Abstract
Interaction between CD40 and its ligand, CD154, has a key function in immune regulation. Recent experimental data support a role of deregulated CD40 signalling in lymphomagenesis. Data from earlier studies that are part of this pooling study implicate a functional polymorphism (−1C>T, rs1883832) in the TNFRSF5 gene encoding CD40 in the etiology of follicular lymphoma. Here, the association of this variant with non-Hodgkin lymphoma (NHL) risk was replicated in a European multicenter study of 855 NHL cases and 1,206 controls. In the combined analysis of 2,617 cases and 3,605 controls, carrying the TT genotype was associated with an increased risk for all NHL (OR = 1.4; p for linear trend = 0.00009), diffuse large B-cell lymphoma (OR = 1.6; p for linear trend = 0.002) and follicular lymphoma (OR = 1.6; p for linear trend = 0.001). These data suggest a possible role of this functional polymorphism in lymphomas originating within the germinal center.
Keywords: lymphoma, TNFRSF5, CD40, polymorphism, epidemiology
Deregulation of immune responses and infectious agents play a role in the etiology of lymphoma.1,2 There is increasing evidence that variants in a number of immune regulatory genes drive interindividual differences in lymphoma susceptibility.3 Recently, Skibola et al. reported that the homozygous variant −1TT genotype of a functional single nucleotide polymorphism (SNP) in the TNFRSF5 gene (−1C>T, rs1883832), located in the Kozak sequence of the 5′UTR,4 was associated with an increased risk of follicular lymphoma (FL).5 TNFRSF5 encodes CD40, a member of the tumor necrosis factor (TNF) receptor family that interacts with its ligand, CD154 encoded by TNFSF5, and orchestrates a multitude of processes that influence B-cell and T-cell biology. The TNFRSF5 −1T risk allele has been associated with reduced CD40 translational efficiency on B cells,6 lower CD40 expression on dendritic cells and reduced circulating levels of soluble CD40 in humans.5 Deregulated CD40 signaling was recently implicated in the initiation of B-cell transformation in mice.7
The objective of our study was to replicate within Epi-Lymph, a European multicenter study, the results of an association between the TNFRSF5 −1TT genotype and risk of FL found in the previous study5 and to conduct a pooled analysis with increased power for subtype-specific analyses.
Materials and Methods
Study populations in Europe and the United States
EpiLymph
The EpiLymph multicenter case–control study consisting of 2,302 lymphoma cases (including non-Hodgkin lymphoma [NHL] and Hodgkin lymphoma) and 2,417 controls was conducted in six countries (Germany, Italy, Spain, Ireland, France and Czech Republic) from 1998 to 2004. Details of the study design have been provided elsewhere.8 Cases were categorized according to the WHO classification.8 Controls were drawn randomly from population registers of the study regions (Germany and Italy) or were recruited from the same hospital as cases (remaining countries). All controls were frequency matched to the cases by age (±5 years), sex and study center. In the hospital-based studies, controls were excluded if the main reason for the hospitalization was cancer, organ transplant and/or systemic infection.
San Francisco Bay Area NHL1 and NHL2 studies
Detailed methods for these two large population-based case–control studies (NHL1: cases = 1,591, controls = 2,515; NHL2: cases = 2,055, controls = 2,081) have been published previously (NHL19,10; NHL25). In brief, rapid case ascertainment and cancer registry data were used to identify incident NHL adult cases diagnosed from 1988 to 1993 (NHL1) and from 2001 to 2006 (NHL2) in six San Francisco Bay Area counties. Controls were identified using random digit dial and frequency matched to cases by 5-year age group, sex and county. Diagnostic materials were rereviewed, and NHL subtypes were classified using the Revised European-American Lymphoma (NHL1) and the WHO classification (NHL2). Biospecimens were collected from eligible participants (NHL1: 63% cases, 66% controls; NHL2: 87% cases, 89% controls).
Informed consent was obtained from all participants before enrolment. The institutional review boards of participating centers approved each study.
