Abstract
The Epstein-Barr virus (EBV)-encoded ZTA protein interacts strongly with and stabilizes the cellular CCAAT/enhancer binding protein α (C/EBPα), leading to the induction of p21-mediated G1 cell cycle arrest. Despite the strong interaction between these two basic leucine zipper (bZIP) family proteins, the ZTA and C/EBPα subunits do not heterodimerize, as indicated by an in vitro cross-linking assay with in vitro-cotranslated 35S-labeled C/EBPα and 35S-labeled ZTA protein. Instead, they evidently form a higher-order oligomeric complex that competes with C/EBPα binding but not with ZTA binding in electrophoretic mobility shift assays (EMSAs). Glutathione S-transferase affinity assays with mutant ZTA proteins revealed that the basic DNA binding domain and the key leucine zipper residues required for homodimerization are all required for the interaction with C/EBPα. ZTA is known to bind to two ZRE sites within the ZTA promoter and to positively autoregulate its own expression in transient cotransfection assays, but there is conflicting evidence about whether it does so in vivo. Examination of the proximal ZTA upstream promoter region by in vitro EMSA analysis revealed two high-affinity C/EBP binding sites (C-2 and C-3), which overlap the ZII and ZIIIB motifs, implicated as playing a key role in lytic cycle induction. A chromatin immunoprecipitation assay confirmed the in vivo binding of both endogenous C/EBPα and ZTA protein to the ZTA promoter after lytic cycle induction but not during the latent state in EBV-infected Akata cells. Reporter assays revealed that cotransfected C/EBPα activated the ZTA promoter even more effectively than cotransfected ZTA. However, synergistic activation of the ZTA promoter was not observed when ZTA and C/EBPα were cotransfected together in either HeLa or DG75 cells. Mutagenesis of either the ZII or the ZIIIB sites in the ZTA promoter strongly reduced C/EBPα transactivation, suggesting that these sites act cooperatively. Furthermore, the introduction of exogenous C/EBPα into EBV-infected HeLa-BX1 cells induced endogenous ZTA mRNA and protein expression, as demonstrated by both reverse transcription-PCR and immunoblotting assays. Finally, double-label immunofluorescence assays suggested that EAD protein expression was activated even better than ZTA expression in latently infected C/EBPα-transfected Akata cells, perhaps because of the presence of a strong B-cell-specific repressed chromatin conformation on the ZTA promoter itself during EBV latency.
Epstein-Barr virus (EBV) is the prototype of the lymphocryptovirus subfamily of herpesviruses, which can cause both B-cell lymphomas and nasopharyngeal carcinoma. In immortalized B cells, usually only limited viral gene expression is associated with the latent state, whereas the induction of the lytic cycle results in viral DNA replication and the release of infectious virions. The exogenous expression of either of two EBV lytic transactivator proteins, ZTA (BZLF1, ZEBRA, Z, or EB1) or RTA (BRLF1 or R), results in a switch from latency to the full lytic cycle (11, 14, 18, 21, 40, 51, 73). Lytic cycle progression can also be triggered by treatment of latently infected B-cell lines with sodium butyrate, phorbol esters (tetradecanoyl phorbol acetate [TPA]), calcium ionophores, or transforming growth factor β or by cross-linking of B-cell receptors (BCRs) with anti-immunoglobulin (IgG) antibodies (1, 23, 37, 58, 75).
