Abstract
Non-Hodgkin lymphoma (NHL) has been associated with immunological defects, chronic inflammatory and autoimmune conditions. Given the link between immune dysfunction and NHL, genetic variants in toll-like receptors (TLRs) have been regarded as potential predictive factors of susceptibility to NHL. Adequate anti-tumoral responses are known to depend on TLR9 function, such that the use of its synthetic ligand is being targeted as a therapeutic strategy. We investigated the association between the functional rs5743836 polymorphism in the TLR9 promoter and risk for B-cell NHL and its major subtypes in three independent case–control association studies from Portugal (1160 controls, 797 patients), Italy (468 controls, 494 patients) and the US (972 controls, 868 patients). We found that the rs5743836 polymorphism was significantly overtransmitted in both Portuguese (odds ratio (OR), 1.85; P = 7.3E–9) and Italian (OR, 1.84; P = 6.0E–5) and not in the US cohort of NHL patients. Moreover, the increased transcriptional activity of TLR9 in mononuclear cells from patients harboring rs5743836 further supports a functional effect of this polymorphism on NHL susceptibility in a population-dependent manner.
Keywords: toll-like receptors, non-Hodgkin lymphoma, single nucleotide polymorphism, TLR9
Introduction
Non-Hodgkin lymphoma (NHL) includes a heterogeneous group of malignant lymphoproliferative diseases whose incidence has substantially increased over the past decades in Western countries.1 Individuals with specific immune deficiencies associated with immune-suppressive therapy after transplantation, HIV infection and congenital conditions are among those with higher incidence rates of lymphoid malignancies. Additionally, clinical and experimental data consistently associated several autoimmune and chronic inflammatory disorders with increased risk of NHL.2,3
Toll-like receptors (TLRs) are widely studied members of the pattern recognition receptor family with a major role in activation and homeostasis of the immune system upon pathogen recognition.4 Among the TLRs that bind nucleic acids, TLR9 recognizes unmethylated CpG DNA motifs (present at a much higher frequency in the genomes of prokaryotes than of eukaryotes) as a ‘danger signal’ that activates the innate immune system. In humans, this receptor is expressed in plasmacytoid dendritic cells and B lymphocytes, known to have a diverse TLR repertoire, where TLR9 predominates.5 A role for TLR9 on viral, fungal, mycobacterial and Helicobacter pylori infections has been described.6–10 TLR9 (and TLR7) has been also involved in the recognition of self-nucleic acids, which is believed to have an important role in the pathogenesis of auto-immune diseases, particularly in systemic lupus erythematosus11 and psoriasis.12
In addition to self-recognition, inappropriate TLR9 activation leading to disease may also involve mechanisms of transcriptional deregulation of the TLR9 gene. Consistently, cells from different lymphoma types (mantle cell, B-cell small lymphocytic, follicular and diffuse large B-cell) have been reported to overexpress TLR9 and to show an increased proliferation upon CpG stimulation, although with a heterogeneous pattern when comparing either different lymphomas or different patients.5,13 More specifically, the highly prevalent rs5743836 polymorphism in the TLR9 gene (1237T>C), has been demonstrated to predispose to Hodgkin lymphoma, as well as to several autoimmune and chronic inflammatory diseases, including asthma and Crohn’s disease.14–18
Given the substantial weight of evidence for immune dysfunction as the underlying basis of lymphomagenesis, a genetic variant unbalancing the regulation and expression of TLR9-associated mechanisms may have a major impact upon host anti-tumoral responses and therefore, an important role in NHL pathogenesis. Epidemiological and genetic studies have shown that inflammatory and infectious conditions, particularly bacterial and viral infections, are associated with increased NHL risk,3,19–22 suggesting that chronic immune stimulation has a role in NHL development.3,21,23–26
Given the role of TLR9 in bacterial and viral recognition, and the link of the TLR9 rs5743836 polymorphism with Hodgkin lymphoma, we hypothesized that rs5743836 could also influence NHL risk. We therefore analyzed whether this polymorphism was associated with susceptibility to B-cell-derived NHL in a large-scale, case–control study.
