Abstract
Acetaminophen is a leading cause of acute liver failure (ALF). Genetic differences might predispose some individuals to develop ALF. In this exploratory study, we evaluated genotype frequency differences among patients enrolled by the ALF Study Group who had developed ALF either intentionally from a single-time-point overdose of acetaminophen (n = 78), unintentionally after chronic high doses of acetaminophen (n = 79), or from causes other than acetaminophen (n = 103). The polymorphisms evaluated included those in genes encoding putative acetaminophen-metabolizing enzymes (UGT1A1, UGT1A6, UGT1A9, UGT2B15, SULT1A1, CYP2E1, and CYP3A5) as well as CD44 and BHMT1. Individuals carrying the CYP3A5 rs776746 A allele were overrepresented among ALF patients who had intentionally overdosed with acetaminophen, with an odds ratio of 2.3 (95% confidence interval, 1.1–4.9; P = 0.034) compared with all other ALF patients. This finding is consistent with the enhanced bioactivation of acetaminophen by the CYP3A5 enzyme. Persons homozygous for the CD44 rs1467558 A allele were also overrepresented among patients who had unintentionally developed ALF from chronic acetaminophen use, with an odds ratio of 4.0 (1.0–17.2, P = 0.045) compared with all other ALF subjects. This finding confirms a prior study that found elevated serum liver enzyme levels in healthy volunteers with the CD44 rs1467558 AA genotype who had consumed high doses of acetaminophen for up to 2 weeks. However, both genetic associations were considered relatively weak, and they were not statistically significant after adjustment for multiple comparisons testing. Nevertheless, both CYP3A5 rs776746 and CD44 rs1467558 warrant further investigation as potential genomic markers of enhanced risk of acetaminophen-induced ALF.
Introduction
Acetaminophen is one of the most widely available and commonly used pain-relieving and fever-reducing drugs. Unfortunately, this drug is also one of the leading causes of acute liver failure (ALF) in this country, accounting for approximately half of all ALF cases identified by the Acute Liver Failure Study Group in the United States (Larson et al., 2005). In most cases, liver injury results from an intentional single-time-point overdose of acetaminophen; but in a substantial number of cases, the injury was unintentional, and the acetaminophen was consumed over a more prolonged time period.
Genetic variability could predispose some individuals to a higher risk of acetaminophen-induced liver injury than would be predicted based on the dose ingested, leading to ALF in a subset of these patients (Patel et al., 1992; Court et al., 2001; Rauchschwalbe et al., 2004; Zhao and Pickering, 2011). Candidate genes for this genetic variability would include those encoding enzymes important in the clearance of acetaminophen, including the UDP-glucuronosyltransferases (UGT) and sulfotransferases (SULT), and also the cytochrome P450 enzymes (CYP) responsible for converting acetaminophen to the hepatotoxic N-acetylparaquinoneimine (NAPQI). In a recent study, we identified a polymorphism in the UGT1A gene (rs8330) associated with an increased rate of acetaminophen detoxification by hepatic glucuronidation and a decreased risk of unintentional acetaminophen-induced liver failure (Court et al., 2013). This polymorphism is likely to have a broad impact on drug glucuronidation because it is located in the 3′ untranslated region of the UGT1A gene that is shared through differential gene splicing by all nine transcribed UGT1A isoforms, including all the hepatic isoforms that have been shown to glucuronidate acetaminophen (UGT1A1, 1A6, and 1A9).
Here, we have used DNA samples from patients enrolled by the Acute Liver Failure Study Group to evaluate associations of other polymorphisms in genes encoding the acetaminophen detoxification enzymes UGT1A1, UGT1A6, UGT1A9, UGT2B15, and SULT1A1, as well as the putative NAPQI-forming enzymes CYP2E1 and CYP3A5. We also evaluated a common nonsynonymous polymorphism in the human CD44 gene (I479T) that was associated with increased serum liver injury biomarker levels in healthy volunteers who chronically consumed acetaminophen (Watkins et al., 2006; Harrill et al., 2009), and also a common nonsynonymous polymorphism in the human betaine-homocysteine methyltransferase 1 (BHMT1) gene (R239Q) because the closely related murine Bhmt2 gene was previously identified as protective against acetaminophen-induced hepatotoxicity in a mouse genomics study (Liu et al., 2010).
Materials and Methods
ALF Patient DNA Samples.
