Abstract
DNA vaccines expressing the envelope (Env) of human immunodeficiency virus type 1 (HIV-1) have been relatively ineffective at generating high-titer, long-lasting immune responses. Oligomeric or trimeric (gp140) forms of Env that more closely mimic the native proteins on the virion are often more effective immunogens than monomeric (gp120) envelopes. In this study, several forms of Env constructed from the HIV-1 isolate YU-2 (HIV-1YU-2) were tested for their immunogenic potential: a trimeric form of uncleaved (−) Env stabilized with a synthetic trimer motif isolated from the fibritin (FT) protein of the T4 bacteriophage, sgp140YU-2(−/FT), was compared to sgp140YU-2(−) without a synthetic trimerization domain, as well as to monomeric gp120YU-2. DNA plasmids were constructed to express Env alone or fused to various copies of murine C3d (mC3d). BALB/c mice were vaccinated (day 1 and week 4) with DNA expressing a codon-optimized envelope gene insert, alone or fused to mC3d. Mice were subsequently boosted (week 8) with the DNA or recombinant Env protein. All mice had high anti-Env antibody titers regardless of the use of mC3d. Sera from mice vaccinated with DNA expressing non-C3d-fused trimers elicited neutralizing antibodies against homologous HIV-1YU-2 virus infection in vitro. In contrast, sera from mice inoculated with DNA expressing Env-C3d protein trimers elicited antibody that neutralized both homologous HIV-1YU-2 and heterologous HIV-1ADA, albeit at low titers. Therefore, DNA vaccines expressing trimeric envelopes coupled to mC3d, expressed in vivo from codon-optimized sequences, elicit low titers of neutralizing antibodies against primary isolates of HIV-1.
Human immunodeficiency virus type 1 (HIV-1) envelope (Env) on the native virion most likely forms a heterologous trimer (10, 22, 34, 55, 60, 62). Oligomeric or trimeric forms of Env that more closely mimic the native protein structure on the viral membrane elicit low to moderate levels of neutralizing antibodies (3, 17, 21, 35, 44, 58). The reason for these disappointing results may be due in part to the inability of these immunogens to remain as a trimer upon inoculation. However, recent attempts have been successful at producing soluble, stabilized Env trimers which contain the gp120 exterior envelope glycoprotein and the ectodomain of gp41 (22, 51, 52, 63-65, 67). Yang et al. have recently stabilized HIV-1YU-2 Env trimers by the addition of synthetic trimeric domains (63, 65). Most recently, Env glycoproteins with the trimeric motif from the T4 bacteriophage fibritin (FT) [sgp140YU-2(−/FT)] have been shown to be more stable in vitro than the previously described glycoproteins with the eukaryotic GCN4 transcription factor motif [sgp140YU-2(−/GCN4)] (65). However, both synthetic trimers exhibited similar patterns of antibody recognition to neutralizing and nonneutralizing antibodies in vitro (65). To date, only sgp140YU-2(−/GCN4) has been tested for immunogenicity and the induction of neutralizing antibodies in mice (66). Mice inoculated with gp140YU-2(−/GCN4) trimerized protein immunogens neutralized both X4- and R5-tropic HIV-1 strains (66).
Since DNA vaccines are comparatively easy to develop and manufacture and are likely to not require a cold chain for worldwide distribution, DNA vaccines provide a promising avenue for the development of new vaccination strategies. These genetic vaccinations consist of eukaryotic expression plasmids that are inoculated into target cells and translated into proteins (16). DNA vaccinations induce protective immunity against a variety of pathogens (37, 48). DNA vaccinations effectively induce both humoral and cellular immune responses to immunogens from diverse infectious agents. DNA vaccines targeting the gp120 subunit of HIV-1 Env have elicited transient antibody titers and have been less successful at generating neutralizing antibodies against HIV-1 (29, 41, 44, 47). This inability to elicit high-titer, cross-clade antibodies may be due to a variety of factors, including the long period of maturation that is required for Env-specific antibodies (11).
The poorly immunogenic nature of Env has made the development of an effective vaccine for HIV challenging. Two novel approaches may provide the ability to overcome some of the previous shortcomings of antibody-based vaccines for Env. Recent studies in our laboratory, as well as others, have shown that the fusion of C3d, a component of the innate immune system, can act as a molecular adjuvant to enhance immunogenicity (30, 31, 38, 49, 50, 57). The addition of three copies of murine C3d (mC3d) to a soluble form of the poorly immunogenic gp120 Env accelerated both the onset and the avidity maturation of antibody in vaccinated mice and enhanced neutralizing antibody titers compared to responses in mice vaccinated with antigen alone (30, 50). The precise mechanism of C3d enhancement is unclear; however, C3d may enhance signaling through CD19 after cross-linking with CD21 on the B-cell surface. Increased signaling through CD19 may increase proliferation of B cells and provide a more rapid development of germinal centers in the spleens and lymph nodes, resulting in an earlier presence of mature plasma cells (14).
Another possible mechanism for overcoming the poor immunogenicity of Env is the use of codon-optimized gene sequences in order to increase protein expression (13, 32, 59, 68). Vaccinations by DNA vaccines elicit lower levels of immune responses than vaccinations with recombinant proteins and viral vector vaccines (44, 45). DNA plasmids expressing codon-optimized gene sequences significantly increase antibody titer (>4-fold) and cellular responses compared to DNA expressing wild-type sequences (1, 6, 12, 59, 68). The use of codon-optimized DNA and codon-optimized gene inserts increases the number of immunostimulatory sequences found in the modified bacterial DNA defined as CpG motifs. The increased number of CpG motifs either in optimized plasmid DNA or in synthetic oligodeoxynucleotides directly prime T-helper cells and induce cytotoxic T-lymphocyte responses (1, 6, 12, 68). Therefore, the use of DNA expressing codon-optimized Env sequences enhances both humoral and cell-mediated responses.
In this study, mice were inoculated with three doses of DNA expressing a codon-optimized Env sequence (monomers or trimers) fused in frame to one, two, or three copies of mC3d. The anti-Env antibody responses elicited with three doses of DNA were similar to the antibody responses in animals primed with DNA (two doses) and boosted with purified recombinant Env (rEnv; one dose). Mice vaccinated with DNA expressing codon-optimized Env genes elicited higher antibody titers than those elicited with DNA expressing Env-mC3d3 from wild-type Env gene sequences. Although DNA expressing codon-optimized Env gene sequences primed the titers of anti-Env antibodies in the absence of C3d, conjugating C3d to trimerized Env did elicit antibodies that were able to neutralize heterologous virus compared to non-C3d-conjugated forms of Env.
MATERIALS AND METHODS
Plasmid DNA.
pTR600, a eukaryotic expression vector, has been described previously (49, 50). Briefly, the vector was constructed to contain the cytomegalovirus (CMV) immediate-early promoter plus intron A for initiating transcription of eukaryotic inserts and the bovine growth hormone polyadenylation [poly(A)] signal for termination of transcription. The vector contains the Col E1 origin of replication for prokaryotic replication and the kanamycin resistance gene (Kanr) for selection in antibiotic media.
The Env gene from the HIV-1 isolate YU-2 (HIV-1YU-2) was modified from the full-length, gp160 precursor to a truncated gp140 (nucleotides [nt] 1 to 1995) version with the following modifications: (i) the cleavage site between the gp120 surface protein and the gp41 transmembrane portion was altered to prevent proteolytic cleavage of the molecule (−), and (ii) the wild-type gp41 trimerization domain was replaced with a synthetic trimerization domain derived from the T4 bacteriophage FT (after amino acid residue K683), previously described (Fig. 1C) (63, 65).
FIG. 1.
