Abstract
Hepatitis E virus (HEV) RNA replication occurred in seven of nine primate cell cultures transfected with in vitro transcripts of an infectious cDNA clone. Cell-to-cell spread did not occur in cell cultures, but rhesus monkeys inoculated with lysates of HEV-transfected PLC/PRF/5 and Huh-7 cells became infected with HEV. A replicon with the ORF2 and ORF3 genes deleted and replaced with the green fluorescent protein gene also replicated in the same primate cells that supported the replication of the full-length genome. Fluorescence-activated cell sorter analysis confirmed that the 7mG cap structure was critical for efficient infectivity, although replication could be initiated at a very low level in its absence. HEV virions were also able to infect a limited number of cells of certain lines.
Hepatitis E virus (HEV), the prototype Hepevirus, and hepatitis A virus (HAV) together are the major etiological agents of enterically transmitted hepatitis (4, 8, 23). HEV has a higher mortality rate, especially in pregnant women, but the reason for this is unknown. Otherwise, the disease caused by the one virus is clinically indistinguishable from that caused by the other, and neither progresses to chronicity. Both viruses have nonenveloped capsids, and both contain a single stranded RNA genome of the positive sense which serves as an mRNA to initiate infection.
In spite of the similarities, HEV and HAV have very different epidemiologies. HAV age-related seroprevalence patterns are those expected for a virus that is transmitted by the fecal-oral route, whereas those of HEV are not, even though fecal contamination is the major source of transmission. In countries in which the virus is endemic, anti-HAV antibodies are generally acquired before the age of 5 years, whereas the major rise in anti-HEV seroprevalence occurs later, in young adults (3). HAV has been found only in humans and in some nonhuman primates. HEV, on the other hand, has been isolated from humans and swine (7, 9, 20): additionally, antibodies reactive with capsid protein from human strains of HEV have been found in many animals, including nonhuman primates (2) and multiple species of rodents including rats (6, 11). A genotype 3 strain of HEV naturally infecting swine has been passed experimentally to monkeys, and a genotype 3 strain infecting humans has been passed to swine (18). However, attempts to transmit other human strains to swine have failed (19). The question of whether HEV is a zoonosis is still open (17), but a recent cluster of hepatitis E cases in Japan was traced to ingestion of raw deer meat, suggesting that this may be the case (26).
The molecular biology of HEV replication is not well understood, mainly because HEV does not grow efficiently in cell culture. Sequence analyses have demonstrated that the HEV genome is 7.2 kb long and contains three open reading frames (ORFs) with a short (∼27- to 35-nucleotide [nt]) 5′ noncoding region and a longer (∼65- to 74-nt) 3′ noncoding region terminating in a poly(A) tract (4, 24). ORF1 encodes putative nonstructural proteins; ORF2 encodes the major, if not the only, capsid protein; and ORF3 encodes a short protein (only 123 amino acids) of unknown function.
Much of the scant knowledge concerning HEV at the molecular level has been obtained through the overexpression of recombinant proteins in vitro. In addition to identification of an active viral RNA-dependent RNA polymerase, (1), such studies have led to the demonstration of guanylyltransferase and methyltransferase activities (15), two enzymatic activities required to synthesize a 7mG cap structure found at the 5′ terminus of most eukaryotic mRNAs. A cap was identified at the 5′ end of the HEV genome through antibody capture and molecular techniques (12, 33). Although a cap was required for infectivity of recombinant genomes in vivo (5), it has been reported that uncapped genomes could replicate in HepG2 cells in culture (22). Therefore, there is controversy over whether the 5′ cap is critical for HEV replication.
Northern blots of livers from monkeys infected with HEV have identified two coterminal subgenomic RNAs believed to encode ORF2 and ORF3 proteins, respectively (24, 31). It is not known whether these subgenomic RNAs are also capped, but since in vitro transcripts of a full-length cDNA clone are infectious for nonhuman primates, the subgenomic RNAs are not required to initiate an infection, and they must be synthesized as part of the replication process.
An efficient cell culture system for HEV would be extremely useful. A group in China has reported the propagation of HEV in cultures of A549 human lung cells, but these results have not been confirmed elsewhere (10, 30). HEV has been shown to infect primary cynomolgus hepatocytes and PLC/PRF/5 cells, but replication in these systems is so inefficient that reverse transcription-PCR (RT-PCR) is required to document it (16, 25). In the present study, we transfected various cells with transcripts from an HEV cDNA clone of known infectivity for animals or with a replicon derived from it in order to identify cells permissive for HEV genome replication. We then attempted to infect permissive cells with a virus pool that had been titered in nonhuman primates.
MATERIALS AND METHODS
Cell cultures.
The following cell lines were purchased from the American Type Culture Collection (Manassas, Va.): Caco-2 (HTB 37), HepG2/C3A (CRL-10741), PLC/PRF/5 (CRL-8024), BRL3A (3RL-1442), Hepa 1-6 (CRL-1830), PK(15) (CCL-33), ST (CRL-1746), A549 (CCL-185), and HS27 (CR6-1634). The 11-1 cells are a subclone of fetal rhesus kidney cells that were cloned by S. Emerson and are highly permissive for HAV. Vero cells, African green monkey kidney cells (passage 5), and tamarin kidney cells (passage 5) were obtained from L. Potash, Novavax, Rockville, Md. PAMD, bovine lung, and bovine thymus cells were obtained from S. Bolin (National Animal Disease Center). Huh-7 cells were originally isolated in Japan (21).
The majority of the cell lines were cultured in Dulbecco's modified Eagle's medium containing 5% fetal bovine serum (ultra-low immunoglobulin G [IgG]; Invitrogen) that had been heat inactivated at 56°C for 30 min. The serum concentration was increased to 10% for Hepa 1-6 cells and to 20% for Caco-2 cells. For PLC/PRF/5 cells, nonessential amino acids (0.1 mM) were added. HepG2/3CA, BRL3, and PK(15) cells were grown in minimal essential medium with Hanks salts, 1.5 g of sodium bicarbonate/liter, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 10% ultra-low-IgG calf serum, with the serum concentration increased to 20% for the PK(15) cells. Cell stocks were grown at 37°C in tightly closed flasks.
Transcription and transfection.