Results and discussion
All cases were histologically confirmed and coded according to the World Health Organization (WHO) classification. Genotype frequencies of Portuguese control individuals (n = 1160) for rs5743836 were in Hardy–Weinberg equilibrium (P>0.05). A comparison of genotype distribution between the NHL patients (n = 797, including cases of follicular lymphoma (FL) n = 240, 30.1%; diffuse large B-cell lymphoma (DLBCL) n = 200, 25.1%, marginal zone lymphoma n = 125, 15.7%, chronic lymphoid leukemia/small lymphocytic lymphoma, (CLL/SLL) n = 85, 10.7%, mantle cell lymphoma n = 82, 10.3%; B-lymphoblastic lymphoma, n = 55, 6.9%, and Burkitt lymphoma, n = 10; 1.2%) and controls revealed a positive association between the presence of rs5743836 and risk of NHL (Table 1), using the dominant (P = 7.3E–9, odds ratio (OR) = 1.85, 95% confidence interval (CI), (1.50–2.27)) or the additive (P = 1.1E–7, OR = 1.66) genetic models. Accordingly, significant associations were also observed for the major NHL subtypes, such as FL (P = 2.9E–4; OR = 1.77; 95% CI, 1.30–2.42), DLBCL (P = 7.2E–6; OR = 2.07; 95% CI, 1.50–2.85) and CLL/SLL (P = 7.06E–6; OR = 2.76; 95% CI, 1.74–4.35), thus pointing to a broad impact of rs5743836 in B-cell NHL, independently of its subtypes.
Table 1.
Analysis of rs5743836 genotype frequencies in B-cell non-Hodgkin lymphoma patients and controls
| Portuguese population |
N (% frequency)a,b
|
Dominant modelc
|
Additive modelc
|
|||||
|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | P-value* | OR (95% CI) | P-value** | OR (95% CI) | ||
| Controls | 1160 | 934 (80.5) | 217 (18.7) | 9 (0.8) | ||||
| DLBCL | 200 | 140 (70.0) | 60 (30.0) | 0 (0.0) | 7.5E–04 | 1.77 (1.27–2.48) | 3.0E–03 | 1.70 |
| FL | 240 | 168 (70.0) | 72 (30.0) | 0 (0.0) | 2.9E–04 | 1.77 (1.30–2.42) | 1.4E–03 | 1.70 |
| CLL/SLL | 85 | 51 (60.0) | 32 (37.6) | 2 (2.4) | 7.1E–06 | 2.76 (1.74–4.35) | 5.8E–06 | 2.42 |
| MCL | 82 | 64 (78.0) | 18 (22.0) | 0 (0.0) | 5.9E–01 | 0.76 (0.04–13.25) | 7.3E–01 | 1.14 |
| MZL | 125 | 77 (61.6) | 48 (38.4) | 0 (0.0) | 9.3E–07 | 2.58 (1.75–3.80) | 7.8E–06 | 2.40 |
| B-lymphoblastic lymphoma | 55 | 43 (78.2) | 12 (21.8) | 0 (0.0) | 6.7E–01 | 1.15 (0.60–2.22) | 7.9E–01 | 1.14 |
| Burkitt lymphoma | 10 | 8 (80.0) | 2 (20.0) | 0 (0.0) | 9.7E–01 | 1.03 (0.22–4.90) | 9.9E–01 | 1.02 |
| B-cell NHL | 797 | 551 (69.1) | 244 (30.6) | 2 (0.3) | 7.3E–09 | 1.85 (1.50–2.27) | 1.1E–07 | 1.66 |
| Italian population | ||||||||
| Controls | 468 | 379 (81.0) | 81 (17.3) | 8 (1.7) | ||||
|
| ||||||||
| DLBCL (n = 55) | 55 | 37 (67.3) | 14 (25.5) | 4 (7.2) | 1.7E–02 | 2.07 (1.13–3.81) | 4.1E–03 | 2.09 |