Schematic representation of the gene inserts used to construct each of the vaccine plasmids. Plasmids expressing HIV-1YU-2 envelope gene inserts sgp120YU-2 (A), sgp140YU-2 (B), and sgp140YU-2(−/FT) (C), were constructed, and a representative of how the individual Envs assemble as either monomers or trimers is shown. A schematic representation of the addition of one, two, and three copies of mC3d fused to the 3′ end of each HIV-1YU-2 envelope gene insert, sgp120YU-2 (A), sgp140YU-2 (B), or sgp140YU-2(−/FT) (C), is shown. Each of the gene inserts was cloned into the previously described eukaryotic vaccine vector pTR600 (30, 50, 51). The vector contains the CMV immediate-early promoter (CMV promoter) plus intron A for efficiently initiating transcription of eukaryotic inserts and the bovine growth hormone polyadenylation terminator for efficient termination of transcription. This vector also contains the Col E1 origin of replication for prokaryotic replication as well as the kanamycin resistance (Kanr) gene for selection in antibiotic media.
The synthetic stabilized, soluble gp140YU-2(−) [sgp140YU-2(−/FT)] construct efficiently secretes trimers of sgp140(−) (65). A plasmid expressing a monomeric, soluble gp120YU-2 (sgp120YU-2) was constructed by PCR amplification of an env fragment that encodes a soluble form of gp120YU-2 (Fig. 1A). In addition, an uncleaved sgp140YU-2 without the synthetic trimer domain was constructed by the addition of a BamHI site directly 5′ to the synthetic FT domain. The plasmid was digested with BamHI and ligated without the synthetic FT domain (Fig. 1B). In each construct the first 32 amino acids have been deleted from the N terminus of the EnvYU-2 gene and replaced with a leader sequence from the trypsin plasminogen activator (tpA). In addition, a His6 tag was engineered at the C terminus of each Env gene insert to facilitate purification of recombinant antigens.
Fusion constructs were generated by the addition of one, two, and three copies of mC3d to the C terminus of each env gene insert (Fig. 1). For the addition of two and three copies of C3d, a linker domain was added between each pair of tandem copies, as previously described (30, 31, 49, 50).
The plasmids were amplified in Escherichia coli strain DH5α, purified using anion-exchange resin columns (Qiagen, Valencia, Calif.), and stored at −20°C in distilled water. Plasmids were verified by appropriate restriction enzyme digestion and gel electrophoresis. Purity of DNA preparations was determined by the OD reading at 260 and 280 nm. Each DNA vaccine inoculation contained >50 fg of endotoxin/μg per DNA inoculation.
Animals and immunizations.
Six- to 8-week-old BALB/c mice (Harlan Sprague Dawley, Indianapolis, Ind.) were used for inoculations. Mice, housed with free access to food and water, were cared for under U.S. Department of Agriculture guidelines for laboratory animals. Mice were anesthetized with 0.03 to 0.04 ml of a mixture of 5 ml of ketamine HCl (100 mg/ml) and 1 ml of xylazine (20 mg/ml). Gene gun immunizations were performed on shaved abdominal skin using the hand held Bio-Rad gene delivery system as described previously (33, 42, 43). Mice were immunized with two gene gun doses containing 1 μg of DNA per 0.5 mg of approximately 1-μm gold beads (Bio-Rad, Hercules, Calif.) at a helium pressure setting of 400 lb/in2. Protein inoculations were administrated at week 8 as a boost, in which 10 μg of recombinant protein corresponding to either sgp120YU-2, sgp140YU-2(−), or the trimerized sgp140YU-2(−/FT) was inoculated by intraperitoneal injections of 100 μl at each of two sites in a solution containing monophosphoryl-lipid A plus trehalose dicorynomycolate adjuvant (Sigma, St. Louis, Mo.).
In vitro expression of DNA vaccines.
The human embryonic kidney cell line 293T (5 × 105 cells/transfection mixture) was transfected with 2 μg of DNA using 12% Lipofectamine (Life Technologies, Grand Island, N.Y.) according to the manufacturer's guidelines. Supernatants were collected and stored at −20°C. Cell lysates were collected in 300 μl of radioimmunoprecipitation lysis buffer (0.05 M Tris-HCl [pH 8.0], 0.1% sodium dodecyl sulfate [SDS], 1.0% Triton X-100, 2 mM phenylmethylsulfonyl fluoride, 0.15 M NaCl) and stored at −20°C. Expression of vaccine constructs was quantified by coating enzyme-linked immunosorbent assay (ELISA) plates with 1.5% of the total supernatant from 293T cells transiently transfected as described above. Plates were blocked with 5% nonfat dry milk in phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBS-T) (1 h) at 25°C and extensively washed with PBS-T, and a 1:5,000 dilution of polyclonal human HIV-infected patient antiserum (HIV immunoglobulin [HIV-Ig]; National Institutes of Health [NIH] AIDS Research and Reference Reagent Program) was added (1 h) at 25°C. Plates were extensively washed, a 1:8,000 dilution of horseradish peroxidase-conjugated goat anti-human antiserum (Bio-Rad) was added (1 h) at 25°C and extensively washed, and tetramethylbenzidine (TMB) substrate (Sigma) was added (1 h) at 25°C for detection. The approximate level of expression was determined by serially diluting commercially purified gp120YU-2 (ImmunoDiagnostics, Inc., Woburn, Mass.) to each plate and extrapolating the concentration based on optical density (OD).
Purification of rEnv.
Recombinant protein corresponding to each of the envelopes was purified from supernatants of 293T cells (105) transiently transfected with plasmid DNA. Supernatants were pooled, concentrated, and equilibrated in the sodium phosphate buffer (10 mM Na2HPO4 · 2H2O, 10 mM NaH2PO4 · H2O, and 500 mM NaCl; pH 7.4) using a single-step buffer exchange spin column (VivaSpin, Inc., Edgewood, N.Y.). The samples were then purified using a HiTrap chelating nickel column (Amersham Biosciences, Piscataway, N.J.). Briefly, the samples were loaded in a total volume of 5 ml and washed with phosphate buffer containing 10 mM imidazole. The samples were eluted by using a gradient from 50 to 300 mM imidazole in phosphate buffer. The fractions were evaluated for purity after visualization by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and quantified using a bicinchoninic acid protein determination kit (Pierce Biotechnology, Inc., Rockford, Ill.).
Virus production and infection.
HIV-1YU-2 was produced in transiently transfected 293T cells as described above. Briefly, 293T cells (105) were transiently transfected with 2 μg of the HIV-1YU-2 proviral DNA plasmid for 48 h. The viral supernatants were harvested, pooled, and aliquoted, and viral infectivity of the HIV-1YU-2 supernatants was determined using increasing concentrations of supernatant to determine the appropriate amount of virus needed (200 μl). The infectivity of HIV-1ADA (NIH AIDS Research and Reference Reagent Program) (26-28, 61) viral stocks was determined using increasing concentrations of virus to determine the amount of virus needed (1,300 50% tissue culture infective doses). Infection of both viruses was performed using GHOST (3) Hi-5 cells (GHOST R5) (NIH AIDS Research and Reference Reagent Program), expressing human CD4 (hCD4) and human CCR5 (hCCR5) containing the HIV-2 LTR-GFP reporter (40). GHOST R5 cells were cultured in Dulbecco's modified Eagle's medium (90%) and supplemented with 10% fetal calf serum, G418-sulfate (500 μg/ml), hygromycin (100 μg/ml), puromycin (1 μg/ml), and penicillin-streptomycin. Infectivity was based on the ability to infect the maximal number of cells in vitro without inducing cell death. GHOST R5 cells were thawed and passaged (two times) and then plated (6.5 × 104 cells) in a 24-well plate. Virus was incubated with cells in complete, nonselective medium (1 to 3 h) at 37°C in the presence of Polybrene (Sigma) (20 μg/ml) in order to enhance infectivity. Cells were harvested 48 h later by aspirating the culture medium, washing (once) with 1 ml of PBS, and gently shaking in EDTA (5 mM)-PBS (15 min) at 25°C. Dislodged cells were placed in paraformaldehyde (4%). Cells were fixed (1 h), pelleted, resuspended in PBS supplemented with 2% fetal calf serum, and analyzed by FACScan using Cell Quest software (Becton Dickinson, Mountain View, Calif.).