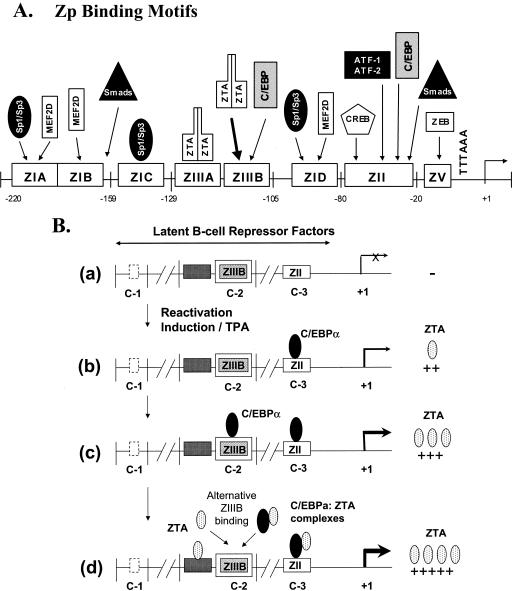
ZTA is a 245-amino-acid DNA binding basic leucine zipper (bZIP) family transcription factor that recognizes both classical AP1 sites and specific ZRE motifs found in downstream target viral genes. It also both positively autoregulates its own promoter and is essential for EBV ori-Lyt-mediated DNA replication (8, 19-21, 40). A variety of cellular factors that can either transcriptionally activate or repress the ZTA promoter (Zp) have also been identified. In particular, a region from −221 to +12 of the Zp contains several sets of cis-acting elements that are important both for maintaining low-level basal expression during latency and for inducing high levels of activation during the lytic cycle (15, 23). The first domain, known as ZI, includes four motifs (ZIA to ZID) that bind to cellular transcription factor MEF2D and are essential for both TPA- and BCR-mediated induction (3, 4, 27, 29, 44). Binding sites for Sp1 and Sp3 in the region may also play a role (43). The second-most-proximal domain, ZII, contains a variant CREB/AP1 site (TGACATCA) that binds to ATF1, ATF2, CREB, and c-JUN as well as an SRE-like site that binds to Smad3/Smad4 and mediates induction of the Zp by transforming growth factor β (37, 42, 45, 54, 68). A third domain, ZIII, is divided into ZIIIA and ZIIIB elements, both of which contain consensus ZRE motifs that can be recognized by the ZTA protein itself and are essential for both BCR- and TPA-mediated induction as well as for positive autoregulation (3, 21, 40). A powerful negative element, ZV, has also been described at positions −17 to −12, and a B-cell-specific cellular repressor protein, ZEB, has been reported to bind to this region (3, 32, 33).
The upregulation of ZTA expression during the onset of the lytic cycle involves two stages. The initial activation of the Zp, which is mediated through the ZI, ZII, and ZIIIB domains, results in low-level basal expression of ZTA after both TPA and anti-IgG antibody treatments (3, 21); the fully activated expression of the Zp involves additional ZIIIA- and ZIIIB-mediated effects thought to involve direct binding of the ZTA protein to the two ZRE sites (3, 21, 29). However, very high levels of ZTA protein may have some inhibitory effects, perhaps through binding at the cap site (40).
The lytic cycle of herpesviruses preferentially takes place in host cells arrested in G1 (24). For EBV, this process requires a function of ZTA that leads to the upregulation of cell cycle kinase inhibitors, such as p21 and p27, and the downregulation of c-MYC (6, 7, 24, 53). Wu et al. recently reported that the ZTA protein binds strongly to the cellular transcription factor CCAAT/enhancer binding protein α (C/EBPα) and that ZTA-induced G1 cell cycle arrest correlates with the induction of both C/EBPα and the p21 protein (71). Importantly, ZTA is unable to induce cell cycle arrest in C/EBPα-null cells, confirming that C/EBPα is essential for this process (71). Furthermore, the ZTA protein evidently both stabilizes C/EBPα and increases transactivation of the C/EBPα and p21 promoters in cooperation with C/EBPα. ZTA has also been found to be associated with C/EBPα promoter DNA in a C/EBPα-dependent manner by endogenous chromatin immunoprecipitation (ChIP) assays with lytically induced EBV-infected lymphoblast cell lines (71).