Plasmids were linearized with BglII (Invitrogen), and 5 μg of DNA was transcribed in a 50-μl reaction volume containing 10 μl of 5× transcription buffer (Promega), 5 μl of 100 mM dithiothreitol (Promega), 2 μl of 40-U/ml RNasin (Promega), 5 μl of nucleoside triphosphates (5 mM [each] ATP, CTP, and UTP; 0.5 mM GTP), 5 μl of 5 mM 7mGTP (Ambion), and 2 μl of 20-U/μl T7 polymerase (Promega). A negative-control sample, run in parallel, was identical to the reaction mixture described above except that T7 polymerase was omitted and replaced with water. A 5-μl volume of 5 mM GTP was substituted for 7mGTP to produce uncapped transcripts. The mixtures were incubated at 37°C for 2 h, with an additional 2 μl of polymerase added after the first hour. Each transcription mixture was cooled on ice and used “as is” for transfection.
Prior to transfection, cells were washed twice with Optimem (Gibco). Mixtures of 20 μl of DMRIE-C (Invitrogen)/ml of Optimem or 30 μl of Lipofectamine 2000 (Invitrogen)/ml of Optimem were prepared. For 12-well plates or 2-well slides, a 10-μl transcription mixture was mixed with 200 μl of an Optimem-liposome mixture, the entire amount was added to one well, and the culture was incubated at 34.5°C for 5 h. Volumes were doubled for a six-well plate. After the 5-h incubation, the mixture was aspirated, fresh medium was added, and the cultures were incubated at 34.5°C. Unless cells were already in chamber slides, cells were trypsinized at the appropriate time and replated on chamber slides prior to immunostaining.
Immune fluorescence microscopy.
Both polyclonal and monoclonal antibodies were used for immune fluorescence microscopy. Chimpanzee 1313 had been infected with HEV in a stool pool from a hepatitis outbreak in Pakistan and later was inoculated with the Mex-14 strain of HEV; the resulting immune serum had an enzyme-linked immunosorbent assay (ELISA) titer of 1:105 against ORF2 recombinant protein spanning amino acids 112 to 607, whereas the preinoculation serum had a titer of <1:20 (limit of detection) against ORF2. The chimpanzee immune serum did not react with cells in which a replicon lacking ORF2 and ORF3 was replicating, indicating that either it did not contain detectable antibodies to ORF1 proteins or the amount of ORF1 proteins synthesized was below the limit of detection (data not shown). Mouse monoclonal antibodies to ORF2 (a gift from GlaxoSmithKline) and to ORF3 (a gift from Abbott Laboratories) were also used. In a radioimmune precipitation assay of in vitro-translated, 35S-labeled ORF2 (amino acids 112 to 607) and ORF3 proteins, anti-ORF2 precipitated only ORF2 from a mixture of ORF2 and ORF3, whereas anti-ORF3 precipitated only ORF3, the chimpanzee immune serum precipitated only ORF2, and the chimpanzee preimmune serum precipitated neither (data not shown).
Cells on slides were rinsed in phosphate-buffered saline (PBS), fixed with acetone, and air dried. Cells were then overlaid with a 1:1 mixture of 10% bovine serum albumin and PBS containing either chimpanzee 1313 serum, a mouse monoclonal antibody, or a mixture of the chimpanzee serum and one of the mouse monoclonal antibodies. After 20 min at room temperature, the slides were rinsed in PBS and overlaid with either Alexa Fluor 488-conjugated goat anti-human IgG (Molecular Probes), an Alexa Fluor 568-conjugated F(ab′)2 fragment of goat-anti-mouse IgG (Molecular Probes), or a mixture of the two. After 20 min at room temperature, slides were washed in PBS, Vectashield (Vector Laboratories) was added, and slides were viewed by indirect immune fluorescence microscopy using a fluorescein isothiocyanate (FITC) filter set for Alexa Fluor 488 (green), a rhodamine filter set for Alexa Fluor 568 (red), and the 25× objective of a Zeiss fluorescence photomicroscope.
For detection of green fluorescent protein (GFP), most of the medium was removed from the cells, and the damp cells were examined for intrinsic fluorescence by fluorescence microscopy using the FITC filter set and the 25× objective of the same microscope used for immunofluorescence. Confocal microscopy was performed by Owen Schwartz, Head of the Biological Imaging Facility at the National Institute of Allergy and Infectious Diseases.
Infections.
A titered 10% stool pool (10−1 dilution) containing 104.5 50% monkey infectious doses (MID50) of Sar55 per 0.5 ml was centrifuged for 2 min at 14,000 rpm in a 5415 C Eppendorf centrifuge, and the supernatant was used to inoculate cell monolayers that had been rinsed once with Optimem. After 3.5 h at 34.5°C, the inoculum was removed, and the cells were washed once before being overlaid with a medium containing antibiotics. Cultures were incubated at 34.5°C. The number of viral genomes remaining in the medium was quantified by Taqman PCR as previously described (5).
Washed monolayers of PLC/PRF/5 cells and Huh-7 cells transfected with transcripts from the full-length capped genome were harvested by trypsinization at 14 and 12 days posttransfection, respectively, mixed with Dulbecco's modified Eagle medium lacking serum and antibiotics, and freeze-thawed three times. Cell debris was removed by centrifugation for 5 min at 10,000 rpm in a 5415 C Eppendorf centrifuge, and the supernatant was stored at −80°C.
One rhesus monkey (Macaca mulatta) each was inoculated intravenously with lysates from the PLC/PRF/5 cells and Huh-7 cells, respectively, and serum samples were collected weekly and monitored for viral genomes, serum alanine aminotransferase (ALT) activity, and anti-HEV antibodies as described previously (5). The housing, maintenance, and care of the animals met or exceeded all requirements for primate husbandry.
Replicon construction.
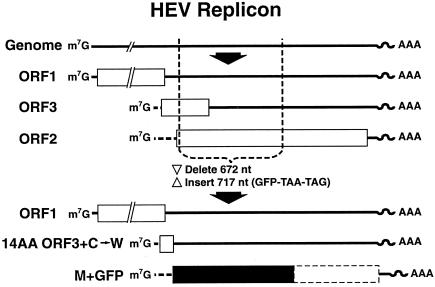
The infectious cDNA clone of the Sar55 strain, pSK-HEV-2 (GenBank accession no. AF444002), has been described previously. The GFP replicon was constructed by synthesizing a PCR fusion product containing the 3′ region of ORF1 fused to the GFP gene and substituting it for the SfiI-to-EcoRI region in the infectious cDNA clone (Fig. 1). The ORF1 region from nt 3883 to 5147 was amplified from the infectious cDNA clone by using the forward primer f E/SFI 3963 (5′ TGAATTAACAGATATTGTGCATTG) and the reverse primer r ORF1/GFP (5′ CATGGTCGCGAACCCATGGGC). The GFP gene was amplified from pIRES-EGFP (Clontech) from nt 292 to 1008 by using the forward primer f ORF2/GFP (5′ GCCCATGGGTTCGCGACCATGGTGAGCAAG GGCGAGGAGCTGTTC) and the reverse primer r EcoRI/GFP (5′ GCGAATTCCTATTACTTGTACAGC TCGTCCATGC) (the termination codons introduced are underlined). Both PCR products were purified with a MinElute PCR Purification kit (Qiagen); then they were mixed together, and fusion PCR was performed by using the forward primer f E/SFI 3963 and the reverse primer r EcoRI/GFP. The fusion product was purified by QIAquick gel extraction (Qiagen), digested with SfiI-EcoRI, and ligated into the large fragment of the infectious HEV clone from which the SfiI-to-EcoRI region had been deleted. The sequence of the entire replicon was verified by sequencing DNA from the Maxiprep preparation used throughout for transfection.