Expression analysis.
For determination of efficient expression of each DNA vaccine plasmid, Western blot analysis was performed. Supernatant (1.5%) from transfected 293T cells was diluted in SDS sample buffer (Bio-Rad) and loaded onto an SDS-polyacrylamide gel (6%) under either (i) denaturing (0.1% SDS)-reducing (5% β-mercaptoethanol [BME]) conditions, (ii) denaturing (0.1% SDS)-nonreducing (0% BME) conditions, or (iii) nondenaturing (0% SDS)-nonreducing (0% BME) conditions. The resolved proteins were transferred onto a nitrocellulose membrane (Bio-Rad) and incubated with a 1:5,000 dilution of HIV-Ig (NIH AIDS Research and Reference Reagent Program) in PBS-T containing 5% nonfat dry milk. After extensive washing, bound human antibodies were detected using a 1:8,000 dilution of horseradish peroxidase-conjugated goat anti-human antiserum (Bio-Rad) and enhanced chemiluminescence (Pierce Biotechnology, Inc.).
ELISA for detection of anti-Env antibodies.
An endpoint ELISA was performed to assess the titers of anti-Env IgG in immune-phase serum using recombinant HIV-1 gp120YU-2 to coat plates as described previously (45). Briefly, plates were coated (25 ng) overnight at 4°C, blocked with 5% nonfat dry milk in PBS-T (1 h) at 25°C, and extensively washed with PBS-T, serial dilutions of mouse antisera from vaccinated mice were allowed to bind (1 h), and the plates were thoroughly washed. Subsequently, the primary antisera were detected by anti-mouse IgG conjugated to horseradish peroxidase (Bio-Rad). The reaction was detected using TMB substrate (Sigma) (1 h) at 25°C. Endpoint titers that were twofold higher than those of vector-only control sera were considered positive.
In vitro neutralization.
Virus neutralization by antisera was determined as follows: virus was mixed with antisera from naïve or vaccinated mice (1 h) at 37°C, and the virus-cell mixture was added to GHOST R5 cells in a total volume of 250 μl containing 20 μg of Polybrene/ml. The neutralizing capacity of antisera was measured by comparing the reduction in the number of infected cells per sample to (i) prebleed sera and (ii) sera from age-matched naïve mice. Inhibition of virus was assessed by the additional reduction in infectivity beyond the background of naïve-mouse and prebleed antisera. Inhibition was measured by 50% inhibition of virus infection (beyond nonspecific inhibition). Samples were run in duplicate assays on the same day and averaged, and data are presented as the average per group.
Statistics.
For statistical analysis, Student's t test was employed. The difference between DNA vaccines expressing Env fused to multiple copies of mC3d was compared to DNA expressing Env alone. Differences in titers were considered statistically significant when the P value was less than 0.05.
RESULTS
Construction and in vitro expression.
Vaccine plasmids expressing secreted forms of Env (monomeric or oligomeric) from HIV-1YU-2 were constructed using the previously described pTR600 vector (Fig. 1) (49, 50). DNA plasmids expressing a soluble gp120YU-2 env gene segment (nt 1 to 1452) encode the entire surface domain but exclude the oligomerization, transmembrane, and cytoplasmic regions of the Env protein. DNA expressing the sgp140YU-2(−) gene segment (nt 1 to 1995) encoded the entire gp120 surface domain of Env along with the ectodomain of gp41, excluding the transmembrane domain. In addition, the proteolytic cleavage site between the gp120 surface protein and the gp41 portion was mutated (nt 1478 and 1487) to prevent proteolytic cleavage of the molecule. Trimerized sgp140YU-2(−) proteins were constructed by the addition of the T4 bacteriophage FT gene sequence at the 3′ end of the env gene that encodes a trimerization motif (nt 2012 to 2105). A second set of DNA vaccines was constructed that encoded either sgp120YU-2, sgp140YU-2(−), or sgp140YU-2(−/FT) fused to one, two, or three copies of mC3d at the carboxyl terminus (Fig. 1).
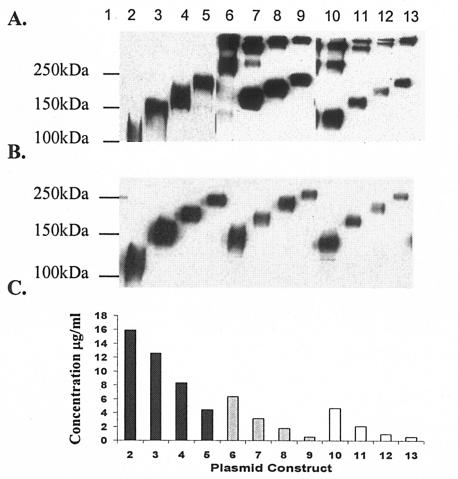
Each DNA plasmid encoding either Env or Env-C3d efficiently expressed proteins that were secreted from transfected cells (Fig. 2). Multiple species of Env were observed in the supernatant of cells transfected with DNA vaccines expressing sgp140YU-2(−) proteins without a synthetic trimerization domain. Soluble gp140YU-2(−) migrated predominately as dimers and higher-molecular-weight aggregates (Fig. 2A, lane 6). The addition of multiple copies of C3d to the carboxyl terminus of sgp140YU-2(−) resulted in the production of predominately monomers and dimers (Fig. 2A, lanes 7 to 9). In contrast, the addition of the synthetic FT trimerization domain to the sgp140YU-2(−) carboxyl terminus stabilized the protein as trimers (Fig. 2A, lane 10). The appearance of monomers and dimers was due to the boiling and denaturing conditions, as confirmed by native polyacrylamide gels (data not shown). In contrast to the non-FT-stabilized sgp140YU-2(−), the addition of multiple copies of mC3d to sgp140YU-2(−/FT) did not destabilize the Env trimerization (Fig. 2A, lanes 11 to 13). However, under reducing conditions, both stabilized and unstabilized sgp140YU-2(−) proteins migrated as monomers (Fig. 2B).
FIG. 2.
Expression of vaccine plasmids was visualized by Western blot analysis. Nonreducing (A) and reducing (B) SDS-PAGE analysis demonstrated that all constructs are efficiently expressed in vitro and FT-stabilized versions of sgp140YU-2(−) form trimers even after boiling and under denaturing conditions with and without the addition of one, two, or three copies of mC3d. Human embryonic kidney cells were transiently transfected with 2 μg of plasmid DNA from each vaccine plasmid. Supernatants were collected, and 1.5% of the total volume was subjected to electrophoresis. Each blot was probed with HIV-Ig followed by anti-human IgG conjugated to horseradish peroxidase and detected with enhanced chemiluminescence. (C) Expression ELISA data to determine the relative level of expression of codon-optimized DNA vaccine plasmids were quantified using 1.5% of the total supernatant from transiently transfected 293T cells and comparing relative amounts to recombinant gp120YU-2 (ImmunoDiagnostics) as a standard and are expressed as micrograms per milliliter of Env secreted into the supernatant. Lane 1, molecular mass marker; lane 2, sgp120YU-2; lane 3, sgp120YU-2 mC3d1; lane 4, sgp120YU-2-mC3d2; lane 5, sgp120YU-2-mC3d3; lane 6, sgp140YU-2(−); lane 7, sgp140YU-2(−)-mC3d1; lane 8, sgp140YU-2(−)-mC3d2; lane 9, sgp140YU-2(−)-mC3d3; lane 10, sgp140YU-2(−/FT); lane 11, sgp140YU-2(−/FT)-mC3d1; lane 12, sgp140YU-2(−/FT)-mC3d2; lane 13, sgp140YU-2(−/FT)-mC3d3.