| FL (n = 150) | 150 | 108 (72.0) | 39 (26.0) | 3 (2.0) | 1.9E–02 | 1.66 (1.08–2.53) | 3.2E–02 | 1.43 |
| CLL/SLL (n = 242) | 242 | 167 (69.0) | 69 (28.5) | 6 (2.5) | 3.3E–04 | 1.91 (1.34–2.73) | 7.3E–04 | 1.61 |
| MCL (n = 22) | 22 | 16 (72.7) | 5 (22.7) | 1 (4.6) | 3.4E–01 | 1.60 (0.61–4.20) | 2.6E–01 | 1.61 |
| MZL (n = 18) | 18 | 12 (66.7) | 6 (33.3) | 0 (0.0) | 1.3E–01 | 2.13 (0.78–5.83) | 2.4E–01 | 1.87 |
| MALT (n = 7) | 7 | 5 (71.4) | 2 (28.6) | 0 (0) | 5.2E–01 | 1.70 (0.33–8.92) | 6.4E–01 | 1.56 |
| B-cell NHL | 494 | 345 (69.8) | 135 (27.3) | 14 (2.9) | 6.0E–05 | 1.84 (1.36–2.48) | 1.1E–04 | 2.00 |
| US population | ||||||||
| Controls (n = 972) | 972 | 674 (69.3) | 275 (28.3) | 23 (2.4) | ||||
|
| ||||||||
| DLBCL (n = 252) | 252 | 175 (69.4) | 70 (27.8) | 7 (2.8) | 1.0E+00 | 1.00 (0.73–1.34) | 9.3E–01 | |
| FL (n = 190) | 190 | 143 (75.3) | 43 (22.6) | 4 (2.1) | 1.2E–01 | 0.75 (0.52–1.06) | 1.3E–01 | |
| CLL/SLL (n = 196) | 196 | 140 (71.4) | 55 (28.1) | 1 (0.5) | 6.1E–01 | 0.91 (0.64–1.27) | 3.2E–01 | |
| MCL (n = 29) | 29 | 21 (72.4) | 8 (27.6) | 0 (0.0) | 8.4E–01 | 0.87 (0.36–1.93) | 5.8E–01 | |
| MZL (n = 76) | 76 | 56 (73.7) | 19 (25.0) | 1 (1.3) | 5.2E–01 | 0.81 (0.47–1.38) | 3.8E–01 | |
| Other (n = 58) | 58 | 44 (75.9) | 14 (24.1) | 0 (0.0) | 3.8E–01 | 0.73 (0.38–1.31) | 2.0E–01 | |
| B-cell NHL | 801 | 579 (72.3) | 209 (26.1) | 13 (1.6) | 1.9E–01 | 0.867 (0.71–1.07) | 1.3E–01 | |
Abbreviations: CI, confidence intervals; DLCBL, diffuse large B-cell lymphoma; FL, follicular cell lymphoma; MALT, mucosa-associated lymphoid tissue lymphoma; MCL, mantle cell lymphoma; MZL, marginal zone lymphoma; NHL, non-Hodgkin lymphoma; OR, odds ratio; SLL/CLL, small lymphocytic lymphoma/chronic lymphocytic leukemia.
Portuguese and Italian patients had a mean age of 60.4±15.8 (51.7% males, 48.3% females) and 62.8±12.4 (58.9% males, 41.1% females), respectively. Controls were unrelated healthy blood donors frequency-matched to cases by gender and age. Patients and controls with history of transplantation, hematological malignancy or HIV infection were excluded from the study. USA patients derived from a study conducted in the San Francisco Bay Area (California, USA) that included incident cases diagnosed from 2001 through 2006. Details of the process, criteria for subject selection and full description of patients have been described elsewhere.38 Note that the number of patients in the table is smaller than that stated in the text as a result of individuals with undetermined genotype. All study participants provided written informed consent.