DNA samples were obtained with appropriate consent from patients enrolled by the Acute Liver Failure Study Group, a consortium of U.S. liver centers established in 1998 to better define causes and outcomes of ALF (Ostapowicz et al., 2002). Standard entry criteria for ALF were employed. All patients were deemed to have had an acute hepatic injury of less than 26 weeks duration and demonstrated an international normalized ratio (INR) of ≥1.5 accompanied by any degree of hepatic encephalopathy as a result. Samples analyzed included those from 260 white patients meeting the above definition, of which 79 patients were considered to have unintentional acetaminophen overdoses and 78 patients to have intentional overdoses, while the remaining 103 patients had developed ALF from a variety of other causes.
The definitions for these patient categories have previously been established (Schiodt et al., 1997; Larson et al., 2005; Khandelwal et al., 2011). In brief, assigning acetaminophen as the cause of ALF required meeting clinical criteria including a history of ingesting more than 4 g of acetaminophen per day within 7 days of admission, the presence of any level of acetaminophen in serum, and significantly elevated alanine aminotransferase (ALT) levels of ≥ 1000 IU/l, with two of three criteria qualifying for inclusion. Exclusion of other potential causes of ALF (listed below) was also required. The definition used to determine an unintentional ingestion is that acetaminophen was taken over days, with a specific cause of pain elicited and denial of suicidal intent (Schiodt et al., 1997). By contrast, patients who were considered to have taken a suicidal (intentional) overdose had taken the acetaminophen at one time point, denied a cause for pain, and admitted to intent of self-harm. Etiologies for the other 103 patients varied greatly, including indeterminate (27), drug-induced liver injury (23), shock/ischemia (15), hepatitis B (11), autoimmune hepatitis (8), other viruses (3), acute fatty liver of pregnancy (2), Budd-Chiari syndrome (2), hepatitis A (2), hepatitis E (1), Wilson’s disease (1), mushroom ingestion (1), and other causes (7).
Genotyping.
Methods for genotyping the UGT polymorphisms have been described previously elsewhere, including UGT1A1 -53(TA) × 5, 6, 7, or 8 (Girard et al., 2005), UGT1A6 S7A (rs6759892), T181A (rs2070959), and R184S (rs1105879) (Krishnaswamy et al., 2005), UGT1A9 -275T>A (rs6714486) (Girard et al., 2004), UGT2B15*2 (rs1902023, D85Y) (Court et al., 2004), and UGT1A-3′UTR c.2042C>G (rs8330) (Court et al., 2013). Taqman allele discrimination assays were used to genotype DNA samples on a ABI 7300 (Applied Biosystems, Foster City, CA) real-time polymerase chain reaction (PCR) instrument for the CYP3A5*3 (rs776746; ABI assay C_26201809-30), CD44 I479T (rs1467558; ABI assay C_2143203_10), and BHMT1 R239Q (rs3733890; ABI assay C_11646606_20) single-nucleotide polymorphisms (SNPs). The CYP2E1 copy number variation (CYP2E1*1x2) was also determined on a ABI 7300 real-time PCR instrument with the Taqman copy number variation assay (ABI assay Hs00231786_cn) using RNase P (ABI assay 4403326) as the reference gene.
Because reference allele frequency data were not available for CYP2E1*1x2, DNA samples were obtained with appropriate consent from 48 U.S. white liver-bank donors (donor details given in Court, 2010) were assayed in addition to the ALF study subject samples. The CYP2E1*1D tandem repeat polymorphism (six repeats in the C allele and eight repeats in the D allele) was assayed by PCR with agarose gel sizing according to the method described previously elsewhere (Hu et al., 1999) with modified PCR primers including 5′-TGG TAC ATT GTG AGA CAG TG-3′ (Pri-745) and 5′-ATA CGG GAA CAC CTC GTT TG-3′ (Pri-746). The PCR product sizes for the C and D alleles were 610 bp and 700 bp, respectively.
The SULT1A1*2 polymorphism (rs9282861, R213H) was assayed by the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method described previously elsewhere (Wang et al., 2002) with modified primers including 5′-TCA GTA ATC CGA GCC TCC AC-3′ (Pri-741) and 5′-GTT TGA TTC GCA CAC TCC CT-3′ (Pri-742). HaeII digestion of PCR product yielded 309 bp and 393 bp fragments for the *1 and *2 alleles, respectively. The genotypes of representative samples were confirmed by direct sequencing of the PCR product.
