Abstract
This report summarizes a symposium sponsored by the American Society for Pharmacology and Experimental Therapeutics at Experimental Biology held April 20-24 in Boston, MA. Presentations discussed the status of cytochrome P450 (P450) knowledge, emphasizing advances and challenges in relating structure with function and in applying this information to drug design. First, at least one structure of most major human drug-metabolizing P450 enzymes is known. However, the flexibility of these active sites can limit the predictive value of one structure for other ligands. A second limitation is our coarse-grain understanding of P450 interactions with membranes, other P450 enzymes, NADPH–cytochrome P450 reductase, and cytochrome b5. Recent work has examined differential P450 interactions with reductase in mixed P450 systems and P450:P450 complexes in reconstituted systems and cells, suggesting another level of functional control. In addition, protein nuclear magnetic resonance is a new approach to probe these protein/protein interactions, identifying interacting b5 and P450 surfaces, showing that b5 and reductase binding are mutually exclusive, and demonstrating ligand modulation of CYP17A1/b5 interactions. One desired outcome is the application of such information to control drug metabolism and/or design selective P450 inhibitors. A final presentation highlighted development of a CYP3A4 inhibitor that slows clearance of human immunodeficiency virus drugs otherwise rapidly metabolized by CYP3A4. Although understanding P450 structure/function relationships is an ongoing challenge, translational advances will benefit from continued integration of existing and new biophysical approaches.
Introduction
In humans, the diverse cytochrome P450 (P450) superfamily consists of 57 genes and 58 distinct mono-oxygenase enzymes with roles ranging from foreign compound metabolism to the oxidation of key endogenous compounds, including vitamins, eicosanoids, steroids, and fatty acids. A single xenobiotic-metabolizing P450 may oxidize hundreds of chemically distinct substrates, while those involved in endogenous pathways often act on a few structurally similar substrates. These membrane proteins are found in either the endoplasmic reticulum or mitochondrial membranes, and require electron delivery via NADPH–cytochrome P450 reductase (CPR) or adrenodoxin, respectively. Additionally, the small heme protein cytochrome b5 (b5) can modulate catalysis. Understanding P450 function therefore requires interrogating the structural features that control interactions among P450s, redox partner proteins, b5, substrates, and the membrane with an ever-expanding set of biophysical techniques. The resulting knowledge is key for progress in comprehending and predicting drug metabolism and manipulating human physiology in many disease states.
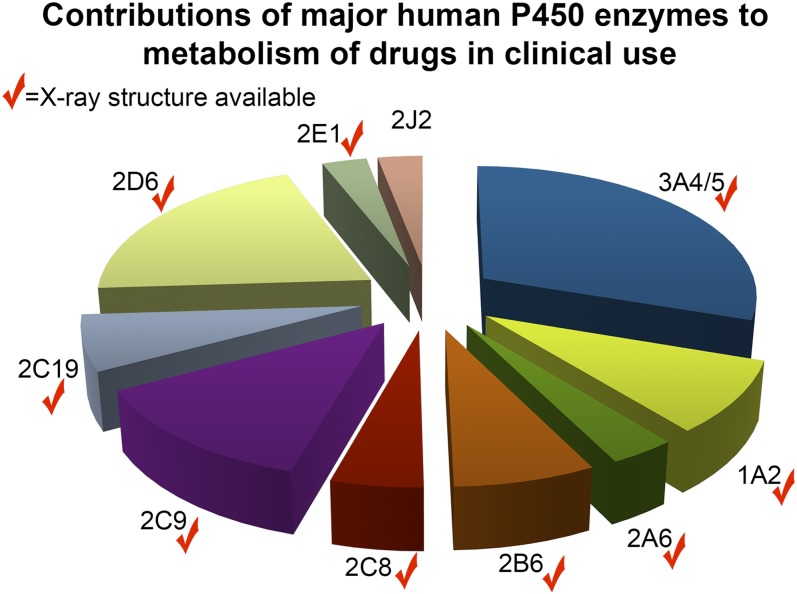
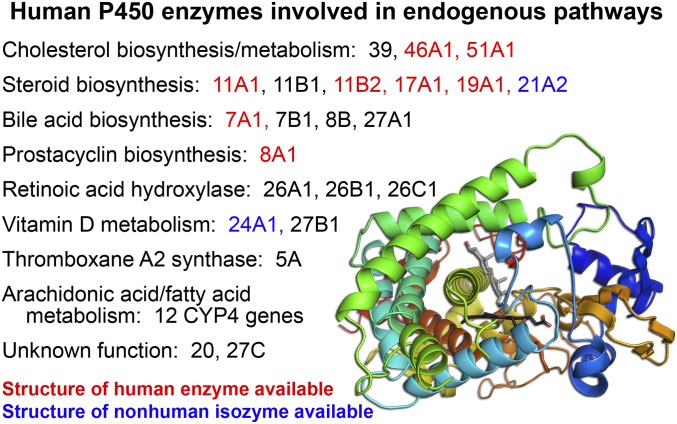
From a structural perspective, we probably know most about specific small-molecule ligand interactions with P450 enzymes garnered largely from structures determined by X-ray crystallography. Building on approaches used to determine the first structure of a membrane cytochrome P450 in 2000 (Williams et al., 2000), structures are now available for most major human drug-metabolizing P450 enzymes (Fig. 1). A synthesis of our basic understanding of common and divergent elements among these enzymes was presented by Eric Johnson. It has become obvious that the flexibility of many xenobiotic-metabolizing P450 active sites results in very different interactions with different ligands. It is frequently true that the structure of a particular P450/ligand complex is of limited utility in understanding the binding mode(s) of structurally distinct ligands to the same P450. There is a significant bottleneck in the time- and labor-intensive process of generating each complex structure de novo experimentally that does not match the pace of drug development, but computational approaches can be confounded by the same protein flexibility. One might expect that P450 enzymes involved in the oxidation of a much more restricted range of endogenous substrates (Fig. 2) might have less flexibility, and thus, these structures may be more useful for approaches like in silico docking and structure-based drug design. However, structures of many of these enzymes are not yet available (Fig. 2).
Fig. 1.
Major human drug-metabolizing P450 enzymes by percentage of clinically used drugs that each metabolizes. Data from (Zanger and Schwab, 2013). Checkmarks indicate enzymes for which at least one structure is present in the Protein Data Bank.
Fig. 2.
Numerous human endogenous pathways include one or more cytochrome P450 enzymes. Those for which structures of the human enzyme are known are in red; those with structures of mammalian homologs are in blue; while those without structures are in black. Lower right inset, structure of CYP17A1 with the prostate cancer drug abiraterone (gray sticks) above the heme prosthetic group (black sticks) displays the common P450 structural elements colored from the N-terminus (blue) to the C-terminus (red).
We know much less about human P450 enzyme interactions with other relevant human proteins. Although structures are available for some P450 enzymes, for human CPR (Xia et al., 2011), and for human microsomal b5 individually (PDB 2I96), no structures are available for human P450 enzymes as a complex with CPR or b5. In vivo, CPR concentrations are typically far below those of induced P450 proteins (Estabrook et al., 1971; Peterson et al., 1976), suggesting that P450 enzymes might compete for available CPR. A presentation from the Backes group further demonstrated that CYP1A2 and CYP2B4 can form heterocomplexes that alter their respective catalytic activities, presumably by modulating their relative binding of CPR. Such P450/P450 complex formation has been validated using crosslinking and bioluminescence energy transfer (BRET) in both simple reconstituted lipid systems and in cells. P450 interactions with cytochrome b5 are complex as well, variously resulting in inhibition, no effect, or increases in P450 catalysis depending on the P450 enzyme, reaction conditions, and substrate. A third presentation from the Scott laboratory elucidated the advantages of protein-detected nuclear magnetic resonance (NMR) as a relatively new approach to investigate membrane P450 interactions with ligands and other proteins. Such studies have great potential to map ligand or protein binding anywhere on the P450 and to probe P450 conformational dynamics. Early studies illustrate part of this potential, having probed the interactions of b5 with human CYP17A1 (Estrada et al., 2013) and with CYP2B4 (Ahuja et al., 2013), confirming residues involved in the interface and revealing that ligands in the P450 active site alter its interactions with other proteins.
To be able to exploit P450 enzymes clinically, the pharmaceutical industry is interested in being able to use structure-function information to predict drug metabolism and to design inhibitors of certain cytochrome P450 enzymes. The most well known and clinically successful examples of P450 enzymes as drug targets involve biosynthetic pathways. Aromatase (CYP19A1) inhibitors are powerful clinical tools to halt estrogen production in hormone-responsive breast cancer (Chumsri et al., 2011). Azole inhibitors of 14α-demethylase (CYP51) block ergosterol synthesis necessary for fungal membrane stability (Kelly et al., 2001). Newer to the market is a CYP17A1 inhibitor that blocks androgen production in prostate cancer patients (Fig. 2). However, there are numerous additional opportunities to target not only P450 enzymes involved in many biosynthetic pathways (Fig. 2), but also P450 enzymes involved in xenobiotic metabolism—either to prevent procarcinogen activation or manipulate drug metabolism. Although many drug companies routinely discontinue the development of compounds that inhibit CYP3A4 because of this enzyme’s involvement in the metabolism of numerous drugs (Fig. 1) and thus its high potential liability for drug-drug interactions, others have sought to take advantage of this capability. The final presentation summarized herein highlighted the design, development, and testing of a novel CYP3A4 inhibitor by Gilead Sciences. Compound development focused on ablating the original human immunodeficiency virus (HIV) protease inhibition while optimizing CYP3A4-selective inhibition to generate cobicistat. This pharmacoenhancer effectively slows the clearance of HIV drugs otherwise rapidly metabolized by CYP3A4. Thus, P450 enzymes involved in either endogenous or xenobiotic metabolism can be effective drug targets, with therapeutic approaches benefitting as we learn more about the relationships between structure and function in these fascinating human enzymes.
Cytochrome P450 Structure: Common Themes and Variations on the Theme
(Eric F. Johnson).
Cytochrome P450s exhibit a conserved and distinctive tertiary structure (Poulos et al., 1987; Hasemann et al., 1995; Williams et al., 2000; Sirim et al., 2010). Human P450s that typically mediate metabolic clearance of drugs are targeted to the endoplasmic reticulum by an N-terminal signal sequence that spans the membrane bilayer with the catalytic domain residing in the cytosol and partially embedded into the membrane surface. As the membrane-targeting leader sequences contribute to aggregation and are not integral parts of the catalytic domain, almost all X-ray crystal structures of microsomal P450s have been determined for proteins expressed without their signal anchor sequence (Johnson and Stout, 2013).
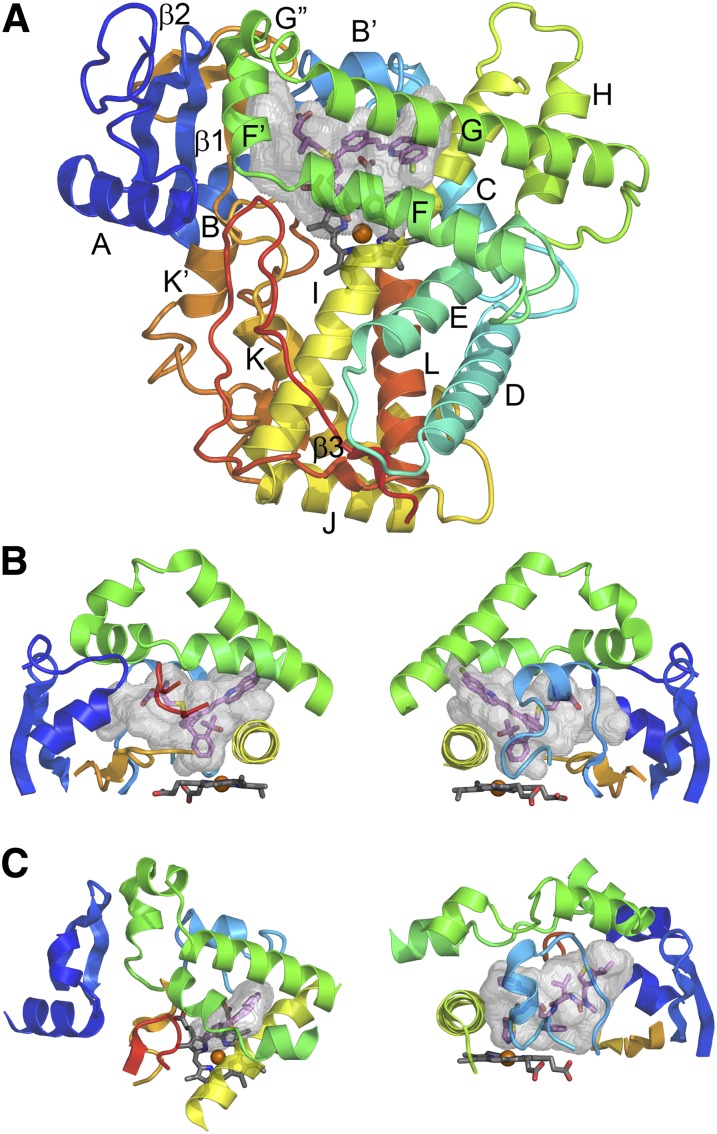
Overall, 12 helices, which are sequentially designated by letters A to L from the N-terminus and three β-sheets are commonly observed for P450 structures, with individual P450s typically exhibiting additional sheets and helices that are less conserved (Fig. 3A). P450s exhibit a triangular prism shape that reflects a V-shaped structural core. Helix I passes through the interior of the protein and packs with helices C, D, E, and H to form a helix bundle along one edge of the protein. Helices J and K form the base of the V. The other side of the V is formed by β-sheet 1, which consists of strands formed by the N-terminal portion of the protein as well as portions of the polypeptide chain that follow helix K. The heme prosthetic group resides between the two sides of the V, above a highly conserved structural motif that contains a conserved cysteine that binds to the heme iron. Molecular oxygen binds to the distal axial position of the heme iron, where sequential reduction and protonation lead to cleavage of the oxygen double bond with expulsion of a water molecule and formation of a reactive iron oxo porphyrin cation radical intermediate that reacts with substrates (Rittle and Green, 2010). The major electron donor for reduction of the heme iron is NADPH cytochrome P450 oxidoreductase, which is also targeted to the endoplasmic reticulum by an N-terminal signal anchor sequence. The reductase exhibits two domains that resemble flavodoxin reductases and flavodoxins, respectively, and a third domain that links the other two domains (Wang et al., 1997). The reduced flavodoxin domain transfers electrons to the heme iron of the P450 by binding to the triangular surface of the protein proximal to the heme.
