Abstract
Idiosyncratic hepatotoxicity has been associated with the oral tyrosine kinase inhibitor lapatinib, which is used in metastatic breast cancer therapy. Lapatinib is extensively metabolized by cytochrome P450 3A4/5 to yield an O-debenzylated metabolite, which can undergo further oxidation to a reactive quinone imine. A recent clinical study reported that concomitant use of lapatinib with dexamethasone increased the incidence of hepatotoxicity in metastatic breast cancer patients treated with lapatinib, and so we hypothesized that induction of CYP3A enhances the bioactivation of lapatinib to reactive intermediates that contribute to hepatotoxicity. Therefore, we examined the effect of CYP3A4 induction on the cytotoxicity and metabolism of lapatinib in the HepaRG human hepatic cell line. Differentiated HepaRG cells were pretreated with dexamethasone (100 μM) or the prototypical CYP3A4 inducer rifampicin (4 μM) for 72 hours, followed by incubation with lapatinib (0–100 μM) for 24 hours. Cell viability was monitored using WST-1 assays, and metabolites were quantified by liquid chromatography coupled to tandem mass spectrometry. Induction of CYP3A4 by dexamethasone or rifampicin enhanced lapatinib-induced cytotoxicity, compared with treatment with lapatinib alone. A direct comparison of the cytotoxicity of lapatinib versus O-debenzylated lapatinib demonstrated that the O-debenzylated metabolite was significantly more cytotoxic than lapatinib itself. Furthermore, pretreatment with 25 μM l-buthionine sulfoximine to deplete intracellular glutathione markedly enhanced lapatinib cytotoxicity. Cytotoxicity was correlated with increased formation of O-debenzylated lapatinib and cysteine adducts of the putative quinone imine intermediate. Collectively, these data suggest that CYP3A4 induction potentiates lapatinib-induced hepatotoxicity via increased reactive metabolite formation.
Introduction
Lapatinib is an orally active dual tyrosine kinase inhibitor of the epidermal growth factor receptor (Erb1) and human epidermal receptor 2 (Erb2) (Rusnak et al., 2001; Moy et al., 2007). Lapatinib is currently approved for use in combination with capecitabine or letrozole for the treatment of advanced or metastatic breast cancer in patients whose tumors overexpress human epidermal receptor 2 and who have received prior therapy with anthracycline, a taxane, and trastuzumab (Geyer et al., 2006; Kroep et al., 2010).
The US Food and Drug Administration recently issued a black-box warning against idiosyncratic hepatotoxicity associated with lapatinib, which has been observed in a small proportion of patients (<1%) in clinical trials and postmarket surveillance (Gomez et al., 2008). Severe and fatal cases of liver injury have been reported with lapatinib use (Moy et al., 2009; Cristofanilli et al., 2013). A recent pharmacogenetic investigation of a subset of metastatic breast cancer patients treated with lapatinib demonstrated that the human leukocyte antigen (HLA) allelic variant DQA1*02:01 was associated with elevations in alanine aminotransferase (Spraggs et al., 2011, 2012). This finding suggests that lapatinib-induced hepatotoxicity has at least a partial immune basis; however, the mechanism(s) remains unclear.
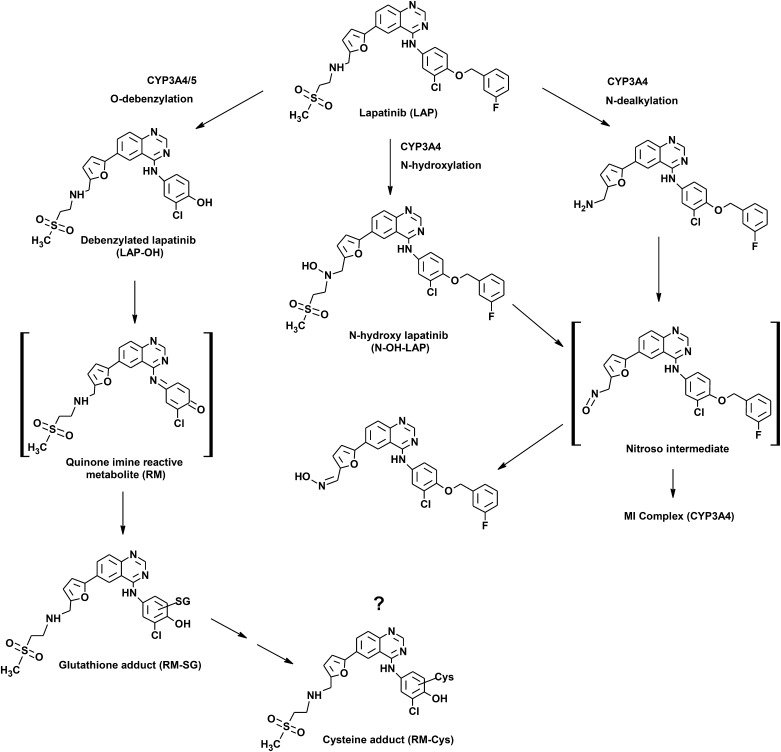
Lapatinib is primarily metabolized by cytochrome P450 (P450) 3A4/5 (GlaxoSmithKline, 2007) through three main pathways: O-dealkylation, N-dealkylation, and N-hydroxylation (Takakusa et al., 2011; Castellino et al., 2012) (Fig. 1). The O-dealkylated metabolite of lapatinib can undergo further oxidization to a reactive quinone imine intermediate (Teng et al., 2010). Glutathione (GSH) adducts of the putative quinone imine have been reported after incubation of lapatinib or its O-dealkylated metabolite with human liver microsomes and individual P450s (Teng et al., 2010; Chan et al., 2012). Previous studies have shown that lapatinib is a time-dependent inhibitor of CYP3A4 (Teng et al., 2010) through formation of a metabolic-intermediate complex via a nitroso intermediate (Takakusa et al., 2011; Barbara et al., 2013). On the other hand, Chan et al. (2012) reported that inactivation of CYP3A5 by lapatinib may proceed through adduction of the quinone imine to the apoprotein. On the basis of these findings, it seems likely that CYP3A-mediated metabolic activation of lapatinib may be involved in initiating lapatinib-induced hepatotoxicity.
Fig. 1.
Proposed bioactivation pathway of lapatinib. MI, metabolic-intermediate.
Concurrent usage of lapatinib with CYP3A4 inducers represents a potentially important drug–drug interaction that may lead to increased drug toxicity. Dexamethasone, a synthetic glucocorticoid, is a CYP3A4 inducer commonly used in metastatic cancer therapy to manage the symptoms of peritumoral edema associated with brain metastasis. A recent retrospective clinical study demonstrated that concomitant use of dexamethasone with lapatinib markedly increased the risk of hepatotoxicity in metastatic breast cancer patients (Teo et al., 2012). Patients receiving the combination of lapatinib plus dexamethasone were significantly more likely to develop clinically important elevations in alanine aminotransferase compared with patients taking lapatinib alone (Teo et al., 2012). In addition, Teo et al. (2012) demonstrated that treatment of transforming growth factor-α mouse hepatocyte cells with lapatinib plus dexamethasone resulted in increased cytotoxicity compared with treatment with lapatinib alone. However, direct evidence that links CYP3A4 induction with metabolic activation and toxicity of lapatinib is lacking.