FIG. 1.
Diagram of HEV replicon expressing GFP. The exact location of the 5′ end of each subgenomic RNA is unknown. Open rectangles, HEV coding regions; solid rectangle, GFP coding region; dashed-line rectangle, residual ORF2 gene (not translated). ORF1 is intact; ORF3 mRNA encodes the first 14 amino acids of ORF3 followed by a tryptophan and a termination codon; ORF2 mRNA encodes GFP followed by two termination codons.
The GDD motif in the polymerase gene was mutated by fusion PCR. The template for PCR was the GFP replicon. The first fragment was amplified with the forward primer f E/SFI 3963, described above, and the reverse primer r JE G66 (5′ CTATCGAATCCGCACCTTTAA AGGCAG). The second fragment was amplified with the forward primer f JE G59 (5′ CTGCCTTTAAAGGTGCGG ATTCGATAG) and the reverse primer r EcoRI/GFP, described above. The rest of the procedure was identical to that used to construct the GFP replicon. The sequence of the entire replicon was determined by automated sequencing. The GAD mutation was introduced into the full-length clone by isolating the SfiI/PauI fragment from the replicon and substituting it for the homologous fragment in the full-length clone. The final clone was fully sequenced and was identical to pSK-HEV-2 except for the GAD mutation.
FACS analysis.
Huh-7 cells that had been transfected with transcripts synthesized in vitro from a GFP replicon encoding the wild-type viral RNA polymerase or encoding the polymerase mutated at the GDD motif to GAD were analyzed on a Becton Dickinson (San Jose, Calif.) FACScan by using a 530/30BP filter to detect GFP and a 585/42BP filter to detect autofluorescence. Autofluorescence was identified based on two negative controls. The first consisted of untreated Huh-7 cells. The second control was a cell culture transfected with a mock transcription mixture that included everything but the T7 polymerase so that RNA was not synthesized. The gate for the fluorescence-activated cell sorter (FACS) was set to exclude virtually all autofluorescent cells in the T7-negative control. No more than 19 cells were gated in any of the T7-negative samples. For each sample, 100,000 cells were counted.
RESULTS
Two human hepatoma cell lines transfected with recombinant HEV genomes produced infectious virions.
An efficient cell culture system for HEV has been very difficult to establish. The inability of HEV virions to infect cells in culture could reflect a number of factors, including the absence of a receptor, failure of the genome to replicate, or deficiencies in morphogenesis or egress. Transfection would bypass potential receptor or uncoating deficiencies. Therefore, as a first step toward sorting out the block(s) to in vitro replication, human hepatoma cells (Huh-7 cells and PLC/PRF/5 cells) were transfected with recombinant full-length capped viral transcripts that were known to infect rhesus monkeys and chimpanzees.
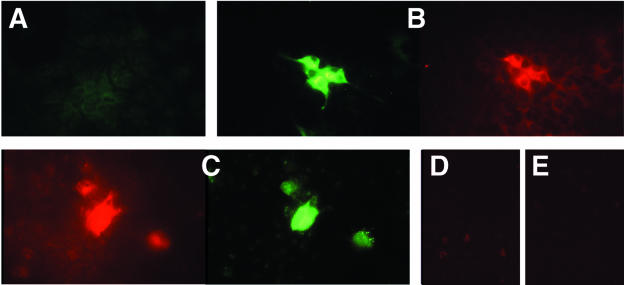
Viral antigens were detected by immunofluorescence microscopy of transfected cells incubated either with a monoclonal antibody specific for ORF2 or ORF3 protein or with a chimpanzee convalescent-phase anti-HEV serum. Stained cells were detected with each antibody in each cell line but not in mock-transfected cells, suggesting that the transfected viral genome may have replicated in the liver-derived cells. Examples of stained Huh-7 cells are shown in Fig. 2. Positive cells were first detected around day 4, and the intensity of the staining increased until around day 7.
FIG. 2.
Immune fluorescence microscopy of Huh-7 cells 14 days after transfection with the infectious HEV genome (A to C) or of mock-transfected Huh-7 cells (D and E). Cells were stained or costained with either chimpanzee 1313 preinfection serum (A), chimpanzee 1313 immune serum (green) and a monoclonal anti-ORF2 antibody (red) (B), a monoclonal anti-ORF3 antibody (red) and chimpanzee 1313 immune serum (green) (C), a monoclonal anti-ORF2 antibody (red) (D), or a monoclonal anti-ORF3 antibody (red) (E).
In order to confirm that production of detectable levels of viral antigen reflected genomic replication, the full-length genome was mutated to convert the conserved GDD motif of the viral RNA-dependent-RNA polymerase to GAD. This mutation has been shown to abolish replication of rubella virus (29), a virus that has a polymerase related to that of HEV (13).
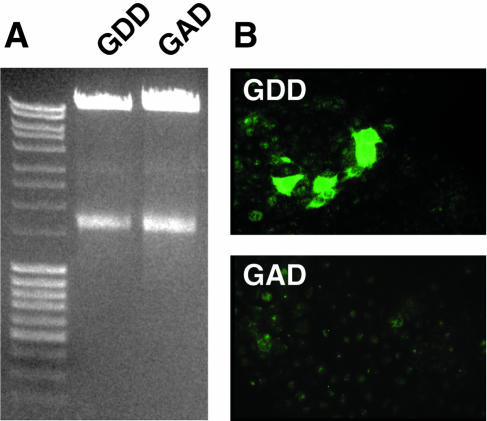
Huh-7 cells were transfected in parallel with similar amounts of transcripts from cDNA clones containing the wild-type and mutated polymerases, respectively (Fig. 3A). Cells were examined by immune fluorescence microscopy on days 7, 12, and 16 posttransfection. Viral antigen was detected on all three days in many cells transfected with the wild-type genome; in contrast, not a single stained cell was detected at any time in cells transfected with the mutant polymerase (Fig. 3B). Therefore, the presence of detectable viral antigen was dependent on genomic replication, confirming that immune fluorescence microscopy provided a valid method for determining whether the viral genome had replicated.