Fig. 3.
P450 secondary and tertiary structure. (A) Topological features of a microsomal P450 as illustrated by the Protein Data Bank:2NNI structure of human microsomal 2C8 colored from blue at the N-terminus to red at the C-terminus. The active site cavity is shown as a transparent surface. The bound substrate, montelukast (violet carbons), and the heme prosthetic group (gray carbons) are shown as stick figures. Twelve helices designated by letters A–L and β-sheets 1 and 2 are highly conserved. Additional helices are evident that are named by letters with prime or double prime designations. (B) Two views of structural components that form the sides of the substrate binding site of 2C8. The F–G region (green) forms the top of the cavity and is cantilevered over helix I (yellow) which forms one side. The opposite side is formed by connections (orange) between helix K and β1–3 and between β1–4 and helix K' near the surface of the heme, and by the N-terminal region (dark blue) that includes helix A and β-1. The gaps under the helix F–G region between helix I and the N-terminal region are filled by the C-terminal loop (red orange) as shown in the left panel and by the B–C loop (light blue) as shown in the right panel. (C) A view of the helix F–G side of human 1A2 α-napthoflavone complex, PDB:2HI4 (left) and of the B–C loop side of the human 3A4 ritonavir complex, Protein Data Bank:3NXU (right) illustrates differences in the topologies of the active sites and the secondary and tertiary structures of the three proteins. This figure was originally published in Johnson EF and Stout CD (2013) Structural diversity of eukaryotic membrane cytochrome P450s. J Biol Chem 288:17082–17090. ©The American Society for Biochemistry and Molecular Biology.
Substrates bind in a cavity between helix I and the opposite corner of the triangle on the distal side of the heme (Poulos et al., 1987; Johnson and Stout, 2013) (Fig. 3, A–C). Substrate oxygenation is initiated by abstraction of a hydrogen atom from the substrate by the oxo atom of the reactive intermediate, followed by recombination of the resulting hydroxyl and substrate radicals (Groves et al., 1978). The hydroxylated product formed by recombination may rearrange to yield more stable products. Inherent differences in the reactivity of substrate atoms (Korzekwa et al., 1990; Yin et al., 1995; Jones et al., 2002) and the probability for close proximity of the atom to the reactive intermediate are also determinants of sites and rates of reaction (Cruciani et al., 2005). The presence of the substrate in close proximity to the heme iron can influence rates of reduction and reduce the formation of alternative products of oxygen reduction (Sligar, 1976). As drug metabolism often reflects the metabolism of new chemical entities that bind to the enzymes in suboptimal ways, these reactions are often poorly coupled and produce multiple metabolites that are likely to reflect different binding orientations and/or dynamic motion within the substrate binding cavity, as suggested by molecular dynamics studies (Collins and Loew, 1988; Bass et al., 1992). Despite these inefficiencies, the overall rates of metabolic clearance are an important determinant of drug dosage, safety, and efficacy. Moreover, compounds that bind tightly to drug-metabolizing enzymes can lead to unsafe levels of another drug as a result of inhibition of the other drug's metabolism (O'Brien and de Groot, 2005; de Groot, 2006; Sun and Scott, 2010).
The substrate-binding cavity is formed by several loop-like structures beginning and ending in close proximity that often contain regular secondary structures (Fig. 3, A–C). The helix F–G region forms a loop that begins and ends in close proximity between helices E and H. The F–G region arches over helix I and the substrate-binding cavity. This region often exhibits four helices: F, F', G', and G. The C-terminal loop projects into the active site under helix F between helix I and helix A to define a side of the cavity. The C-terminal loop begins after helix L and ends near the C-terminus, where the beginning and end of the loop form antiparallel strands of a β-sheet 3 (Fig. 3A). This loop may also contain an antiparallel β-sheet close to the hairpin turn that resides in the active site. A third loop that begins after helix B and ends in close proximity before helix C fills the gap between helix I, β-sheet 1, and helix G and forms the third side of the triangle. The B–C loop often exhibits a B' helix.
Dynamic relationships between the loop-like structures that form the substrate binding cavity enable adaptations that open and close solvent access channels to facilitate access to and egress from the active site for solvent molecules, substrates, inhibitors, and products (Cojocaru et al., 2007) and that facilitate adaptations to accommodate structurally diverse drugs (Wester et al., 2003; Ekroos and Sjogren, 2006; Halpert, 2011). These adaptations can be important for positioning the substrate near the reactive intermediate and prolongation of residency during the reaction cycle. Conversely, X-ray crystallography, which has been used to determine the atomic structures of drug-metabolizing P450s, requires that the protein crystallizes in ways that restrain the internal dynamics of the protein, as motion would lead to diffuse scattering of X-rays. Fortunately, some aspects of conformational dynamics can be inferred from structural changes that occur when the proteins are crystallized with diverse ligands and under different conditions.
Six P450s—3A4, 2C9, 2D6, 1A2, 2C19, and 2C8—are hepatic enzymes that contribute most often to metabolic clearance of the most frequently prescribed therapeutic drugs (Zanger et al., 2008). Comparisons of structures for the six enzymes reveal distinct active site topologies and chemical properties that contribute to differential substrate recognition, as shown in Fig. 3, B and C for P450s 2C8, 1A2, and 3A4. Of these six P450s, 1A2 exhibits the smallest and narrowest active site (Sansen et al., 2007). This architecture is conserved in structures of P450s 1A1 (Walsh et al., 2013) and 1B1 (Wang et al., 2011), and the narrow hydrophobic cavity is consistent with the roles of these enzymes in metabolism of polycyclic aromatic hydrocarbons. This narrow cavity is reinforced by the passage of helix F below helix G, and this redirection of the F helix is associated with a distortion of the F helix as it passes over the active site cavity (Fig. 3C). The active site of P450 2C8 is narrow near the heme iron but is broader than P450 1A2 in the upper portions of the cavity (Schoch et al., 2004; Schoch et al., 2008), as also seen for P450s 2C9, 2C19, and 2D6. This narrow portion of the cavity near the heme iron sequesters an aromatic group of montelukast close to the reactive intermediate in the structure of P450 2C8 (Fig. 3B). In contrast, the active site cavity of P450 3A4 exhibits a much larger volume near the heme iron, which reflects smaller amino acid side chains and a change in conformation for the connector between helix K and β-sheet 1 (Fig. 3C). The helix F–G region of P450 3A4 also differs from the family 1 and 2 enzymes in that the portions of the F–G loop that cross over the large active site do not exhibit a regular secondary structure (Williams et al., 2004; Yano et al., 2004). Comparisons of different P450 3A4 structures indicate that this region flexes to accommodate different substrates and inhibitors (Ekroos and Sjogren, 2006; Sevrioukova and Poulos, 2012).
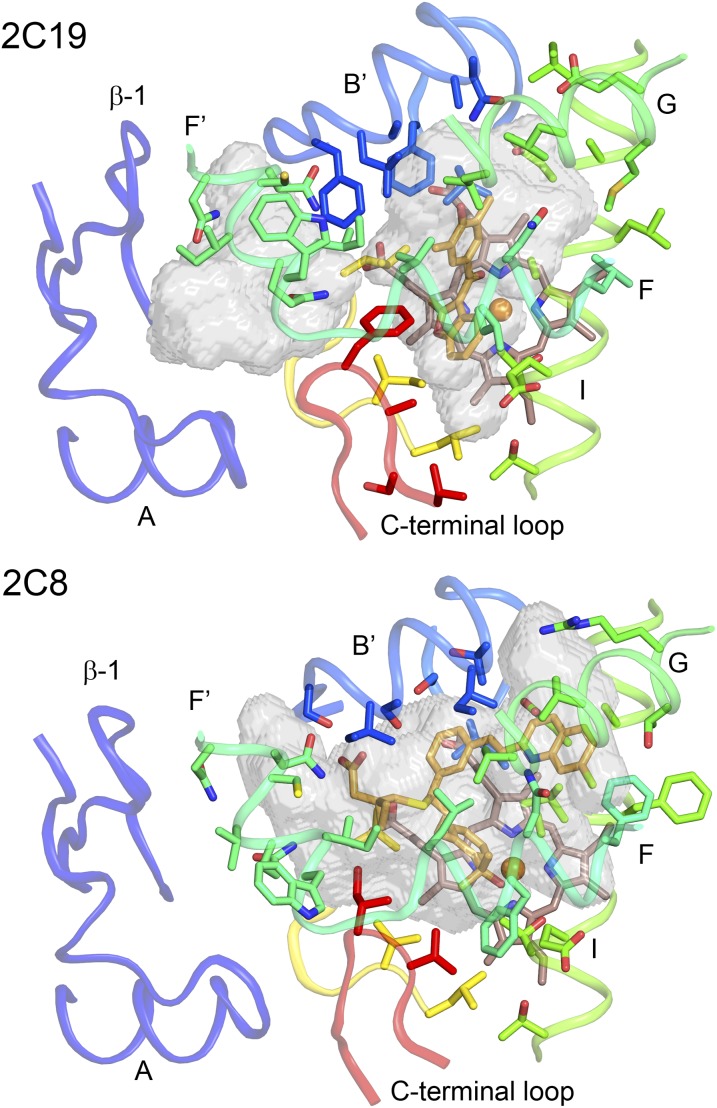
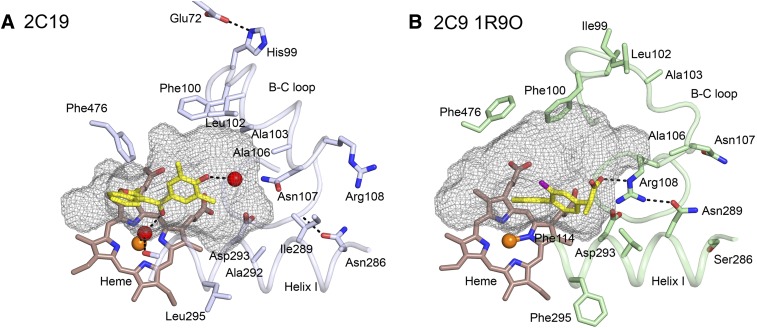
Three of the six enzymes discussed here are in the 2C subfamily and have much higher amino acid sequence identities with each other than with P450s 1A2, 2D6, or 3A4. P450 2C8 exhibits 75% amino acid sequence identity with P450s 2C19 and 2C9, whereas P450s 2C9 and 2C19 exhibit >90% sequence identity. Although this sequence conservation leads to a higher degree of conservation for the structural cores of the enzymes, their active sites differ. While P450s 2C8 and 2C19 exhibit highly similar backbone conformations, 30 of the 50 amino acid side chains in the active site of P450 2C8 differ from those of P450 2C19 (Reynald et al., 2012). Moreover, the presence of larger aromatic residues in the P450 2C19 active site partitions the cavity into a smaller active site and an antechamber under helix F' that is likely to serve as a substrate entrance channel (Fig. 4). The PDB 1R9O structure of the P450 2C9–flurbiprofen complex exhibits an active site that is similar to that of P450 2C19 (Reynald et al., 2012). A major difference is seen for the conformation of the helix B–C loop that allows Arg108, which is outside the active site of P450 2C19, to move into the active site cavity and form an ionic bond with the carboxylate of the substrate flurbiprofen (Wang et al., 2012) (Fig. 5). These distinct conformations of the helix B–C loop also lead to differences in the shapes of the active site cavities of the two enzymes. Mutagenesis studies suggest that these conformational differences reflect amino acid differences for residues 286 and 289 that influence the conformation of the helix B–C loop (Ibeanu et al., 1996; Jung et al., 1998; Klose et al., 1998; Tsao et al., 2001).