A human-relevant hepatocyte model is essential to characterize the relationship between reactive metabolite (RM) formation and lapatinib-induced hepatotoxicity. HepaRG cells are an immortalized human liver progenitor cell line that has recently emerged as a useful model for evaluating metabolism-mediated drug toxicity (McGill et al., 2011). These cells are derived from human hepatocellular carcinoma and can be differentiated into hepatocyte-like and biliary epithelial-like cells (Aninat et al., 2006; Guillouzo et al., 2007). Differentiated HepaRG cells are capable of expressing many of the major drug metabolizing enzymes (e.g., CYP3A4) and transporters at levels comparable to primary human hepatocytes (Aninat et al., 2006; Guillouzo et al., 2007). In addition, HepaRG cells have been shown to respond to prototypical P450 inducers (Kanebratt and Andersson, 2008; Anthérieu et al., 2010). The advantage of HepaRG cells over primary hepatocytes is their ready availability, high reproducibility, and well characterized complement of gene products relevant to absorption, distribution, metabolism, and excretion (Andersson et al., 2012).
The objective of the current investigation was to use HepaRG cells as a model to characterize the role of CYP3A4-mediated metabolic activation in lapatinib-induced hepatotoxicity. An important aim was to establish the link between CYP3A4 induction and RM formation by directly testing whether increased CYP3A4 activity resulted in elevated RM formation and enhanced cytotoxicity. Presumably the RM-glutathione conjugate would be further metabolized through the mercapturic acid pathway, and this has been explored in the HepaRG model.
Materials and Methods
Lapatinib (free base) was purchased from LC Laboratories (Woburn, MA). The O-dealkylated metabolite of lapatinib, debenzylated lapatinib, was chemically synthesized from lapatinib, as described previously (Teng et al., 2010). D4-debenzylated lapatinib was supplied by Concert Pharmaceuticals Inc. (Lexington, MA). Stock solutions of lapatinib and lapatinib metabolites were prepared in dimethylsulfoxide (DMSO). Midazolam and d4-1′-hydroxymidazolam were purchase from Cerilliant (Rock Round, TX). l-glutathione (reduced) was purchased from Sigma Aldrich (St. Louis, MO). Optima liquid chromatography (LC)/mass spectrometry (MS)-grade water and Optima LC/MS-grade acetonitrile were purchased from Fisher Scientific (Pittsburgh, PA). All other chemicals and reagents were of analytical grade and were purchased from commercial sources.
HepaRG Cells.
All cell incubation studies were carried out at 37°C in a humidified atmosphere, 5% CO2. Cryopreserved differentiated and undifferentiated HepaRG cells were supplied by BioPredic International (Rennes, France). Undifferentiated cells were terminally differentiated in-house according to the supplier’s protocols, as described previously (Gripon et al., 2002). Briefly, cells were first grown for 2 weeks to confluence in culture medium followed by culturing for 2 weeks in culture medium supplemented with 2% DMSO. Differentiated cells were plated at a density of approximately 70,000 cells/well in collagen-coated 96-well microtiter plates according to the manufacturer’s instructions and maintained in general purpose medium composed of Williams’ E medium with GlutaMAX-I supplemented with HepaRG Thaw, Seed, and General Purpose Supplement 670 (HPRG670; Life Technologies, Carlsbad, CA).
To induce CYP3A4, HepaRG cells were incubated with serum-free induction medium composed of Williams’ E medium with GlutaMAX-I supplemented with HepaRG Serum-Free Induction Medium Supplement 650 (HPRG650; Life Technologies) and containing dexamethasone (100 μM) or rifampicin (4 μM). Cells were incubated for a total of 72 hours in induction medium, with medium changed every 24 hours. Control incubations were conducted with DMSO (0.1%). Initial experiments were carried out to determine maximal induction of CYP3A4 using a range of concentrations of dexamethasone (0.1–500 μM). To note, dexamethasone treatment alone was not cytotoxic to HepaRG cells over a range of concentrations tested. After the induction period, CYP3A4 activity in HepaRG cells was measured using midazolam as a probe substrate. Cells were incubated with serum-free incubation medium containing midazolam (3 μM) for 1 hour. The reaction was quenched by addition of an equal volume of ice-cold methanol containing d4-1′-hydroxymidazolam (100 ng/ml), and the supernatant was collected for analysis of 1′-hydroxymidazolam by LC coupled to tandem MS (LC-MS/MS), as described previously (Kirby et al., 2006). Treatment with 100 μM dexamethasone was found to yield maximal induction (approximately 7-fold) of midazolam 1′-hydroxylation activity, compared with the control treatment, and thus 100 μM dexamethasone was used for all subsequent induction studies. After the 72-hour induction period, cell medium was replaced with cytotoxicity medium composed of Williams’ E medium with GlutaMAX-I supplemented with HepaRG Tox medium supplement 630 (HPRG630; Life Technologies). Cells were incubated with lapatinib (10–100 μM) or O-debenyzlated lapatinib (10–100 μM) for 24 hours. Test compounds were dissolved in DMSO/acetonitrile (4:1), and added into HepaRG cell medium to a final concentration of 0.4% DMSO, 0.1% acetonitrile. For CYP3A4 inhibition, cells were coincubated with ketoconazole (4 μM) for 24 hours. In a preliminary range-finding study, incubation of HepaRG cells with 4 μM ketoconazole was found to inhibit CYP3A4 activity by >90%, as measured by midazolam 1′-hydroxylation.
Primary Human Hepatocytes.
Cryopreserved plateable human hepatocytes from two adult donors, one male (lot no. Hu4246) and one female (lot no. Hu1389), were generously provided by Life Technologies (Carlsbad, CA). Hepatocyte donors were fully characterized by the supplier and selected based on relative extent of CYP3A4 induction, as characterized by the supplier. Cryopreserved hepatocytes were thawed and plated according to the supplier’s protocol. Briefly, cells were thawed at 37°C for 60–90 seconds followed by dilution into 50 ml of warmed Cryopreserved Hepatocyte Recovery Medium (Life Technologies). The cell suspension was centrifuged at 100g for 10 minutes. Cells were plated in Williams’ E medium (no phenol red) containing the Hepatocyte Plating Supplement Pack (Life Technologies) on 96-well collagen-coated Geltrex plates (Life Technologies) at a seeding density of 0.5–0.7 × 105 cells/well. After 6 hours, the medium was replaced with incubation medium containing Williams’ E Medium and the Hepatocyte Maintenance Supplement Pack (serum-free). Primary hepatocyte cultures were maintained in incubation medium for 48 hours, followed by incubation with lapatinib (100 μM) for 24 hours.
Cell Viability and Metabolite Formation.
Cell viability in differentiated HepaRG cells and primary human hepatocytes was assessed using WST-1 cell viability assays (Clontech Laboratories, Inc., Mountain View, CA) according to the manufacturer’s instructions. Control incubations were conducted by incubation of cells with vehicle control (0.4% DMSO, 0.1% acetonitrile). Briefly, 10 µl of the premixed WST-1 cell proliferation reagent was added to each well of the 96-well plate (1:10 final dilution), and the plate was incubated for 30 minutes at 37°C, followed by shaking at room temperature for 1 minute. Absorbance was measured at 440 nm using a Tecan Infinite M200 microplate reader (Tecan Systems, Inc., San Jose, CA). The reference wavelength was 690 nm. Cell viability after treatment with test compounds was quantified by calculating the percent viability compared with cell incubations with vehicle control (untreated).
To determine the extent of lapatinib metabolite formation after incubations in HepaRG cells, 100 μl of ice-cold acetonitrile containing 100 ng/ml d4-debenzylated lapatinib (internal standard) was added to each well in a 96-well microtiter plate. Cells were scraped, and the supernatant was transferred to a 1.7-ml centrifuge tube, vortexed and sonicated for 1 minute, followed by centrifugation at 13,000 rpm for 10 minutes at 4°C. The supernatant was transferred to a separate vial and dried under N2 gas at 37°C using a Biotage TurboVap (Biotage, Charlotte, NC). Samples were reconstituted in 100 μl of an acetonitrile/water (1:1) mixture and transferred to LC/MS vials for analysis. For detection and analysis of reactive metabolite glutathione (RM-SG) and cysteine (RM-Cys) adducts, the supernatant from cell extracts was dried under N2 gas as described above, and samples were reconstituted in 50 μl of an acetonitrile/water (3:7) mixture for LC-MS/MS analysis.