Statistical Analyses.
The genotype frequencies of each ALF patient group (unintentional acetaminophen, intentional acetaminophen, or other causes) were compared with the combined genotype frequencies for the other two ALF groups by chi-square test with one degree of freedom (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl, accessed June 25, 2013). Both recessive (combining homozygous reference and heterozygote genotype numbers) and dominant (combining heterozygote and homozygous variant genotype numbers) genetic models were evaluated to enhance the statistical power. Allele frequency differences between ALF patient groups were also evaluated in a similar manner by comparing the allele frequency of one group with the combined allele frequency of the other two ALF groups. P < 0.05 was considered statistically significant. For statistical analysis purposes, the UGT1A1 promoter repeat expansion polymorphism consisting of either 5, 6, 7, or 8 TA repeats (i.e., 4 possible alleles) was reduced to two effective allele groups of either 5 or 6 repeats (fast metabolizer alleles) or 7 or 8 repeats (slow metabolizer alleles).
Results and Discussion
The number of ALF patients with each of the assayed genotypes is given in Table 1, grouped by ALF etiology. None of the genotype frequencies evaluated were found to deviate from the expected Hardy-Weinberg equilibrium values. Allele frequencies for these polymorphisms are shown in Fig. 1 to allow for comparisons with published or database allele frequencies for a race/ethnicity-matched population. We elected to analyze the acetaminophen-induced ALF patients as either one group (versus the nonacetaminophen ALF patients) or as two separate groups based upon intent (intentional or unintentional hepatotoxicity).
TABLE 1.
Comparison of genotype frequencies in 260 patients who developed acute liver failure either unintentionally from chronic acetaminophen use (n = 79), intentionally from acute acetaminophen overdose (n = 78), or from causes other than acetaminophen (n = 103)
| Genotype | Acetaminophen |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Unintentional |
Intentional |
Other causes |
||||||||
| n | (%) | P valuea | n | (%) | P valuea | n | (%) | P valuea | ||
| UGT1A-3′UTR | C/C | 56 | (71) | 0.027b 0.98 | 42 | (54) | 0.13, 0.48 | 60 | (58) | 0.50, 0.53 |
| rs8330 | C/G | 20 | (25) | 32 | (41) | 40 | (39) | |||
| G/G | 3 | (4) | 4 | (5) | 3 | (3) | ||||
| UGT1A1 (TA)n | 56/56 | 39 | (50) | 0.28, 0.71 | 42 | (54) | 0.064, 0.33 | 35 | (35) | 0.0066b 0.56 |