DNA plasmids expressing lower-molecular-weight proteins expressed higher concentrations of Env (Fig. 2C). DNA plasmids expressing sgp120YU-2 had higher protein expression (16 μg/ml) than DNA expressing sgp140YU-2(−) (4 to 6 μg/ml). DNA expressing EnvYU-2 from wild-type gene sequences had a ∼100-fold decrease in protein expression (data not shown) compared to Env expressed from codon-optimized sequences; however, similar to Env-C3d expressed from wild-type genes, conjugating C3d to any of the Envs (monomers or oligomers) progressively reduced protein expression (Fig. 2C).
Purification of envelope proteins.
Envelope proteins were purified and samples were analyzed under denaturing conditions. Recombinant proteins were subjected to boiling conditions (100°C) and resolved by SDS-6% PAGE. Under these conditions, sgp140YU-2(−) oligomers were destabilized, and therefore both higher- and lower-molecular-weight forms were detected; however, the addition of the synthetic stabilization motif FT resulted in the secretion of soluble, higher-molecular-weight Env proteins, as previously described (data not shown).
Each purified protein maintained a native structure. Purified proteins were screened against a panel of anti-Env monoclonal antibodies (data not shown). Monoclonal antibodies (IgG1b12, IgG2G12, and F105), which recognize epitopes in the gp120 surface protein, efficiently detected each purified Env protein [sgp120YU-2, sgp140YU-2(−), and sgp140YU-2(−/FT)]. The monoclonal antibody 2F5, which maps to a linear sequence in gp41, recognized sgp140YU-2(−) and sgp140YU-2(−/FT) but did not detect purified sgp120YU-2 (which lacks the gp41 ectodomain). Therefore, each Env protein was expressed and purified in a native conformation similar to the gp160 glycoprotein on the virion surface.
Envelope monomers elicit similar high-titer anti-Env antibody as envelope oligomers.
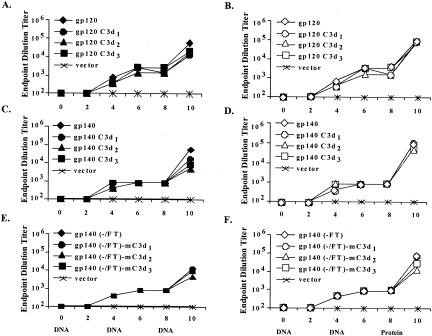
BALB/c mice were vaccinated on day 1 and boosted at week 4 with gold particles coated with DNA expressing codon-optimized env gene inserts (2 μg) and administered via gene gun. Anti-Env antibodies were present (<1:103) in sera from mice vaccinated with DNA plasmids expressing any of the Env proteins (monomers or oligomers) at week 4 (Fig. 3). Mice vaccinated with the pTR600 vector elicited no anti-Env antibody. Mice primed with DNA expressing codon-optimized gene inserts elicited significantly higher titer antibodies to Env with fewer inoculations compared to wild-type gene inserts (30, 50) (data not shown). After the second inoculation, the titer of anti-Env antibodies was boosted (>1:103) in all mice vaccinated with plasmids expressing any Env protein.
FIG. 3.
Anti-Env IgG raised by sgp120YU-2, sgp140YU-2(−), and sgp140YU-2(-/FT) inoculations was determined by an endpoint dilution titer. BALB/c mice (n = 10 per group) were primed at day 0 with each vaccine construct indicated, sgp120YU-2, sgp140YU-2(−), or sgp140YU-2(−/FT) with or without the addition of one, two, or three copies of mC3d, using 2 μg of DNA inoculated via gene gun and boosting at week 4 with an additional 2 μg of DNA via gene gun. At week 8, each group was split into two and either received a third inoculation of 2 μg of DNA or was boosted with 10 μg of recombinant protein via intraperitoneal injection at two sites of 100 μl with RIBI (Sigma) as adjuvant. Solid symbols represent mice receiving DNA as a third inoculation (A, C, and E); open symbols represent mice receiving recombinant protein as a third inoculation (B, D, and F). Sera were collected prior to and 2 weeks after each inoculation as indicated, and each mouse was assayed for specific IgG levels by ELISA. The 96-well plates were coated with 25 ng of gp120YU-2 recombinant protein. Data are represented as the average of five mice. Preimmune-phase sera from mice had no detectable specific IgG. Endpoint dilution titers were determined by diluting the sera until OD values reached background levels. As shown, all of the constructs elicited similar high-titer Env-specific antibodies regardless of the vaccination regimen used.
At week 8, mice were divided into two groups: one group was given a third inoculation of codon-optimized DNA plasmid (2 μg), and the second group was administered purified, rEnv protein (10 μg). Mice vaccinated with DNA expressing sgp120YU-2 were boosted with Envgp120, and mice vaccinated with DNA expressing sgp140YU-2(−) were boosted with Envgp140. Mice vaccinated with a third inoculation of DNA or boosted with recombinant protein had similar anti-Env titers at week 10 (titers ≥ 1:25,000) (Fig. 3A to D). Mice vaccinated with DNA expressing sgp120YU-2 and boosted with DNA or Env protein at week 8 had anti-Env titers greater than 1:105 (Fig. 3A and B). Overall, monomers of Env appeared to elicit anti-Env titers similar to those elicited by oligomers of Env (compare Fig. 3A to D). In addition, mice vaccinated with DNA expressing codon-optimized env gene inserts elicited the same high-titer anti-Env antibodies as rEnv protein (three vaccinations) (data not shown).
sgp140 stabilized by (−/FT) elicits lower levels of anti-Env antibodies.
Mice vaccinated with DNA expressing sgp140YU-2(−/FT) had consistently lower titers of anti-Env antibodies than mice vaccinated with nonstabilized sgp140YU-2(−) (Fig. 3E). After three inoculations of DNA, sera from mice vaccinated with nonstabilized sgp140YU-2(−) DNA had anti-Env IgG titers that were 1 log higher (1:105 compared to ≤1:104) than sera from mice inoculated with stabilized sgp140YU-2(−) DNA. At week 8, half of the mice previously inoculated with DNA plasmids (day 1 and week 4) were vaccinated with trimerized recombinant sgp140YU-2(−) protein. Sera from these mice had higher titers of antibodies to Env than sera from mice vaccinated with three inoculations of DNA (Fig. 3F).
Multiple copies of mC3d elicit similar high-titer anti-Env antibodies.
A second set of DNA vaccine plasmids was constructed to express Env proteins fused to various copies of mC3d (Fig. 3). Mice were vaccinated with DNA plasmids expressing either sgp120YU-2 or sgp140YU-2(−) fused to one, two, or three copies of mC3d. After the third inoculation, DNA vaccines expressing sgp140YU-2(−)-mC3d2 elicited the lowest titers of anti-Env antibodies (∼1:6,400) compared to DNA vaccines expressing sgp140YU-2(−) (>1:104) (Fig. 3C). However, mice primed (day 1 and week 4) with DNA expressing sgp140YU-2(−) or sgp140YU-2(−) conjugated to mC3d (one, two, or three copies) and then boosted with recombinant gp140YU-2(−) had similar levels of anti-Env antibodies (>1:25,000) at week 10 regardless of the DNA used to prime the mice (Fig. 3D). In contrast, sera from mice vaccinated with DNA expressing sgp120YU-2 conjugated to mC3d and then boosted with either the same DNA plasmid or recombinant sgp120YU-2 had similar high-titer anti-Env antibody at week 10 (Fig. 3A and B). These results indicate that the relative expression of Env-C3d vaccines is directly related to the titer of anti-Env antibody raised in these mice. Mice primed with DNA expressing Env-C3d proteins were boosted to titers greater than 1:50,000 by the administration of a single boost of rEnv protein, regardless of the number of copies of mC3d fused to monomeric forms of Env (Fig. 3B).