C/EBPα is also a member of the bZIP family of DNA binding nuclear transcription factors, which includes c-JUN, c-FOS, ATF, and CREB as well as ZTA (8, 34). C/EBPα and its close relatives C/EBPβ and CHOP-10 play important roles in cellular differentiation (16, 35, 67, 74). The C/EBPα gene encodes two predominant isoforms, namely, a 42-kDa full-length form that has antimitotic activity and a 30-kDa truncated form that is made from an alternative translational initiation site and that lacks any antimitotic activity (5, 41, 49). C/EBPα can positively autoregulate its own gene promoter (13, 59, 60), and the overexpression of C/EBPα causes G1 cell cycle arrest through the induction of p21 as well as through the direct inhibition of both cdk2 and E2F (28, 57, 61, 62, 64).
Because both C/EBPα and ZTA are bZIP family proteins, it seemed plausible that the two may heterodimerize with each other, like c-JUN and c-FOS. The replication-associated protein (RAP, or ORF-K8) encoded by Kaposi's sarcoma-associated herpesvirus (KSHV) is a highly diverged evolutionary and positional analogue of EBV ZTA that also retains a bZIP-like domain but does not bind to either ZRE or AP1 sites and does not act as a direct transcriptional activator of KSHV genes. However, like ZTA, RAP does interact strongly with C/EBPα and can induce C/EBPα- and p21-mediated G1 cell cycle arrest (69). Nevertheless, RAP, despite also being a distant member of the bZIP family, neither interacts with ZTA nor heterodimerizes with C/EBPα (72).
Wang et al. recently demonstrated that C/EBPα is able to reciprocally transactivate both KSHV RAP and RTA promoters and that the introduction of exogenous C/EBPα induces mRNA and protein expression from both of these immediate-early genes in endogenous latent genomes in KSHV-infected PEL cell lines (65, 66). Therefore, we examined here whether C/EBPα may also transactivate the EBV Zp. Kouzarides et al. (31) originally reported that the ZRE site in the ZIIIB domain of the Zp is also a binding site for C/EBPα, and they used DNase I footprinting assays to reveal another nonconsensus C/EBPα-interacting region in the ZII domain. In this study, we evaluated the functional contributions of these two C/EBP binding sites to C/EBPα activation and Zp autoregulation in transient reporter gene cotransfection assays and confirmed that both ZTA and C/EBPα associate specifically with the Zp in lytically infected cells. We also examined the nature of the strong specific protein-protein interaction between C/EBPα and ZTA and some of the effects of this interaction on DNA recognition in electrophoretic mobility shift assays (EMSAs). Finally, we also evaluated both the ability of ZTA to induce endogenous cellular C/EBPα expression and the ability of C/EBPα to induce ZTA and EAD expression in cell lines latently infected with EBV.
MATERIALS AND METHODS
Cell lines.
Human HeLa cells, EBV-positive HeLa-BX1 cells, and Vero cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum in a humidified 5% CO2 incubator at 37°C. EBV-positive Akata cells and EBV-negative DG75 cells were grown in RPMI 1640 medium containing 10% fetal bovine serum in a humidified 5% CO2 incubator at 37°C.
Expression plasmids and in vitro transcription template plasmids.
Plasmid pRTS21 is a mammalian expression vector carrying a genomic version of the EBV ZTA gene driven by the simian virus 40 promoter-enhancer region (SV2) in the plasmid pSG5 background (Stratagene) (55). Plasmid pLMP2A is an expression vector for EBV LMP2A in the pSG5 background. Plasmid pSEW-C01 is a mammalian expression vector for intact C/EBPα (codons 1 to 358) driven by the human cytomegalovirus promoter-enhancer region in the pcDNA background; plasmid pSEW-C02 is similar, except for the addition of an in-frame 5′ Flag epitope tag motif. Plasmid pYNC172a is a black beetle virus leader region (BBV) T7 in vitro transcription-translation vector encoding full-length C/EBPα. Plasmid pSEW-C05 encodes a fusion of full-length glutathione S-transferase (GST) and C/EBPα (codons 1 to 358).