| 5or6->7or8 | 56/78 | 30 | (38) | 30 | (38) | 54 | (53) | |||
| 78/78 | 9 | (12) | 6 | (8) | 12 | (12) | ||||
| UGT1A6 | T/T | 32 | (41) | 0.13, 0.87 | 28 | (36) | 0.58, 0.67 | 26 | (27) | 0.050, 0.58 |
| rs6759892 | T/G | 35 | (44) | 38 | (49) | 55 | (56) | |||
| G/G | 12 | (15) | 11 | (14) | 17 | (17) | ||||
| UGT1A6 | A/A | 40 | (51) | 0.16, 0.38 | 36 | (47) | 0.57, 0.35 | 36 | (37) | 0.061, 0.94 |
| rs2070959 | A/G | 30 | (38) | 36 | (47) | 53 | (54) | |||
| G/G | 9 | (11) | 5 | (6) | 9 | (9) | ||||
| UGT1A6 | A/A | 37 | (47) | 0.23, 0.39 | 35 | (45) | 0.38, 0.19 | 33 | (34) | 0.049b 0.68 |
| rs1105879 | A/C | 32 | (41) | 37 | (48) | 54 | (55) | |||
| C/C | 10 | (13) | 5 | (6) | 11 | (11) | ||||
| UGT1A9 | T/T | 72 | (91) | 0.44 | 66 | (85) | 0.16 | 93 | (90) | 0.55 |
| rs6714486 | T/A | 7 | (9) | 12 | (15) | 10 | (10) | |||
| UGT2B15 | G/G | 21 | (30) | 0.62, 0.45 | 19 | (25) | 0.58, 0.06 | 27 | (28) | 0.95, 0.29 |
| rs1902023 | G/T | 36 | (51) | 34 | (45) | 52 | (53) | |||
| T/T | 14 | (20) | 23 | (30) | 19 | (19) | ||||
| SULT1A1 | G/G | 36 | (46) | 0.87, 0.64 | 34 | (44) | 0.89, 0.94 | 46 | (45) | 0.97, 0.61 |
| rs9282861 | G/A | 35 | (44) | 36 | (47) | 49 | (48) | |||
| A/A | 8 | (10) | 7 | (9) | 8 | (8) | ||||
| CYP2E1*1D | 6/6 | 75 | (96) | 0.83 | 77 | (99) | 0.21 | 98 | (95) | 0.32 |
| 6->8 repeats | 6/8 | 3 | (4) | 1 | (1) | 5 | (5) | |||
| CYP2E1*1x2 | 1/1 | 74 | (94) | 0.94, 0.13 | 74 | (95) | 0.65, 0.51 | 96 | (93) | 0.73, 0.42 |
| 1->2 copies | 1/2 | 4 | (5) | 4 | (5) | 7 | (7) | |||
| 2/2 | 1 | (1) | 0 | (0) | 0 | (0) | ||||
| CYP3A5 | G/G | 74 | (94) | 0.082 | 64 | (82) | 0.034b | 92 | (89) | 0.73 |
| rs776746 | G/A | 5 | (6) | 14 | (18) | 11 | (11) | |||
| CD44 | G/G | 51 | (65) | 0.21, 0.045b | 59 | (76) | 0.19, 0.75 | 72 | (70) | 0.98, 0.11 |
| rs1467558 | G/A | 23 | (29) | 17 | (22) | 30 | (29) | |||
| A/A | 5 | (6) | 2 | (3) | 1 | (1) | ||||
| BHMT1 | G/G | 34 | (43) | 0.56, 0.41 | 31 | (40) | 0.20, 0.10 | 54 | (52) | 0.081, 0.43 |
| rs3733890 | G/A | 37 | (47) | 33 | (42) | 38 | (37) | |||
| A/A | 8 | (10) | 14 | (18) | 11 | (11) | ||||
P values are given for chi-square test of both dominant (left value) and recessive (right value) genetic models comparing differences in genotype numbers for the indicated ALF group with the summed genotype numbers for the other two ALF groups. Only one P value is listed for the UGT1A9, CYP2E1*1D, and CYP3A5 polymorphisms because no homozygous variant genotype individuals were found, so only the dominant model could be tested.
P < 0.05.
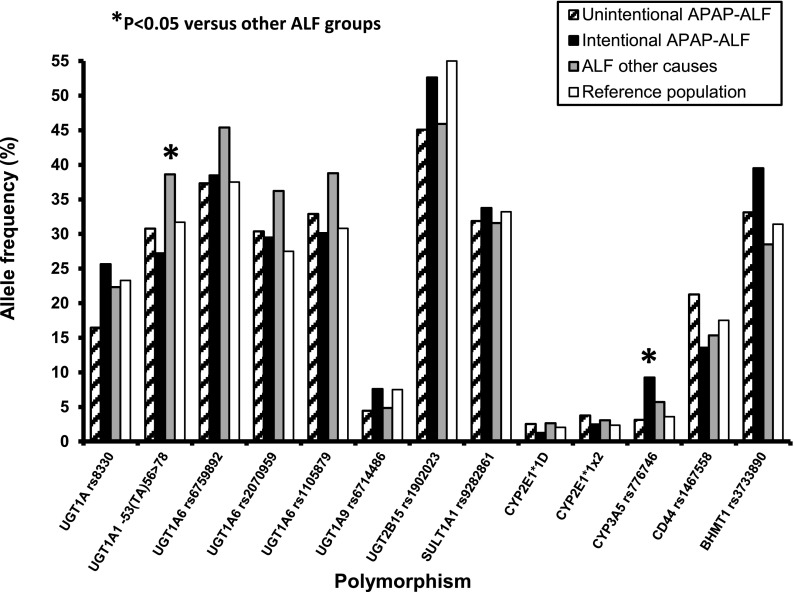
Fig. 1.