Genotyping
Genotyping of TNFRSF5 −1C>T SNP (rs1883832) was performed by Pyrosequencing™, Qiagen, Hilden, Germany and TaqMan®, Applied Biosystems™, Darmstadt, Germany.5
Statistical analysis
Data for the TNFRSF5 −1C>T SNP (rs1883832) for five Epi-Lymph study centers (Spain, France, Italy, Ireland and the Czech Republic) were used to replicate results from an earlier analysis including the German component of the same study and the San Francisco Bay Area NHL1 and 2 studies5 and to conduct a pooled analysis of the combined data to increase power for analyses by NHL subtype. Statistical analyses were conducted using SAS version 9 (SAS Institute, Cary, NC) and the rmeta package in R version 2.14.
Analyses were restricted to HIV-negative, non-Hispanic whites to diminish potential confounding effects because of underlying population structure (2,617 cases, 3,605 controls; 4 cases and 5 controls without genotype information, see Supporting Information Table 2). Unconditional logistic regression models adjusted for age as a continuous variable, sex and individual study centers (six EpiLymph, two San Francisco Bay Area) were used to obtain odds ratios (OR) as estimates of relative risk (hereafter called risk). The contribution of individual studies to the overall effect estimates was assessed in sensitivity analyses. Genotype was coded as an ordinal variable based on the number of rare alleles (0, 1 and 2) and as a binary variable assuming dominant inheritance. The referent group for genotype analyses was the most common homozygous genotype in controls. Adjusted logistic regression was used to determine linear trend in genotype odds ratios when genotype was included in the model as an ordinal variable.
Results and Discussion
A total of 855 NHL cases and 1,206 controls were available for the replication study. The TNFRSF5 −1TT genotype was positively associated with risk of NHL (OR = 1.6, p value for linear trend = 0.001; Table 1). Risk was increased for diffuse large B-cell lymphoma (DLBCL; OR = 2.1, p value for linear trend = 0.001) and FL (OR = 1.9, p value for linear trend = 0.04; Table 1), but not for chronic lymphocytic leukemia (OR = 0.78, p value for linear trend = 0.84).
Table 1.
Odds ratios (OR) and 95% confidence intervals (CI) for risk of non-Hodgkin lymphoma (NHL) and NHL by histologic subtype groups associated with the single nucleotide polymorphism (SNP) at position −1 of the Kozak consensus sequence of CD40 gene (rs1883832, C>T), restricted to HIV-negative non-Hispanic white participants
| NHL | Genotype | Replication set1 | Pooled set2 | ||
|---|---|---|---|---|---|
| Cases/controls | OR3 (95%CI) | Cases/controls | OR3 (95%CI) | ||
| All NHL | CC | 420/661 | 1.0 | 1,343/1,992 | 1.0 |
| CT | 351/460 | 1.2 (1.0–1.5) | 1,041/1,366 | 1.1 (1.0–1.3) | |
| TT | 84/85 | 1.6 (1.2–2.3) | 229/242 | 1.4 (1.2–1.8) | |
| CT/TT | 435/545 | 1.3 (1.1–1.5) | 1,270/1,608 | 1.2 (1.1–1.3) | |
| p trend | 0.001 | 0.00009 | |||
| DLBCL | CC | 125/661 | 1.0 | 391/1,992 | 1.0 |
| CT | 116/460 | 1.3 (1.0–1.8) | 311/1,366 | 1.2 (0.98–1.4) | |
| TT | 31/85 | 2.1 (1.3 –3.3) | 72/242 | 1.6 (1.2–2.1) | |
| CT/TT | 147/545 | 1.4 (1.1–1.9) | 383/1,608 | 1.2 (1.0–1.4) | |
| p trend | 0.001 | 0.002 | |||
| FL | CC | 44/661 | 1.0 | 244/1,992 | 1.0 |
| CT | 43/460 | 1.4 (0.91–2.2) | 213/1,366 | 1.3 (1.1–1.6) | |
| TT | 11/85 | 1.9 (0.96–3.9) | 46/242 | 1.6 (1.1–2.2) | |
| CT/TT | 54/545 | 1.5 (0.99–2.3) | 259/1,608 | 1.3 (1.1–1.6) | |
| p trend | 0.04 | 0.001 | |||
| CLL/SLL | CC | 119/661 | 1.0 | 263/1,992 | 1.0 |
| CT | 87/460 | 1.1 (0.79–1.5) | 202/1,366 | 1.1 (0.92–1.4) | |
| TT | 12/85 | 0.78 (0.41–1.5) | 38/242 | 1.2 (0.81–1.7) | |
| CT/TT | 99/545 | 1.0 (0.76–1.4) | 240/1,608 | 1.1 (0.94–1.4) | |
| p trend | 0.84 | 0.19 | |||
EpiLymph centers except Germany (Spain, France, Italy, Ireland and Czech Republic).