Plasmids pYNC100 and pCJC514 are BBV T7 in vitro transcription-translation vectors carrying cDNA genes for wild-type ZTA (245 amino acids) and wild-type KSHV RAP (237 amino acids), respectively (8, 72). Plasmid pFYW04 is a BBV T7 in vitro transcription-translation vector encoding the ZTA-(RAP lz) fusion protein, which contains the N-terminal activation and basic domains from ZTA (codons 1 to 196) and the C-terminal leucine zipper domain from KSHV RAP (codons 188 to 237). This plasmid was constructed by joining a 600-bp BamHI/SacI PCR fragment made from pYNC100 as a template (primer LGH3172, 5′-CTATGGATCCGTCTTCGCTGAAGATGATG-3′ [underlining indicates restriction sites], and primer LGH3173, 5′-CTATGAGCTCCTTAAACTTGGCCCGGCA-3′) to a 160-bp SacI/EcoRI PCR fragment made from pCJC514 as a template (primer LGH3106, 5′-CTATGAGCTCCAGCAGGCATTAGAAGAA-3′, and primer LGH3102, 5′-CTATGGATTCCTAACATGGTGGGAGTGG-3′) in a BamHI/EcoRI-cleaved pYNC100 background. Plasmid pFYW44 is a BBV T7 in vitro transcription-translation vector carrying a 740-bp BamHI/EcoRI ZTA cDNA fragment containing a 178E/179E/180L mutation, obtained by PCR amplification of plasmid pGL28 (38) with primers LGH4902 (5′-CAGTGGATCCAATGATGGACCCAAACTCG-3′) and LGH4903 (5′-GACTGAATTCTTAGAAATTTAAGAGATCC-3′). Plasmids pFYW46, pFYW47, pFYW48, and pFYW49 are BBV T7 in vitro transcription-translation vectors carrying 740-bp BamHI/EcoRI ZTA cDNA fragments harboring separate 197K/200S, 204D, 205R/206D, and 214R/218R mutations, respectively (22).
Reporter genes.
Plasmid pHC41 is a chloramphenicol acetyltransferase (CAT) reporter containing an insertion of a 260-kb HindIII/XbaI fragment from positions −221 to +39 in the 5′-upstream flanking region within the EBV Zp (Zp-CAT). Zp-2 M-CAT, Zp-3 M-CAT, and Zp-2/3 M-CAT are pHC41 derivatives containing nucleotide substitutions that destroy the C-2 site, the C-3 site, and both the C-2 and the C-3 sites, respectively, in the Zp (see Fig. 8A). Zp 2 M-CAT (pFYW33), Zp 3 M-CAT (pFYW42), and Zp 2/3 M-CAT (pFYW43) were all generated by ligating together two separate PCR-amplified fragments. For the C-2/ZIIIB mutant, primers LGH3208 (5′-CGCAAGCTTGATGAATGTCTGCTGCATGC-3′) and LGH4224 (5′-GATCCCGCTCGAGGTACATTAGCGGAGCCTGTGGCTCATGCATAGT-3′ [bold type indicates mutated nucleotides]) were used to generate a 110-bp fragment from pHC41 flanked by a 5′ HindIII site and a 3′ XhoI site, and primers LGH4222 (5′-GATCCCGCTCGAGGACACACCTAAATTTAGCAC-3′) and LGH3207 (5′-CATTCTAGACTTCAGCAAAGATAGCAAAGG-3′) were used to generate a 150-bp fragment from pHC41 flanked by a 5′ XhoI site and a 3′ XbaI site. For the C-3/ZII mutant, primers LGH3208 and LGH4324 (5′-GATCCGCGGATCCATGTCATGGTTTGGGACGTG-3′) were used to generate a 160-bp fragment from pHC41 flanked by a 5′ HindIII site and a 3′ BamHI site, and primers LGH4325 (5′-GATCCGCGGATCCGGAGGCTGGTGCCTTGGCTT-3′) and LGH3207 were used to generate a 100-bp fragment from pHC41 flanked by a 5′ BamHI site and a 3′ XbaI site. For the C-2-C-3 double mutant, primers LGH3208 and LGH4324 were used to generate a 160-bp fragment from pFYW33 flanked by a 5′ HindIII site and a 3′ BamHI site, and primers LGH4325 and LGH3207 were used to generate a 100-bp fragment flanked by a 5′ BamHI site and a 3′ XbaI site.