Allele frequencies for candidate genes potentially involved in the pathogenesis of acetaminophen-induced acute liver failure (APAP-ALF). Shown are the frequencies determined for 261 patients who had developed ALF either unintentionally from chronic acetaminophen use (n = 79), intentionally from a single-time-point acetaminophen overdose (n = 78), or from causes other than acetaminophen (n = 103). Chi-square analysis showed a higher allele frequency (*P < 0.05) for the CYP3A5 fast-metabolizer rs776746 G allele in patients who had a single-time-point acetaminophen overdose, and for UGT1A1 slow glucuronidation -53(TA)7 or 8 alleles in the patients with ALF from causes other than acetaminophen when compared with the other two ALF groups. Also shown for comparison are allele frequencies obtained from the National Center for Biotechnology Information (NCBI) database for a race/ethnicity-matched white population (60 Utah residents with northern and western European ancestry), except for the CYP2E1*1x2 copy number variation, which were determined in this study using DNA from 42 white individuals (see the Materials and Methods section).
As we reported previously (Court et al., 2013), although the two ALF groups consumed essentially the same total dose of acetaminophen, the intentional group did so at a single time point whereas the unintentional group consumed the acetaminophen over a period of days to weeks such that the daily dose was lower and there was an opportunity for adaptive changes. Consequently, a distinct but potentially overlapping set of genes could influence risk of hepatotoxicity in these ALF groups.
As reported in preliminary form previously (Court et al., 2013), the UGT1A-3′UTR rs8330 variant carrier genotype frequency was significantly lower in the unintentional acetaminophen hepatotoxicity group (29%) as compared with the other ALF subgroups (44%), with an odds ratio (95% confidence interval [CI]) of 0.53 (0.30–0.94; P = 0.027). This finding is consistent with a protective effect of the variant rs8330 G allele through enhancement of acetaminophen glucuronidation and detoxification, as demonstrated by a series of in vitro studies (Court et al., 2013). Conversely, it would imply that noncarriers of this allele (i.e., rs8330 CC genotype) would be at increased risk of unintentional acetaminophen hepatotoxicity. The only other polymorphism associated with the unintentional acetaminophen-induced ALF group was CD44 rs1467558, with patients homozygous for the CD44 variant A allele being overrepresented with an odds ratio of 4.0 (95% CI, 1.0–17.2; P = 0.045). This finding is consistent with a prior study that showed an association of this same polymorphism with elevated serum alanine aminotransferase levels (a biomarker of hepatocellular injury) in healthy volunteers who consumed the maximum recommended dose of acetaminophen for up to 2 weeks (Watkins et al., 2006; Harrill et al., 2009). However, because acetaminophen dosing was stopped in that study once significant liver enzyme elevations were detected in the affected subjects, it is not clear whether those subjects would have proceeded to develop fulminant hepatic failure with continued drug exposure.
Given that both UGT1A-3′UTR rs8330 and CD44 rs1467558 might additively contribute to the risk for acetaminophen-induced ALF, we also determined the number of subjects who had both of the risk genotypes (rs8330 CC and rs1467558 TT). Interestingly, 5 of 156 acetaminophen-induced ALF patients (3 of 79 unintentional, and 2 of 78 intentional) had both of these risk alleles, but none of the 103 nonacetaminophen ALF patients had both these alleles. Unfortunately, the relatively small number of patients with both genotypes precluded any definitive statistical analysis.
The only other polymorphism that appeared to influence the risk for acetaminophen-induced hepatotoxicity was CYP3A5 rs776746, with the A allele being more frequently observed in the intentional acetaminophen-induced ALF patients relative to the patients with ALF from other causes, with an odds ratio of 2.3 (95% CI, 1.1–4.9; P = 0.034). This association is consistent with enhanced formation of NAPQI from acetaminophen by CYP3A5 in individuals carrying the rs776746 A allele, in contrast with individuals who have the G allele but lack active CYP3A5 enzyme because of aberrant gene splicing (Kuehl et al., 2001). Although the capacity of CYP3A5 to oxidize acetaminophen has not been reported, a recent study showed that recombinant CYP3A4 had the highest acetaminophen-bioactivation capacity out of nine human hepatic CYPs tested (other than CYP3A5), including CYP2E1 and CYP1A2 (Laine et al., 2009). Given the high degree of overlap in substrate selectivity between CYP3A enzymes, it is likely that CYP3A5 could oxidize acetaminophen, and this hypothesis should be tested in future studies.