Fig. 4.
Comparison of the active site cavities of 2C19 complexed with (2-methyl-1-benzofuran-3-yl)-(4-hydroxy-3,5-dimethylphenyl)methanone (Protein Data Bank:4GQS) and 2C8 complexed with montelukast (Protein Data Bank:2NNI). Differences in the sizes, shapes, and polarity of the amino acid side-chains lining the two cavities lead to changes in the shapes of the cavities and to the presence of two cavities in 2C19. Carbons are colored from blue at the N-terminus to red at the C-terminus for the proteins. Ligand and heme carbons are colored orange and brown, respectively. Oxygen, nitrogen, chlorine, sulfur, and iron are colored red, blue, green, yellow, and orange, respectively. The active sites and antechamber are shown as transparent surfaces.
Fig. 5.
The active site cavities (mesh surface) of the P450 2C19 complex (A) with (2-methyl-1-benzofuran-3-yl)-(4-hydroxy-3,5-dimethylphenyl)methanone (Protein Data Bank:4GQS) and of the 2C9 complex (B) with flurbiprofen (Protein Data Bank:1R9O). Amino acid residues that contribute to differential substrate selectivity and/or that directly shape the distal portions of the cavity are displayed as sticks with carbons colored light blue and pale green, respectively. Ligand and heme carbons are colored yellow and brown, respectively. Oxygen, nitrogen, fluorine, and iron are colored red, blue, purple, and orange. Water molecules are represented as a red spheres; hydrogen bonds are represented by dashed lines. This figure was originally published in Reynald RL, Sansen S, Stout CD, and Johnson EF (2012) Structural characterization of human cytochrome P450 2C19: Active site differences between P450's 2C8, 2C9 and 2C19. J Biol Chem 287:44581–44591. ©The American Society for Biochemistry and Molecular Biology.
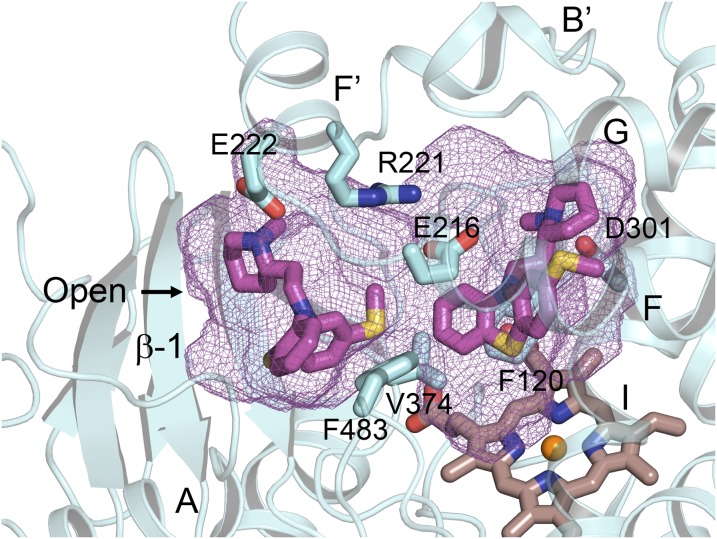
Conversely, P450 2D6 contributes significantly to metabolic clearance of substrates with positive charges, and this role reflects the presence of acidic amino acid side-chains, Glu216, and Asp301 in the active site (Wang et al., 2009). Although Glu216 is unique to P450 2D6, Asp301 is conserved in P450s 2C8, 2C9, and 2C19, as well as 1A2. Asp301 in P450 2D6 exhibits greater accessibility to solvent and substrates in P450 2D6; this reflects in part an insertion of four additional amino acids relative to the 2C P450s in the connector between the helix B' and helix C connector. This is illustrated by the binding of thioridazine in P450 2D6, where thioridazine forms an ionic bond with Asp301 in crystal structures of the complex (Fig. 6). This complex has been crystallized in two crystal lattices. In the conformation of the protein seen in one lattice, thioridazine is bound in a closed active site with an adjacent antechamber similar to that seen for the P450 2D6 prinomastat complex (Wang et al., 2012) and for P450 2C19 (Reynald et al., 2012). In the second crystalline form, the protein exhibits an open conformation that is stabilized by the binding of a second molecule, thioridazine, in the antechamber adjacent to the active site (Fig. 6). As a result, the entrance channel opens at the juncture of the first turn in β-sheet 1, helix A, and helix F', creating a continuous cavity from the active site to the surface of the enzyme. Interestingly, the thioridazine molecule in the access channel interacts with Glu222 near the entrance. This amino acid side chain may facilitate initial binding of charged substrate molecules in the entrance channel for translocation to the active site for metabolism, where Glu216 or Asp301 would stabilize binding of the charged substrate for metabolism.
Fig. 6.
The substrate binding site of 2D6 complexed with two molecules of thioridazine (PDB:3TBG). Simultaneous binding of two molecules leads to an open active site cavity with one thioridazine molecule bound close to the heme with its charged, protonated nitrogen forming an ionic bond with Asp301 on helix I. The second thioridazine molecule is bound in the open entrance channel with the protonated nitrogen forming an ionic bond with Asp222. Two additional charged amino acids in the 2D6 active site, Glu216 and Arg221, exhibit favorable ionic interactions in this complex. Protein, ligand, and heme carbons are colored cyan, magenta, and brown, respectively. Heteroatoms are colored as described in the previous figure legends.
This access channel in P450 2D6 corresponds to that first described for the prokaryotic P450 102A1 (Ravichandran et al., 1993), which closes when a substrate is bound (Li and Poulos, 1997). The presence of a second substrate molecule in the similarly expanded active site cavities has been reported also for the adrenal steroid 21-hydroxylase, P450 21A2, with progesterone, P450s 2B4 and 2B6 with amlodipine, and P450 2A13 with nicotine-derived nitrosamine ketone molecules bound in the active site. In contrast to the soluble P450 102A1, the entrance channels for P450 2D6 and other drug-metabolizing P450s are likely to be buried in the surface of the membrane (Berka et al., 2011; Cojocaru et al., 2011).
Examination of the crystal lattice for P450 2D6 thioridazine complex indicated that the entrance to the substrate-binding site was accessible from a solvent channel in the crystal. Addition of potent inhibitors to the mother liquor of P450 2D6 crystallized with thioridazine led to replacement of both molecules of thioridazine and the binding of single molecules of the inhibitors, prinomastat or ajmalicine, in the active site. Moreover, binding of the new ligands was accompanied by adaptive changes in the active site structure to enhance nonbonded interactions between the protein and ligand.
This brief review has highlighted structural differences among six of the most prominent P450s in hepatic drug clearance. Additionally, determination of multiple structures for individual P450 enzymes provides a basis for understanding substrate binding and conformational adaptations that can occur when different substrates bind. One of these multiple structures may be best suited for in silico modeling of a specific substrate complex. Hopefully, molecular dynamics simulations can better define the energetic contributions of conformational flexibility of these P450s to substrate binding and provide a means to improve estimates of binding affinity.
Physical Interactions among NADPH-Cytochrome P450 Reductase, CYP1A2, and CYP2B4 in the Endoplasmic Reticulum
(Patrick Connick, James R. Reed, and Wayne L. Backes).
Enzymes of the cytochrome P450 system exist as integral membrane-bound proteins on the cytoplasmic side of the endoplasmic reticulum. The high protein to lipid ratio of the endoplasmic reticulum (Singer, 1975) leads to protein crowding and facilitates the formation of protein:protein complexes. These complexes are not only observed among P450s and their redox partners: NADPH–cytochrome P450 reductase and cytochrome b5 (Bridges et al., 1998; Estrada et al., 2013), but also occur between multiple P450 enzymes (Davydov, 2011; Reed and Backes, 2012). Both homomeric (Davydov et al., 1992; Szczesna-Skorupa et al., 2000; Ozalp et al., 2005; Jamakhandi et al., 2007; Davydov et al., 2010; Hu et al., 2010; Reed et al., 2012) and heteromeric (Yamazaki et al., 1997; Backes et al., 1998; Hazai and Kupfer, 2005; Subramanian et al., 2009; Reed et al., 2010; Subramanian et al., 2010) P450-P450 complexes have been reported among several different P450 enzymes.
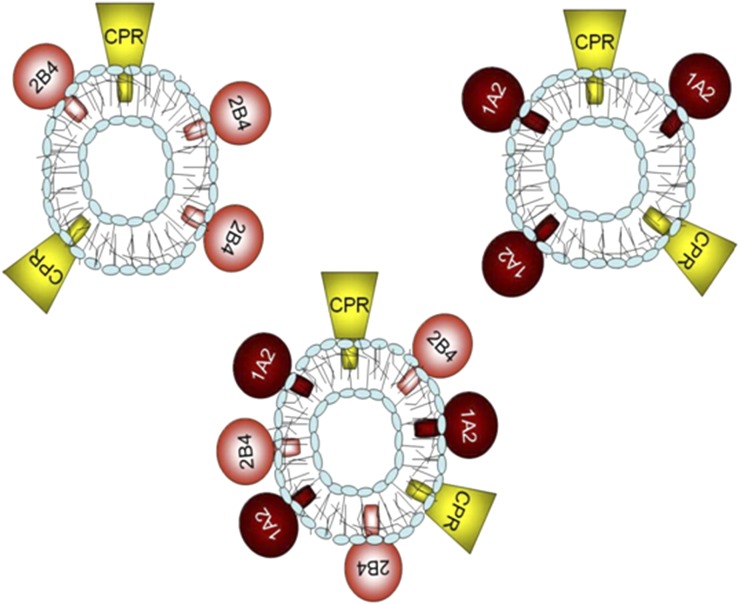
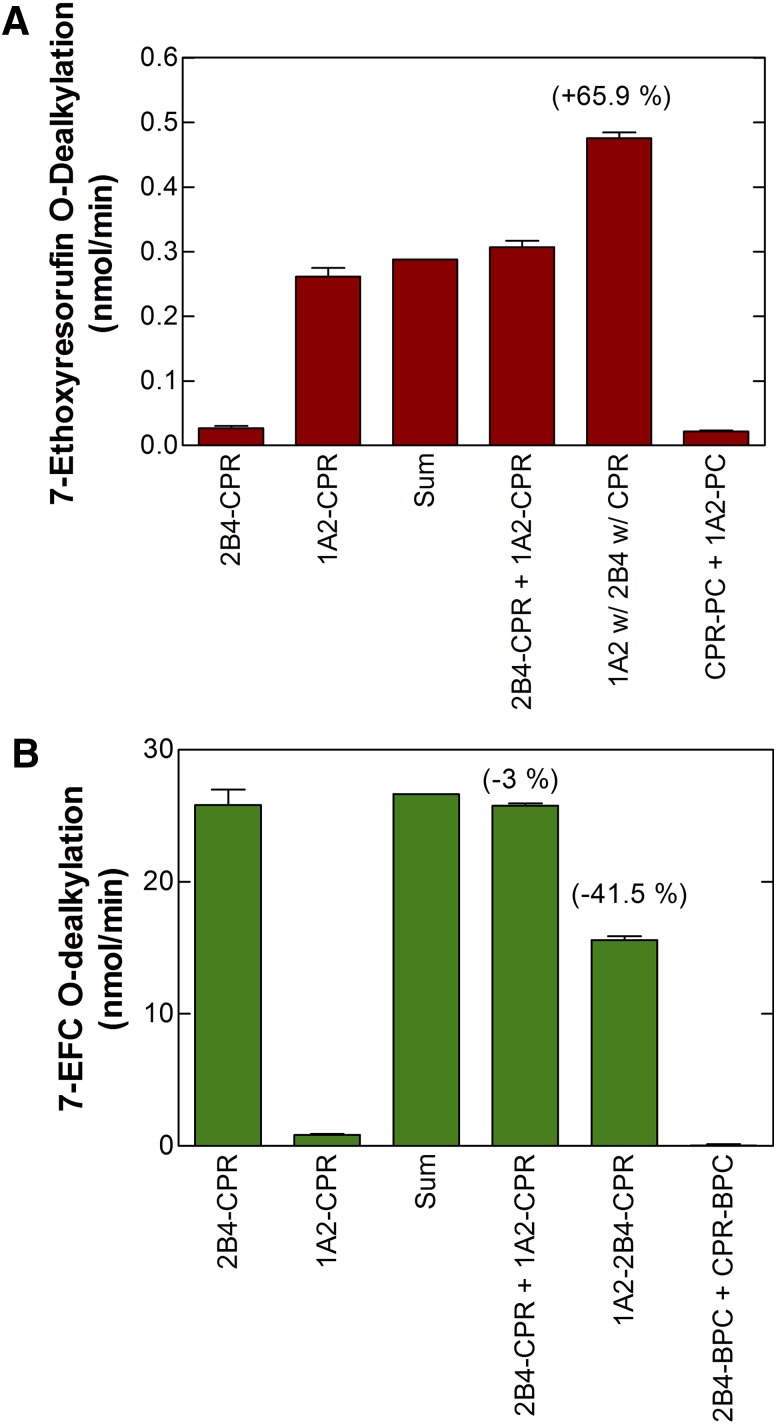
Although many different P450 enzymes have been reported to affect the function of other P450 enzymes, the current report is focused on continuing our characterization of the interactions between CYP1A2 and CYP2B4. Our work has shown that CYP1A2 and CYP2B4 form complexes that affect CPR binding when the P450s are reconstituted in phosphatidylcholine vesicles at subsaturating levels of CPR. This was shown using the following strategy: we prepared simple reconstituted systems containing a single P450 (either CYP1A2 or CYP2B4) and subsaturating CPR, as well as reconstituted systems containing both P450 enzymes at equimolar concentrations and CPR at the same subsaturating CPR:P450 ratio (Fig. 7). Next, the metabolism of a CYP1A2-selective substrate was measured in each of the systems. If the proteins were organized in the same manner in the simple and mixed P450-reconstituted systems, we would expect that the activities in the mixed reconstituted system to be the sum of the rates from the simple reconstituted systems. However, any changes in the ability of CPR to associate with either CYP would be expected to lead to either inhibition or stimulation as compared with the sum of the simple activities. This can be seen with the CYP1A2-selective substrate 7-ethoxyresorufin (Fig. 8A), where in the mixed reconstituted system, 7-ethoxyresorufin activity is greater than the sum of the rates from the simple systems. Conversely, when the CYP2B4-selective substrate 7-ethoxy-4-trifluoromethylcoumarin was tested, an inhibition of the activity was observed with a mixed reconstituted system (Fig. 8B). These results are consistent with a model where CYP1A2 and CYP2B4 form a complex whose CYP1A2 moiety has a dramatically increased affinity for CPR, as illustrated in Fig. 9. Consequently, at subsaturating CPR, a common situation in the endoplasmic reticulum, much of the CPR will be sequestered by CYP1A2. This conclusion is supported by kinetic studies showing that the data cannot be explained by simple competition between CYP1A2 and CYP2B4 for the limiting levels of CPR (Backes et al., 1998).