FIG. 3.
Transfection of cells with full-length genomes containing a wild-type (GDD) or a mutated (GAD) polymerase. (A) Agarose gel of 5 μl of each transcription reaction product used for transfection. (B) Immune fluorescence micrographs of representative cell monolayers stained with chimpanzee 1313 immune serum 14 days after transfection with wild-type (GDD) or mutated (GAD) genomes.
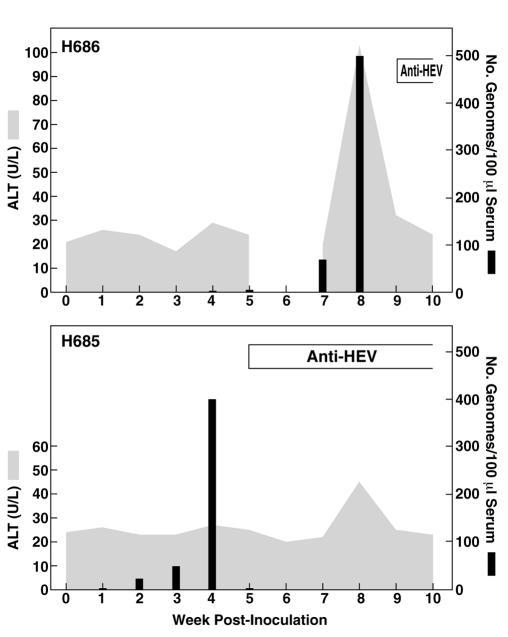
The percentage of cells in which viral antigen could be detected remained stable at less than 10%, suggesting that either infectious virions were not being assembled or, if they were, they were unable to infect the remaining cells. In order to determine if infectious virus was being produced in these transfected cells, cells were harvested by trypsinization on day 14 (PLC/PRF/5) or day 12 (Huh-7) posttransfection and lysed by three cycles of freeze-thawing. The lysate from each cell line was inoculated intravenously into a rhesus macaque, which was then monitored for evidence of HEV infection. The animal inoculated with the PLC/PRF/5 lysate developed a characteristic course of hepatitis E as evidenced by elevated levels of liver enzymes in serum, viremia, and seroconversion (week 9) to anti-HEV ORF2 (Fig. 4A). The HEV genome was amplified by RT-PCR from the feces of rhesus H686 at week 7 postinoculation, and the consensus sequence spanning nt 130 to the poly(A) tail was determined. The sequence was identical to that of the cDNA clone except for a single nucleotide change at position 586 (C to T), which did not change the amino acid. Two unique silent mutations (nucleotide positions 286 and 4396) present in the cDNA clone used for transfection of the PLC/PRF/5 cells were present in the virus recovered from the macaque, demonstrating that the virus had originated from the recombinant genome.
FIG. 4.
Course of HEV infection in rhesus macaques inoculated intravenously with cell lysates from transfected cell cultures. H686, lysate from PLC/PRF/5 cells. No sample was available for week 6. H685, lysate from Huh-7 cells. The ALT peak in the H685 graph was not at the correct time or high enough to be considered indicative of hepatitis.
The monkey inoculated with the lysate of transfected Huh-7 cells experienced viremia and seroconverted to anti-HEV at week 5 postinoculation but did not develop overt hepatitis (Fig. 4B). Because the severity of hepatitis E is related to the viral dose as well as to biological variation in the monkeys, the absence of disease was not unexpected. Short regions spanning nt 286 and 4396 were amplified from serum and sequenced, and the presence of the two unique silent mutations confirmed that this virus also originated from the recombinant genome.
Replication of the HEV genome in different cell lines.
The detection of viral antigen and the recovery of infectious virus from transfected PLC/PRF/5 and Huh-7 cells demonstrated that the viral genome had replicated in these cells and that assembly of infectious virions had occurred. In order to determine the degree of permissiveness of various cell lines for HEV genomic replication, additional lines were transfected with the full-length genome. The chimpanzee immune serum was used to monitor replication because it was more sensitive than the monoclonal antibodies for detecting positive cells by immunofluorescence microscopy. A monoclonal antibody to ORF2 or ORF3 was used to confirm results. A number of human and nonhuman primate cell lines, but not all, were able to support HEV replication (Table 1). Numbers of stained cells were different in different cell lines but were consistently less than 15%. There was no indication of cell-to-cell virus spread. The two human liver cell lines PLC/PRF/5 and Huh-7 and the human intestinal cell line Caco-2 produced the highest number of stained cells and the most intensely stained cells. Some primate cell lines such as the tamarin kidney cells and human foreskin cultures (HS27) were negative for HEV replication. The rhesus kidney subclone 11-1 was negative when DMRIE-C was the transfection facilitator but was positive when Lipofectamine 2000 was used, suggesting that inefficient transfection rather than an inability to replicate HEV accounted for the negative results in these cells. However, the tamarin kidney cells remained negative even when Lipofectamine 2000 was used. Viral antigen (indicating replication of the HEV genome) was not detected in any nonprimate cell tested.
TABLE 1.
Transfection of cultured cells with HEV transcripts
| Cell line | Resultsa of transfectionb with:
|
Species | Tissue | |
|---|---|---|---|---|
| pSK7ac | GFPd | |||
| PLC/PRF/5 | ++++ (D) | +++ (D) | Human | Liver |
| Huh-7 | ++++ (D) | ++++ (D) | Human | Liver |
| Caco-2 | ++++ (D) | +++ (D) | Human | Intestine |
| HepG2/3CA | ++ (D), + (L) | − (D), ++++ (L) | Human | Liver |
| AGMK (p5) | ND | + (D) | African green monkey | Kidney |
| FKRhK (11-1) | − (D), ++ (L) | + (D), ++ (L) | Rhesus monkey | Kidney |
| Vero | + (D), − (L) | ++ (D) | African green monkey | Kidney |
| HS27 | − (D) | − (L) | Human | Foreskin |
| A549 | −/+ (D) | − (D, L) | Human | Lung |
| Tamarin | − (D, L) | − (D, L) | Tamarin | Kidney |
| PK(15) | − (D, L) | − (D, L) | Pig | Kidney |
| ST | ND | − (D) | Pig | Testis |
| PamD | − (D) | − (D) | Pig | Macrophage |
| Bovine lung | − (D) | − (D) | Cow | Lung |
| Bovine thymus | − (D) | − (D) | Cow | Thymus |
| BRL-3A | ND | − (D) | Rat | Liver |
| Hepa 1-6 | ND | − (D, L) | Mouse | Liver |
+, positive for HEV antigen or GFP; ++ to ++++, increasing numbers of positive cells; −, negative for HEV antigen or GFP; ND, not done.