Analysis and Quantitation of Metabolites by LC-MS/MS.
Drug metabolites from cell incubations were analyzed using a high-performance LC system consisting of two Shimadzu LC-10AD pumps with a gradient controller and a Shimadzu SIL-10ADvp autoinjector (Shimadzu Scientific Instruments Inc., Columbia, MD) coupled to a Waters Micromass Quattro Micro II triple quadrupole mass spectrometer (Waters Corporation, Milford, MA), as described previously (Takakusa et al., 2011). A 30-μl aliquot of each sample was injected into the equilibrated high-performance LC system. Solvents A and B were Optima LC/MS-grade water (Fisher Scientific) with 0.1% (v/v) trifluoroacetic acid and Optima LC/MS-grade acetonitrile (Fisher Scientific) with 0.1% (v/v) trifluoroacetic acid, respectively. Analyte separation was achieved with an Agilent Zorbax SB-C18 column (5 μm, 2.1 mm × 150 mm) (Agilent Technologies, Santa Clara, CA) at a flow rate of 0.3 ml/min. The gradient program for analysis of lapatinib metabolites was as follows: isocratic gradient at 15% B (0–1.5 minutes), linear gradient from 15% to 95% B (1.5–5 minutes), isocratic at 95% B (5–7 minutes), and returned to 15% B (7.1 minutes). The eluent was introduced directly into the mass spectrometer via electrospray ionization in positive ion mode. The MS conditions were as follows: capillary voltage, 3.5 kV; cone voltage, 60 V; source temperature, 120°C; desolvation temperature, 350°C; ionization mode, electrospray ionization in the positive ion mode; and analyzer, V mode. The MS data were acquired in MS/MS mode utilizing multiple reaction monitoring (MRM) with collision energy 30 V. The following LC-MS/MS MRM method was developed to permit the detection and quantitation of lapatinib and lapatinib metabolites based on structurally specific fragmentation obtained from collision-induced dissociation: lapatinib (LAP) m/z 581 → 365, retention time 6.8 minutes; O-debenzylated lapatinib (LAP-OH) m/z 473 → 350, retention time 5.2 minutes; RM-SG adduct m/z 778 → 655, retention time 4.9 minutes; and RM-Cys adduct m/z 592 → 382, retention time 4.8 minutes; internal standard, d4-debenzylated lapatinib m/z 477 → 352, retention time 5.2 minutes. The MS spectral data were analyzed and deconvoluted by using MassLynx software (version 4.1; Waters Corporation).
Glutathione Depletion.
HepaRG cells were pretreated with l-buthionine sulfoximine (BSO) (25 μM) for 24 hours, followed by incubation with lapatinib (10, 50, and 100 μM) or vehicle control for 24 hours. BSO treatment alone was not cytotoxic to HepaRG cells over the range of concentrations tested (25–200 μM). Cell viability was monitored using WST-1 assays as described above.
Statistical Analyses.
All cell incubations were performed in triplicate in one to three independent experiments (as indicated). The mean and standard deviation (S.D.) or standard error of the mean (S.E.M.) for each experiment were determined using GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA). Treatment groups were compared with their respective controls using the t test for unpaired data. P values were calculated by two-tailed analysis, and differences at P < 0.05 were considered significant.
Results
Cytotoxicity of Lapatinib in HepaRG Cells and Primary Human Hepatocytes.
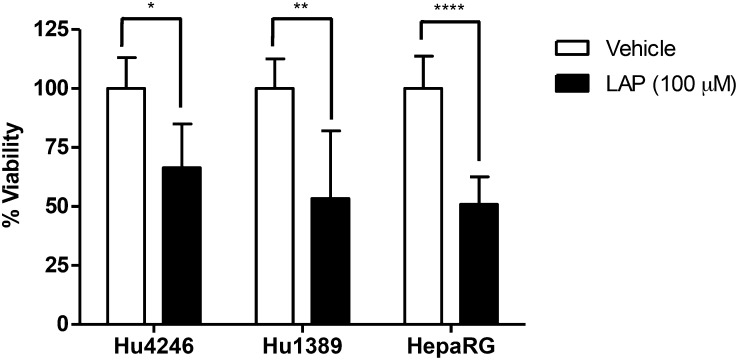
The cytotoxic effects of lapatinib were examined in HepaRG cells and compared with primary human hepatocytes to evaluate HepaRG cells as a model to study lapatinib-induced hepatotoxicity. Fig. 2 shows the percent viability of HepaRG cells treated with lapatinib (100 μM) for 24 hours in three independent experiments performed in triplicate and the percent viability of primary human hepatocytes from two donors (Hu4246 and Hu1389) treated with lapatinib (100 μM) for 24 hours in triplicate for each donor. Incubation of HepaRG cells with lapatinib resulted in 50.9% ± 11.7% viability (mean ± S.D.) (P < 0.0001), compared with the control. Similarly, treatment of primary human hepatocytes Hu4286 and Hu1389 with lapatinib for 24 hours resulted in 66.4% ± 18.5% (P = 0.0151) and 53.4% ± 28.6% viability (P = 0.0095), respectively, compared with the control (Fig. 2). There was no statistically significant difference between the viability of primary human hepatocytes versus HepaRG cells treated with lapatinib. These data demonstrate that lapatinib induces cytotoxicity to a similar extent in primary human hepatocytes and HepaRG cells.
Fig. 2.
Cytotoxicity of lapatinib in primary human hepatocytes and HepaRG cells. Cryopreserved plated human hepatocytes from two donors (Hu4246 and Hu1389) and HepaRG cells were treated with lapatinib (100 μM) for 24 hours. Cell viability was monitored using WST-1 assays, and viability is expressed as the percent viability compared with vehicle treatment. Data represent the mean ± S.D. of triplicate values for Hu4246 and Hu1389, and the mean ± S.D. of triplicate values from three independent experiments for HepaRG cells. *P < 0.05; **P < 0.01; ****P < 0.0001 compared with vehicle (unpaired t test, two-tailed P values).
Effect of CYP3A4 Induction on the Cytotoxicity of Lapatinib in HepaRG Cells.
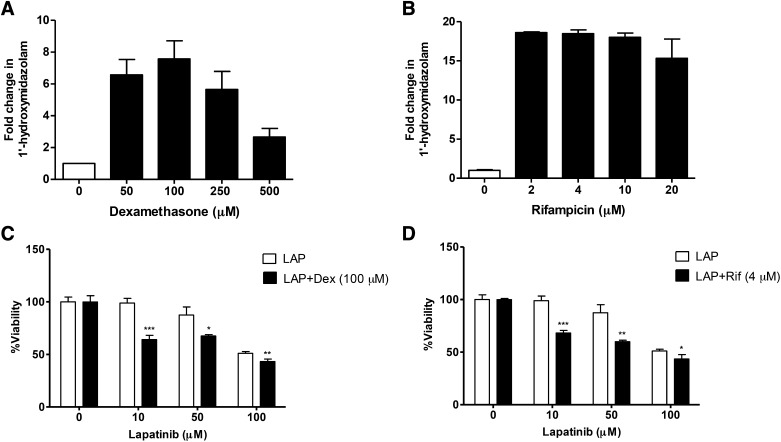
To further probe the role of CYP3A4-mediated metabolism as a mechanism in lapatinib hepatotoxicity, we sought to evaluate the effect of dexamethasone on CYP3A4 activity and lapatinib-induced cytotoxicity using HepaRG cells as an in vitro model. First, the extent to which dexamethasone induces CYP3A4 activity in HepaRG cells was monitored using midazolam as a CYP3A4 probe substrate. Maximal induction of CYP3A4-dependent midazolam hydroxylation (approximately 7-fold) was achieved with 100 μM dexamethasone (Fig. 3A), compared with the vehicle control. In addition, CYP3A4 activity was monitored in cells treated with the prototypical CYP3A4 inducer rifampicin. Treatment of cells with 2–4 μM rifampicin resulted in an 18.5-fold increase in midazolam 1′-hydroxylase activity, and midazolam 1′-hydroxylation remained significantly elevated up to 20 μM rifampicin (Fig. 3B).