Fig. 7.
Illustration of the experimental protocol for examining the interactions between multiple P450 enzymes. Vesicles contain CYP1A2 and CPR, CYP2B4 and CPR, or both CYP1A2 and CYP2B4 in the same vesicle as CPR. Reproduced from Reed et al., 2010.
Fig. 8.
Stimulation of CYP1A2-selective activities and inhibition of CYP2B4-selective activities upon coreconstitution of both CYP1A2 and CYP2B4 with subsaturating CPR. Both simple reconstituted systems containing a single P450 and subsaturating CPR, and mixed systems containing both CYP1A2 and CYP2B4 and subsaturating CPR were prepared in bovine phosphatidylcholine. The metabolism of CYP1A2-selective (panel A) and CYP2B4-selective (panel B) substrates were then examined. (A) Metabolism of the CYP1A2 substrate 7-ethoxyresorufin was synergistically stimulated when CYP1A2, CYP2B4, and CPR were coreconstituted in the same vesicle (1A2 w/2B4 w/CPR). Such stimulation was not observed when CYP1A2/CPR vesicles were mixed with CYP2B4/CPR vesicles (CPR-CYP1A2 + CPR-CYP2B4). Controls include simple systems containing CYP2B4 and CPR (2B4 + CPR), CYP1A2 and CPR (1A2 + CPR). An additional control is shown where CYP1A2 and CPR were reconstituted into separate vesicles (CPR-PC + 1A2-PC), demonstrating that both proteins must be in the same vesicle to function. (B) Metabolism of the CYP2B4 substrate 7-ethoxy-4-trifluoromethylcoumarin was inhibited in mixed reconstituted systems containing CPR and both P450s (1A2-2B4-CPR), but not when the proteins were present in separate vesicles (2B4-CPR + 1A2-CPR). The following reconstituted systems were: simple system containing 2B4 and CPR (2B4-CPR), simple system containing 1A2 and CPR (1A2-CPR), the arithmetic sum of the rates from the 1A2-CPR + 2B4-CPR systems (SUM), the mixing of the two simple reconstituted systems (2B4-CPR + 1A2-CPR), reconstitution of CPR, CYP1A2, and CYP2B4 in the same vesicles (1A2-2B4-CPR), and mixing of a CPR in one vesicle (CPR-PC) and CYP2B4 (2B4-PC) in another vesicle. Adapted from Reed et al., 2010.
Fig. 9.
A model that is consistent with the kinetic data supporting the concept of the formation of P450-P450 complexes. When CPR is at subsaturating concentrations, it selectively associates with the CYP1A2 moiety of the CYP1A2•CYP2B4 complex.
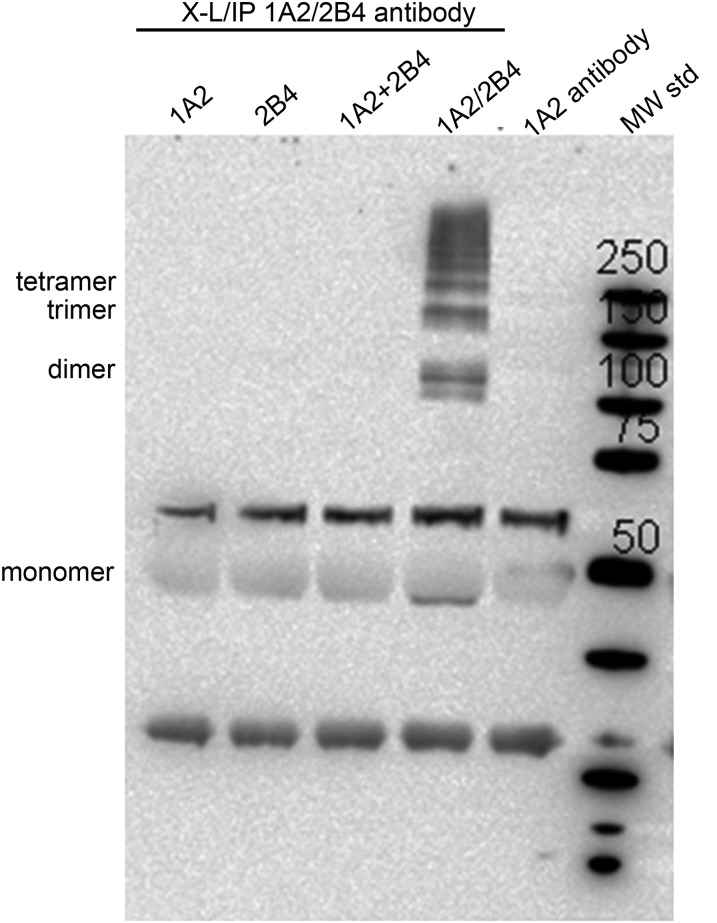
To further support the existence of CYP1A2 and CYP2B4 as heteromeric complexes, the potential for formation of physical complexes between the P450s was examined by crosslinking followed by immunoprecipitation. Mixed reconstituted systems containing both CYP1A2 and CYP2B4 in the same membrane, as well as simple membrane systems containing only a single P450, were crosslinked using the water-soluble bifunctional agent bis(sulfosuccinimidyl) suberate, which reacts with amine groups. After crosslinking, the samples were solubilized, immunoprecipitated with anti-CYP1A2, and blotted with anti-CYP2B4 (Fig. 10). In the lane labeled 1A2+2B4, these proteins were reconstituted into separate membranes and then combined, whereas the proteins shown in the lane labeled 1A2/2B4 were coreconstituted into the same membrane (Fig. 10). These data demonstrate that only when both proteins were present in the same membranes were they capable of forming heteromeric CYP1A2•CYP2B4 complexes. Samples containing CYP1A2 were immunoprecipitated but not recognized by anti-2B4 antibody. Samples containing only CYP2B4 could not be immunoprecipitated. Thus no heteromeric complexes were observed in the simple systems. These results show that CYP1A2 and CYP2B4 were capable of forming physical complexes only when both proteins were coreconstituted in the same membranes, and that in addition to dimers, larger molecular weight complexes were obtained.
Fig. 10.
Demonstration of heteromeric CYP1A2•CYP2B4 complexes in reconstituted systems. Four distinct reconstituted systems were generated, two simple systems containing either CYP1A2 or CYP2B4, a combination of the separate vesicle systems (1A2 + 2B4), or a mixed reconstituted system containing equimolar concentrations of CYP1A2 and CYP2B4 in the same vesicles. These systems were first cross-linked with bis(sulfosuccinimidyl) suberate, then immunoprecipitated with CYP1A2 antibody, and finally immunoblotted with anti-CYP2B4 antibody. Higher molecular weight complexes (labeled as dimer, trimer, and tetramer) were observed only when both P450 enzymes were present in a common membrane. The “1A2 antibody” lane is a control generated without any reconstituted system. The molecular weight standard (MW std) has the masses of the bands indicated. Adapted from Reed et al., 2010.
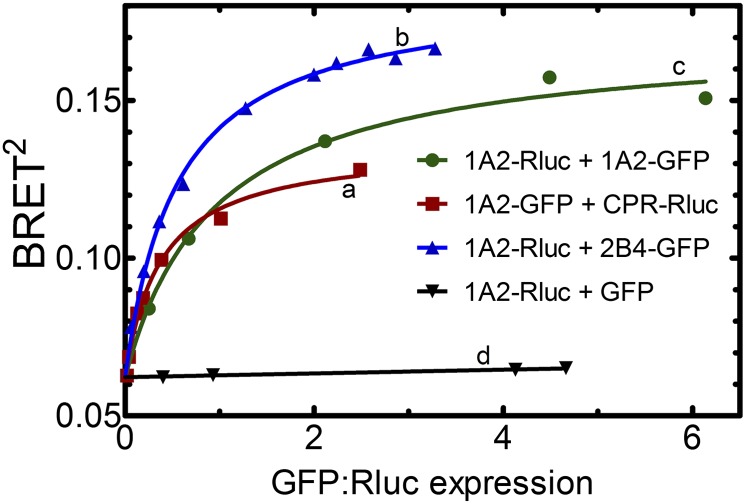
The potential of these P450/P450 complexes to form in living cells was examined using bioluminescence energy transfer (BRET). In these studies, vectors were constructed for the expression of fusion proteins consisting of CYP1A2, CYP2B4, or CPR, each with either green fluorescent protein (GFP) or Renilla luciferase (Rluc) fused to the C-terminus. Various combinations of P450-Rluc and P450-GFP constructs were cotransfected into human embryonic kidney 293T cells at different ratios of the Rluc- and GFP-containing DNA. BRET is based on the ability of Rluc, which emits light at 410 nm, to transfer the energy to GFP, which emits at 510 nm. The BRET ratio is defined as the ratio of 510 nm emission to 410 nm emission, and so it serves as a measure of the amount of energy transfer occurring. This energy transfer is highly dependent on the proximity of the GFP and Rluc chromophores, so the presence of P450-GFP•P450-Rluc complexes can be detected as a BRET signal. Specific complexes are expected to generate a hyperbolic BRET response as the ratio of P450-GFP to P450-Rluc expression varies (Fig. 11, curves a–c), whereas the BRET response of nonspecific interactions is expected to be independent of the expression ratio (Fig. 11, curve d). Using BRET, we were able to detect the interaction between CYP1A2 and CPR (Fig. 11, curve a). Data in Fig. 11 curve b demonstrate that CYP1A2 forms homomeric complexes in cells based on the significant BRET response. Finally, we transfected CYP1A2-Rluc and CYP2B4-GFP constructs into human embryonic kidney 293T cells to determine if heteromeric complexes are formed (Fig. 11, curve c). The BRET signal of this system clearly demonstrates that specific complexes between CYP1A2 and CYP2B4 are generated in cellular systems.
Fig. 11.