For the Portuguese and Italian studies, genotyping was performed using bi-directional PCR amplification of specific alleles (Bi-PASA) as described.39 Accurate genotyping was validated by direct sequencing randomly selected DNA samples. Concordant results were obtained for 100% of the samples in blind analysis. In addition, as routine quality control, each Bi-PASA genotyping set comprised randomly selected replicates of previously typed samples and two negative controls (water). For the US study, DNA was isolated from peripheral blood mononuclear cells using the QIAamp DNA Blood Maxi Kit protocol (Qiagen, Valencia, CA, USA) and quantified using PicoGreen dsDNA Quantitation kits (Invitrogen, Carlsbad, CA, USA) according to the manufacturers’ specifications. Genotyping was performed using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA, USA) on the ABI Prism 7700 Sequence Detection System. Replicate, blinded quality control samples were included to assess reproducibility of the above genotyping procedures. Ambiguous genotypes were regenotyped as necessary. All plates contained positive (all three genotypes) and negative (water) controls. Reactions were done with the following protocol: 95 °C for 10 min, then 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Probes and primer sets used were as follows: forward primer, 5′-GCCTTGGGATGTGCTGTTC-3′; reverse primer, 5′-CAGAGACATAATGGAGGCAAAGGA-3′; T probe, 5′-CCTGAAAACTCCC-3′; and C probe, 5′-CTGGAAACTCCC-3′.
Association tests were conducted using a dominant and an additive model (Cochran–Armitage trend test) tests. For the Portuguese and Italian studies, Fisher’s exact test and Pearson’s χ2-test were used to compare genotype frequencies between patients and controls. Unconditional logistic regression was used to compute odds ratios (ORs) and corresponding 95% confidence intervals (CI) adjusted for age and sex. For the US study, association tests were conducted using the PLINK 1.04 software. ORs and 95% CI were calculated by median-unbiased estimation using the mid-p method from the epitools R package (http://sites.google.com/site/medepi/epitools). P-values were calculated regarding dominant
or the additive
genetic model.
No significant associations were found when considering other features of clinical value including international prognostic indexes (IPI and FLIPI), clinical stage of the disease or overall survival (data not shown).
To validate our findings, two independent Caucasian populations were used: an Italian cohort (467 NHL cases, including cases of CLL/SLL, n = 242, 49.0%; FL, n = 150, 30.4%; DLBCL, n = 55, 11.1%; mantle cell lymphoma, n = 22, 4.5%; marginal zone lymphoma, n = 18, 3.6% and mucosa-associated lymphoid tissue lymphoma, n = 7, 1.4%, and 468 controls) and a US population of European ancestry (821 NHL cases, including cases of CLL/SLL, n = 201, 24.5%; FL, n = 194, 23.6%; DLBCL, n = 255, 31.1%; mantle cell lymphoma, n = 31, 3.8%; marginal zone lymphoma, n = 80, 9.7%, other B-NHL cases, n = 60, 7.3%, and 1001 controls).27 In both populations, rs5743836 frequencies for controls were in Hardy–Weinberg equilibrium (P>0.05). Whereas the association with overall NHL (with either the dominant (P = 6.0E–5; OR = 1.84; 95% CI, 1.36–2.48) or the additive (P = 1.1E–4; OR = 1.60) genetic models) or its major subtypes FL (P = 1.9E–2; OR = 1.66; 95% CI, 1.08–2.53), DLBCL (P = 1.7E–2; OR = 2.07; 95% CI, 1.13–3.81) and CLL/ SLL (P = 3.3E–4; OR = 1.91; 95% CI, 1.34–2.73) was confirmed in the Italian cohort, no association between NHL susceptibility and the presence of rs5743836 was found in the US cohort (Table 1).