Fig. 3.
Induction of CYP3A4 activity by dexamethasone and rifampicin, and the effect of CYP3A4 induction on the cytotoxicity of lapatinib in HepaRG cells. HepaRG cells were pretreated with varying concentrations of dexamethasone (A) or rifampicin (B) or vehicle for 72 hours, followed by incubation with midazolam (3 μM) for 1 hour. CYP3A4 activity was assessed by measurement of 1′-hydroxymidazolam using LC-MS/MS. Fold change in 1′-hydroxymidazolam was calculated by comparison with pretreatment with vehicle control (DMSO). Data represent mean ± S.E.M. of three to four values. For cytotoxicity studies, HepaRG cells were pretreated with 100 μM dexamethasone (C) or 4 μM rifampicin (D) or vehicle for 72 hours, followed by incubation with LAP (10, 50, 100 μM) or vehicle control for 24 hours. Cell viability was monitored using WST-1 assays. For (C), LAP + Dex was compared with incubation with LAP alone at each concentration. For (D), LAP + Rif was compared with incubation with LAP alone at each concentration. Data represent the mean ± S.D. of triplicate values. *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired t test, two-tailed P values). Dex, dexamethasone; Rif, rifampicin.
Next, the effect of CYP3A4 induction on the cytotoxicity of lapatinib was evaluated. The results from WST-1 cell viability assays are shown in Fig. 3C. Dexamethasone potentiated the cytotoxicity of lapatinib at each concentration tested (10 μM, P = 0.0006; 50 μM, P = 0.0114; and 100 μM, P = 0.0093) compared with cells treated with lapatinib alone. Rifampicin was used as a positive control inducer to verify the effect of CYP3A4 induction on lapatinib’s cytotoxicity (Fig. 3D). Similarly, pretreatment of HepaRG cells with rifampicin (4 μM) resulted in a significant decrease in cell viability compared with treatment with lapatinib alone (Fig. 3D).
Cytotoxicity of Lapatinib versus Lapatinib Metabolites.
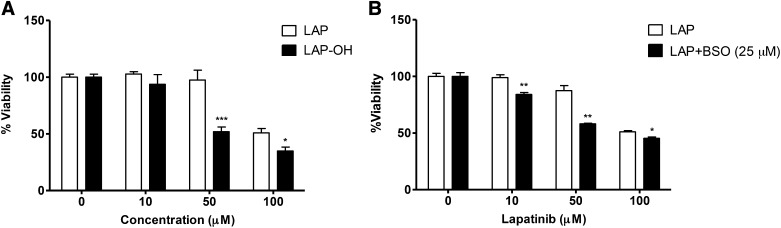
The increase in lapatinib’s cytotoxicity observed with CYP3A4 induction is consistent with the toxicity arising from a metabolite rather than the parent drug alone. To better address this possibility, a direct comparison was made between the cytotoxicity of LAP versus its O-debenzylated metabolite (LAP-OH). Fig. 4A shows the results from WST-1 cell viability assays after treatment of HepaRG cells with LAP or LAP-OH for 24 hours. The viability of cells treated with 50 μM LAP was 97.5% ± 8.70% viability compared with 51.90% ± 4.17% viability for cells treated with 50 μM LAP-OH (P = 0.0002). Cell viability was 50.0% ± 3.36% for cells treated with 100 μM LAP compared with 34.9% ± 3.49% viability for cells treated with 100 μM LAP-OH (P = 0.0067) (Fig. 4A). This demonstrates that LAP-OH is significantly more cytotoxic to HepaRG cells than lapatinib itself. We also compared the cytotoxicity of lapatinib versus the N-hydroxy lapatinib metabolite (N-OH-LAP); however, N-OH-LAP was not cytotoxic over the range of concentrations tested (data not shown). Taken together, these data suggest that LAP-OH is likely a precursor metabolite to the putative cytotoxic species.
Fig. 4.
Cytotoxicity of lapatinib versus O-debenzylated lapatinib in HepaRG cells and the effect of glutathione depletion by BSO on lapatinib-induced cytotoxicity. (A) HepaRG cells were incubated with LAP or LAP-OH (10, 50, 100 μM) or vehicle control for 24 hours. Cell viability was monitored using WST-1 assays. (B) HepaRG cells were pretreated with 25 μM BSO for 24 hours, prior to incubation with LAP (10, 50, 100 μM) or vehicle control for 24 hours. For (A), LAP-OH was compared with incubation with LAP at each concentration. Data represent means ± S.E.M. of triplicate values from three independent experiments. For (B), LAP + BSO was compared with incubation with LAP alone at each concentration. Data represent the mean ± S.D. of triplicate values. *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired t test, two-tailed P values).
Effect of Glutathione Depletion on the Cytotoxicity of Lapatinib.
Previous studies have shown that LAP-OH can undergo further metabolism to an electrophilic quinone imine intermediate (Teng et al., 2010). Quinone imines can readily react with cellular nucleophiles, such as GSH or cysteine residues of proteins, potentially leading to toxicities (Park et al., 2005). On the basis of this observation, we tested whether depletion of intracellular GSH could modulate lapatinib cytotoxicity. BSO is an inhibitor of γ-glutamylcysteine synthetase, which catalyzes the rate-limiting step in glutathione synthesis, and thus BSO was used experimentally to deplete intracellular GSH stores. In control incubations, treatment with BSO (25 μM) alone was not cytotoxic to the cells. However, pretreatment with BSO sensitized the cells to cytotoxicity by lapatinib at subtoxic doses (10 μM) and augmented the toxicity of lapatinib at higher doses (50 μM and 100 μM) compared with cells treated with lapatinib alone (Fig. 4B). This synergistic effect suggests that detoxification of reactive metabolites by GSH conjugation may play an important role in preventing cellular injury from electrophilic intermediates derived from lapatinib.
Effect of CYP3A4 Induction on the Metabolism of Lapatinib in HepaRG Cells.
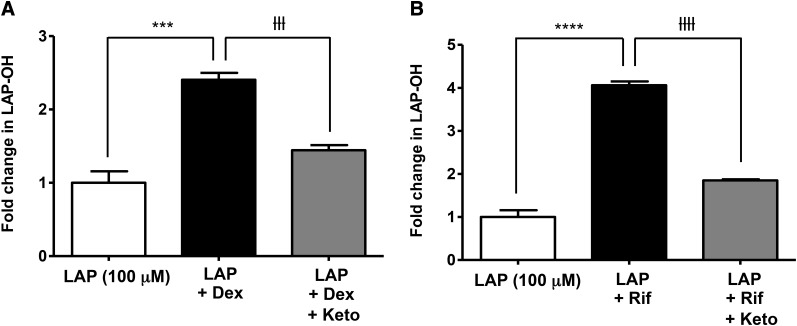
After establishing that both CYP3A4 induction and GSH depletion potentiate the cytotoxicity of lapatinib, the effect of CYP3A4 induction on the formation of lapatinib metabolites was monitored to evaluate the correlation between cytotoxicity and metabolism. Dexamethasone increased LAP-OH levels by 2.4-fold (P = 0.0002) (Fig. 5A) compared with treatment with lapatinib alone, and rifampicin increased LAP-OH formation by 4-fold (P < 0.0001) (Fig. 5B). Coincubation with the CYP3A4 inhibitor ketoconazole (4 μM) attenuated the effect of dexamethasone (P = 0.0001) and rifampicin (P < 0.0001) on the formation of LAP-OH.