D, transfected by use of DMRIE-C; L, transfected by use of Lipofectamine 2000.
Full-length transcripts of infectious clone.
Transcripts from GFP replicon.
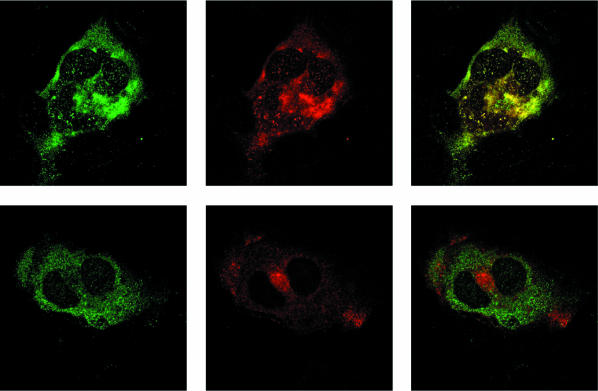
Huh-7 cells were doubly stained with chimpanzee convalescent-phase serum and a monoclonal antibody to either ORF2 or ORF3 and were examined by confocal microscopy. The chimpanzee antibodies and the antibody to ORF3 often stained different regions of the cytoplasm, especially at later times posttransfection (Fig. 5). In contrast, the monoclonal antibody to ORF2 always appeared to colocalize with the chimpanzee antibody, in agreement with the ability of this serum to immunoprecipitate ORF2 protein but not ORF3 protein.
FIG. 5.
Confocal microscopy of Huh-7 cells stained with anti-HEV at day 7 posttransfection. (Top left) Chimpanzee 1313 immune serum; (top center) anti-ORF2; (top right) merge. (Bottom left) Chimpanzee 1313 immune serum; (bottom center) anti-ORF3; (bottom right) merge. Magnification, ×126.
Construction of an HEV replicon expressing GFP.
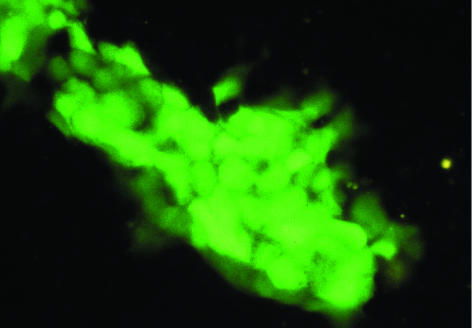
A segment of the HEV genome containing most of the ORF3 gene and the first half of the ORF2 gene downstream of the first methionine was removed, and the GFP gene was inserted in frame with the ORF2 methionine initiation codon (Fig. 1). The replication-negative control genome containing the polymerase GAD mutation was similarly modified to encode GFP. Thus, only the first 14 amino acids of ORF3 and none (excepting the first methionine) of ORF2 would be translated, and GFP would be translated from the “ORF2” subgenomic mRNA. Capped transcripts from the construct containing the wild-type polymerase resulted in the appearance of green fluorescent cells beginning at 2 to 4 days posttransfection when cells were viewed directly with the FITC filter set under a fluorescence microscope. In contrast, green fluorescent cells were not observed even by day 14 following transfection with capped transcripts encoding the mutant polymerase. Since a functional polymerase was needed for the synthesis of GFP to be detected, the wild-type genome in which the GFP gene replaced much of the ORF2 and ORF3 genes must have been replication competent. Results from transfection of various cell lines with the competent GFP replicon were comparable to those obtained with the full-length genome and confirmed that some cells but not others were able to support HEV replication (Table 1). For each cell line in general, similar numbers of cells were positive when transfected either with the full-length genome or with the GFP replicon, and positive cells appeared at about the same time regardless of whether antibody staining (full-length genome assay) or intrinsic fluorescence (replicon assay) was the detection method. At day 3 to 4 following transfection, GFP-positive cells were first observed as single isolated cells, but at later times doublets or small clusters of GFP-positive cells were often observed, suggesting that the transfected cells were dividing. The transfected Caco-2 cells appeared to divide more frequently, thus producing large colonies of GFP-positive cells by day 10 (Fig. 6).
FIG. 6.
Fluorescence microscopy of Caco-2 cells producing GFP. Cell division has resulted in a colony containing more than 30 GFP-producing cells. Magnification, ×25.
FACS analysis of GFP-producing cells.
FACS analysis of cells transfected with the replicon was undertaken to determine if enough GFP per cell was produced to make this procedure useful for studying HEV genome replication. Huh-7 cells were chosen for these experiments because they were more efficiently transfected and were easier to passage and disperse than were PLC/PRF/5, HepG2/3CA, or Caco-2 cells. Cells in four wells of a six-well plate were transfected with the same transcription mixture, and cells from one well each were harvested on consecutive days (days 1 through 4) posttransfection and subjected to FACS analysis. A very few cells contained enough GFP to be sorted on day 1, but the number increased to 11.6% by day 4 (data not shown). Therefore, it appeared that FACS analysis could be used to quantify the number of transfected cells.
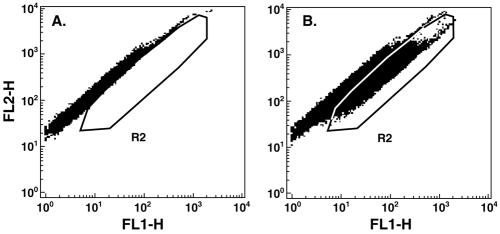
The FACS analysis was repeated to examine later time points, to determine the effect of omitting the cap structure, and to confirm the deleterious effect of mutating the polymerase active site from GDD to GAD. Four wells of Huh-7 cells in a six-well plate were each transfected with a different sample, and one well was left untreated. The next day, the cells were trypsinized and aliquoted into four new wells of a six-well plate for each sample. One well each was harvested for FACS on days 4, 6, and 8 posttransfection. Cells in the remaining well for each sample were split 1 to 8 on day 8 and again on day 19 and were harvested for FACS on day 25. An example of the FACS distribution is shown in Fig. 7.
FIG. 7.
Dot plot of FACS analysis. Cells, depicted as squares, are plotted as autofluorescence (FL2-H) versus fluorescence intensity in the FITC channel (FL1-H). (A) The pattern from control transfected cells was used to set the R2 window to exclude autofluorescent cells and to include cells containing GFP. (B) FACS distribution of cells 8 days after transfection with the GFP replicon. Only those cells that fell within the R2 area were scored as GFP positive.