UGT1A1 -53TA(n) also appeared to differ between ALF groups in that there was a higher frequency of carriers of the slow glucuronidation UGT1A1 alleles (7 or 8 TA repeats) in the nonacetaminophen ALF patients (66 of 101 patients; 65%) as compared with patients with acetaminophen-induced ALF (75 of 157 patients; 48%) with an odds ratio of 2.0 (95% CI, 1.2–3.4; P = 0.0066). We recently reported a UGT1A1 slow-metabolizer genotype frequency of 48% (20 of 44 subjects) for white subjects within our human liver bank samples (Yasar et al., 2013), suggesting that the acetaminophen-induced ALF patients probably do not differ in the frequency of this polymorphism from a race- and ethnicity-matched population, whereas the nonacetaminophen ALF patients probably have an unexpectedly higher frequency of slow UGT1A1 glucuronidation carriers. Further stratification of the nonacetaminophen ALF cases by etiology showed the highest frequencies of the UGT1A1 poor-metabolizer genotypes in patients with ALF from shock (12 of 15 patients; 80%), from indeterminate causes (18 of 26 patients; 69%), and from drug-induced hepatitis (14 of 22 patients; 64%).
Slower glucuronidation by UGT1A1 of drugs (other than acetaminophen) administered to these patients may have contributed to the pathogenesis of ALF in these patients. However, a review of the drugs implicated as a cause ALF in the slow metabolizer patients identified only one drug known to be cleared by glucuronidation (valproate), and this drug was used in only one slow and one fast UGT1A1 metabolizer patient. It is also possible that slower glucuronidation of bilirubin by UGT1A1 (the only enzyme capable of conjugating bilirubin) in carriers of the slow-metabolizer UGT1A1 genotypes might also have enhanced the development of symptoms of jaundice in the nonacetaminophen ALF patients.
There are several limitations to the current study that should be pointed out. We were unable to use a classic case-control study design because we did not have access to DNA samples from a control population who had received a similar acetaminophen dose (acute or chronic) as the ALF patients but had not developed ALF. Consequently, our approach may have limited the statistical power to discern genotype frequency differences. The number of DNA samples studied was also relatively small for a genetic association study of this type, although we used all the available ALF patient DNA samples. Furthermore, we tested different genotypes for frequency differences between groups, which would tend to increase the type I (false-positive) error rate, so P = 0.05 is probably not stringent enough for statistical significance. Consequently, our results should be considered exploratory and will need to be confirmed by analysis of other larger patient populations.
In conclusion, we have identified polymorphisms in the CD44 (rs1467558) and CYP3A5 (rs776746) genes that may contribute to risk for acetaminophen-induced ALF, in addition to the previously discovered UGT1A-3′UTR variant (rs8330). These SNPs warrant further investigation as potential genomic markers of enhanced risk of acetaminophen-induced ALF.
Acknowledgments
Members of the Acute Liver Failure Study Group who contributed as coauthors to this paper include William M. Lee (the study principal investigator), Anne M. Larson, Iris Liou, University of Washington, Seattle, WA; Oren Fix, University of California, San Francisco; Michael Schilsky, Mount Sinai School of Medicine, New York, NY (current address: Yale University, New Haven, CT); Timothy McCashland, University of Nebraska, Omaha, NE; Natalie Murray, Baylor University Medical Center, Dallas, TX; A. Obaid S. Shaikh, University of Pittsburgh, Pittsburgh, PA; Daniel Ganger, Northwestern University, Chicago, IL; Atif Zaman, University of Oregon, Portland, OR; Steven H.B. Han, University of California, Los Angeles, CA; Robert Fontana, University of Michigan, Ann Arbor, MI; Brendan McGuire, University of Alabama, Birmingham, AL; Raymond T. Chung, Massachusetts General Hospital, Boston, MA; Alastair Smith, Duke University Medical Center, Durham, NC; Robert Brown, Cornell/Columbia University, New York, NY; Jeffrey Crippin, Washington University, St. Louis, MO; Edwyn Harrison, Mayo Clinic, Scottsdale, AZ; Adrian Reuben, Medical University of South Carolina, Charleston, SC; Santiago Munoz, Albert Einstein Medical Center, Philadelphia, PA; Rajender Reddy, University of Pennsylvania, Philadelphia, PA; R. Todd Stravitz, Virginia Commonwealth University, Richmond, VA; Lorenzo Rossaro, University of California at Davis, Sacramento, CA; Raj Satyanarayana, Mayo Clinic, Jacksonville, FL; Grace Samuel, Carla Pezzia, Corron Sanders, Nahid Attar, and Linda S. Hynan of the University of Texas Southwestern Administrative Group; and Valerie Durkalski, Catherine Dillon, Holly Battenhouse, and Wenle Zhao of the Medical University of South Carolina Data Coordination Unit. Other members of the Acute Liver Failure Study Group during 1998–2011 who also contributed significantly to this work include Timothy Davern, University of California, San Francisco, CA (current address: California Pacific Medical Center, San Francisco, CA); Andres Blei, Northwestern University, Chicago, IL (deceased); J. Eileen Hay, Mayo Clinic, Rochester, MN; Tarek Hassanein, University of California, San Diego, CA; Ezmina Lalani, University of Texas Southwestern Administrative Group; and Tomoko Goddard, Medical University of South Carolina Data Coordination Unit.