Fig. 5.
Effect of CYP3A4 induction and inhibition on the formation of O-debenzylated lapatinib in HepaRG cells. HepaRG cells were incubated with 100 μM dexamethasone (A) or 4 μM rifampicin (B) for 72 hours, followed by incubation with LAP (100 μM) for 24 hours. For CYP3A4 inhibition, HepaRG cells were coincubated with ketoconazole (4 μM) + LAP (100 μM). Formation of LAP-OH was quantified by LC-MS/MS utilizing MRM. Data represent the mean ± S.D. of triplicate values. ***P < 0.001 and ****P < 0.0001 for comparison of LAP versus LAP + Dex and LAP versus LAP + Rif; ƚƚƚP < 0.001 and ƚƚƚƚP < 0.0001 for comparison of LAP + Dex versus LAP + Dex + Keto and LAP + Rif versus LAP + Rif + Keto (unpaired t test, two-tailed P values). Dex, dexamethasone; Keto, ketoconazole; Rif, rifampicin.
Detection of Reactive Metabolite Glutathione Adducts.
The generation of reactive metabolite thiol adducts was investigated as a surrogate to examine conversion of LAP-OH to the electrophilic quinone imine intermediate. GSH adducts of the putative quinone imine were previously identified in GSH-trapping studies in human liver microsomes incubated with lapatinib or LAP-OH (Teng et al., 2010). The predicted precursor ion of the RM-SG adduct is m/z 778. Fig. 6A shows the enhanced product ion spectrum of the GSH adduct (m/z 778) obtained from incubation of LAP-OH (100 μM) in human liver microsomes supplemented with NADPH and GSH (50 mM) for 1 hour. Collision-induced dissociation of m/z 778 yielded the characteristic fragmentation of the RM-SG adduct with neutral loss of 123 atomic mass units (amu) to yield the major product ion m/z 655 [M + H – 123]+. This corresponds to fragmentation of the lapatinib portion of the molecule, and neutral loss of 129 amu corresponds to loss of the pyroglutamic acid from the GSH adduct, as described previously (Evans et al., 2004), to yield the secondary product ion m/z 526 [M + H – 252]+ (Fig. 6A). This product ion spectrum is consistent with previous reports from LC-MS/MS analysis of the RM-SG adduct formed in human liver microsomes and recombinantly expressed CYPs (Teng et al., 2010; Chan et al., 2012).
Fig. 6.
Identification of reactive metabolite GSH and cysteine adducts. (A) Enhanced product ion scans of the putative quinone imine reactive metabolite-GSH adduct (RM-SG) (m/z 778) formed in pooled human liver microsomes incubated with O-debenzylated lapatinib (LAP-OH) (100 μM) for 1 hour and supplemented with NADPH and GSH (50 mM). (B) MRM chromatogram of RM-SG from incubation of LAP-OH (100 μM) in HepaRG cells for 24 hours. (C) Enhanced product ion scans of the putative quinone imine RM-Cys adduct (m/z 592). (D) MRM chromatogram of RM-Cys from incubation of LAP-OH (100 μM) in HepaRG cells for 24 hours.
Lapatinib-derived thiol adducts have not been reported in intact cells. Therefore, we determined whether such RM-SG adducts were formed in HepaRG cells after incubation with lapatinib or LAP-OH. For these studies, HepaRG cells were incubated with lapatinib (100 μM) or LAP-OH (50 μM to 100 μM) for 24 hours, and cell media supernatant was analyzed by LC-MS/MS. An MRM method was developed based on the characteristic precursor to product fragmentation of the GSH adduct m/z 778 to m/z 655, described above. The levels of GSH adducts in HepaRG cells treated with lapatinib (100 μM) were near the limit of detection; however, GSH adducts were readily detectable in cells treated with LAP-OH (100 μM). Fig. 6B shows a representative LC-MS/MS MRM chromatogram of the RM-SG adduct (retention time 4.87 minutes) detected in HepaRG cells incubated with LAP-OH for 24 hours.
Identification and Characterization of Reactive Metabolite Cysteine Adducts.
The tripeptide portion of GSH S-conjugates can undergo subsequent metabolism to yield cysteinyl-glycine and cysteine S-conjugates via the sequential actions of γ-glutamyl transferase and dipeptidases (Hinchman and Ballatori, 1994). Teng et al. (2010) previously described the identification of the cysteinyl-glycine conjugate of the quinone imine intermediate of lapatinib in human liver microsomal incubations with lapatinib or LAP-OH that were supplemented with GSH. In the present study, we evaluated the formation of cysteine S-conjugates of the quinone imine (RM-Cys), which would be generated as downstream detoxication products in HepaRG cells treated with lapatinib or LAP-OH.
The predicted precursor ion of the RM-Cys adduct is m/z 592 when analyzed by LC/MS in positive ion mode. Cysteine adducts were readily detectable in HepaRG cells after 24-hour incubation with lapatinib or LAP-OH (100 μM). The enhanced product ion spectrum of the RM-Cys adduct in culture media from HepaRG cells treated with LAP-OH for 24 hours is shown in Fig. 6C. The product ion spectrum reveals the expected fragmentation pattern of RM-Cys adducts resulting from neutral loss of 123 amu to yield the product ion m/z 469 [M + H – 123]+. This corresponds to fragmentation of the drug portion of the molecule and secondary loss of 87 amu corresponding to retro-Michael fragmentation of the Cys moiety to yield the major product ion, m/z 382 [M + H – 210]+ (Fig. 6C). On the basis of these findings, an LC-MS/MS MRM method was developed for RM-Cys with the major precursor to product ion transition m/z 592 → m/z 382. Fig. 6D shows a representative LC-MS/MS chromatogram from analysis of RM-Cys adducts in the cell supernatant after incubation of HepaRG cells with LAP-OH (100 μM) for 24 hours. A chromatographic peak at 4.83 minutes was detected from MRM analysis of RM-Cys (Fig. 6D).
After identifying the GSH and cysteine adducts in HepaRG cells, the time course of their formation was investigated. HepaRG cells were incubated with LAP-OH (50 μM) over a 24-hour period. Whereas both RM-SG and RM-Cys were detected in HepaRG cells treated with LAP-OH, the levels of Cys adducts increased over the 24-hour period with a corresponding decrease in GSH adduct levels, suggesting sequential metabolism of GSH adducts to Cys adducts. RM-Cys adducts were also detected in the cell supernatant of primary human hepatocytes incubated with lapatinib for 24 hours (data not shown). Collectively, these data suggest that Cys adducts downstream thiol metabolites generated from the detoxification of the putative quinone imine reactive intermediate of lapatinib. This represents the first identification of RM-Cys adducts generated from lapatinib in human-derived hepatic cells, supporting the use of this conjugate as a stable biomarker for the formation of lapatinib reactive metabolites in hepatic cell cultures.