FIG. 8.
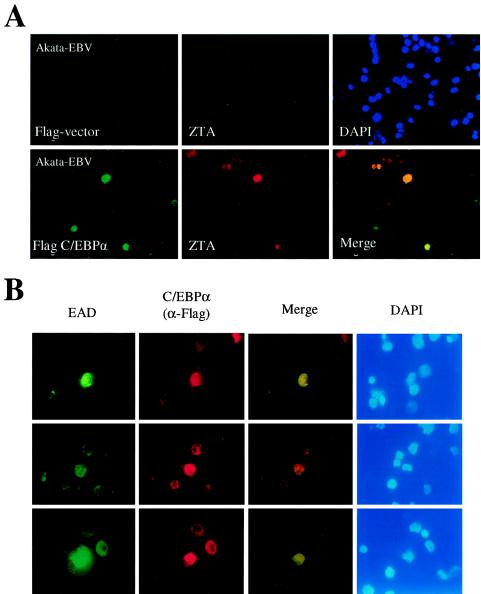
Induction of endogenous ZTA and EAD protein expression by C/EBPα exogenously introduced into a B-lymphoblast cell line latently infected with EBV. (A) Introduction of Flag-C/EBPα encoded by pSEW-C02 into electroporated EBV-infected Akata cells. (Upper panel) Double-label IFA showing the lack of Flag-protein expression from empty vector control DNA, detected with an anti-Flag MAb (FITC, green) (left); the absence of enhanced ZTA protein expression in the same cell population, detected with an anti-ZTA PAb (rhodamine, red) (middle); and stained nuclear DNA in the whole-cell population (DAPI, blue) (right). (Lower panel) Double-label IFA showing the expression of Flag-C/EBPα from plasmid pSEW-C02, detected with an anti-Flag MAb (FITC, green) (left); the activation of ZTA protein expression in two out of four visible C/EBPα-positive cells, detected with an anti-ZTA PAb (rhodamine, red) (middle); and a merge of the two frames (yellow) (right). Induced ZTA expression was found to occur in 28% of Flag-C/EBPα-positive cells. (B) Activation of EBV EAD (PPF or BMLF1) protein expression by the introduction of exogenous Flag-C/EBPα plasmid DNA (pSEW-C02) into EBV-infected Akata cells by electroporation. All three sets of panels show examples of induced EAD expression detected by double-label IFA: EAD-expressing cells, detected with a mouse anti-EAD MAb (FITC, green) (first set of panels); C/EBPα-expressing cells, detected with a rabbit anti-Flag PAb (rhodamine, red) (second set of panels); a merge of those two sets of panels (yellow) (third set of panels); and staining of all nuclei in the same fields (DAPI, blue) (fourth set of panels). Induced EAD expression was found to occur in 85% of Flag-C/EBPα-positive cells.
DNA transfection and CAT assays.