Pooled data include also Germany center of EpiLymph and San Francisco Bay Area NHL1 and NHL2 study data.5
Odds ratios and 95% confidence intervals are adjusted for age (continuous), sex and individual study center.
Abbreviations: DLBCL: diffuse large B-cell lymphoma; FL: follicular lymphoma; CLL/SLL: chronic lymphocytic leukemia/small lymphocytic lymphoma.
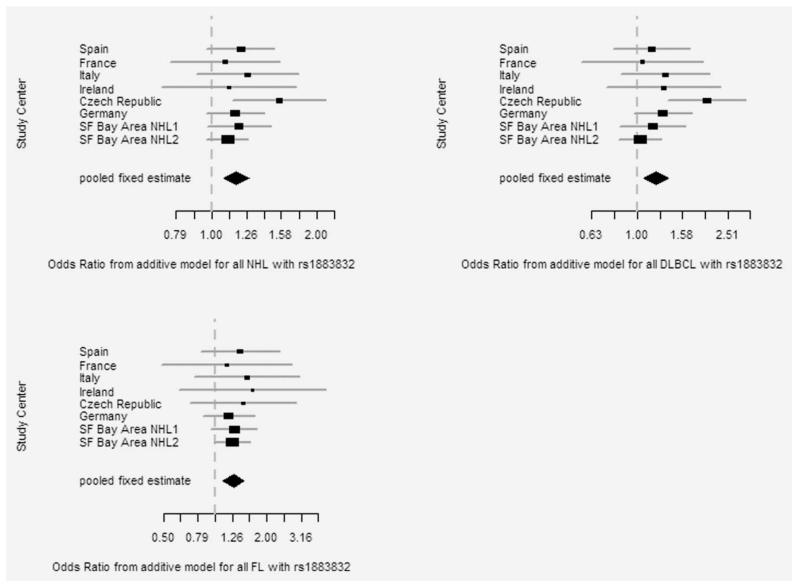
In the pooled study population of 2,617 NHL cases and 3,605 controls, carriers of the TNFRSF5 −1TT genotype showed a 1.4-fold increase in risk of NHL (p for trend = 0.00009). The excess risk was seen for DLBCL and FL, where the −1TT genotype conferred a 1.6-fold increase in risk (p value for linear trend = 0.002 and 0.001, respectively; Table 1). Risk for chronic lymphocytic leukemia was not increased in association with the −1TT genotype. Study-specific and pooled-adjusted risk estimates and 95% confidence intervals from fixed-effect models for TNFRSF5 −1C>T for all NHL, DLBCL and FL are depicted in Figure 1. In sensitivity analyses, none of the study altered the observed OR estimate by >10% (Fig. 1). Results from a sensitivity analysis excluding hospital controls were similar to results from the analysis including hospital controls (risk for NHL per allele, OR = 1.15, 95% CI = 1.04–1.26; OR = 1.18, 95% CI = 1.08–1.27; respectively).
Figure 1.
Odds ratios (ORs) and 95% confidence intervals (CIs) for risk per allele for the rs13883832 polymorphism associated with risk of all non-Hodgkin lymphoma (NHL), diffuse large B-cell (DLBCL) and follicular lymphoma (FL) obtained from age- and sex-adjusted unconditional logistic regression models restricted to HIV-negative non-Hispanic whites by study center. Rectangular shapes represent the study-specific ORs, the diamond shape represents the pooled OR with the size of the rectangles proportional to the weight (sample size) of the individual study in the pooled estimate (diamond). Study centers are identified on the y-axis and ORs and 95% CIs along the x-axis. The broken vertical line represents the OR = 1.0 (null effect).