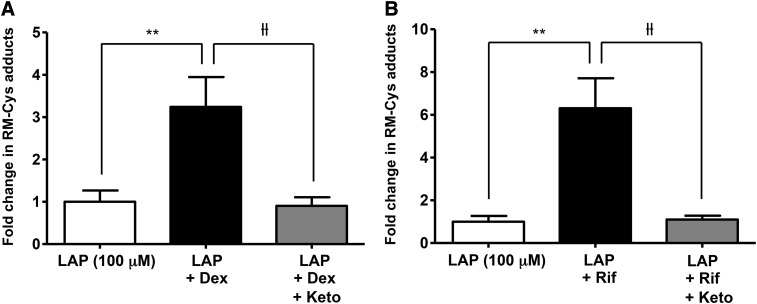
To further elucidate the role of CYP3A4 in the bioactivation of lapatinib, the effect of CYP3A4 induction on the formation of RM-Cys adducts was evaluated. Relative quantitation of RM-Cys adducts in the cell supernatant was achieved by LC-MS/MS analysis utilizing the MRM method described above. As shown in Fig. 7, pretreatment with dexamethasone increased the levels of RM-Cys adducts approximately 3-fold (P = 0.0068) in HepaRG cells compared with cells treated with lapatinib alone (Fig. 7A), and induction by rifampicin (4 μM) increased the levels of RM-Cys adducts approximately 6-fold (P = 0.0030) (Fig. 7B). Coincubation with ketoconazole (4 μM) attenuated the effect of dexamethasone (P = 0.0053) and rifampicin (P = 0.0031) on RM-Cys adduct formation. Collectively, these findings provide direct evidence that CYP3A4 induction enhances the generation of reactive metabolites of lapatinib.
Fig. 7.
Effect of CYP3A4 induction and inhibition on formation of RM-Cys adducts in HepaRG cells. HepaRG cells were incubated with 100 μM dexamethasone (A) or 4 μM rifampicin (B) for 72 hours, followed by incubation with lapatinib (100 μM) with or without coincubation with ketoconazole (4 μM) for 24 hours. Relative levels of RM-Cys adducts were quantified by LC-MS/MS MRM. Data represent the mean ± S.D. of triplicate values. **P < 0.01 for comparison of LAP versus LAP + Dex and LAP versus LAP + Rif; ƚƚP < 0.01 for comparison of LAP + Dex versus LAP + Dex + Keto and LAP + Rif versus LAP + Rif + Keto (unpaired t test, two-tailed P values). Dex, dexamethasone; Keto, ketoconazole; Rif, rifampicin.
Discussion
In this study, we investigated the role of metabolic activation in the cytotoxicity of lapatinib using HepaRG cells. The HepaRG cell line is a bipotent progenitor cell line that has emerged as a useful in vitro tool to evaluate human drug metabolism and toxicity (Andersson et al., 2012). Differentiated HepaRG cells retain most liver-specific functions, including stable expression of nuclear receptors, drug metabolizing enzymes, and drug transporters (Guillouzo et al., 2007; Anthérieu et al., 2010; Andersson et al., 2012). Notably, high levels of CYP3A4 expression and activity are maintained in differentiated HepaRG cells over long periods in culture. To evaluate HepaRG cells as a model, the cytotoxicity of lapatinib was compared between HepaRG cells and primary human hepatocytes, which are the gold standard model for human drug metabolism and toxicity studies. Treatment of HepaRG cells with lapatinib (100 μM) for 24 hours resulted in an approximately 50% decrease in cell viability, and a similar extent of cell death was observed in primary human hepatocytes treated with lapatinib (100 μM) under the same conditions (Fig. 2). Thus, the cytotoxic effect of lapatinib appears comparable in HepaRG cells and primary human hepatocytes.
Unlike other hepatic cell lines, expression of CYP3A4 is inducible in HepaRG cells using prototypical CYP3A4 inducers (Kanebratt and Andersson, 2008; Anthérieu et al., 2010). Indeed, dexamethasone (100 μM) significantly induced CYP3A4 activity in HepaRG cells in the present investigation. Treatment of HepaRG cells with dexamethasone for 72 hours induced CYP3A4 activity by 7-fold compared with the control, as measured by midazolam 1′-hydroxylation (Fig. 3A). It should be noted that the concentrations of dexamethasone to elicit this effect are well above the therapeutic levels, which are in the low nanomolar to submicromolar range (McCune et al., 2000). As proposed by Pascussi et al., (2001), these low concentrations activate the glucocorticoid receptor and increase pregnane X receptor (PXR) expression leading to transactivation of CYP3A4 gene expression, whereas supramicromolar concentrations cause direct activation of PXR. The dexamethasone-lapatinib interaction noted by Teo et al. (2012) may reflect CYP3A4 mRNA induction as a secondary glucocorticoid receptor–mediated response. On the other hand, the concentrations of dexamethasone used in HepaRG cells here are consistent with direct activation of PXR to produce maximal CYP3A4 induction, which allowed us to specifically probe the effects of CYP3A4-mediated metabolism on the cytotoxicity of lapatinib.
Importantly, we found that induction of CYP3A4 potentiated the cytotoxicity of lapatinib in parallel with increased formation of lapatinib metabolites on the proposed bioactivation pathway (Fig. 1). Pretreatment of HepaRG cells with either dexamethasone or rifampicin, followed by incubation with lapatinib (100 μM), significantly elevated lapatinib-induced cytotoxicity (Fig. 3, C and D), in conjunction with increased formation of the O-debenzylated metabolite of lapatinib (LAP-OH) (Fig. 5). Coincubation of HepaRG cells with lapatinib and ketoconazole attenuated the levels of metabolite formation observed with both dexamethasone and rifampicin (Fig. 5). These data are consistent with the view that metabolic activation by CYP3A4 plays a causative role in lapatinib-induced cytotoxicity. Previous studies have shown that drug-mediated nuclear receptor activation and P450 induction leading to increased metabolic activation can increase drug-induced toxicity. Cheng et al. (2009) demonstrated that treatment of mice humanized for PXR and CYP3A4 with acetaminophen and rifampicin enhanced acetaminophen-induced liver injury compared with mice treated with acetaminophen alone. In addition, Zhang et al. (2002) showed that phenobarbital, a well known activator of the constitutive androstane receptor (CAR), induced expression of CYP1A2 and CYP3A11 mRNAs in wild-type CAR mice and markedly enhanced acetaminophen toxicity, whereas CAR-knockout mice were resistant to acetaminophen toxicity.
A direct comparison of the cytotoxicity of lapatinib versus LAP-OH was also performed. The finding that LAP-OH was significantly more cytotoxic to HepaRG cells than lapatinib itself (Fig. 4A) strongly suggests that the toxicity is due, at least in part, to conversion of the parent drug to metabolite(s). LAP-OH is a significant metabolite in humans after oral administration of lapatinib (Castellino et al., 2012). In a recent study on the human metabolism of lapatinib, LAP-OH was found to be primarily excreted in feces and was not detected in plasma (Castellino et al., 2012). The median value reported for this metabolite was 4% of the dose, but it may represent up to 19.2% of the excreted dose in humans (Castellino et al., 2012).
The formation of reactive drug metabolites has been proposed as an initial step in the development of drug-induced liver injury (Park et al., 2005). In vitro investigations have shown that LAP-OH can be further oxidized to a reactive quinone imine intermediate, which can covalently adduct GSH and potentially other cellular nucleophiles (Teng et al., 2010). Therefore, we tested the hypothesis that depletion of GSH may increase the cellular accumulation of reactive metabolites of lapatinib, rendering HepaRG cells more susceptible to injury. Treatment of cells with BSO to deplete GSH resulted in potentiation of lapatinib-induced cytotoxicity (Fig. 4B). It is postulated that reactive metabolites of lapatinib may cause cellular damage by direct adduction to cellular proteins and/or disruption of cellular redox balance (Teng et al., 2010; Castellino et al., 2012). Thus, environmental or genetic factors that compromise the detoxication and antioxidant defense pathways involved in the inactivation of toxic metabolites could be expected to contribute to individual susceptibility to idiosyncratic lapatinib-induced hepatotoxicity.