DG75 cells were transfected by the electroporation method as described previously (66). A total of 107 cells were mixed with 5 to 10 μg of plasmid DNA in 0.5 ml of RPMI 1640 medium and electroporated at 300 V and 950 μF by using a GenePulser (Bio-Rad, Hercules, Calif.). Lipofectamine (Invitrogen) DNA transfection was performed for Vero or HeLa cells after replating at 5 × 105 cells/ml. Transfection was done with a total of 2.5 μg of DNA (adjusted with vector plasmid as a carrier) per well in a six-well plate (0.5 μg of CAT reporter gene, 1 μg of C/EBPα, and/or 0.1 μg of ZTA effector DNA), and the cells were harvested at 48 h after transfection. After pelleting and freezing-thawing, 10 μl of the supernatant was added to CAT assay solution (0.47 M Tris-HCl [pH 7.9], 0.5 mM acetyl coenzyme A, 15 μCi of [14C]chloramphenicol) to reach a final volume of 150 μl, and the mixture was incubated for 45 min at 37°C. Ethyl acetate (1 ml) was added to each tube, and the mixture was vortexed vigorously for 15 s. The upper, organic phase of the mixture was transferred to a new microtube and vacuum dried for 1 h. The samples were resuspended in 20 μl of ethyl acetate and spotted onto thin-layer chromatography plates, and the plates were placed in a tank containing 200 ml of methanol-chloroform (1:19) solution for 80 min to measure acetylation activity. The CAT assay products on the thin-layer chromatography plates were quantitated with an Instant Imager.
EBV lytic cycle induction and indirect IFAs.
Transfection of Akata cells latently infected with EBV was performed by the electroporation method as described previously (66). At 48 h after transfection, the cell pellet was gently washed with phosphate-buffered saline (PBS), resuspended in 200 μl of PBS, and plated on polylysine glass slides as previously described (70). Indirect immunofluorescence assays (IFAs) and fluorescence microscopy were performed as described elsewhere (70). Secondary donkey or goat fluorescein isothiocyanate (FITC)- or rhodamine-conjugated IgG (Jackson Pharmaceuticals, West Grove, Pa.) was used to detect primary antibodies. Mounting solution with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Shield) was used to visualize cellular DNA. Primary antibodies included mouse M2 anti-Flag monoclonal antibody (MAb) and rabbit anti-Flag polyclonal antibody (PAb) (Sigma), rabbit anti-C/EBPα PAb (69), mouse anti-ZTA MAb (Argene, North Massapequa, N.Y.), rabbit anti-ZTA PAb (1:800) (38), mouse anti-EAD (PPF or BMLF1) MAb (Argene), and rat anti-LANA1 MAb (LN53; Advanced Biotechnologies Inc., Columbia, Md.).
Extraction of mRNA, RT-PCR, and Western immunoblotting.
HeLa-BX1 cells (9) were transfected either with empty vector or SV2-C/EBPα, and mRNA was extracted 40 h after transfection by using a GenElute Direct mRNA Miniprep kit (Sigma). Cells were harvested and resuspended by vortexing in 0.5 ml of lysis solution containing proteinase K (0.2 mg/ml), followed by incubation at 65°C for 10 min. Next, 32 μl of 5 M NaCl and 25 μl of oligo(dT) beads were added to the solution, which was allowed to stand at room temperature for 10 min. The oligo(dT)-mRNA complex was pelleted by microcentrifugation for 5 min at maximum speed, washed once with 350 μl of wash solution, and washed twice with 350 μl of low-salt wash solution. The poly(A) mRNA was eluted at 65°C in 100 μl of elution solution. For reverse transcription (RT), the following Promega reagents were used: 20 U of avian myeloblastosis virus (AMV) reverse transcriptase, 10 μl of AMV 5× RT buffer, 1 μl of RNasin RNA inhibitor, 4 μl of 5 mM deoxynucleoside triphosphates, and 0.5 μg of random primers. These reagents were mixed and incubated at 42°C for 1 h with 30 μl of mRNA and H2O to a final volume of 50 μl. The synthesized cDNAs were used as templates for ZTA PCRs with primers LGH2617 (5′-ACATCTGCTTCAACAGGAGG-3′) and LGH2618 (5′-AGCAGACATTGGTGTTCCAC-3′); the PCR mixtures contained 2 μl of cDNA template, 0.25 μg of each primer, 3 μl of 25 mM MgCl2, 4 μl of 2.5 mM deoxynucleoside triphosphates, 3.5 μl of dimethyl sulfoxide, 2.5 U of Taq DNA polymerase (Promega), 5 μl of 10× buffer (Promega), and H2O to a final volume of 50 μl. The PCR conditions were 94°C for 5 min for 1 cycle; 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 30 cycles; and 72°C for 10 min for 1 cycle. The PCR products were analyzed on 2% agarose gels. HeLa-BX1 cell lysates for Western immunoblot analyses were prepared as described elsewhere (71), and anti-ZTA MAb was used at a 1:1,000-fold dilution for the detection of ZTA protein.