A third set of DNA vaccines was constructed to express sgp140YU-2(−), stabilized by FT, and fused to one, two, or three copies of mC3d. The expressed proteins resulted in a trimerized sgp140YU-2(−) complex fused to three, six, or nine copies of mC3d upon trimerization (Fig. 1C). Mice vaccinated with DNA expressing sgp140 YU-2(−/FT)-mC3d elicited lower titers of anti-Env antibody than sgp140YU-2(−)-mC3d DNA. However, once again, boosting mice with trimerized sgp140YU-2(−) recombinant protein enhanced the levels of antibodies to similar anti-Env titers as in mice vaccinated with sgp140YU-2(−) DNA.
Neutralizing antibodies elicited by Env oligomers conjugated to multiple copies of C3d.
Sera from vaccinated mice (week 10) were tested for the ability to neutralize homologous HIV-1YU-2 in vitro. Mice primed with DNA expressing sgp120YU-2-mC3d3 and boosted with sgp120YU-2 protein had antibodies that neutralized viral infection of permissive cells (1:20 dilution); however, sera from mice vaccinated with three doses of DNA plasmid expressing these same proteins had little neutralizing antibody (Table 1). In contrast, neutralizing antibodies were elicited in mice vaccinated with DNA expressing nonstabilized sgp140YU-2(−)-mC3d3 (1:20) and then boosted with either the same DNA plasmid or recombinant sgp140YU-2(−) protein. However, mice vaccinated with any of the other sgp140YU-2(−) constructs (sgp140YU-2(−), sgp140 YU-2(−)-mC3d1, or sgp140 YU-2(−)-mC3d2) had no detectable neutralizing antibodies, regardless of whether the mice were boosted at week 8 with DNA plasmid or recombinant protein (Table 1). Interestingly, mice vaccinated with DNA expressing the trimerized sgp140YU-2(−/FT) proteins, with or without the addition of mC3d, and then boosted with either trimerized recombinant sgp140YU-2(−) or DNA expressing sgp140YU-2(−/FT) with or without mC3d had detectable neutralizing antibodies (1:20). In addition, sera from mice primed with DNA expressing sgp140YU-2(−/FT)-mC3d2 or sgp140YU-2(−/FT)-mC3d3 and boosted with trimerized recombinant sgp140YU-2(−) neutralized viral infection at a dilution of 1:40 (Table 1). Mice vaccinated with three inoculations of DNA expressing sgp140YU-2(−/FT)-mC3d1 also inhibited HIV-1YU-2 (1:40). Therefore, all DNA vaccine plasmids expressing stabilized trimers of sgp140YU-2(−/FT) with and without the addition of one, two, and three copies of mC3d elicited neutralizing antibodies (1:20).
TABLE 1.
Neutralizing antibody titers in vaccinated mice
| Vaccine plasmid | Antibody titer toa:
|
|||
|---|---|---|---|---|
| HIV-1YU-2
|
HIV-1ADA
|
|||
| DNA boost | Protein boost | DNA boost | Protein boost | |
| gp120YU-2 | — | — | — | — |
| gp120YU-2 C3d1 | — | — | — | — |
| gp120YU-2 C3d2 | — | — | — | — |
| gp120YU-2 C3d3 | — | 1:20 | — | — |
| gp140YU-2(−) | — | — | — | — |
| gp140YU-2(−)-mC3d1 | — | — | — | — |
| gp140YU-2(−)-mC3d2 | — | — | — | — |
| gp140YU-2(−)-mC3d3 | 1:20 | 1:20 | — | — |
| gp140(−/FT) | 1:20 | 1:20 | — | — |
| gp140(−/FT)-mC3d1 | 1:40 | 1:20 | — | — |
| gp140(−/FT)-mC3d2 | 1:20 | 1:40 | — | 1:20 |
| gp140(−/FT)-mC3d3 | 1:20 | 1:40 | — | 1:20 |
Sera collected at week 10 were analyzed for the presence of neutralizing antibodies against homologous HIV-1YU-2 virus and heterologous HIV-1ADA virus at dilutions of 1:20 and 1:40 for the ability to inhibit 50% of virus infection. Inhibition of virus was assessed by the additional reduction in infectivity beyond the background of naïve-mouse and prebleed sera. Samples were run in duplicate, and data are presented per group, where the ability to inhibit 50% of infection at the indicated dilution is shown. A − indicates the lack of inhibition of infection at the lowest dilution of serum.
Sera from vaccinated mice (week 10) were also tested for the ability to neutralize heterologous HIV-1ADA in vitro. Only mice primed with DNA expressing sgp140YU-2-mC3d2 and sgp140YU-2-mC3d3 and boosted with trimerized recombinant sgp140YU-2(−) protein had antibodies that neutralized viral infection of permissive cells (1:20) (Table 1).
DISCUSSION
The development of an effective vaccine against HIV-1 has been challenging due to the poorly immunogenic nature of Env. Inoculations of the gp120 subunit of Env elicit transient titers of neutralizing antibody (53). Oligomeric or trimeric forms of Env, which more closely mimic the native protein on the virion, elicit low to moderate levels of antibodies that neutralize viral infection (2, 9, 15, 21, 44, 45, 58, 66). These oligomeric forms often do not remain as distinct trimers following purification or administration in vivo; however, they are more effective immunogens than monomeric forms of Env (4, 9, 21, 52, 58, 66). Several strategies have been employed to stabilize Envgp140 in a trimer conformation (22, 51, 63-65, 67). Env molecules, stabilized using synthetic trimerization domains, induce antibodies that neutralize both R5 and X4 HIV-1 isolates (66).
Previous studies from our laboratory demonstrated that the levels of neutralizing antibodies to HIV-1 can be enhanced using C3d fused to monomeric gp120 (sgp120-mC3d) (30, 50). Rodents inoculated with DNA vaccines expressing HIV-1IIIB gp120 fused to three copies of murine or human C3d had enhanced anti-Env IgG titers and accelerated affinity maturation of antibody, as well as higher levels of neutralizing antibodies compared to that with Env DNA (27). The goal of this study was to determine if C3d, acting as a molecular adjuvant, could improve the neutralizing antibodies elicited by codon-optimized gene sequences expressing oligomeric envelopes.
In this study, DNA plasmids expressing codon-optimized gene sequences encoding sgp120YU-2, sgp140YU-2, and sgp140YU-2(−/FT) alone or fused to mC3d were all efficiently secreted from cells in vitro (Fig. 2). The conjugation of increasing numbers of mC3d gene segments led to a progressive decrease in the overall amount of protein secreted into the supernatant of transfected cells compared to non-C3d-fused Envs (Fig. 2C) (30, 31, 50). DNA plasmids expressing sgp140YU-2(−) without the synthetic trimerization domain were secreted predominately as dimers and higher-molecular-weight aggregates (Fig. 2A), as previously described for uncleaved Envgp140 proteins (4, 17, 51, 52). The addition of mC3d shifted the formation of Env to monomers and dimers (Fig. 2A). In contrast, plasmids expressing sgp140YU-2(−), stabilized by the addition of FT, were efficiently secreted as trimers of Env (Fig. 2A). Unlike DNA expressing sgp140YU-2(−), the addition of mC3d to sgp140YU-2(−/FT) did not dissociate Env trimers (Fig. 2A). This lack of dissociation is due to the addition of the FT carboxyl-terminal domain that stabilizes triple-helical coiled-coil proteins (25, 56, 66). Residues within each of the three carboxyl-terminal FT domains exert extensive hydrophobic and polar intersubunit interactions that result in trimerization (56).