Skibola et al.5 recently showed that healthy controls and lymphoma cases with the TNFRSF5 −1TT genotype had lower circulating soluble CD40 levels and reduced cell surface expression of CD40 on dendritic cells. This is consistent with previous studies that have reported lower CD40 expression in B cells of −1T versus −1C carriers in B-cell lines derived from patients with Grave’s disease and in transfected Rat-2 fibroblasts.6,11,12 Mice and humans that lack TNFRSF5 or TNFSF5 gene expression have reduced antibody production and Ig class switching and are unable to mount effective responses against infectious agents.13
Interestingly, the TNFRSF5 −1TT genotype was associated with an elevated risk of the two major NHL subtypes, DLBCL and FL, neoplasms that develop mainly in the germinal center (GC). CD40-CD154 ligation plays a pivotal role in centrocyte differentiation in the GC by downregulation of BCL6.14 Low BCL6 activity permits differentiation of centrocytes into plasma cells or memory cells, allowing B cells to exit the GC. This process may be attenuated by the TNFRSF5 −1TT genotype where low CD40 expression could hinder B- and T-cell interactions, allowing B cells to linger in GCs and undergo further somatic hypermutation and proliferation. This may increase the likelihood of B cells with preneoplastic lesions to progress to malignancy.
The strength of our study is its large sample size that allows the investigation of the main effects of TNFRSF5 −1C>T in relation to the risk of major NHL subtypes. However, small numbers limited the analysis of rarer lymphoma subtypes and subgroups stratified by age and sex.
In conclusion, this pooled analysis highlights the important role of the functional TNFRSF5 −1C>T variant in the etiology of DLBCL and FL. Future studies with careful molecular characterization of GC lymphomas are needed to assess risk related to BCL6 and BCL2 mutation status and further clarify mechanisms that contribute to lymphomagenesis.
Supplementary Material
Acknowledgements
We thank Ms. Aurélie Meunier (IARC, Lyon), Ms. Evelin Deeg (DKFZ, Heidelberg) and Ms. Zita Primusz-Kuesel (DKFZ, Heidelberg) for data management, and Ms. Marlen Auer and Ms. Bettina Ehret for performing genotyping. This work was supported by The German José Carreas Leukemia Foundation (DJCLS_R04/08 and R07/26f) (to AN); Federal Ministry of Education and Research (BMBF 01 EO 0803) (to AN); the EC 5th Framework Program Quality of Life grant No. QLK4-CT-2000-00422 (to PB, PBo); EC 6th Framework Program Food grant No. FOOD-CT-2006-023103 (to PB, P Bo); the Federal Office for Radiation Protection grants No. StSch4261 and StSch4420 (Germany) (to NB); the Spanish Ministry of Health grants CIBERESP (06/02/0073), FIS 08-1555 and Marato TV3 (051210) (Spain) (to SS); La Fondation de France (to MM); Compagnia di San Paolo di Torino, Programma Oncologia 2001 (to PC); and National Cancer Institute grants R01-CA45614, R01-CA89745 and R01-CA87014 (to EAH) and R01-CA104862, R01-CA122663 (to CSF).