Demonstration of the existence of P450-P450 complexes in living cells using bioluminescence resonance energy transfer. Vectors were created containing CYP1A2, CYP2B4, or CPR cDNA upstream of either GFP or Rluc so that the resultant fusion proteins contained a C-terminal tag. These vectors (one -Rluc and one -GFP) were transfected into human embryonic kidney 293T cells to coexpress the fusion proteins at a variety of GFP-to-Rluc ratios. A hyperbolic increase in BRET signal (BRET2, measured as the ratio of the 510 nm GFP fluorescence/410 nm Rluc luminescence) is indicative of specific complexes between the proteins. Interaction between CYP1A2 and CPR was detected as a BRET signal after the cotransfection of CYP1A2-GFP and CPR-Rluc (curve a). Formation of homomeric CYP1A2 complexes is shown by the BRET response generated by cotransfection of CYP1A2-Rluc and CYP1A2-GFP (curve b). Formation of the heteromeric CYP1A2•CYP2B4 complex is shown by cotransfection of CYP1A2-Rluc with CYP2B4-GFP (curve c). A control curve showing a lack of complex formation was generated by cotransfecting CYP1A2-Rluc and GFP (without a P450 attached) (curve d). Curves b and d are reproduced from Reed et al., 2012.
These results demonstrate that CYP1A2 and CYP2B4 interact in membranes and this interaction has a significant effect on the function of these enzymes. The interactions between these P450s are mediated through the formation of heteromeric complexes, and the proteins must be in the same membrane to interact. Finally, CYP1A2 and CYP2B4 have the ability to form these complexes in living cells, capable of generating both homomeric CYP1A2 and CYP2B4 complexes, as well as heteromeric CYP1A2•CYP2B4 complexes. Taken together, these data support the idea that P450s normally exist in the endoplasmic reticulum as complexes, and that changes in the relative amounts of individual P450s can affect metabolism not only due to their own metabolic capabilities, but also by conformationally affecting the behavior of surrounding P450 enzymes.
Investigations of Human Cytochrome P450 Enzymes with Solution NMR
(D. Fernando Estrada, Jennifer S. Laurence, and Emily E. Scott).
Major challenges exist in understanding the interactions of cytochrome P450 enzymes with diverse small molecules and with protein partners CPR and b5. Although substantial progress has been made in determining the X-ray structures of a number of human cytochrome P450 enzymes with various ligands, it is frequently true that the structure of a particular P450/ligand complex can be of limited utility in understanding the binding mode(s) of structurally distinct ligands to even the same P450. However, generating each desired P450/ligand cocrystal structure is a laborious effort that is inconsistent with feedback required in the drug development process. Advances have been made in the timeline to generate crystals of CYP2D6 complexes (vide supra), but this solution is at the moment unique to CYP2D6. In addition, no structures exist to date for membrane P450 enzymes in complex with either b5 or CPR to guide our understanding of the structural elements of electron transfer and/or allosteric effects. Mutagenesis (Geller et al., 1999; Naffin-Olivos and Auchus, 2006; Im and Waskell, 2011), crosslinking (Bridges et al., 1998; Gao et al., 2006; Zhao et al., 2012; Peng and Auchus, 2013), and protein docking approaches (Bridges et al., 1998; Gao et al., 2006; Im and Waskell, 2011) have provided insights into these protein/protein interactions, supplemented recently by the first structure of the bacterial soluble P450cam with its redox partner, putidaredoxin (Hiruma et al., 2013; Tripathi et al., 2013), but much is lacking in our current understanding of these aspects of P450 systems. Protein-detected NMR is a high-resolution structural technique that has been little applied to P450 systems, but has the potential to provide significant new and orthogonal information on human P450 interactions with both ligands and other proteins in solution, as well as on protein dynamics that can only be inferred from current X-ray structure information. Once individual residues are assigned to particular resonances in the NMR spectrum, NMR can be used to probe such interactions on a rapid time-scale commensurate with drug discovery processes. NMR complements crystallography because it has the advantages of dealing well with ligands of moderate affinity (like many P450 substrates), with ligands that bind at a distant location from the heme iron (in contrast to binding studies based on spin-shift changes at the heme), and it provides valuable dynamic information. Straightforward analyses of changes in spectral line shape and resonance position inform understanding of such interactions (Skinner and Laurence, 2008). NMR may also have an advantage in evaluating interactions mediated by electrostatics, such as those thought to play an important role in electron transfer encounter complexes. While the high salt environment often required for crystallization can render the P450 electron donor complex difficult to capture, NMR conditions employ low ionic strength and may facilitate the characterization of interactions with redox partner proteins that regulate P450 function.
The basic utility and feasibility of using solution NMR to probe P450/redox partner interactions and P450/ligand interactions has already been demonstrated effectively for the soluble bacterial P450cam (Rui et al., 2006; Asciutto et al., 2012), but this approach is in its infancy with respect to membrane-bound human cytochrome P450 enzymes. Membrane P450 enzymes typically express in recombinant systems at very low levels, and isotopic enrichment required for NMR analysis can further depress yield. They also aggregate and/or precipitate under NMR-compatible experimental conditions much more readily than their soluble counterparts. However, efforts already invested in generating highly purified stable, monodisperse membrane P450 protein in sufficient quantities for crystallography studies can be directly applied to generating P450 protein for solution NMR studies. If these technical issues can be surmounted, the cytochrome P450 community and the pharmaceutical industry stand to gain much from application of NMR to P450 systems.
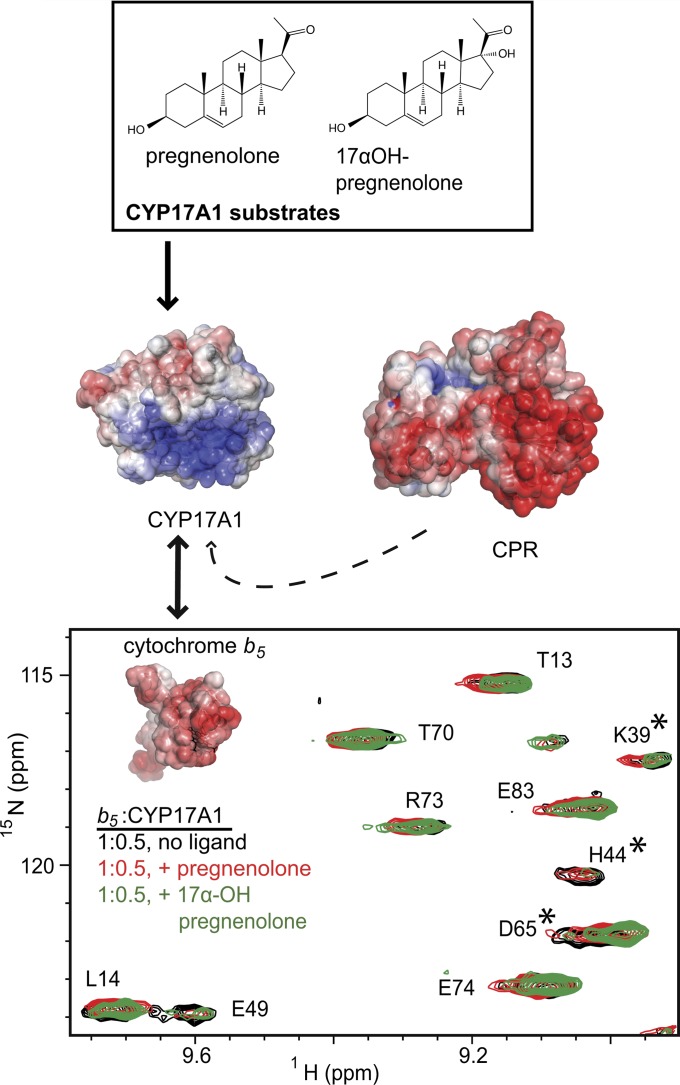
Current work in our laboratory focuses on using NMR to probe the interactions between P450, b5, and CPR. While CPR is essential for electron delivery to P450, the effects of b5 are less well understood, as it is able to variously inhibit, facilitate, or have no effect on P450 catalysis depending on the enzyme, substrate, and reaction conditions. Ideas put forth in the literature regarding the basis of such effects include providing the second electron in the catalytic cycle, decreasing uncoupling, and allosteric effects on P450. The steroidogenic human cytochrome P450 17A1 (CYP17A1) provides an excellent system for probing b5 effects on P450 function. CYP17A1, also called CYPc17, performs two reactions. One is hydroxylation of pregnenolone or progesterone to the corresponding 17α-hydroxy product; the second is a 17,20-lyase reaction cleaving the acyl group from 17α-hydroxypregnenolone to generate the C19 androgen dehydroepiandrosterone. Cytochrome b5 has a selective effect on these two reactions, with <2-fold effect on the hydroxylation reaction, but mediating a 10-fold increase in the lyase reaction (Akhtar et al., 2005).
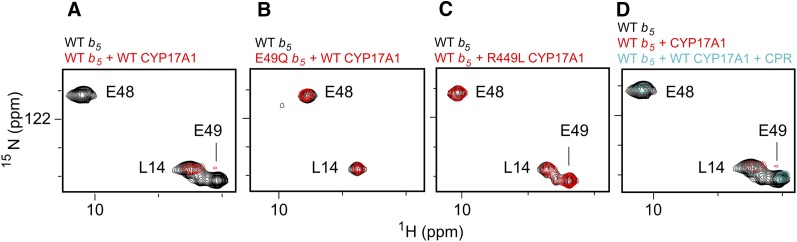
The experimental system used to probe these protein/protein interactions consists of a catalytically active version of human P450 CYP17A1 missing the N-terminal transmembrane helix (DeVore and Scott, 2012), the soluble domain of human b5 (residues 1–107), and full-length rat CPR (Shen et al., 1989). As the resonances for b5 have already been assigned (Nunez et al., 2010), changes in the spectrum when CYP17A1 is added are readily translated back to individual b5 amino acids and surfaces. Titration of uniformly labeled 15N-b5 with increasing concentrations of unlabeled CYP17A1 led to substantial broadening for select b5 residues (Estrada et al., 2013). Specifically, the resonances of b5 backbone amides for the sequential G47, E48, E49, and V50 residues in or near the α2 helix are either completely absent or significantly reduced upon the addition of an equimolar concentration of CYP17A1 (e.g., Fig. 12A). The b5 mutations E48G and E49G are known to decrease the ability of b5 to stimulate the CYP17A1 lyase activity without negatively affecting the b5-insensitive hydroxylase reaction (Naffin-Olivos and Auchus, 2006). To further examine if these residues are important in the physical interactions between the two proteins, b5 proteins incorporating these same mutations were examined using NMR. In this case, although the NMR spectra of the b5 mutant proteins were very similar to those of wild-type, showing that the b5 backbone structure was not altered by mutation, either b5 E48Q or E49Q was sufficient to disrupt its interaction with CYP17A1, as evidenced by the lack of line broadening in the b5 NMR spectrum when CYP17A1 was added (e.g., Fig. 12B). Residues are also known on the proximal surface of CYP17A1 that do not alter the hydroxylase activity but do strongly reduce the lyase activity, primarily identified from clinical patients (Lee-Robichaud et al., 2004). On CYP17A1, the mutations R347H, R449L, or R358Q similarly disrupted the P450/b5 interaction as observed by the absence of NMR line broadening (e.g., Fig. 12C). Thus, there is a direct, physical interaction between CYP17A1 and b5 that is mediated by acidic residues in the b5 α2 helix and basic residues R347, 449, and R358 on the proximal face of CYP17A1.
Fig. 12.
Examples of NMR spectra establishing contact surfaces between CYP17A1 and b5 and the mutually exclusive nature of b5 and CPR binding. (A) Comparison between the HSQC of uniformly 15N-labeled b5 alone (black) and a 1:1 mixture with unlabeled CYP17A1 (red) demonstrates that selected resonances in/near the b5 α helix two are broadened and disappear (e.g., E48 and E49, but not L14), suggesting the glutamic acid residues are involved in the CYP17A1/b5 interaction. (B) When b5 residues involved in the interaction with CYP17A1 are mutated (e.g., E49Q, red spectrum), the same resonances for residues in the b5 α helix two are not broadened compared with the 15N-b5 spectrum alone (black), suggesting the CYP17A1/b5 complex does not form. (C) If residues on the CYP17A1 proximal surface known to impair b5 facilitation of the CYP17A1 lyase reaction are mutated (e.g., CYP17A1 R449L, red spectrum), the mutant CYP17A1 also does not cause line broadening of b5 residues (compared with the b5 spectum, black). (D) Although the spectrum of wild type b5 alone (black) has selective resonances line broadened upon addition of CYP17A1 (red spectrum), the subsequent addition of CPR (cyan spectrum) causes these resonances (e.g., E48 and E49) to reappear, suggesting that CPR disrupts CYP17A1 interaction with b5. Adapted from (Estrada et al., 2013).
Since b5 has a differential effect on the two reactions of CYP17A1, we were interested in how the binding of different substrates and inhibitors in the CYP17A1 active site might impact the CYP17A1/b5 interaction. In the presence of the type-II iron-coordinating inhibitor abiraterone, the hydroxylase substrate pregnenolone, or the lyase substrate 17α-hydroxypregnenolone, the CYP17A1/b5 complex still formed, but the effects are different for all three. The CYP17A1 (abiraterone)/b5 spectra are most similar to CYP17A1(no ligand)/b5 spectra, while titrations in the presence of either substrate result in additional changes. Observations (Fig. 13) include differential shifts of D65 and line broadening of additional resonances, including the backbone amides of K39 and H44 preceding the residues of b5 α2 that were observed to be involved in the b5/CYP17A1 interaction in the absence of CYP17A1 ligand. H44 is one of two histidines coordinate to the heme iron in b5. In addition, it was observed that the locations of a number of other resonances were affected in a way unique to each substrate. Thus, not only does the presence of substrate in the CYP17A1 active site significantly alter the CYP17A1/b5 interaction, but the interaction of CYP17A1 with b5 is different depending on the substrate identity, consistent with the differential effects of b5 on the hydroxylase and lyase reactions.