There are several possible reasons for the discrepant results obtained for the association of rs5743836 with NHL. It is noteworthy that several other studies have found minor allele control frequencies of rs5743836, similar to those assessed in our European populations.8,28,29 This suggests that, despite the fact that the populations enrolled in our study shared the same population bottleneck expanding out of Europe, the decreased minor allele frequency detected in the Portuguese and Italian cohorts likely reflects a greater genetic drift in Europe.30 On the other hand, familial aggregation and twin studies for lymphoproliferative disorders have ruled out the role of highly penetrant genes in affecting risk of the majority of NHL cases.31 It is therefore likely that the additive effects of genetic variants and interactions with environmental and infectious agents may have a significant role in the differential relevance of the rs5743836 polymorphism in the risk for NHL. This hypothesis is in agreement with epidemiological and genetic studies, showing that inflammatory and infectious conditions are associated with increased NHL risk, suggesting that chronic immune stimulation has a role in NHL development.3,20–22
The rs5743836 polymorphism in TLR9 has been consistently associated with increased transcriptional activity.9,32,33 Indeed, we have also found that unstimulated mononuclear cells isolated from both healthy controls and NHL patients bearing the rs5743836 polymorphism display increased TLR9 mRNA expression (Figure 1), thereby supporting the notion that rs5743836 triggers an enhanced TLR9 function. Therefore, it is possible to predict that the complex immunological response of various cell types to CpG, involving both direct and indirect effects of TLR9, is likely deregulated in individuals carrying the rs5743836 polymorphism. In fact, it has been shown that increased B-cell proliferation upon CpG stimulation is a common feature of different NHL subtypes, even though the proliferation pattern varies among NHL subtypes and individual patients.13 Signal transduction initiated by TLR9 activation results in nuclear translocation of nuclear factor-κB34 that may enhance lymphocyte neoplastic transformation by promoting proliferation and survival of mutated cells.35 In this regard, it may be worthwhile to investigate whether the rs5743836 polymorphism displays a preferential association with lymphoma subtypes with constitutive activation of nuclear factor-κB pathways. The neoplastic process leading to the development of NHL may be usurping impairments in TLR signaling pathways to advance cancer progression, which suggests that targeting these pathways may open novel therapeutic avenues. CpG molecules are now regarded as highly promising for cancer therapy, mostly due to their direct effect on TLR9 activation on immune cell subpopulations that have an important role in anti-tumor immunity, including B cells.36 However, therapeutic applications of these CpG should be individually tailored, as individual genetic variations may affect the outcome of the TLR9 signaling pathway.
Figure 1.
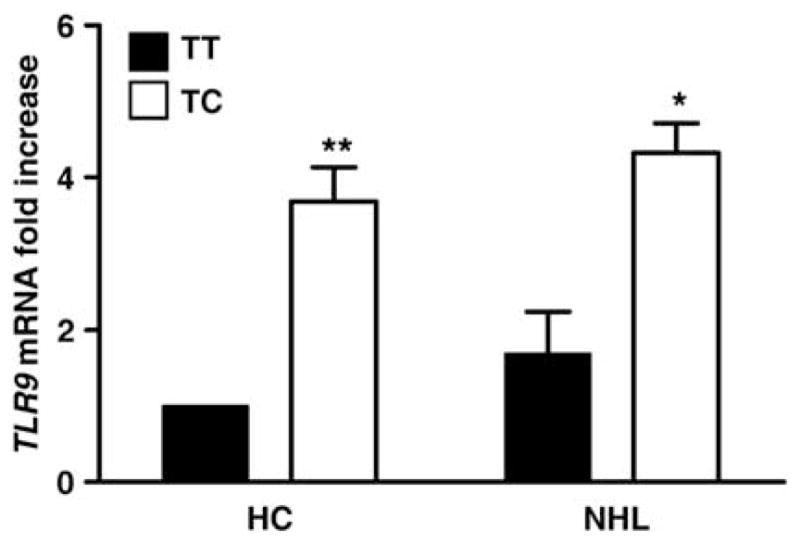
Relative expression of TLR9 in unstimulated mononuclear cells derived from healthy controls (HC) and NHL patients bearing distinct rs5743836 genotypes (TT, wild-type and TC). Shown is data pooled from 16 HC and 16 NHL individuals (eight of each genotype) and expressed as mean±s.d. Samples from patients with FL (n = 4), DLBCL (n = 2) and CLL/SLL (n = 2) histologies, after diagnosis and before initiation of treatment, were included in the NHL category. No significant differences in TLR9 expression were observed among the NHL subtypes. *P<0.05 and ** P <0.01 by Student’s t-test.