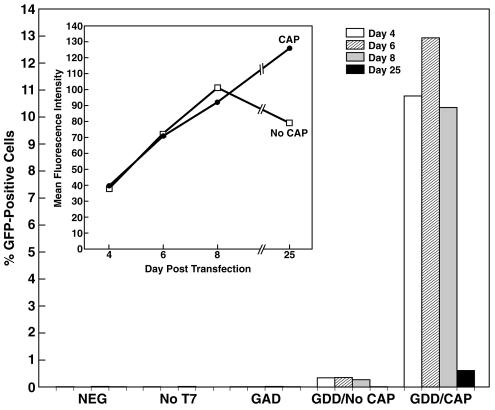
The percentage of GFP-positive cells (or gated cells for the negative controls) for each sample for each day is shown in Fig. 8. Cell cultures transfected with the negative-control transcription mixture lacking the T7 polymerase or with the capped polymerase mutant (GAD) transcripts were virtually identical to untreated Huh-7 cultures, with 0.02% or fewer GFP-positive cells in each case. In contrast, the number of GFP-positive cells following transfection with capped transcripts of the replicon encoding the wild-type polymerase ranged between 10.78 and 12.93% of total cells on days 4 through 8. When the cap was omitted from the replicon transcripts, positive cells were still detected, but the number (0.27 to 0.35%) was 32 to 38 times lower than that for the corresponding capped sample. The number of GFP-positive cells on day 25 dropped dramatically for both the capped and uncapped samples (to 0.61 and 0.01%, respectively).
FIG. 8.
FACS analysis of untransfected Huh-7 cells (NEG) or of Huh-7 cells transfected either with a transcription mixture from which polymerase had been omitted (No T7) or in which the GDD motif of the polymerase had been mutated to GAD (GAD) or with uncapped (GDD/No CAP) or capped (GDD/CAP) transcripts from the replicon containing the wild-type polymerase. A total of 100,000 cells were counted for each sample at each time. (Inset) Mean fluorescence intensity of GFP-positive cells from capped (circles) or uncapped (squares) sample.
The mean fluorescence intensities of the GFP-positive cells were almost identical in the capped and uncapped samples for each day, except for the day-25 uncapped sample (Fig. 8, inset). The fluorescence intensity increased with time, as would be expected if GFP was still accumulating. Although very few GFP-positive cells were detected on day 25, those that were detected retained a high level of fluorescence.
In agreement with the FACS data, microscopic examination of the cell monolayer on day 25 prior to harvesting revealed a small number of green fluorescent cells in both the capped and uncapped samples. In each case, some positive cells were in minicolonies consisting of 10 or more green cells in close proximity, indicating that the cells were dividing and suggesting that GFP was still being produced (data not shown).
Infection of cells.
Four cell lines (PLC/PRF/5, Huh-7, Caco-2, and HepG2/3CA) that replicated the transfected genome efficiently and A549 cells, which had been reported to support HEV infection (10, 30), were inoculated with a titered stool pool of Sar55 virions. Cells in a 12-well plate at about 50% confluency were inoculated with 12,600 MID50 for an approximate multiplicity of infection of 1 MID50 per 20 cells. After adsorption for 3.5 h, the inoculum was removed, and the number of genomes remaining in the inoculum was quantified by Taqman RT-PCR.
Ninety-four percent of the genomes in the input inoculum were still present in the medium of the Caco-2 cells after the adsorption period, suggesting that the virus was unable to bind efficiently to these cells under the conditions used. This was in contrast to the results with the other four cell lines, in which approximately 20% of the input virus remained in the medium, suggesting that almost 80% of the virus bound to each of these cell lines.
Cells were passed to four-well chamber slides and stained for immunofluorescence microscopy with chimpanzee 1313 immune serum on day 8. Positively stained cells were not detected for the Caco-2, PLC/PRF/5, or A549 cell line. Therefore, efficient binding to cells did not necessarily lead to infection. Positive cells were found in the Huh-7 and HepG2/3CA cultures, but fewer than 10 positive cells were found per well. The HepG2/3CA culture contained more positive cells than did the Huh-7 culture. However, after dilution and passage to T25 flasks, positive cells were no longer detected in either the HepG2/3CA or the Huh-7 culture.
Caco-2 and HepG2/3CA cells did not efficiently attach to slides following trypsinization, and it was not clear whether infected cells were being selectively lost during preparation for microscopy. Therefore, the infection was repeated with the same Sar55 inoculum but with cells in eight-well chamber slides coated with collagen in an effort to promote cell attachment. Cells were not confluent at the time of infection, but all except the PLC/PRF/5 cells were confluent when the slides were fixed on day 6 postinfection. After the cells were stained with chimpanzee 1313 immune serum for immune fluorescence microscopy, the entire well was manually scanned and the number of positive cells was counted. Only a very low number of positive cells was found in the Huh-7 (4 cells), Caco-2 (4 cells), A549 (15 cells), or PLC/PRF/5 (7 cells) cell line. In contrast, 202 positive cells were detected in the HepG2/3CA cell line.
DISCUSSION
Following transfection into seven of nine primate cell lines, synthetic genomes transcribed from an infectious cDNA clone of HEV were able to produce enough ORF2 and ORF3 proteins to be clearly visible by immunofluorescence microscopy. Unfortunately, we have not yet been able to increase the number of antigen-positive cells to a level that would allow direct detection of replicating RNA; this most likely is due to inefficient capping in vitro coupled with the requirement for a cap and the inability of the virus to spread to other cells in the culture. However, two independent lines of evidence suggested that the viral genome had replicated in these cells. First, mutation of the viral polymerase at a highly conserved site in the catalytic domain abolished the immunostaining of cells, indicating that the synthesis of ORF2 and ORF3 proteins was dependent on viral RNA replication. This conclusion is in agreement with the currently held belief that ORF2 and ORF3 proteins are each synthesized from subgenomic RNAs which would have to be generated and amplified by the viral RNA-dependent RNA polymerase. Second, virus infectious for rhesus macaques was recovered from both cell lines tested, thus demonstrating that the transfected genomes had successfully initiated and completed the replication cycle.
FACS analysis of cells transfected with a replicon expressing GFP confirmed that production of virally encoded proteins was dependent on genomic replication. A mutant replicon expressing an impaired polymerase produced few if any cells containing enough GFP to be detected, whereas the replicon containing the authentic polymerase caused 10% or more of the cells to be counted as positive.