Fig. 13.
Distinct ligands differentially affect the interaction of CYP17A1 with cytochrome b5. Depending on the presence and identity of ligands in the enclosed active site on the distal side of the heme, differential effects are observed in the interactions between the CYP17A1 proximal surface and b5. In the example shown, residues K39, H44, and D65 of b5 (asterisks) are line-broadened or perturbed in a way that is unique to either pregnenolone-bound (red spectrum) or 17α-hydroxypregnenolone-bound (green spectrum) CYP17A1 compared with the unliganded spectrum (black). The addition of rat CPR (dashed arrow), and its ability to displace bound b5 provides an additional tool with which to examine differential ligand effects on the CYP17A1/b5 interaction. Adapted from Estrada et al., 2013.
When the b5/CYP17A1 (unliganded) complex is formed, the b5 G47-V50 residues are broadened and disappear in the NMR spectrum (Fig. 12A), but subsequent addition of CPR results in the restoration of these resonances (Fig. 12D), suggesting the binding of CPR and b5 on CYP17A1 are mutually exclusive. The simplest explanation is partially overlapping binding sites on the CYP17A1 proximal face, consistent with mutagenesis studies probing CYP2B4/CPR interactions (Im and Waskell, 2011). Again, the ability of CPR to free b5 from its interaction with CYP17A1 is modulated by the CYP17A1 liganded state. While b5 is completely liberated by CPR when the CPR concentration is equimolar with CYP17A1 and b5 when CYP17A1 is unliganded, it takes an additional 0.5 molar of CPR equivalent to do so when the lyase substrate 17α-hydroxypregnenolone is in the CYP17A1 active site, and even more CPR is needed to return the free b5 spectrum when the hydroxylase substrate pregnenolone is present. Thus the presence and identity of ligands in the buried CYP17A1 active site appear to alter protein/protein interactions on the proximal surface.
In addition to the new information about specific protein/protein interactions and their modulation by ligands with CYP17A1, there are many additional opportunities to use NMR to investigate other membrane P450 systems. This would be important because of the variable effects of b5 on different P450 enzymes and because cross-linking studies suggest different surfaces might be involved in b5 interactions for different P450 enzymes (Gao et al., 2006; Sulc et al., 2012). Overall this work illustrates a significant advantage of NMR—a single P450 of interest may be analyzed in the presence of multiple protein and small-molecule components. (Fig. 13) Using only 2-dimensional experiments, we were able to leverage the mutually exclusive nature of the CYP17A1/b5 and CYP17A1/CPR interactions, combined with the presence or absence of ligand (a fourth component) to describe a complex set of relationships. The permutations available for study by NMR rapidly increase by expanding this experimental template to include additional substrates and inhibitors. Interpretation of these data in the context of the known component structures can rapidly produce valuable information about the complex in the absence of a high-resolution structure of the entire assembly.
In addition, once resonances are assigned in a particular P450, ligand-binding sites throughout the protein and their affinities can be readily determined using a similar NMR approach, which would have substantial utility for deciphering allosteric interactions. The challenge here is resonance assignment in membrane P450 enzymes. Although the process of relating NMR resonances to individual amino acids is relatively straightforward for small proteins that express well in recombinant systems under isotopic labeling conditions, human cytochrome P450 enzymes have neither of these advantages. At ∼55 kDa and ∼500 amino acids, uniformly labeled NMR spectra are very crowded and require the application of selective amino acid labeling strategies and/or deuteration. Human P450 forms engineered to remove the single transmembrane helix are still localized in the membrane but have the benefits of increased expression in bacterial systems, increased solubility under solution NMR conditions, can often be maintained in a monomeric state, and are catalytically active. However, even truncated versions of P450 enzymes have reduced expression yields in the expensive media and growth conditions needed to generate the 15N, 13C, deuterated samples required for traditional resonance assignment approaches. Once the appropriate sample is generated, however, methodologies are already in place to make residue assignments. Such information will be invaluable not just for examining xenobiotic-metabolizing cytochrome P450 enzymes, for many of which we already have some structural information in place with at least one ligand, but even more so for human cytochrome P450 enzymes, for which no structural information is currently available but which participate in physiologically important processes that make them drug targets.
Novel Pharmacoenhancer Cobicistat: Discovery and Development of a CYP3A Inhibitor
(Lianhong Xu and Manoj C. Desai).
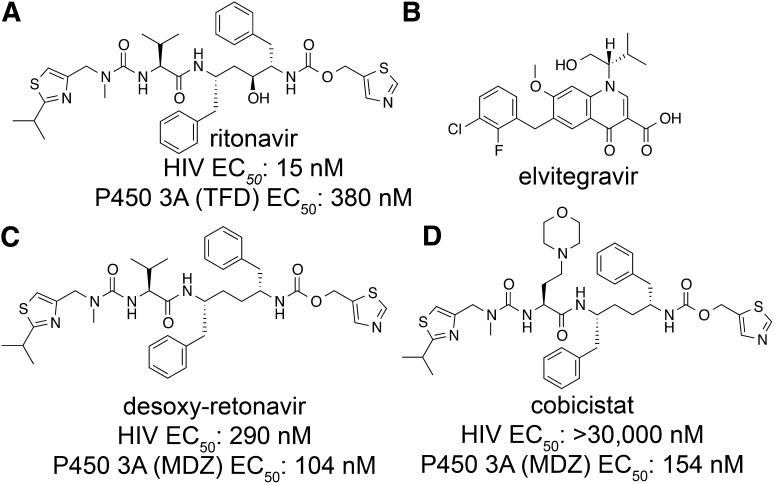
Cytochrome P450 3A4 is the most abundant P450 enzyme in the human liver and plays a key role in both the detoxification of xenobiotics and the metabolism of endobiotic signaling molecules (Nelson et al., 1996; Rendic and Di Carlo, 1997; Wrighton et al., 2000; Rendic, 2002). Most importantly, it metabolizes >50% of all approved drugs and, in many cases, limits the ability to achieve a sufficient steady-state minimum drug concentration required for durable efficacy. All approved HIV protease inhibitors (PIs) in particular are metabolized primarily by the CYP3A subfamily (CYP3A4 and CYP3A5) in the liver and intestine, and have unfavorable pharmacokinetic profiles, including poor and/or variable oral bioavailability and rapid metabolic degradation with relative short plasma elimination half-lives.1 As a result, early regimens containing a PI were typically complex, requiring frequent dosing with high pill burdens, and were associated with significant side effects and undesirable toxicities. Ritonavir (Fig. 14A), an approved PI (600 mg twice daily), is also a potent mechanism-based inhibitor of CYP3A4. Utilization of ritonavir at a dose that is subtherapeutic as a PI (100 or 200 mg daily) results in improved pharmacokinetics of concomitant PIs, thereby improving effectiveness while simplifying the regimen. This unique clinical practice reduces the pill burden and enhances the adherence of the coadministered PIs—a strategy that has become a cornerstone of PI-containing highly active antiretroviral therapy (Gallant, 2004). Ritonavir therefore serves a critical function in the development of PI-based highly active antiretroviral therapy regimens and in the chronic management of HIV infection. Without the boosting effect of ritonavir, achieving convenient dosing regiments for the large peptidomimetic PIs would not have been possible. In fact, ritonavir has been used as a pharmacoenhancer for more than 15 years. With the exception of predictable drug interactions, there has been no strong evidence to demonstrate significant side effects arising from sustained CYP3A4 inhibition by long-term use of ritonavir as a booster.
Fig. 14.
Structures of small-molecule compounds. (A) Structure of ritonavir. EC50 is inhibition of terfenadine (TFD) hydroxylase (CYP3A) activity in human liver microsomes. (B) Structure of elvitegravir. (C) Structure and activity of desoxy-ritonavir. (D) Structure and activity of cobicistat. For (C) and (D) EC50 is inhibition of midazolam (MDZ) 1′-hydroxylase by CYP3A.
Elvitegravir (Fig. 14B) is an HIV integrase strand transfer inhibitor with potent antiretroviral activity against wild-type and laboratory strains resistant to nucleoside/nucleotide reverse transcriptase inhibitors, nonnucleoside reverse transcriptase inhibitors, and protease inhibitors (Shimura et al., 2008). However, it is extensively metabolized primarily by CYP3A4 in the liver and intestine in vivo, requiring high-dose twice-daily dosing. Similar with that observed for PIs, coadministration of ritonavir with elvitegravir can significantly improve the pharmacokinetic profile of elvitegravir, enabling its use as a once-daily drug. Nonetheless, the use of a subtherapeutic dose of ritonavir may have the potential to select for PI-resistant viruses in a non-PI-containing regimen, thus limiting the use of ritonavir-boosted elvitegravir to only treatment-experienced patients or a PI-containing regimen. In addition, ritonavir is associated with side effects and complications, including causing lipid disorders and triggering undesired drug interactions as an inducer of drug-metabolizing enzymes such as cytochrome P450, p-glycoprotein, and uridine 5′-diphospho-glucuronosyltransferases. Therefore pharmacoenhancers were developed that maintain ritonavir’s boosting efficacy but overcome its liabilities.
Despite many studies, the precise mechanism of CYP3A4 inhibition by ritonavir remains unclear. It is likely that the overall inhibitory effects of ritonavir arise from a combination of both type II binding to the heme iron and mechanism-based inactivation. The type II interaction enables ritonavir binding to CYP3A to occur with fast kinetics, high affinity, and a very slow disassociation rate. Although the direct interaction between ritonavir and the P450 heme as a type II ligand may play a role in the CYP3A inhibition in vivo, the clinical observation of ritonavir’s sustained pharmacoenhancing effect beyond the persistence of ritonavir in plasma is consistent with a mechanism-based interaction (Xu and Desai, 2009).
Chemical structure-activity relationships were used as the basis to design novel CYP3A4 inhibitors. Since ritonavir (Fig. 14A) is a unique pharmacoenhancer that exerts sustained pharmacological effects with a long-term safety record in the clinical setting, it was used as a starting point, with the goal of eliminating its anti-HIV activity while maintaining its potent inhibition of CYP3A and mode of action. Initial attempts to eliminate the antiviral activity of ritonavir were focused on removal of the key hydroxyl group that mimics the transition state of amide hydrolysis through the formation of hydrogen-bonds to the oxygen atoms of the catalytic Asp25 and Asp25′ residues in the HIV protease active site (Kempf et al., 1995). Desoxy-ritonavir (Fig. 14C) was about 20 times less potent than ritonavir in a cell-based antiviral assay, but retained full inhibitory activity against CYP3A. After extensive structure activity relationship studies with desoxy-ritonavir, cobicistat (Fig. 14C) was identified as a potent, selective, and orally bioavailable inhibitor of CYP3A (Xu et al., 2010).
Extensive in vitro studies of cobicistat and ritonavir side by side have shown that cobicistat is a potent and selective CYP3A inhibitor that lacks significant anti-HIV activity. Cobicistat shares a spectrum of CYP3A substrate specificity similar to that of ritonavir, retains the characteristic of mechanism-based inhibition of CYP3A, and is equipotent to ritonavir in inhibiting CYP3A metabolism of a group of CYP3A substrates. Cobicistat is also more selective for CYP3A than ritonavir, with much reduced inhibitory activity toward CYP2D6, CYP2C8, and CYP2C9. In addition, in vitro studies suggest that cobicistat may have a lower potential to cause undesired drug-drug interactions and lipid disorders than ritonavir (Xu et al., 2010).
Cobicistat was also investigated in clinical studies versus ritonavir as a pharmacoenhancer for drugs metabolized by CYP3A to improve their systemic exposure. In a randomized, placebo-controlled, double-blind, multicenter, 48-week phase 3 study, cobicistat demonstrated comparable efficacy to ritonavir as a pharmacoenhancer for the PI atazanavir. The study found that an HIV regimen containing cobicistat-boosted atazanavir was not inferior to a regimen containing ritonavir-boosted atazanavir at 48 weeks of therapy (Gallant et al., 2012). Currently, cobicistat is under regulatory review. If approved, cobicistat may be an effective option for boosting the potency of HIV regimens that are based on PIs.