In summary, our results suggest a potential role for the host genetic background in B-cell NHL susceptibility. Altogether, and despite the fact that B-cell lymphomas are known for their ability to trigger disparate responses to TLR9 agonists,13 the genetic associations found herein were not, for the most part, subtype-specific. This suggests that altered TLR9 signaling pathways may underlie shared mechanisms of susceptibility to B-cell NHL, particularly to the subtypes where the findings were stronger, such as CLL/SLL. As a matter of fact, apoptosis of human CLL B-cells has been found to be intimately related with TLR9 activation,37 thereby suggesting that genetic variation of this receptor may also impact the outcome of TLR9 agonist therapy. However, deeper insights into the pathogenetic mechanisms underlying TLR9-mediated mechanisms in NHL are needed. Ultimately, functional studies dissecting the role of the rs5743836 may allow the identification of potential therapeutic targets.
Acknowledgments
AC, NSO, MTC and AJA were financially supported by a fellowship from Fundação para a Ciência e Tecnologia, Portugal. MS is a Ciência 2007 fellow. This study was supported by Fundação para a Ciência e Tecnologia, Portugal (PIC/IC/83313/2007) and by Fundação Calouste Gulbenkian, Serviço de Saúde e Desenvolvimento Humano, Portugal (Grant Number:Proc/60666-MM/734). CFS, PB and LC were supported by National Institutes of Health (NIH) grants CA122663 and CA104682, and PB also by NIH grants CA45614 and CA89745. We are grateful to Paulo Vieira, Cecília Leão, Manuel T Silva, Nuno Sousa, Jorge Correia-Pinto, Joana Palha, Margarida Correia-Neves, Margarida Lima and Matthew Berry for all their input throughout these studies and critical reading of the manuscript. We are grateful to the patients who joint this study, as well as to all members of the Life and Health Sciences Research Institute and School of Health Sciences, University of Minho, who contributed in any way to the development of this work.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 2.Grulich AE, Vajdic CM, Cozen W. Altered immunity as a risk factor for non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2007;16:405–408. doi: 10.1158/1055-9965.EPI-06-1070. [DOI] [PubMed] [Google Scholar]
- 3.Smedby KE, Askling J, Mariette X, Baecklund E. Autoimmune and inflammatory disorders and risk of malignant lymphomas—an update. J Intern Med. 2008;264:514–527. doi: 10.1111/j.1365-2796.2008.02029.x. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 5.Bourke E, Bosisio D, Golay J, Polentarutti N, Mantovani A. The toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 2003;102:956–963. doi: 10.1182/blood-2002-11-3355. [DOI] [PubMed] [Google Scholar]
- 6.Berrington WR, Hawn TR. Mycobacterium tuberculosis, macrophages, and the innate immune response: does common variation matter? Immunol Rev. 2007;219:167–186. doi: 10.1111/j.1600-065X.2007.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho A, Cunha C, Carotti A, Aloisi T, Guarrera O, Di Ianni M, et al. Polymorphisms in Toll-like receptor genes and susceptibility to infections in allogeneic stem cell transplantation. Exp Hematol. 2009;37:1022–1029. doi: 10.1016/j.exphem.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho A, Pasqualotto AC, Pitzurra L, Romani L, Denning DW, Rodrigues F. Polymorphisms in toll-like receptor genes and susceptibility to pulmonary aspergillosis. J Infect Dis. 2008;197:618–621. doi: 10.1086/526500. [DOI] [PubMed] [Google Scholar]