Grant sponsor: German José Carreas Leukemia Foundation; Grant number: DJCLS_R04/08 and R07/26f; Grant sponsor: Federal Ministry of Education and Research (BMBF); Grant number: 01 EO 0803; Grant sponsor: EC 5th Framework Program Quality of Life; Grant number: QLK4-CT-2000-00422; Grant sponsor: EC 6th Framework Program Food; Grant number: FOOD-CT-2006-023103; Grant sponsor: Federal Office for Radiation Protection; Grant number: StSch4261 and StSch4420 (Germany); Grant sponsor: Spanish Ministry of Health grants CIBERESP; Grant numbers: 06/02/0073, FIS 08-1555 and Marato TV3 (051210) (Spain); Grant sponsor: La Fondation de France; Grant sponsor: Compagnia di San Paolo di Torino, Programma Oncologia 2001; Grant sponsor: National Cancer Institute; Grant numbers: R01-CA45614, R01-CA89745, R01-CA87014, R01-CA104862 and R01-CA122663
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Grulich AE, Vajdic CM, Cozen W. Altered immunity as a risk factor for non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2007;16:405–8. doi: 10.1158/1055-9965.EPI-06-1070. [DOI] [PubMed] [Google Scholar]
- 2.Engels EA. Infectious agents as causes of non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2007;16:401–4. doi: 10.1158/1055-9965.EPI-06-1056. [DOI] [PubMed] [Google Scholar]
- 3.Skibola CF, Curry JD, Nieters A. Genetic susceptibility to lymphoma. Haematologica. 2007;92:960–9. doi: 10.3324/haematol.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomer Y, Concepcion E, Greenberg DA. A C/T single-nucleotide polymorphism in the region of the CD40 gene is associated with Graves’ disease. Thyroid. 2002;12:1129–35. doi: 10.1089/105072502321085234. [DOI] [PubMed] [Google Scholar]
- 5.Skibola CF, Nieters A, Bracci PM, Curry JD, Agana L, Skibola DR, Hubbard A, Becker N, Smith MT, Holly EA. A functional TNFRSF5 gene variant is associated with risk of lymphoma. Blood. 2008;111:4348–54. doi: 10.1182/blood-2007-09-112144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobson EM, Concepcion E, Oashi T, Tomer Y. A Graves’ disease-associated Kozak sequence single-nucleotide polymorphism enhances the efficiency of CD40 gene translation: a case for translational pathophysiology. Endocrinology. 2005;146:2684–91. doi: 10.1210/en.2004-1617. [DOI] [PubMed] [Google Scholar]
- 7.Homig-Holzel C, Hojer C, Rastelli J, Casola S, Strobl LJ, Muller W, Quintanilla-Martinez L, Gewies A, Ruland J, Rajewsky K, Zimber-Strobl U. Constitutive CD40 signaling in B cells selectively activates the noncanonical NF-kappaB pathway and promotes lymphomagenesis. J Exp Med. 2008;205:1317–29. doi: 10.1084/jem.20080238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Sanjose S, Benavente Y, Nieters A, Foretova L, Maynadié M, Cocco PL, Staines A, Vornanen M, Boffetta P, Becker N, Alvaro T, Brennan P. The Effect of Regular Use of Hair Dye on Lymphoid Neoplasm in Europe. Am J Epidemiol. 2006;164:47–55. doi: 10.1093/aje/kwj187. [DOI] [PubMed] [Google Scholar]
- 9.Holly EA, Lele C, Bracci PM, McGrath MS. Case-control study of non-Hodgkin’s lymphoma among women and heterosexual men in the San Francisco Bay Area, California. Am J Epidemiol. 1999;150:375–89. doi: 10.1093/oxfordjournals.aje.a010017. [DOI] [PubMed] [Google Scholar]
- 10.Holly EA, Lele C. Non-Hodgkin’s lymphoma in HIV-positive and HIV-negative homosexual men in the San Francisco Bay Area: allergies, prior medication use, and sexual practices. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:211–22. doi: 10.1097/00042560-199707010-00005. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson EM, Huber AK, Akeno N, Sivak M, Li CW, Concepcion E, Ho K, Tomer Y. A CD40 Kozak sequence polymorphism and susceptibility to antibody-mediated autoimmune conditions: the role of CD40 tissue-specific expression. Genes Immun. 2007;8:205–14. doi: 10.1038/sj.gene.6364375. [DOI] [PubMed] [Google Scholar]
- 12.Park JH, Chang HS, Park CS, Jang AS, Park BL, Rhim TY, Uh ST, Kim YH, Chung IY, Shin HD. Association analysis of CD40 polymorphisms with asthma and the level of serum total IgE. Am J Respir Crit Care Med. 2007;175:775–82. doi: 10.1164/rccm.200609-1286OC. [DOI] [PubMed] [Google Scholar]
- 13.Grewal IS, Flavell RA. CD40 and CD154 in cell-mediated immunity. Annu Rev Immunol. 1998;16:111–35. doi: 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- 14.Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, Pernis A, Pasqualucci L, la-Favera R. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–92. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.