Cobicistat has favorable physicochemical properties, especially high aqueous solubility, allowing it to be formulated as a tablet and coformulated with other drugs. Cobicistat was coformulated with the nucleoside/nucleotide reverse transcriptase inhibitors tenofovir disoproxil fumarate and emtricitabine, in addition to the integrase inhibitor elvitegravir, in a once-daily single-tablet regimen known as Stribild (cobicistat 150 mg, tenofovir disoproxil fumarate 300 mg, emtricitabine 200 mg, and elvitegravir 150 mg). Stribild is the first and only integrase inhibitor–containing single-tablet regimen. Two independent, fully powered phase 3 noninferiority trials have compared Stribild with two current standard-of-care regimens for initial HIV treatment; these studies confirmed that Stribild has high efficacy and a good tolerability profile and is statistically noninferior to the two standard-of-care regimens (DeJesus et al., 2012; Sax et al., 2012). The US Food and Drug Administration approved Stribild to treat HIV-1 infection in treatment-naive adults on August 27, 2012. It is expected that Stribild will become an important complete regimen option for adult HIV-infected subjects.
In summary, drug-drug interactions as a result of CYP3A enzyme inhibition are often regarded as a liability. In the case of cobicistat and ritonavir, however, associated drug-drug interactions have proven to be an asset in the treatment of life-threatening HIV infection. The second-generation pharmacoenhancer cobicistat was discovered by extensive structure-activity relationship studies. Cobicistat maintains the CYP3A4-inhibition potency without any antiviral activity and possesses significantly improved physicochemical properties, and finds broader use with its improved overall profile. The first integrase inhibitor-based, once-daily single-tablet highly active antiretroviral therapy regimen Stribild has demonstrated promising clinical results. The potent, persistent CYP3A inhibition properties and improved physicochemical properties associated with cobicistat enable elvitegravir to be a once-daily drug. As our knowledge of CYP3A inhibition improves and our experience with pharmacoenhancers continues to increase, it is expected that a clean and safe pharmacoenhancer will also have broader utility against other life-threatening diseases, such as cancer, albeit with close monitoring to avoid unfavorable drug-drug interactions.
Summary
An increasing variety of diverse techniques and biophysical approaches are being used to probe the complex relationships between cytochrome P450 structure and their function. The strategies presented herein focus both on P450 interactions with ligands and partner proteins. This work and other efforts provide a partial basis for the continuing challenge to understand drug and endogenous compound metabolism.
Abbreviations
- b5
cytochrome b5
- BRET
bioluminescence energy transfer
- CPR
NADPH–cytochrome P450 reductase
- GFP
green fluorescent protein
- HIV
human immunodeficiency virus
- NMR
nuclear magnetic resonance
- P450
cytochrome P450
- PI
protease inhibitor
- Rluc
Renilla luciferase
Authorship Contributions
Participated in research design: Johnson, Connick, Reed, Backes; Xu, Desai, Estrada, Laurence, Scott.
Conducted experiments: Connick, Reed, Estrada.
Performed data analysis: Johnson, Connick, Reed, Backes, Estrada, Laurence, Scott.
Wrote or contributed to the writing of the manuscript: Johnson, Connick, Backes, Xu, Desai, Estrada, Laurence, Scott.
Footnotes
This work was supported in part by the National Institutes of Health [Grants R01 GM031001, F32 GM103069, R01 GM076343, R01 ES004344, and P42 ES013648] and by Gilead Sciences, Inc.
This report is a summary of a session at Experimental Biology 2013 sponsored by the Drug Metabolism Division of the American Society for Pharmacology and Experimental Therapeutics.
- 1) Gerber, JG (2000) Using pharmacokinetics to optimize antiretroviral drug-drug interactions in the treatment of human immunodeficiency virus infection Clin Infect Dis 30 Suppl 2:S123-9.
- 2) Moyle, GJ and Black, D (2001) Principles and practice of HIV-protease inhibitor pharmacoenhancement. HIV Med 2(2):105-13.
- 3) Cooper CL, van Heeswijk RP, Gallicano K, Cameron DW (2003) A review of low-dose ritonavir in protease inhibitor combination therapy. Clin Infect Dis 36(12):1585-92.
- 4) Becker SL (2003) The role of pharmacological enhancement in protease inhibitor-based highly active antiretroviral therapy. Expert Opin Investig Drugs 12(3):401-12.
- 5) Gallant JE (2004) Protease-inhibitor boosting in the treatment-experienced patient. AIDS Rev 6(4):226-33.
- 6) Youle M (2007) Overview of boosted protease inhibitors in treatment-experienced HIV-infected patients. J Antimicrob Chemother 60(6):1195-205.
- 7) Busse, KH and Penzak, SR (2008) Pharmacological enhancement of protease inhibitors with ritonavir: an update. Expert Review of Clinical Pharmacology 1:533-545.
- 8) Xu L, Desai MC (2009) Pharmacokinetic enhancers for HIV drugs. Curr Opin Investig Drugs 10(8):775-86.
References
- Ahuja S, Jahr N, Im SC, Vivekanandan S, Popovych N, Le Clair SV, Huang R, Soong R, Xu J, Yamamoto K, et al. (2013) A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data. J Biol Chem 288:22080–22095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar MK, Kelly SL, Kaderbhai MA. (2005) Cytochrome b(5) modulation of 17alpha hydroxylase and 17-20 lyase (CYP17) activities in steroidogenesis. J Endocrinol 187:267–274 [DOI] [PubMed] [Google Scholar]
- Asciutto EK, Young MJ, Madura J, Pochapsky SS, Pochapsky TC. (2012) Solution structural ensembles of substrate-free cytochrome P450(cam). Biochemistry 51:3383–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes WL, Batie CJ, Cawley GF. (1998) Interactions among P450 enzymes when combined in reconstituted systems: formation of a 2B4-1A2 complex with a high affinity for NADPH-cytochrome P450 reductase. Biochemistry 37:12852–12859 [DOI] [PubMed] [Google Scholar]
- Bass MB, Paulsen MD, Ornstein RL. (1992) Substrate mobility in a deeply buried active site: analysis of norcamphor bound to cytochrome P-450cam as determined by a 201-psec molecular dynamics simulation. Proteins 13:26–37 [DOI] [PubMed] [Google Scholar]
- Berka K, Hendrychová T, Anzenbacher P, Otyepka M. (2011) Membrane position of ibuprofen agrees with suggested access path entrance to cytochrome P450 2C9 active site. J Phys Chem A 115:11248–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. (1998) Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J Biol Chem 273:17036–17049 [DOI] [PubMed] [Google Scholar]
- Chumsri S, Sabnis GJ, Howes T, Brodie AM. (2011) Aromatase inhibitors and xenograft studies. Steroids 76:730–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru V, Balali-Mood K, Sansom MS, Wade RC. (2011) Structure and dynamics of the membrane-bound cytochrome P450 2C9. PLOS Comput Biol 7:e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru V, Winn PJ, Wade RC. (2007) The ins and outs of cytochrome P450s. Biochim Biophys Acta 1770:390–401 [DOI] [PubMed] [Google Scholar]
- Collins JR, Loew GH. (1988) Theoretical study of the product specificity in the hydroxylation of camphor, norcamphor, 5,5-difluorocamphor, and pericyclocamphanone by cytochrome P-450cam. J Biol Chem 263:3164–3170 [PubMed] [Google Scholar]
- Cruciani G, Carosati E, De Boeck B, Ethirajulu K, Mackie C, Howe T, Vianello R. (2005) MetaSite: understanding metabolism in human cytochromes from the perspective of the chemist. J Med Chem 48:6970–6979 [DOI] [PubMed] [Google Scholar]
- Davydov DR. (2011) Microsomal monooxygenase as a multienzyme system: the role of P450-P450 interactions. Expert Opin Drug Metab Toxicol 7:543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov DR, Knyushko TV, Hoa GH. (1992) High pressure induced inactivation of ferrous cytochrome P-450 LM2 (IIB4) CO complex: evidence for the presence of two conformers in the oligomer. Biochem Biophys Res Commun 188:216–221 [DOI] [PubMed] [Google Scholar]
- Davydov DR, Sineva EV, Sistla S, Davydova NY, Frank DJ, Sligar SG, Halpert JR. (2010) Electron transfer in the complex of membrane-bound human cytochrome P450 3A4 with the flavin domain of P450BM-3: the effect of oligomerization of the heme protein and intermittent modulation of the spin equilibrium. Biochim Biophys Acta 1797:378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot MJ. (2006) Designing better drugs: predicting cytochrome P450 metabolism. Drug Discov Today 11:601–606 [DOI] [PubMed] [Google Scholar]
- DeJesus E, Rockstroh JK, Henry K, Molina JM, Gathe J, Ramanathan S, Wei X, Yale K, Szwarcberg J, White K, et al. GS-236-0103 Study Team (2012) Co-formulated elvitegravir, cobicistat, emtricitabine, and tenofovir disoproxil fumarate versus ritonavir-boosted atazanavir plus co-formulated emtricitabine and tenofovir disoproxil fumarate for initial treatment of HIV-1 infection: a randomised, double-blind, phase 3, non-inferiority trial. Lancet 379:2429–2438 [DOI] [PubMed] [Google Scholar]
- DeVore NM, Scott EE. (2012) Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001. Nature 482:116–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekroos M, Sjögren T. (2006) Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci USA 103:13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estabrook RW, Hildebrandt AG, Baron J, Netter KJ, Leibman K. (1971) A new spectral intermediate associated with cytochrome P-450 function in liver microsomes. Biochem Biophys Res Commun 42:132–139 [DOI] [PubMed] [Google Scholar]
- Estrada DF, Laurence JS, Scott EE. (2013) Substrate-modulated cytochrome P450 17A1 and cytochrome b5 interactions revealed by NMR. J Biol Chem 288:17008–17018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant JE, Koenig E, Andrade-Villanueva J, Chetchotisakd P, DeJesus E, Antunes F, Arastéh K, Moyle G, Rizzardini G, Fehr J. (2013) Cobicistat versus ritonavir as a pharmacoenhancer of atazanavir plus emtricitabine/tenofovir disoproxil fumarate in treatment-naive HIV type 1-infected patients: week 48 results. J Infect Dis 208(1):32–9 [DOI] [PubMed] [Google Scholar]
- Gallant JE. (2004) Protease-inhibitor boosting in the treatment-experienced patient. AIDS Rev 6:226–233 [PubMed] [Google Scholar]
- Gao Q, Doneanu CE, Shaffer SA, Adman ET, Goodlett DR, Nelson SD. (2006) Identification of the interactions between cytochrome P450 2E1 and cytochrome b5 by mass spectrometry and site-directed mutagenesis. J Biol Chem 281:20404–20417 [DOI] [PubMed] [Google Scholar]
- Geller DH, Auchus RJ, Miller WL. (1999) P450c17 mutations R347H and R358Q selectively disrupt 17,20-lyase activity by disrupting interactions with P450 oxidoreductase and cytochrome b5. Mol Endocrinol 13:167–175 [DOI] [PubMed] [Google Scholar]
- Groves JT, McClusky GA, White RE, Coon MJ. (1978) Aliphatic hydroxylation by highly purified liver microsomal cytochrome P-450. Evidence for a carbon radical intermediate. Biochem Biophys Res Commun 81:154–160 [DOI] [PubMed] [Google Scholar]
- Halpert JR. (2011) Structure and function of cytochromes P450 2B: from mechanism-based inactivators to X-ray crystal structures and back. Drug Metab Dispos 39:1113–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. (1995) Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure 3:41–62 [DOI] [PubMed] [Google Scholar]
- Hazai E, Kupfer D. (2005) Interactions between CYP2C9 and CYP2C19 in reconstituted binary systems influence their catalytic activity: possible rationale for the inability of CYP2C19 to catalyze methoxychlor demethylation in human liver microsomes. Drug Metab Dispos 33:157–164 [DOI] [PubMed] [Google Scholar]
- Hiruma Y, Hass MA, Kikui Y, Liu WM, Olmez B, Skinner SP, Blok A, Kloosterman A, Koteishi H, Löhr F, et al. (2013) The structure of the cytochrome P450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography. J Mol Biol DOI:10.1124/pr.55.3.4 [published ahead of print]. [DOI] [PubMed] [Google Scholar]
- Hu G, Johnson EF, Kemper B. (2010) CYP2C8 exists as a dimer in natural membranes. Drug Metab Dispos 38:1976–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeanu GC, Ghanayem BI, Linko P, Li L, Pederson LG, Goldstein JA. (1996) Identification of residues 99, 220, and 221 of human cytochrome P450 2C19 as key determinants of omeprazole activity. J Biol Chem 271:12496–12501 [DOI] [PubMed] [Google Scholar]