Trimerized HIV-1 envelopes appear to confer a conformation that more closely mimics the envelope structure on the native virion, which elicits conformationally induced antibodies (7, 17). Previously, oligomers or trimers of Env were constructed to express envelopes with a mutated proteolytic cleavage between gp120/gp41 (7, 17-20, 54, 63-65, 67) or the addition of disulfide bonds (4, 51, 52). Stabilization of uncleaved oligomers of gp140YU-2 by the addition of synthetic trimerization domains to gp41 formed soluble, stable trimers of Env under harsh conditions, such as high temperatures (boiling at 100°C), denaturing conditions, or during the purification process (63-65).
DNA vaccine plasmids expressing codon-optimized env genes alone or conjugated to C3d all elicited high-titer anti-Env antibodies (Fig. 3). Several studies have demonstrated that viral proteins, such as those encoded by HIV-1 env or gag, display a distinct codon bias that results in moderate to low protein expression compared to eukaryotic genes (1, 12, 32, 59, 68). Wild-type env sequences have a preference for adenosine (A) and thymine (T) in the wobble position of each codon, whereas highly expressed mammalian genes preferably encode a guanine (G) or cytosine (C) at this position (1, 32, 59). Construction of synthetic sequences encoding Env that substitute a G or C for an A or T at the wobble position dramatically increases protein expression, which for HIV genes is Rev independent (1, 32, 59). Compared to wild-type sequences expressing Env, codon-optimized sequences expressing Env have the same level of expression using 100-fold less DNA (data not shown).
Previously it has been reported that C3d, when conjugated to an antigen, acts as a molecular adjuvant to enhance poorly immunogenic molecules, such as gp120, when expressed from plasmid DNA in vivo (30, 31, 50). Mice vaccinated with DNA expressing wild-type env sequences (gp120) had little to no anti-Env antibodies (30, 31, 50). However, the addition of three copies of mC3d to Env increased anti-Env antibody titers (1:103 to 1:104). In contrast, plasmids expressing codon-optimized env sequences elicited high-titer anti-Env antibodies, regardless of whether Env was conjugated to C3d (Fig. 3). Codon-optimized sgp120 sequences expressed approximately 20-fold more protein than wild-type sequences (Fig. 2) (30, 50), which directly correlated with an increase in antibody response directed against the envelope (1). The enhancement effect of C3d was observed in mice vaccinated with a reduced dose (0.1 μg) of DNA expressing codon-optimized env sequences. Mice vaccinated with three doses (0.1 μg) of DNA expressing codon-optimized sgp120 elicited antibody titers between 1:100 and 1:1,000; however, mice primed with three doses of DNA expressing codon-optimized sgp120-mC3d3 (0.1 μg) elicited high-titer antibody (>1:104) (unpublished observations). Therefore, both codon optimization of env sequences and C3d conjugation to Env appear to enhance anti-Env antibodies in an independent, but not in a synergistic, manner.
The binding of C3d to CR2 (CD21) on the surface of circulating B cells is one possible mechanism for the enhancement of antibody responses by these C3d-conjugated vaccines (23). Direct stimulation of B cells by C3d leads to a cross-linking of the CR2 with the signaling receptor, CD19 (14, 23, 24). Antigen-specific B cells then become activated and are directly stimulated to produce antibody (14, 23, 24). DNA expressing Env-C3d most likely broadened the height of antibody responses by (i) directly stimulating antibody-producing B cells through CR2 (CD21) and expanding the pool of anti-Env-specific B cells (23, 24); (ii) reducing B-cell apoptosis (36, 46); or (iii) lowering with C3d-conjugated antigen both the concentration threshold and the affinity threshold for B-cell activation (8, 39).
Interestingly, even though similar levels of anti-Env antibodies were elicited in mice vaccinated with plasmids expressing codon-optimized sgp120YU-2 alone or conjugated to C3d, only sera from mice primed with DNA expressing sgp120YU-2-mC3d3 and boosted with rEnv had antibodies that neutralized HIV-1YU-2 viral infection (1:20). These data are in agreement with previous studies in which only mice vaccinated with plasmids expressing monomeric gp120 fused to mC3d had high levels of neutralizing antibodies against homologous virus (30). The present study extends our previous research by demonstrating that multiple copies of mC3d fused to monomeric gp120 from a primary isolate of HIV-1 (HIV-1YU-2) are able to improve neutralizing antibody titers compared to gp120 alone.
Previous studies have suggested that oligomers or trimers of Env may be more effective immunogens than monomeric rEnv (9, 21, 44, 45, 66). This study is the first to examine the use of oligomeric forms of Env fused to mC3d in order to elicit neutralizing antibodies against HIV-1. Unlike DNA plasmids expressing sgp120YU-2-mC3d3 (three doses), mice vaccinated with DNA expressing sgp140YU-2(−)-mC3d3 (three doses) had antibodies that neutralized virus infection (1:20 dilution). The ability of antibodies from mice vaccinated with sgp140YU-2(−)-mC3d3 to neutralize homologous virus is unclear; however, it may be due to the oligomerization of the uncleaved sgp140(−) portion of the sgp140YU-2(−)-mC3d3 molecule. In contrast to DNA expressing monomeric non-C3d forms of Env, plasmids expressing stabilized trimers of sgp140YU-2(−/FT) elicited antibodies that neutralized homologous viral infection (Table 1). The addition of C3d to these stabilized Env immunogens broadened the neutralizing capacity of elicited antibodies to both homologous (HIV-1YU-2) and heterologous (HIV-1ADA) isolates, but only after a single dose of rEnv (Table 1).
DNA priming followed by boosting with recombinant protein is an effective strategy for eliciting high-titer neutralizing antibody (44). Richmond et al. reported significant increases in antibody titer, avidity, and neutralizing antibodies after protein boosting of animals primed with DNA expressing wild-type Env sequences (44). However, in their study, inoculation of three doses of DNA expressing Env from wild-type gene sequences elicited poor antibody responses and, therefore, a robust antibody response was observed following two subsequent recombinant protein boosts (44). In contrast, priming with two doses of DNA expressing codon-optimized env genes followed by recombinant protein boost provided a similar neutralizing titer as that in mice vaccinated with three doses of DNA expressing codon-optimized env genes (Table 1) and, therefore, the effect of the protein boost was not readily observed.
Interestingly, however, in this study increasing numbers of C3d molecules conjugated to sgp140YU-2(−/FT) did not enhance the titers of neutralizing antibody in sera collected from mice vaccinated with only DNA plasmid alone (Table 1). The addition of one, two, or three copies of the gene encoding mC3d resulted in three, six, or nine copies of the mC3d protein conjugated to the stabilized sgp140YU-2(-/FT) molecule (Fig. 1). One possible explanation for the lack of correlation between neutralizing antibody and copies of C3d may be that the terminal C3d molecule of each fusion protein is interacting with CR2 on the surface of B cells. Therefore, even though plasmids encoding sgp140YU-2(−/FT)-mC3d3 have three times the number of mC3d proteins (nine molecules) in the expressed protein compared to sgp140 (−/FT)-mC3d1 (three molecules), only the terminal C3d protein of each molecule may be able to bind to the cell surface receptors. This hypothesis may account for the similar titers of neutralizing antibody elicited by Env monomers [sgp120YU-2 or sgp140YU-2(−)] conjugated to mC3d3 and sgp140YU-2(−/FT)-mC3d1, both fused with three copies of mC3d (Table 1).