GSH conjugates of the putative quinone imine intermediate of lapatinib were previously reported from incubation of lapatinib or LAP-OH in human liver microsomes and recombinantly expressed P450 enzymes (Teng et al., 2010; Chan et al., 2012). In the present study, RM-SG adducts were detected in HepaRG cells incubated with LAP-OH utilizing LC-MS/MS analysis (Fig. 6B). Furthermore, cysteine adducts of the putative quinone imine intermediate were also detected in cells treated with lapatinib and LAP-OH (Fig. 6D). Cysteinyl-glycine conjugates, but not N-acetyl-cysteine conjugates, were also observed in cells treated with LAP-OH; however levels of the cysteinyl-glycine conjugates were very low. During a 24-hour incubation of HepaRG cells with LAP-OH, formation of RM-Cys adducts increased with a corresponding decrease in the levels of RM-SG adducts (data not shown). This suggests that the RM-Cys adduct is a downstream thiol metabolite generated from sequential metabolism of the initial GSH conjugate (Fig. 1). Conversion of GSH conjugates to the corresponding cysteine S-conjugates is likely mediated by the sequential actions of γ-glutamyl transpeptidase and dipeptidase, which catalyze the hydrolysis of glutamate and glycine moieties from the GSH tripeptide (Meister and Anderson, 1983).
On the basis of the above findings, analysis of RM-Cys adducts was used as a biomarker to monitor the formation of reactive metabolites of lapatinib in HepaRG cells. Induction of CYP3A4 markedly increased the formation of RM-Cys adducts in HepaRG cells treated with lapatinib plus dexamethasone or rifampicin compared with treatment with lapatinib alone (Fig. 7). Coincubation with the CYP3A4 inhibitor ketoconazole significantly reduced the levels of RM-Cys adducts. These findings suggest that CYP3A4 plays a key role in the generation of reactive metabolites from lapatinib in human liver.
Recent clinical evidence suggests that lapatinib-induced hepatotoxicity has an underlying immunologic component. Pharmacogenetic analysis of a subset of metastatic breast cancer patients treated with lapatinib revealed a significant association between HLA risk alleles and hepatotoxicity (Spraggs et al., 2011, 2012). Lapatinib patients that carried the HLA variant DQA1*02:01 had a higher incidence of developing autoimmune hepatitis compared with HLA-negative patients, suggesting activation of the adaptive immune system (Spraggs et al., 2012). This situation could arise from covalent binding of reactive metabolites to proteins to form haptens, which are recognized by specific HLA proteins, resulting in inflammatory liver injury (Spraggs et al., 2011). However, more work is needed in this area.
Finally, we note that the peak plasma concentrations of lapatinib after an oral dose of 1250 mg have been reported to be 1.57 to 3.77 μg/ml, which is equivalent to 2.70 to 6.49 μM (GlaxoSmithKline, 2007). This is lower than the nominal hepatotoxic concentrations of lapatinib used in the present study, which were around 100 μM. However, neither the intracellular concentrations of lapatinib achieved in HepaRG cells, nor the concentration of lapatinib in human liver after chronic dosing are known, so direct comparisons cannot be made. A recent preclinical investigation showed that oral administration of [14C]lapatinib in a male rat resulted in the highest levels of radioactivity in the liver when analyzed by whole-body autoradiography, suggesting signification accumulation of lapatinib in liver (Polli et al., 2008). Regardless, the potential discrepancy in lapatinib concentrations that elicit in vitro cytotoxicity and in vivo hepatotoxicity is an important caveat to the conclusions drawn here and a more comprehensive experimental model, perhaps utilizing CYP3A transgenic mice, may be needed to fully understand the contribution of P450-mediated bioactivation to in vivo mechanisms of lapatinib-induced idiosyncratic hepatotoxicity.
In conclusion, this study provides evidence that lapatinib bioactivation by CYP3A4 may play an important role in initiating hepatocellular injury. Induction of CYP3A4 by dexamethasone and rifampicin markedly enhanced the cytotoxicity of lapatinib, which was correlated with increased formation of reactive metabolite cysteine adducts. Additional studies are required to provide further insight into the genetic and environmental factors that contribute to patient susceptibility to lapatinib idiosyncratic hepatotoxicity.
Acknowledgments
The authors thank Susan Grepper (Life Technologies, San Diego, CA) for providing samples of primary human hepatocytes, Concert Pharmaceuticals (Lexington, MA) for providing the deuterated analogue of O-debenzylated lapatinib for use in LC-MS/MS quantitative studies, and Eric Chun Yong Chan (National University of Singapore, Singapore) for valuable discussions.
Abbreviations
- BSO
l-buthionine sulfoximine
- CAR
constitutive androstane receptor
- DMSO
dimethylsulfoxide
- GSH
glutathione
- HLA
human leukocyte antigen
- LAP
lapatinib
- LAP-OH
O-debenzylated lapatinib
- LC
liquid chromatography
- LC-MS/MS
liquid chromatography coupled to tandem mass spectrometry. MRM, multiple reaction monitoring
- MS
mass spectrometry
- P450
cytochrome P450
- PXR
pregnane X receptor
- RM
reactive metabolite
- RM-Cys
cysteine conjugate of the reactive metabolite generated from O-debenzylated lapatinib
- RM-SG
glutathione conjugate of the reactive metabolite generated from O-debenzylated lapatinib
Authorship Contributions
Participated in research design: Hardy, Wahlin, Rettie, Nelson.
Conducted experiments: Hardy, Wahlin, Papageorgiou.
Contributed new reagents or analytic tools: Papageorgiou, Unadkat.
Performed data analysis: Hardy, Wahlin.
Wrote or contributed to the writing of the manuscript: Hardy, Rettie.
Footnotes
This research was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant P01 GM32165]; the National Institutes of Health National Center for Research Resources [Grant UL1-RR025014]; the University of Washington School of Pharmacy Drug Metabolism, Pharmacokinetics, and Transport Research Program; the University of Washington School of Pharmacy Elmer M. Plein Research Award; and the UNCF-Merck Science Initiative.
Current affiliation: Department of Pharmaceutical Sciences, Lipscomb University College of Pharmacy, Nashville, Tennessee.