Since GFP-producing cells could be observed in the original wells used for transfection, it was possible to identify colonies of green fluorescent cells that formed as the transfected cell divided until a confluent cell monolayer was formed. The generation of these colonies suggested that the pool of functional replicons was large enough to tolerate multiple dilutions and still produce detectable GFP as the cell population expanded.
The FACS analysis also emphasized the importance of the 5′ cap structure for infectivity. Previously Emerson et al. provided evidence for a cap structure on the 5′ terminus of natural viral genomes and showed that in vitro-synthesized transcripts of the genome lacking the cap were not infectious for primates whereas those containing a cap were (5). In the present study, capped replicon transcripts were 32 to 38 times more likely to produce GFP-positive cells than were uncapped transcripts. The uncapped transcripts were able on rare occasions to initiate an infection cycle, as indicated by the detection of some cells during FACS analysis and the detection of a low number of green cells by fluorescence microscopy. Since RNAs that normally are capped can be translated in cell-free translation systems in the absence of a cap, the cap is a facilitator of translation rather than an absolute necessity. Therefore, it seems likely that on infrequent occasions, the uncapped RNA was able to interact with ribosomes and thus the ORF1 protein(s), including the polymerase and capping enzymes, was translated. The first genome synthesized de novo by these enzymes should be capped, and the replication cycle should thereafter be identical to one initiated by a capped RNA. Panda et al. previously reported that uncapped synthetic HEV genomes were infectious for HepG2 cells (22). Our data support this conclusion in principle but disagree in one respect. Whereas Panda et al. (22) reported efficient transfection (nearly 20% of the cells) with uncapped RNAs, we found that transfection with uncapped RNAs was very inefficient compared to that with capped RNAs both in vitro (this study) and in vivo (5). Since Panda et al. (22) did not compare capped and uncapped RNAs, the reason for this difference is unclear, and further studies are required.
It was surprising to find that the HEV genome could replicate in so many different primate cell lines. This result suggests that the block to growing HEV in cell cultures of primate cells is probably not due to a lack of cell factors necessary for translation or replication. However, species-specific factors are most likely required, since neither the replicon nor the viral genome was able to replicate in any of the nonprimate cells tested, including liver cells from rats or mice. It should be noted that the inability to detect replication in tamarin cells is in accord with the previous failure to infect tamarins with the Sar55 virus (27).
Attempts to infect cells were not as successful as the transfections, suggesting that most cultured cells may lack the receptor for HEV. There was a significant difference among cell lines in the number of cells that were infected with the Sar55 inoculum. Only the HepG2/3CA cells were infected to a level that made artifactual entry an unlikely explanation. However, infection of HepG2/3CA cells was still inefficient, and viral spread was not observed.
HEV appears not to be very cytopathic, if at all. Following transfection with the entire genome, some ORF2-positive Huh-7 cells were still detected by immune fluorescence microscopy after 162 days in cell culture (data not shown). Replication of the viral genome did not prevent cell division, as evidenced by the detection of ORF2 protein in pairs of cells presumed to have divided recently, and of “colony” formation of GFP-producing cells, especially Caco-2 cells. The decreased percentage of GFP-producing cells at day 25 posttransfection therefore may not result from cell death but more likely reflects overgrowth by non-GFP-producing cells following the two large subculturing steps, which would lead to increased cell division.
Although ORF3 contains only 123 amino acids, it has been postulated to carry out numerous functions. Studies with expressed recombinant ORF3 have demonstrated that it reacts with the cell cytoskeleton as well as with nonglycosylated recombinant ORF2 protein (28, 32). In addition, it has been suggested that it has regulatory functions through binding to multiple cellular proteins containing SH3 domains and through activation of mitogen-activated protein kinase (14). In spite of all these proposed functions, the GFP replicon lacking both the ORF3 and ORF2 proteins was able to replicate in all the same cell lines as the full-length genome that produced both ORF3 and ORF2 proteins. Therefore, neither ORF3 nor ORF2 was essential for replication of the genome in vitro. It remains to be determined if RNA replication is more efficient when ORF2 and/or ORF3 proteins are present, but there was not an obvious difference in time to ORF2 protein versus GFP detection, and the efficiencies of transfection were similar for the full-length genome and the replicon. Although others have reported colocalization of ORF2 and ORF3 when overexpressed from independent vectors (28), we did not regularly detect colocalization of ORF2 and ORF3 when the two proteins were synthesized as part of the normal replication cycle (Fig. 3). ORF3 showed a clear tendency to segregate into large masses apparently devoid of ORF2. Therefore, it will be very interesting to determine if ORF2 and ORF3 protein interactions change during the course of the virus replication cycle. The replication of the genome offers an advantage over the use of independent vector-expressed proteins in that all the viral proteins should be present in the ratios characteristic of the normal viral replication cycle, and therefore, observed interactions may be more representative of those occurring in an actual infection in vivo. Since the RNA transcribed from the cDNA clone used in these experiments is also infectious for primates (5), it should now be possible to compare in vitro and in vivo molecular requirements for HEV replication and to determine which are indeed critical.
Acknowledgments
This work was supported in part by National Institute of Allergy and Infectious Diseases contract 1-AO-02733.
We thank Kevin Holmes for helpful suggestions on presenting the FACS data and Sandra Chang for excellent secretarial assistance.