Overall, priming with DNA expressing trimerized HIV-1 Env from a codon-optimized gene insert fused to multiple copies of C3d, followed by a recombinant trimerized protein boost, can elicit similar high-titer neutralizing antibodies in vaccinated mice as with purified recombinant trimers of these same immunogens (66). Note, however, that while immunization with recombinant trimers of Env yielded antibodies that neutralized heterologous viruses (66), non-C3d-conjugated Envs expressed from DNA were unable to elicit neutralizing antibodies. The levels of neutralizing antibodies seen in the present study were disappointingly low and lacked the significant enhancement effect by C3d that was shown in previous studies (30, 36a, 50). Future studies should use immunogens that are more likely to elicit neutralizing antibodies and, therefore, the enhancement effect by C3d will be more pronounced. A variety of Env immunogens have been shown to elicit neutralizing antibodies against HIV, including (i) the use of modified envelopes, such as deletions in variable loop domains or deglycosylation to expose cryptic sites (2); (ii) a selective mutation(s) in gp41 that enhances neutralizing sensitivity; or (iii) the use of envelopes that elicit cross-clade neutralizing antibodies (15).
The results suggest that other immune responses may be activated by C3d-conjugated antigens. C3d enhances antigen presentation of processed antigens by B lymphocytes (5) and, therefore, in Env-C3d vaccines Env may be more efficiently presented by B cells to T-helper cells. In addition, C3d can activate T cells expressing both major histocompatibility complex class I and II molecules in an antigen-dependent manner (38). Also, recently Liu et al. showed that higher levels of anti-Env gamma interferon-secreting T cells could be elicited by Env-C3d fusion proteins (36a). Therefore, examination of the mechanism of both humoral and cellular immune enhancement mediated by C3d would be quite useful for both understanding the function of complement as well as developing more effective vaccines.
Acknowledgments
This research was supported by National Institutes of Health grant awards AI44325 to T.M.R., A157630 to J.F.B., and AI31783 and AI44328 and by the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, and the late William F. McCarty-Cooper to J.S.
We thank Douglas Fearon for supplying the mC3d constructs. We thank Tom Green and Edwin Franklin for technical assistance. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH: GHOST (3) Hi-5 from Vineet N. KewalRamani and Dan R. Littman; HIV-1Ada-M from Howard Gendelman; and HIV-Ig from NABI and NHLBI.
REFERENCES
- 1.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, P. W., T. J. Gregory, L. Riddle, G. R. Nakamura, M. A. Champe, J. P. Porter, F. M. Wurm, R. D. Hershberg, E. K. Cobb, and J. W. Eichberg. 1990. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature 345:622-625. [DOI] [PubMed] [Google Scholar]
- 4.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boackle, S. A., V. M. Holers, and D. R. Karp. 1997. CD21 augments antigen presentation in immune individuals. Eur. J. Immunol. 27:122-129. [DOI] [PubMed] [Google Scholar]
- 6.Bojak, A., J. Wild, L. Deml, and R. Wagner. 2002. Impact of codon usage modification on T cell immunogenicity and longevity of HIV-1 gag-specific DNA vaccines. Intervirology 45:275-286. [DOI] [PubMed] [Google Scholar]
- 7.Broder, C. C., P. L. Earl, D. Long, S. T. Abedon, B. Moss, and R. W. Doms. 1994. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc. Natl. Acad. Sci. USA 91:11699-11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, R. H., M. O. Spycher, Y. C. Ng, R. Hoffman, and D. T. Fearon. 1988. Synergistic interaction between complement receptor type 2 and membrane IgM on B lymphocytes. J. Immunol. 141:457-463. [PubMed] [Google Scholar]
- 9.Chakrabarti, B. K., W. P. Kong, B. Y. Wu, Z. Y. Yang, J. Friborg, X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 11.Cole, K. S., J. L. Rowles, B. A. Jagerski, M. Murphey-Corb, T. Unangst, J. E. Clements, J. Robinson, M. S. Wyand, R. C. Desrosiers, and R. C. Montelaro. 1997. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J. Virol. 71:5069-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deml, L., A. Bojak, S. Steck, M. Graf, J. Wild, R. Schirmbeck, H. Wolf, and R. Wagner. 2001. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J. Virol. 75:10991-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deml, L., R. Schirmbeck, J. Reimann, H. Wolf, and R. Wagner. 1999. Immunostimulatory CpG motifs trigger a T helper-1 immune response to human immunodeficiency virus type-1 (HIV-1) gp160 envelope proteins. Clin. Chem. Lab. Med. 37:199-204. [DOI] [PubMed] [Google Scholar]
- 14.Dempsey, P. W., M. E. Allison, S. Akkaraju, C. C. Goodnow, and D. T. Fearon. 1996. C3d of complement as a molecular adjuvant: bridging innate and acquired immunity. Science 271:348-350. [DOI] [PubMed] [Google Scholar]
- 15.Dong, M., P. F. Zhang, F. Grieder, J. Lee, G. Krishnamurthy, T. VanCott, C. Broder, V. R. Polonis, X. F. Yu, Y. Shao, D. Faix, P. Valente, and G. V. Quinnan, Jr. 2003. Induction of primary virus-cross-reactive human immunodeficiency virus type 1-neutralizing antibodies in small animals by using an alphavirus-derived in vivo expression system. J. Virol. 77:3119-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnelly, J. J., J. B. Ulmer, and M. A. Liu. 1998. DNA vaccines. Dev. Biol. Stand. 95:43-53. [PubMed] [Google Scholar]
- 17.Earl, P. L., C. C. Broder, D. Long, S. A. Lee, J. Peterson, S. Chakrabarti, R. W. Doms, and B. Moss. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earl, P. L., R. W. Doms, and B. Moss. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earl, P. L., S. Koenig, and B. Moss. 1991. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J. Virol. 65:31-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earl, P. L., and B. Moss. 1993. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res. Hum. Retrovir. 9:589-594. [DOI] [PubMed] [Google Scholar]
- 21.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farzan, M., H. Choe, E. Desjardins, Y. Sun, J. Kuhn, J. Cao, D. Archambault, P. Kolchinsky, M. Koch, R. Wyatt, and J. Sodroski. 1998. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 72:7620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fearon, D. T. 1998. The complement system and adaptive immunity. Semin. Immunol. 10:355-361. [DOI] [PubMed] [Google Scholar]
- 24.Fearon, D. T., and R. H. Carter. 1995. The CD19/CR2/TAPA-1 complex of B lymphocytes: linking natural to acquired immunity. Annu. Rev. Immunol. 13:127-149. [DOI] [PubMed] [Google Scholar]
- 25.Frank, S., R. A. Kammerer, D. Mechling, T. Schulthess, R. Landwehr, J. Bann, Y. Guo, A. Lustig, H. P. Bachinger, and J. Engel. 2001. Stabilization of short collagen-like triple helices by protein engineering. J. Mol. Biol. 308:1081-1089. [DOI] [PubMed] [Google Scholar]
- 26.Gendelman, H. E., L. M. Baca, C. A. Kubrak, P. Genis, S. Burrous, R. M. Friedman, D. Jacobs, and M. S. Meltzer. 1992. Induction of IFN-alpha in peripheral blood mononuclear cells by HIV-infected monocytes. Restricted antiviral activity of the HIV-induced IFN. J. Immunol. 148:422-429. [PubMed] [Google Scholar]
- 27.Gendelman, H. E., J. M. Orenstein, L. M. Baca, B. Weiser, H. Burger, D. C. Kalter, and M. S. Meltzer. 1989. The macrophage in the persistence and pathogenesis of HIV infection. AIDS 3:475-495. [DOI] [PubMed] [Google Scholar]