Abbreviations
- ALF
acute liver failure
- BHMT1
betaine-homocysteine methyltransferase 1
- CYP
cytochrome P450
- NAPQI
N-acetylparaquinoneimine
- PCR
polymerase chain reaction
- SNP
single-nucleotide polymorphism
- SULT
sulfotransferase
- UGT
UDP-glucuronosyltransferase
Authorship Contributions
Participated in research design: Court, Lee, Greenblatt, Rossaro, Munoz, Shaikh, Crippin, McGuire, Chung, Fix, McCashland, Ganger, Fontana, Brown, Schilsky, Satyanarayana, Reuben, Liou, Hynan, Sanders, Durkalski, Dillon, Battenhouse, Attar, Pezzia, Samuel.
Conducted experiments: Court, Vasiadi, Hazarika, Zaman, Rossaro, Munoz, Larson, Murray, Smith, Brown, Satyanarayana.
Contributed new reagents or analytic tools: Greenblatt, Stravitz, Crippin, Harrison, Reddy, Zhao.
Performed data analysis: Court, Vasiadi.
Wrote or contributed to the writing of the manuscript: Court, Lee, Greenblatt, Peter, Rossaro, Han, Smith, Fontana.
Footnotes
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grants R01-GM061834 and R01-GM102130] (to M.H.C.); and by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant U01-DK058369] (to W.M.L. for the Acute Liver Failure Study Group), and [Contract N01-DK-7-0004 / HHSN267200700004C] (to the Liver Tissue and Cell Distribution System, Minneapolis, Minnesota).
References
- Court MH. (2010) Interindividual variability in hepatic drug glucuronidation: studies into the role of age, sex, enzyme inducers, and genetic polymorphism using the human liver bank as a model system. Drug Metab Rev 42:209–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH, Duan SX, von Moltke LL, Greenblatt DJ, Patten CJ, Miners JO, Mackenzie PI. (2001) Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP-glucuronosyltransferase isoforms. J Pharmacol Exp Ther 299:998–1006 [PubMed] [Google Scholar]
- Court MH, Freytsis M, Wang X, Peter I, Guillemette C, Hazarika S, Duan SX, Greenblatt DJ, Lee WM, Acute Liver Failure Study Group (2013) The UDP-glucuronosyltransferase (UGT) 1A polymorphism c.2042C>G (rs8330) is associated with increased human liver acetaminophen glucuronidation, increased UGT1A exon 5a/5b splice variant mRNA ratio, and decreased risk of unintentional acetaminophen-induced acute liver failure. J Pharmacol Exp Ther 345:297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH, Hao Q, Krishnaswamy S, Bekaii-Saab T, Al-Rohaimi A, von Moltke LL, Greenblatt DJ. (2004) UDP-glucuronosyltransferase (UGT) 2B15 pharmacogenetics: UGT2B15 D85Y genotype and gender are major determinants of oxazepam glucuronidation by human liver. J Pharmacol Exp Ther 310:656–665 [DOI] [PubMed] [Google Scholar]
- Girard H, Court MH, Bernard O, Fortier LC, Villeneuve L, Hao Q, Greenblatt DJ, von Moltke LL, Perussed L, Guillemette C. (2004) Identification of common polymorphisms in the promoter of the UGT1A9 gene: evidence that UGT1A9 protein and activity levels are strongly genetically controlled in the liver. Pharmacogenetics 14:501–515 [DOI] [PubMed] [Google Scholar]
- Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, Hao Q, von Moltke LL, Greenblatt DJ, Guillemette C. (2005) UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology 42:448–457 [DOI] [PubMed] [Google Scholar]