REFERENCES
- 1.Agrawal, S., D. Gupta, and S. K. Panda. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282:87-101. [DOI] [PubMed] [Google Scholar]
- 2.Arankalle, V. A., M. V. Joshi, A. M. Kulkarni, S. S. Gandhe, L. P. Chobe, S. S. Rautmare, A. C. Mishra, and V. S. Padbidri. 2001. Prevalence of anti-hepatitis E virus antibodies in different Indian animal species. J. Viral Hepat. 8:223-227. [DOI] [PubMed] [Google Scholar]
- 3.Arankalle, V. A., S. A. Tsarev, M. S. Chadha, D. W. Alling, S. U. Emerson, K. Banerjee, and R. H. Purcell. 1995. Age-specific prevalence of antibodies to hepatitis A and E viruses in Pune, India, 1982 and 1992. J. Infect. Dis. 171:447-450. [DOI] [PubMed] [Google Scholar]
- 4.Emerson, S. U., and R. H. Purcell. 2003. Hepatitis E virus. Rev. Med. Virol. 13:145-154. [DOI] [PubMed] [Google Scholar]
- 5.Emerson, S. U., M. Zhang, X. J. Meng, H. Nguyen, M. St Claire, S. Govindarajan, Y. K. Huang, and R. H. Purcell. 2001. Recombinant hepatitis E virus genomes infectious for primates: importance of capping and discovery of a cis-reactive element. Proc. Natl. Acad. Sci. USA 98:15270-15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favorov, M. O., M. Y. Kosoy, S. A. Tsarev, J. E. Childs, and H. S. Margolis. 2000. Prevalence of antibody to hepatitis E virus among rodents in the United States. J. Infect. Dis. 18:449-455. [DOI] [PubMed] [Google Scholar]
- 7.Garkavenko, O., A. Obriadina, J. Meng, D. A. Anderson, H. J. Benard, B. A. Schroeder, Y. E. Khudyakov, H. A. Fields, and M. C. Croxson. 2001. Detection and characterisation of swine hepatitis E virus in New Zealand. J. Med. Virol. 65:525-529. [PubMed] [Google Scholar]
- 8.Hollinger, F. B., and S. U. Emerson. 2001. Hepatitis A virus, p. 799-840. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 9.Hsieh, S. Y., X. J. Meng, Y. H. Wu, S. T. Liu, A. W. Tam, D. Y. Lin, and Y. F. Liaw. 1999. Identity of a novel swine hepatitis E virus in Taiwan forming a monophyletic group with Taiwan isolates of human hepatitis E virus. J. Clin. Microbiol. 37:3828-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, R., N. Nakazono, K. Ishii, D. Li, O. Kawamata, R. Kawaguchi, and Y. Tsukada. 1995. Hepatitis E virus (87A strain) propagated in A549 cells. J. Med. Virol. 47:299-302. [DOI] [PubMed] [Google Scholar]
- 11.Kabrane-Lazizi, Y., J. B. Fine, J. Elm, G. E. Glass, H. Higa, A. Diwan, C. J. Gibbs, Jr., X. J. Meng, S. U. Emerson, and R. H. Purcell. 1999. Evidence for widespread infection of wild rats with hepatitis E virus in the United States. Am. J. Trop. Med. Hyg. 61:331-335. [DOI] [PubMed] [Google Scholar]
- 12.Kabrane-Lazizi, Y., X. J. Meng, R. H. Purcell, and S. U. Emerson. 1999. Evidence that the genomic RNA of hepatitis E virus is capped. J. Virol. 73:8848-8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin, E. V., A. Gorbalenya, M. A. Purdy, M. N. Rozanov, G. R. Reyes, and D. W. Bradley. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. USA 89:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korkaya, H., S. Jameel, D. Gupta, S. Tyagi, R. Kumar, M. Zafrullah, M. Mazumdar, S. K. Lal, L. Xiaofang, D. Sehgal, S. R. Das, and S. Sahal. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 276:42389-42400. [DOI] [PubMed] [Google Scholar]
- 15.Magden, J., N. Takeda, T. Li, P. Auvinen, T. Ahola, T. Miyamura, A. Merits, and L. Kaariainen. 2001. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J. Virol. 75:6249-6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng, J., P. Dubreuil, and J. Pillot. 1997. A new PCR-based seroneutralization assay in cell culture for diagnosis of hepatitis E. J. Clin. Microbiol. 35:1373-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng, X. J. 2000. Novel strains of hepatitis E virus identified from humans and other animal species: is hepatitis E a zoonosis? J. Hepatol. 33:842-845. [DOI] [PubMed] [Google Scholar]
- 18.Meng, X. J., P. G. Halbur, M. S. Shapiro, S. Govindarajan, J. D. Bruna, I. K. Mushahwar, R. H. Purcell, and S. U. Emerson. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 72:9714-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng, X. J., P. G. Halbur, J. S. Haynes, T. S. Tsareva, J. D. Bruna, R. L. Royer, R. H. Purcell, and S. U. Emerson. 1998. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch. Virol. 143:1405-1415. [DOI] [PubMed] [Google Scholar]
- 20.Meng, X. J., R. H. Purcell, P. G. Halbur, J. R. Lehman, D. M. Webb, T. S. Tsareva, J. S. Haynes, B. J. Thacker, and S. U. Emerson. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 94:9860-9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 22.Panda, S. K., I. H. Ansari, H. Durgapal, S. Agrawal, and S. Jameel. 2000. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J. Virol. 74:2430-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell, R. H., and S. U. Emerson. 2001. Hepatitis E virus, p. 3051-3061. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Tam, A. W., M. M. Smith, M. E. Guerra, C. C. Huang, D. W. Bradley, K. E. Fry, and G. R. Reyes. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam, A. W., R. White, E. Reed, M. Short, Y. Zhang, T. R. Fuerst, and R. E. Lanford. 1996. In vitro propagation and production of hepatitis E virus from in vivo-infected primary macaque hepatocytes. Virology 215:1-9. [DOI] [PubMed] [Google Scholar]
- 26.Tei, S., N. Kitajima, K. Takahashi, and S. Mishiro. 2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362:371-373. [DOI] [PubMed] [Google Scholar]
- 27.Tsarev, S. A., T. S. Tsareva, S. U. Emerson, A. Z. Kapikian, J. Ticehurst, W. London, and R. H. Purcell. 1993. ELISA for antibody to hepatitis E virus (HEV) based on complete open-reading frame-2 protein expressed in insect cells: identification of HEV infection in primates. J. Infect. Dis. 168:369-378. [DOI] [PubMed] [Google Scholar]
- 28.Tyagi, S., H. Korkaya, M. Zafrullah, S. Jameel, and S. K. Lal. 2002. The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 277:22759-22767. [DOI] [PubMed] [Google Scholar]
- 29.Wang, X., and S. Gillam. 2001. Mutations in the GDD motif of rubella virus putative RNA-dependent RNA polymerase affect virus replication. Virology 285:322-331. [DOI] [PubMed] [Google Scholar]
- 30.Wei, S., P. Walsh, R. Huang, and S. S. To. 2000. 93G, a novel sporadic strain of hepatitis E virus in South China isolated by cell culture. J. Med. Virol. 61:311-318. [DOI] [PubMed] [Google Scholar]
- 31.Xia, X., R. Huang, and D. Li. 2000. Studies on the subgenomic RNAs of hepatitis E virus. Wei Sheng Wu Xue Bao 41:622-627. [PubMed] [Google Scholar]
- 32.Zafrullah, M., M. H. Ozdener, S. K. Panda, and S. Jameel. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 71:9045-9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, M., R. H. Purcell, and S. U. Emerson. 2001. Identification of the 5′ terminal sequence of the SAR-55 and MEX-14 strains of hepatitis E virus and confirmation that the genome is capped. J. Med. Virol. 65:293-295. [DOI] [PubMed] [Google Scholar]