- Im SC, Waskell L. (2011) The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5). Arch Biochem Biophys 507:144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamakhandi AP, Kuzmic P, Sanders DE, Miller GP. (2007) Global analysis of protein-protein interactions reveals multiple CYP2E1-reductase complexes. Biochemistry 46:10192–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EF, Stout CD. (2013) Structural diversity of eukaryotic membrane cytochrome p450s. J Biol Chem 288:17082–17090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JP, Mysinger M, Korzekwa KR. (2002) Computational models for cytochrome P450: a predictive electronic model for aromatic oxidation and hydrogen atom abstraction. Drug Metab Dispos 30:7–12 [DOI] [PubMed] [Google Scholar]
- Jung F, Griffin KJ, Song W, Richardson TH, Yang M, Johnson EF. (1998) Identification of amino acid substitutions that confer a high affinity for sulfaphenazole binding and a high catalytic efficiency for warfarin metabolism to P450 2C19. Biochemistry 37:16270–16279 [DOI] [PubMed] [Google Scholar]
- Kelly SL, Lamb DC, Cannieux M, Greetham D, Jackson CJ, Marczylo T, Ugochukwu C, Kelly DE. (2001) An old activity in the cytochrome P450 superfamily (CYP51) and a new story of drugs and resistance. Biochem Soc Trans 29:122–128 [DOI] [PubMed] [Google Scholar]
- Kempf DJ, Marsh KC, Denissen JF, McDonald E, Vasavanonda S, Flentge CA, Green BE, Fino L, Park CH, Kong XP, et al. (1995) ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc Natl Acad Sci USA 92:2484–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose TS, Ibeanu GC, Ghanayem BI, Pedersen LG, Li L, Hall SD, Goldstein JA. (1998) Identification of residues 286 and 289 as critical for conferring substrate specificity of human CYP2C9 for diclofenac and ibuprofen. Arch Biochem Biophys 357:240–248 [DOI] [PubMed] [Google Scholar]
- Korzekwa KR, Jones JP, Gillette JR. (1990) Theoretical studies on cytochrome P-450 mediated hydroxylation: A predictive model for hydrogen atom abstractions. J Am Chem Soc 112:7042–7046 [Google Scholar]
- Lee-Robichaud P, Akhtar ME, Wright JN, Sheikh QI, Akhtar M. (2004) The cationic charges on Arg347, Arg358 and Arg449 of human cytochrome P450c17 (CYP17) are essential for the enzyme’s cytochrome b5-dependent acyl-carbon cleavage activities. J Steroid Biochem Mol Biol 92:119–130 [DOI] [PubMed] [Google Scholar]
- Li H, Poulos TL. (1997) The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat Struct Biol 4:140–146 [DOI] [PubMed] [Google Scholar]
- Naffin-Olivos JL, Auchus RJ. (2006) Human cytochrome b5 requires residues E48 and E49 to stimulate the 17,20-lyase activity of cytochrome P450c17. Biochemistry 45:755–762 [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, et al. (1996) P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6:1–42 [DOI] [PubMed] [Google Scholar]
- Nunez M, Guittet E, Pompon D, van Heijenoort C, Truan G. (2010) NMR structure note: oxidized microsomal human cytochrome b5. J Biomol NMR 47:289–295 [DOI] [PubMed] [Google Scholar]
- O’Brien SE, de Groot MJ. (2005) Greater than the sum of its parts: combining models for useful ADMET prediction. J Med Chem 48:1287–1291 [DOI] [PubMed] [Google Scholar]
- Ozalp C, Szczesna-Skorupa E, Kemper B. (2005) Bimolecular fluorescence complementation analysis of cytochrome p450 2c2, 2e1, and NADPH-cytochrome p450 reductase molecular interactions in living cells. Drug Metab Dispos 33:1382–1390 [DOI] [PubMed] [Google Scholar]
- Peng HM, Auchus RJ. (2013) The action of cytochrome b(5) on CYP2E1 and CYP2C19 activities requires anionic residues D58 and D65. Biochemistry 52:210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JA, Ebel RE, O’Keeffe DH, Matsubara T, Estabrook RW. (1976) Temperature dependence of cytochrome P-450 reduction. A model for NADPH-cytochrome P-450 reductase:cytochrome P-450 interaction. J Biol Chem 251:4010–4016 [PubMed] [Google Scholar]
- Poulos TL, Finzel BC, Howard AJ. (1987) High-resolution crystal structure of cytochrome P450cam. J Mol Biol 195:687–700 [DOI] [PubMed] [Google Scholar]
- Ravichandran KG, Boddupalli SS, Hasermann CA, Peterson JA, Deisenhofer J. (1993) Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450’s. Science 261:731–736 [DOI] [PubMed] [Google Scholar]
- Reed JR, Backes WL. (2012) Formation of P450 · P450 complexes and their effect on P450 function. Pharmacol Ther 133:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Connick JP, Cheng D, Cawley GF, Backes WL. (2012) Effect of homomeric P450-P450 complexes on P450 function. Biochem J 446:489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Eyer M, Backes WL. (2010) Functional interactions between cytochromes P450 1A2 and 2B4 require both enzymes to reside in the same phospholipid vesicle: evidence for physical complex formation. J Biol Chem 285:8942–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendic S. (2002) Summary of information on human CYP enzymes: human P450 metabolism data. Drug Metab Rev 34:83–448 [DOI] [PubMed] [Google Scholar]
- Rendic S, Di Carlo FJ. (1997) Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab Rev 29:413–580 [DOI] [PubMed] [Google Scholar]
- Reynald RL, Sansen S, Stout CD, Johnson EF. (2012) Structural characterization of human cytochrome P450 2C19: active site differences between P450s 2C8, 2C9, and 2C19. J Biol Chem 287:44581–44591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittle J, Green MT. (2010) Cytochrome P450 compound I: capture, characterization, and C-H bond activation kinetics. Science 330:933–937 [DOI] [PubMed] [Google Scholar]
- Rui L, Pochapsky SS, Pochapsky TC. (2006) Comparison of the complexes formed by cytochrome P450cam with cytochrome b5 and putidaredoxin, two effectors of camphor hydroxylase activity. Biochemistry 45:3887–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2007) Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem 282:14348–14355 [DOI] [PubMed] [Google Scholar]
- Sax PE, DeJesus E, Mills A, Zolopa A, Cohen C, Wohl D, Gallant JE, Liu HC, Zhong L, Yale K, et al. GS-US-236-0102 study team (2012) Co-formulated elvitegravir, cobicistat, emtricitabine, and tenofovir versus co-formulated efavirenz, emtricitabine, and tenofovir for initial treatment of HIV-1 infection: a randomised, double-blind, phase 3 trial, analysis of results after 48 weeks. Lancet 379:2439–2448 [DOI] [PubMed] [Google Scholar]
- Schoch GA, Yano JK, Sansen S, Dansette PM, Stout CD, Johnson EF. (2008) Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J Biol Chem 283:17227–17237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. (2004) Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J Biol Chem 279:9497–9503 [DOI] [PubMed] [Google Scholar]
- Sevrioukova IF, Poulos TL. (2012) Interaction of human cytochrome P4503A4 with ritonavir analogs. Arch Biochem Biophys 520:108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen AL, Porter TD, Wilson TE, Kasper CB. (1989) Structural analysis of the FMN binding domain of NADPH-cytochrome P-450 oxidoreductase by site-directed mutagenesis. J Biol Chem 264:7584–7589 [PubMed] [Google Scholar]
- Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, et al. (2008) Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J Virol 82:764–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SJ. (1975) Architecture and Topography of Biologic Membranes, HP Publishing Co., Inc., New York, New York [Google Scholar]
- Sirim D, Widmann M, Wagner F, Pleiss J. (2010) Prediction and analysis of the modular structure of cytochrome P450 monooxygenases. BMC Struct Biol 10:34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner AL, Laurence JS. (2008) High-field solution NMR spectroscopy as a tool for assessing protein interactions with small molecule ligands. J Pharm Sci 97:4670–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sligar SG. (1976) Coupling of spin, substrate, and redox equilibria in cytochrome P450. Biochemistry 15:5399–5406 [DOI] [PubMed] [Google Scholar]
- Subramanian M, Low M, Locuson CW, Tracy TS. (2009) CYP2D6-CYP2C9 protein-protein interactions and isoform-selective effects on substrate binding and catalysis. Drug Metab Dispos 37:1682–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Tam H, Zheng H, Tracy TS. (2010) CYP2C9-CYP3A4 protein-protein interactions: role of the hydrophobic N terminus. Drug Metab Dispos 38:1003–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulc M, Jecmen T, Snajdrova R, Novak P, Martinek V, Hodek P, Stiborova M, Hudecek J. (2012) Mapping of interaction between cytochrome P450 2B4 and cytochrome b5: the first evidence of two mutual orientations. Neuroendocrinol Lett 33 (Suppl 3):41–47 [PubMed] [Google Scholar]
- Sun H, Scott DO. (2010) Structure-based drug metabolism predictions for drug design. Chem Biol Drug Des 75:3–17 [DOI] [PubMed] [Google Scholar]
- Szczesna-Skorupa E, Chen CD, Kemper B. (2000) Cytochromes P450 2C1/2 and P450 2E1 are retained in the endoplasmic reticulum membrane by different mechanisms. Arch Biochem Biophys 374:128–136 [DOI] [PubMed] [Google Scholar]
- Tripathi S, Li H, Poulos TL. (2013) Structural basis for effector control and redox partner recognition in cytochrome P450. Science 340:1227–1230 [DOI] [PubMed] [Google Scholar]
- Tsao CC, Wester MR, Ghanayem B, Coulter SJ, Chanas B, Johnson EF, Goldstein JA. (2001) Identification of human CYP2C19 residues that confer S-mephenytoin 4′-hydroxylation activity to CYP2C9. Biochemistry 40:1937–1944 [DOI] [PubMed] [Google Scholar]
- Walsh AA, Szklarz GD, Scott EE. (2013) Human cytochrome P450 1A1 structure and utility in understanding drug and xenobiotic metabolism. J Biol Chem 288:12932–12943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Savas U, Hsu MH, Stout CD, Johnson EF. (2012) Crystal structure of human cytochrome P450 2D6 with prinomastat bound. J Biol Chem 287:10834–10843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Savas U, Stout CD, Johnson EF. (2011) Structural characterization of the complex between alpha-naphthoflavone and human cytochrome P450 1B1. J Biol Chem 286:5736–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang LP, Zhang XZ, Huang SQ, Bartlam M, Zhou SF. (2009) New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab Rev 41:573–643 [DOI] [PubMed] [Google Scholar]
- Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. (1997) Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA 94:8411–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester MR, Johnson EF, Marques-Soares C, Dijols S, Dansette PM, Mansuy D, Stout CD. (2003) Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1 A resolution: evidence for an induced fit model of substrate binding. Biochemistry 42:9335–9345 [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. (2000) Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell 5:121–131 [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. (2004) Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science 305:683–686 [DOI] [PubMed] [Google Scholar]
- Wrighton SA, Schuetz EG, Thummel KE, Shen DD, Korzekwa KR, Watkins PB. (2000) The human CYP3A subfamily: practical considerations. Drug Metab Rev 32:339–361 [DOI] [PubMed] [Google Scholar]
- Xia C, Panda SP, Marohnic CC, Martásek P, Masters BS, Kim JJ. (2011) Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency. Proc Natl Acad Sci USA 108:13486–13491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Desai MC. (2009) Pharmacokinetic enhancers for HIV drugs. Curr Opin Investig Drugs 10:775–786 [PubMed] [Google Scholar]
- Xu L, Liu HC, Murray BP, Callebaut C, Lee MS, Hong A, Strickley RG, Tsai LK, Stray KM, Wang Y, Rhodes GR, Desai M.C., et al. (2010) Cobicistat (GS-9350): A potent and selective inhibitor of human CYP3A as a novel pharmacoenhancer. ACS Med Chem Lett 1:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Gillam EM, Dong MS, Johnson WW, Guengerich FP, Shimada T. (1997) Reconstitution of recombinant cytochrome P450 2C10(2C9) and comparison with cytochrome P450 3A4 and other forms: effects of cytochrome P450-P450 and cytochrome P450-b5 interactions. Arch Biochem Biophys 342:329–337 [DOI] [PubMed] [Google Scholar]
- Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem 279:38091–38094 [DOI] [PubMed] [Google Scholar]
- Yin H, Anders MW, Korzekwa KR, Higgins L, Thummel KE, Kharasch ED, Jones JP. (1995) Designing safer chemicals: predicting the rates of metabolism of halogenated alkanes. Proc Natl Acad Sci USA 92:11076–11080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M. (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141 [DOI] [PubMed] [Google Scholar]
- Zanger UM, Turpeinen M, Klein K, Schwab M. (2008) Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal Bioanal Chem 392:1093–1108 [DOI] [PubMed] [Google Scholar]
- Zhao C, Gao Q, Roberts AG, Shaffer SA, Doneanu CE, Xue S, Goodlett DR, Nelson SD, Atkins WM. (2012) Cross-linking mass spectrometry and mutagenesis confirm the functional importance of surface interactions between CYP3A4 and holo/apo cytochrome b(5). Biochemistry 51:9488–9500 [DOI] [PMC free article] [PubMed] [Google Scholar]