol. 72:1411-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roller, R. J., and B. Roizman. 1994. A herpes simplex virus 1 US11-expressing cell line is resistant to herpes simplex virus infection at a step in viral entry mediated by glycoprotein D. J. Virol. 68:2830-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Satoh-Horikawa, K., H. Nakanishi, K. Takahashi, M. Miyahara, M. Nishimura, K. Tachibana, A. Mizoguchi, and Y. Takai. 2000. Nectin-3, a new member of immunoglobulin-like cell adhesion molecules that shows homophilic and heterophilic cell-cell adhesion activities. J. Biol. Chem. 275:10291-10299. [DOI] [PubMed] [Google Scholar]
- 58.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 59.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 60.Takai, Y., and H. Nakanishi. 2003. Nectin and afadin: novel organizers of intercellular junctions. J. Cell Sci. 116:17-27. [DOI] [PubMed] [Google Scholar]
- 61.Terry-Allison, T., R. I. Montgomery, J. C. Whitbeck, R. Xu, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J. Virol. 72:5802-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walther, W., U. Stein, and C. Eder. 1994. RNA analysis using miniprep RNA in reverse transcription PCR. BioTechniques 17:674-675. [PubMed] [Google Scholar]
- 63.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, G., E. Avitabile, G. Campadelli-Fiume, and B. Roizman. 2003. The domains of glycoprotein D required to block apoptosis induced by herpes simplex virus 1 are largely distinct from those involved in cell-cell fusion and binding to nectin1. J. Virol. 77:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]