References
- Andersson TB, Kanebratt KP, Kenna JG. (2012) The HepaRG cell line: a unique in vitro tool for understanding drug metabolism and toxicology in human. Expert Opin Drug Metab Toxicol 8:909–920 [DOI] [PubMed] [Google Scholar]
- Aninat C, Piton A, Glaise D, Le Charpentier T, Langouët S, Morel F, Guguen-Guillouzo C, Guillouzo A. (2006) Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos 34:75–83 [DOI] [PubMed] [Google Scholar]
- Anthérieu S, Chesné C, Li R, Camus S, Lahoz A, Picazo L, Turpeinen M, Tolonen A, Uusitalo J, Guguen-Guillouzo C, et al. (2010) Stable expression, activity, and inducibility of cytochromes P450 in differentiated HepaRG cells. Drug Metab Dispos 38:516–525 [DOI] [PubMed] [Google Scholar]
- Barbara JE, Kazmi F, Parkinson A, Buckley DB. (2013) Metabolism-dependent inhibition of CYP3A4 by lapatinib: evidence for formation of a metabolic intermediate complex with a nitroso/oxime metabolite formed via a nitrone intermediate. Drug Metab Dispos 41:1012–1022 [DOI] [PubMed] [Google Scholar]
- Castellino S, O’Mara M, Koch K, Borts DJ, Bowers GD, MacLauchlin C. (2012) Human metabolism of lapatinib, a dual kinase inhibitor: implications for hepatotoxicity. Drug Metab Dispos 40:139–150 [DOI] [PubMed] [Google Scholar]
- Chan EC, New LS, Chua TB, Yap CW, Ho HK, Nelson SD. (2012) Interaction of lapatinib with cytochrome P450 3A5. Drug Metab Dispos 40:1414–1422 [DOI] [PubMed] [Google Scholar]
- Cheng J, Ma X, Krausz KW, Idle JR, Gonzalez FJ. (2009) Rifampicin-activated human pregnane X receptor and CYP3A4 induction enhance acetaminophen-induced toxicity. Drug Metab Dispos 37:1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofanilli M, Johnston SR, Manikhas A, Gomez HL, Gladkov O, Shao Z, Safina S, Blackwell KL, Alvarez RH, Rubin SD, et al. (2013) A randomized phase II study of lapatinib + pazopanib versus lapatinib in patients with HER2+ inflammatory breast cancer. Breast Cancer Res Treat 137:471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DC, Watt AP, Nicoll-Griffith DA, Baillie TA. (2004) Drug-protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem Res Toxicol 17:3–16 [DOI] [PubMed] [Google Scholar]
- Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, Jagiello-Gruszfeld A, Crown J, Chan A, Kaufman B, et al. (2006) Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med 355:2733–2743 [DOI] [PubMed] [Google Scholar]
- GlaxoSmithKline (2007) Tykerb (lapatinib) product information. Available at: http://us.gsk.com/products/assets/us_tykerb.pdf Accessed on October 8, 2013.
- Gomez HL, Doval DC, Chavez MA, Ang PC, Aziz Z, Nag S, Ng C, Franco SX, Chow LW, Arbushites MC, et al. (2008) Efficacy and safety of lapatinib as first-line therapy for ErbB2-amplified locally advanced or metastatic breast cancer. J Clin Oncol 26:2999–3005 [DOI] [PubMed] [Google Scholar]
- Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. (2002) Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci USA 99:15655–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. (2007) The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact 168:66–73 [DOI] [PubMed] [Google Scholar]
- Hinchman CA, Ballatori N. (1994) Glutathione conjugation and conversion to mercapturic acids can occur as an intrahepatic process. J Toxicol Environ Health 41:387–409 [DOI] [PubMed] [Google Scholar]
- Kanebratt KP, Andersson TB. (2008) HepaRG cells as an in vitro model for evaluation of cytochrome P450 induction in humans. Drug Metab Dispos 36:137–145 [DOI] [PubMed] [Google Scholar]
- Kirby B, Kharasch ED, Thummel KT, Narang VS, Hoffer CJ, Unadkat JD. (2006) Simultaneous measurement of in vivo P-glycoprotein and cytochrome P450 3A activities. J Clin Pharmacol 46:1313–1319 [DOI] [PubMed] [Google Scholar]
- Kroep JR, Linn SC, Boven E, Bloemendal HJ, Baas J, Mandjes IA, van den Bosch J, Smit WM, de Graaf H, Schröder CP, et al. (2010) Lapatinib: clinical benefit in patients with HER 2-positive advanced breast cancer. Neth J Med 68:371–376 [PubMed] [Google Scholar]
- McCune JS, Hawke RL, LeCluyse EL, Gillenwater HH, Hamilton G, Ritchie J, Lindley C. (2000) In vivo and in vitro induction of human cytochrome P4503A4 by dexamethasone. Clin Pharmacol Ther 68:356–366 [DOI] [PubMed] [Google Scholar]
- McGill MR, Yan HM, Ramachandran A, Murray GJ, Rollins DE, Jaeschke H. (2011) HepaRG cells: a human model to study mechanisms of acetaminophen hepatotoxicity. Hepatology 53:974–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A, Anderson ME. (1983) Glutathione. Annu Rev Biochem 52:711–760 [DOI] [PubMed] [Google Scholar]
- Moy B, Kirkpatrick P, Kar S, Goss P. (2007) Lapatinib. Nat Rev Drug Discov 6:431–432 [DOI] [PubMed] [Google Scholar]
- Moy B, Rappold E, Williams L. (2009) Hepatobiliary abnormalities in patients with metastatic cancer treated with lapatinib (Abstract 1043). J Clin Oncol 27(Suppl 15S). [Google Scholar]
- Park BK, Kitteringham NR, Maggs JL, Pirmohamed M, Williams DP. (2005) The role of metabolic activation in drug-induced hepatotoxicity. Annu Rev Pharmacol Toxicol 45:177–202 [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Drocourt L, Gerbal-Chaloin S, Fabre JM, Maurel P, Vilarem MJ. (2001) Dual effect of dexamethasone on CYP3A4 gene expression in human hepatocytes. Sequential role of glucocorticoid receptor and pregnane X receptor. Eur J Biochem 268:6346–6358 [DOI] [PubMed] [Google Scholar]
- Polli JW, Humphreys JE, Harmon KA, Castellino S, O’Mara MJ, Olson KL, John-Williams LS, Koch KM, Serabjit-Singh CJ. (2008) The role of efflux and uptake transporters in [N-3-chloro-4-[(3-fluorobenzyl)oxy]phenyl-6-[5-([2-(methylsulfonyl)ethyl]aminomethyl)-2-furyl]-4-quinazolinamine (GW572016, lapatinib) disposition and drug interactions. Drug Metab Dispos 36:695–701 [DOI] [PubMed] [Google Scholar]
- Rusnak DW, Affleck K, Cockerill SG, Stubberfield C, Harris R, Page M, Smith KJ, Guntrip SB, Carter MC, Shaw RJ, et al. (2001) The characterization of novel, dual ErbB-2/EGFR, tyrosine kinase inhibitors: potential therapy for cancer. Cancer Res 61:7196–7203 [PubMed] [Google Scholar]
- Spraggs CF, Budde LR, Briley LP, Bing N, Cox CJ, King KS, Whittaker JC, Mooser VE, Preston AJ, Stein SH, et al. (2011) HLA-DQA1*02:01 is a major risk factor for lapatinib-induced hepatotoxicity in women with advanced breast cancer. J Clin Oncol 29:667–673 [DOI] [PubMed] [Google Scholar]
- Spraggs CF, Parham LR, Hunt CM, Dollery CT. (2012) Lapatinib-induced liver injury characterized by class II HLA and Gilbert’s syndrome genotypes. Clin Pharmacol Ther 91:647–652 [DOI] [PubMed] [Google Scholar]
- Takakusa H, Wahlin MD, Zhao C, Hanson KL, New LS, Chan EC, Nelson SD. (2011) Metabolic intermediate complex formation of human cytochrome P450 3A4 by lapatinib. Drug Metab Dispos 39:1022–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng WC, Oh JW, New LS, Wahlin MD, Nelson SD, Ho HK, Chan ECY. (2010) Mechanism-based inactivation of cytochrome P450 3A4 by lapatinib. Mol Pharmacol 78:693–703 [DOI] [PubMed] [Google Scholar]
- Teo YL, Saetaew M, Chanthawong S, Yap YS, Chan ECY, Ho HK, Chan A. (2012) Effect of CYP3A4 inducer dexamethasone on hepatotoxicity of lapatinib: clinical and in vitro evidence. Breast Cancer Res Treat 133:703–711 [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang W, Chua SS, Wei P, Moore DD. (2002) Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science 298:422–424 [DOI] [PubMed] [Google Scholar]