- Harrill AH, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, Boorman GA, Russo MW, Sackler RS, Harris SC, et al. (2009) Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res 19:1507–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Hakkola J, Oscarson M, Ingelman-Sundberg M. (1999) Structural and functional characterization of the 5′-flanking region of the rat and human cytochrome P450 2E1 genes: identification of a polymorphic repeat in the human gene. Biochem Biophys Res Commun 263:286–293 [DOI] [PubMed] [Google Scholar]
- Khandelwal N, James LP, Sanders C, Larson AM, Lee WM, Acute Liver Failure Study Group (2011) Unrecognized acetaminophen toxicity as a cause of indeterminate acute liver failure. Hepatology 53:567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy S, Hao Q, Al-Rohaimi A, Hesse LM, von Moltke LL, Greenblatt DJ, Court MH. (2005) UDP glucuronosyltransferase (UGT) 1A6 pharmacogenetics: I. Identification of polymorphisms in the 5′-regulatory and exon 1 regions, and association with human liver UGT1A6 gene expression and glucuronidation. J Pharmacol Exp Ther 313:1331–1339 [DOI] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, et al. (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391 [DOI] [PubMed] [Google Scholar]
- Laine JE, Auriola S, Pasanen M, Juvonen RO. (2009) Acetaminophen bioactivation by human cytochrome P450 enzymes and animal microsomes. Xenobiotica 39:11–21 [DOI] [PubMed] [Google Scholar]
- Larson AM, Polson J, Fontana RJ, Davern TJ, Lalani E, Hynan LS, Reisch JS, Schiødt FV, Ostapowicz G, Shakil AO, et al. Acute Liver Failure Study Group (2005) Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology 42:1364–1372 [DOI] [PubMed] [Google Scholar]
- Liu HH, Lu P, Guo Y, Farrell E, Zhang X, Zheng M, Bosano B, Zhang Z, Allard J, Liao G, et al. (2010) An integrative genomic analysis identifies Bhmt2 as a diet-dependent genetic factor protecting against acetaminophen-induced liver toxicity. Genome Res 20:28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapowicz G, Fontana RJ, Schiødt FV, Larson A, Davern TJ, Han SH, McCashland TM, Shakil AO, Hay JE, Hynan L, et al. U.S. Acute Liver Failure Study Group (2002) Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med 137:947–954 [DOI] [PubMed] [Google Scholar]
- Patel M, Tang BK, Kalow W. (1992) Variability of acetaminophen metabolism in Caucasians and Orientals. Pharmacogenetics 2:38–45 [DOI] [PubMed] [Google Scholar]
- Rauchschwalbe SK, Zühlsdorf MT, Wensing G, Kuhlmann J. (2004) Glucuronidation of acetaminophen is independent of UGT1A1 promotor genotype. Int J Clin Pharmacol Ther 42:73–77 [DOI] [PubMed] [Google Scholar]
- Schiødt FV, Rochling FA, Casey DL, Lee WM. (1997) Acetaminophen toxicity in an urban county hospital. N Engl J Med 337:1112–1117 [DOI] [PubMed] [Google Scholar]
- Wang Y, Spitz MR, Tsou AM, Zhang K, Makan N, Wu X. (2002) Sulfotransferase (SULT) 1A1 polymorphism as a predisposition factor for lung cancer: a case-control analysis. Lung Cancer 35:137–142 [DOI] [PubMed] [Google Scholar]
- Watkins PB, Kaplowitz N, Slattery JT, Colonese CR, Colucci SV, Stewart PW, Harris SC. (2006) Aminotransferase elevations in healthy adults receiving 4 grams of acetaminophen daily: a randomized controlled trial. JAMA 296:87–93 [DOI] [PubMed] [Google Scholar]
- Yasar U, Greenblatt DJ, Guillemette C, Court MH. (2013) Evidence for regulation of UDP-glucuronosyltransferase (UGT) 1A1 protein expression and activity via DNA methylation in healthy human livers. J Pharm Pharmacol 65:874–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Pickering G. (2011) Paracetamol metabolism and related genetic differences. Drug Metab Rev 43:41–52 [DOI] [PubMed] [Google Scholar]