- 28.Gendelman, H. E., J. M. Orenstein, M. A. Martin, C. Ferrua, R. Mitra, T. Phipps, L. A. Wahl, H. C. Lane, A. S. Fauci, D. S. Burke, et al. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham, B. S., and P. F. Wright. 1995. Candidate AIDS vaccines. N. Engl. J. Med. 333:1331-1339. [DOI] [PubMed] [Google Scholar]
- 30.Green, T. D., D. C. Montefiori, and T. M. Ross. 2003. Enhancement of antibodies to the human immunodeficiency virus type 1 envelope by using the molecular adjuvant C3d. J. Virol. 77:2046-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green, T. D., B. R. Newton, P. A. Rota, Y. Xu, H. L. Robinson, and T. M. Ross. 2001. C3d enhancement of neutralizing antibodies to measles hemagglutinin. Vaccine 20:242-248. [DOI] [PubMed] [Google Scholar]
- 32.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 33.Haynes, J. R., D. E. McCabe, W. F. Swain, G. Widera, and J. T. Fuller. 1996. Particle-mediated nucleic acid immunization. J. Biotechnol. 44:37-42. [DOI] [PubMed] [Google Scholar]
- 34.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klaniecki, J., T. Dykers, B. Travis, R. Schmitt, M. Wain, A. Watson, P. Sridhar, J. McClure, B. Morein, J. T. Ulrich, et al. 1991. Cross-neutralizing antibodies in rabbits immunized with HIV-1 gp160 purified from simian cells infected with a recombinant vaccinia virus. AIDS Res. Hum. Retrovir. 7:791-798. [DOI] [PubMed] [Google Scholar]
- 36.Kozono, Y., R. C. Duke, M. S. Schleicher, and V. M. Holers. 1995. Co-ligation of mouse complement receptors 1 and 2 with surface IgM rescues splenic B cells and WEHI-231 cells from anti-surface IgM-induced apoptosis. Eur. J. Immunol. 25:1013-1017. [DOI] [PubMed] [Google Scholar]
- 36a.Liu, F., I. Mboudjeka, S. Shen, T. W. Chou, S. Wang, T. M. Ross, and S. Liu. Vaccine, in press. [DOI] [PubMed]
- 37.Liu, M. A., T. M. Fu, J. J. Donnelly, M. J. Caulfield, and J. B. Ulmer. 1998. DNA vaccines. Mechanisms for generation of immune responses. Adv. Exp. Med. Biol. 452:187-191. [PubMed] [Google Scholar]
- 38.Mitchell, J. A., T. D. Green, R. A. Bright, and T. M. Ross. 2003. Induction of heterosubtypic immunity to influenza A virus using a DNA vaccine expressing hemagglutinin-C3d fusion proteins. Vaccine 21:902-914. [DOI] [PubMed] [Google Scholar]
- 39.Mongini, P. K., M. A. Vilensky, P. F. Highet, and J. K. Inman. 1997. The affinity threshold for human B cell activation via the antigen receptor complex is reduced upon co-ligation of the antigen receptor with CD21 (CR2). J. Immunol. 159:3782-3791. [PubMed] [Google Scholar]
- 40.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mustafa, F., J. F. Richmond, R. Fernandez-Larsson, S. Lu, R. Fredriksson, E. M. Fenyo, M. O'Connell, E. Johnson, J. Weng, J. C. Santoro, and H. L. Robinson. 1997. HIV-1 Env glycoproteins from two series of primary isolates: replication phenotype and immunogenicity. Virology 229:269-278. [DOI] [PubMed] [Google Scholar]
- 42.Pertmer, T. M., M. D. Eisenbraun, D. McCabe, S. K. Prayaga, D. H. Fuller, and J. R. Haynes. 1995. Gene gun-based nucleic acid immunization: elicitation of humoral and cytotoxic T lymphocyte responses following epidermal delivery of nanogram quantities of DNA. Vaccine 13:1427-1430. [DOI] [PubMed] [Google Scholar]
- 43.Pertmer, T. M., and H. L. Robinson. 1999. Studies on antibody responses following neonatal immunization with influenza hemagglutinin DNA or protein. Virology 257:406-414. [DOI] [PubMed] [Google Scholar]
- 44.Richmond, J. F., S. Lu, J. C. Santoro, J. Weng, S. L. Hu, D. C. Montefiori, and H. L. Robinson. 1998. Studies of the neutralizing activity and avidity of anti-human immunodeficiency virus type 1 Env antibody elicited by DNA priming and protein boosting. J. Virol. 72:9092-9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richmond, J. F., F. Mustafa, S. Lu, J. C. Santoro, J. Weng, M. O'Connell, E. M. Fenyo, J. L. Hurwitz, D. C. Montefiori, and H. L. Robinson. 1997. Screening of HIV-1 Env glycoproteins for the ability to raise neutralizing antibody using DNA immunization and recombinant vaccinia virus boosting. Virology 230:265-274. [DOI] [PubMed] [Google Scholar]
- 46.Roberts, T., and E. C. Snow. 1999. Cutting edge: recruitment of the CD19/CD21 coreceptor to B cell antigen receptor is required for antigen-mediated expression of Bcl-2 by resting and cycling hen egg lysozyme transgenic B cells. J. Immunol. 162:4377-4380. [PubMed] [Google Scholar]
- 47.Robinson, H. L. 1997. DNA vaccines for immunodeficiency viruses. AIDS 11:S109-S119. [PubMed] [Google Scholar]
- 48.Robinson, H. L., and T. M. Pertmer. 2000. DNA vaccines for viral infections: basic studies and applications. Adv. Virus Res. 55:1-74. [DOI] [PubMed] [Google Scholar]
- 49.Ross, T. M., Y. Xu, R. A. Bright, and H. L. Robinson. 2000. C3d enhancement of antibodies to hemagglutinin accelerates protection against influenza virus challenge. Nat. Immunol. 1:127-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ross, T. M., Y. Xu, T. D. Green, D. C. Montefiori, and H. L. Robinson. 2001. Enhanced avidity maturation of antibody to human immunodeficiency virus envelope: DNA vaccination with gp120-C3d fusion proteins. AIDS Res. Hum. Retrovir. 17:829-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanders, R. W., M. Vesanen, N. Schuelke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, P. J. Maddon, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spearman, P. 2003. HIV vaccine development: lessons from the past and promise for the future. Curr. HIV Res. 1:101-120. [DOI] [PubMed] [Google Scholar]
- 54.Srivastava, I. K., L. Stamatatos, H. Legg, E. Kan, A. Fong, S. R. Coates, L. Leung, M. Wininger, J. J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2002. Purification and characterization of oligomeric envelope glycoprotein from a primary R5 subtype B human immunodeficiency virus. J. Virol. 76:2835-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tao, Y., S. V. Strelkov, V. V. Mesyanzhinov, and M. G. Rossmann. 1997. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure 5:789-798. [DOI] [PubMed] [Google Scholar]
- 57.Test, S. T., J. Mitsuyoshi, C. C. Connolly, and A. H. Lucas. 2001. Increased immunogenicity and induction of class switching by conjugation of complement C3d to pneumococcal serotype 14 capsular polysaccharide. Infect. Immun. 69:3031-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.VanCott, T. C., J. R. Mascola, R. W. Kaminski, V. Kalyanaraman, P. L. Hallberg, P. R. Burnett, J. T. Ulrich, D. J. Rechtman, and D. L. Birx. 1997. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J. Virol. 71:4319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vinner, L., H. V. Nielsen, K. Bryder, S. Corbet, C. Nielsen, and A. Fomsgaard. 1999. Gene gun DNA vaccination with Rev-independent synthetic HIV-1 gp160 envelope gene using mammalian codons. Vaccine 17:2166-2175. [DOI] [PubMed] [Google Scholar]
- 60.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 61.Westervelt, P., H. E. Gendelman, and L. Ratner. 1991. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc. Natl. Acad. Sci. USA 88:3097-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 63.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, C. W., Y. Chishti, R. E. Hussey, and E. L. Reinherz. 2001. Expression, purification, and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J. Biol. Chem. 276:39577-39585. [DOI] [PubMed] [Google Scholar]
- 68.zur Megede, J., M. C. Chen, B. Doe, M. Schaefer, C. E. Greer, M. Selby, G. R. Otten, and S. W. Barnett. 2000. Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J. Virol. 74:2628-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]