- 9.Ng MT, Van’t Hof R, Crockett JC, Hope ME, Berry S, Thomson J, et al. Increase in NF-kappaB binding affinity of the variant C allele of the toll-like receptor 9 -1237T/C polymorphism is associated with Helicobacter pylori-induced gastric disease. Infect Immun. 2010;78:1345–1352. doi: 10.1128/IAI.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pine SO, McElrath MJ, Bochud PY. Polymorphisms in toll-like receptor 4 and toll-like receptor 9 influence viral load in a seroincident cohort of HIV-1-infected individuals. AIDS. 2009;23:2387–2395. doi: 10.1097/QAD.0b013e328330b489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshak-Rothstein A. Toll-like receptors in systemic auto-immune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 13.Jahrsdorfer B, Muhlenhoff L, Blackwell SE, Wagner M, Poeck H, Hartmann E, et al. B-cell lymphomas differ in their responsiveness to CpG oligodeoxynucleotides. Clin Cancer Res. 2005;11:1490–1499. doi: 10.1158/1078-0432.CCR-04-1890. [DOI] [PubMed] [Google Scholar]
- 14.Hong J, Leung E, Fraser AG, Merriman TR, Vishnu P, Krissansen GW. TLR2, TLR4 and TLR9 polymorphisms and Crohn’s disease in a New Zealand Caucasian cohort. J Gastroenterol Hepatol. 2007;22:1760–1766. doi: 10.1111/j.1440-1746.2006.04727.x. [DOI] [PubMed] [Google Scholar]
- 15.Lachheb J, Dhifallah IB, Chelbi H, Hamzaoui K, Hamzaoui A. Toll-like receptors and CD14 genes polymorphisms and susceptibility to asthma in Tunisian children. Tissue Antigens. 2008;71:417–425. doi: 10.1111/j.1399-0039.2008.01011.x. [DOI] [PubMed] [Google Scholar]
- 16.Lazarus R, Klimecki WT, Raby BA, Vercelli D, Palmer LJ, Kwiatkowski DJ, et al. Single-nucleotide polymorphisms in the Toll-like receptor 9 gene (TLR9): frequencies, pairwise linkage disequilibrium, and haplotypes in three U.S. ethnic groups and exploratory case-control disease association studies. Genomics. 2003;81:85–91. doi: 10.1016/s0888-7543(02)00022-8. [DOI] [PubMed] [Google Scholar]
- 17.Mollaki V, Georgiadis T, Tassidou A, Ioannou M, Daniil Z, Koutsokera A, et al. Polymorphisms and haplotypes in TLR9 and MYD88 are associated with the development of Hodgkin’s lymphoma: a candidate-gene association study. J Hum Genet. 2009;54:655–659. doi: 10.1038/jhg.2009.90. [DOI] [PubMed] [Google Scholar]
- 18.Torok HP, Glas J, Tonenchi L, Bruennler G, Folwaczny M, Folwaczny C. Crohn’s disease is associated with a toll-like receptor-9 polymorphism. Gastroenterology. 2004;127:365–366. doi: 10.1053/j.gastro.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 19.Ivanovski M, Silvestri F, Pozzato G, Anand S, Mazzaro C, Burrone OR, et al. Somatic hypermutation, clonal diversity, and preferential expression of the VH 51p1/VL kv325 immunoglobulin gene combination in hepatitis C virus-associated immunocytomas. Blood. 1998;91:2433–2442. [PubMed] [Google Scholar]
- 20.Muller AM, Ihorst G, Mertelsmann R, Engelhardt M. Epidemiology of non-Hodgkin’s lymphoma (NHL): trends, geographic distribution, and etiology. Ann Hematol. 2005;84:1–12. doi: 10.1007/s00277-004-0939-7. [DOI] [PubMed] [Google Scholar]
- 21.Pagano JS. Viruses and lymphomas. N Engl J Med. 2002;347:78–79. doi: 10.1056/NEJMp020056. [DOI] [PubMed] [Google Scholar]
- 22.Schollkopf C, Melbye M, Munksgaard L, Smedby KE, Rostgaard K, Glimelius B, et al. Borrelia infection and risk of non-Hodgkin lymphoma. Blood. 2008;111:5524–5529. doi: 10.1182/blood-2007-08-109611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerhan JR, Ansell SM, Fredericksen ZS, Kay NE, Liebow M, Call TG, et al. Genetic variation in 1253 immune and inflammation genes and risk of non-Hodgkin lymphoma. Blood. 2007;110:4455–4463. doi: 10.1182/blood-2007-05-088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lecuit M, Abachin E, Martin A, Poyart C, Pochart P, Suarez F, et al. Immunoproliferative small intestinal disease associated with Campylobacter jejuni. N Engl J Med. 2004;350:239–248. doi: 10.1056/NEJMoa031887. [DOI] [PubMed] [Google Scholar]
- 25.Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, Isaacson PG. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet. 1991;338:1175–1176. doi: 10.1016/0140-6736(91)92035-z. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto K. Pathogenesis of Sjogren’s syndrome. Autoimmun Rev. 2003;2:13–18. doi: 10.1016/s1568-9972(02)00121-0. [DOI] [PubMed] [Google Scholar]
- 27.Skibola CF, Bracci PM, Halperin E, Conde L, Craig DW, Agana L, et al. Genetic variants at 6p21. 33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009;41:873–875. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamann L, Glaeser C, Hamprecht A, Gross M, Gomma A, Schumann RR. Toll-like receptor (TLR)-9 promotor polymorphisms and atherosclerosis. Clin Chim Acta. 2006;364:303–307. doi: 10.1016/j.cca.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Lammers KM, Ouburg S, Morre SA, Crusius JB, Gionchett P, Rizzello F, et al. Combined carriership of TLR9-1237C and CD14-260T alleles enhances the risk of developing chronic relapsing pouchitis. World J Gastroenterol. 2005;11:7323–7329. doi: 10.3748/wjg.v11.i46.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skibola CF, Curry JD, Nieters A. Genetic susceptibility to lymphoma. Haematologica. 2007;92:960–969. doi: 10.3324/haematol.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lange NE, Zhou X, Lasky-Su J, Himes BE, Lazarus R, Soto-Quiros M, et al. Comprehensive genetic assessment of a functional TLR9 promoter polymorphism: no replicable association with asthma or asthma-related phenotypes. BMC Med Genet. 2011;12:26. doi: 10.1186/1471-2350-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novak N, Yu CF, Bussmann C, Maintz L, Peng WM, Hart J, et al. Putative association of a TLR9 promoter polymorphism with atopic eczema. Allergy. 2007;62:766–772. doi: 10.1111/j.1398-9995.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 34.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 35.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 36.Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 37.Liang X, Moseman EA, Farrar MA, Bachanova V, Weisdorf DJ, Blazar BR, et al. Toll-like receptor 9 signaling by CpG-B oligodeoxynucleotides induces an apoptotic pathway in human chronic lymphocytic leukemia B cells. Blood. 2010;115:5041–5052. doi: 10.1182/blood-2009-03-213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skibola CF, Bracci PM, Halperin E, Nieters A, Hubbard A, Paynter RA, et al. Polymorphisms in the estrogen receptor 1 and vitamin C and matrix metalloproteinase gene families are associated with susceptibility to lymphoma. PLoS One. 2008;3:e2816. doi: 10.1371/journal.pone.0002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvalho A, Marques A, Maciel P, Rodrigues F. Study of disease-relevant polymorphisms in the TLR4 and TLR9 genes: a novel method applied to the analysis of the Portuguese population. Mol Cell Probes. 2007;21:316–320. doi: 10.1016/j.mcp.2007.03.005. [DOI] [PubMed] [Google Scholar]