Radiolabeled in vitro-translated proteins.
In vitro-translated wild-type C/EBPα or ZTA protein used in these studies was made by using a TNT quick coupled transcription-translation system (Promega) with 2.0 μg of pSEW-C01 or pYNC100 template DNA, respectively, in 40 μl of TNT quick master mix-1 μl of RNase inhibitor-2 μl of cold methionine. The mixture was incubated at 30°C for 90 min and was stored subsequently at −80°C. For labeling and verification of protein expression, 2 μl of [35S]methionine (Amersham) was used, and the proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and detected by autoradiography.
EMSAs.
For EMSAs, 2 μl of in vitro-translated protein was added to a 19-μl reaction mixture containing binding buffer A (10 mM HEPES [pH 7.5], 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1% Triton X-100, 5% glycerol) and 2 μg of poly-dI/dC (Sigma). After incubation for 5 min at room temperature, 1 μl of 32P-labeled dCTP oligonucleotide (50,000 cpm) was added, and the mixture was incubated for 1 h. For supershift experiments, the reaction mixture was incubated at 20°C for 30 min, 0.5 μl of anti-C/EBPα PAb or 1 μl of anti-ZTA MAb was added, and the mixture was incubated for 30 min.
Oligonucleotides were purchased from the Invitrogen Primer Synthesis Facility. Underlining represents known or expected recognition sequences. LGH4246(5′-GATCCATGCCATGCATATTTCAACTGGGCTGTCTAAA-3′) andLGH4247 (5′-GATCAATAGACAGCCCAGTTGAAATATGCATGGCATG-3′) were annealed to form the C-1 probe. LGH4248 (5′-GATCAGCCACAGGCATTGCTAATGTACCTCATAGACA-3′) and LGH4249 (5′-GATCTGTCTATGAGGTACATTAGCAATGCCTGTGGCT-3′) were annealed to form the C-2 probe. LGH4309 (5′-GATCACGTCCCAAACCATGACATCACAGAGGAGGCTG-3′) and LGH4310 (5′-GATCCAGCCTCCTCTGTGATGTCATGGTTTGGGACGT-3′) were annealed to form the C-3 probe. LGH4252 (5′-GATCGAGGCTGGTGCCTTGGCTTTAAAGGGGAGATGT-3′) and LGH4253 (5′-GATCACATCTCCCCTTTAAAGCCAAGGCACCAGCCTC-3′) were annealed to form the C-4 probe. LGH4258 (5′-GATCAGCCACAGGCTCCGCTAATGTACCTCATAGACA-3′) and LGH4259 (5′-GATCTGTCTATGAGGTACATTAGCGGAGCCTGTGGCT-3′) were annealed to form the Zp-2 M probe. LGH436 (5′-GATCCTCACCTTGCGCAATTTGGTCTAGAA-3′) and LGH437 (5′-GATCTTCTAGACCAAATTGCGCAAGGTGAG-3′) were annealed to form the C/EBP(R) probe. LGH291 (5′-GATCCTCACCATGTGCAAATTGGTCTAGAA-3′) and LGH292 (5′-GATCTTCTAGACCAATTTGCACATGGTGAG-3′) were annealed to form the ZRE(5) probe (40). LGH4276(5′-GATCGAGGCGGTGGGCGTTGCGCCGCGGCCTGCCTGG-3′) andLGH4277 (5′-GATCCCAGGCAGGCCGCGGCGCAACGCCCACCGCCTC-3′) were annealed to form the C/EBPα-P probe. LGH4232 (5′-GATCGAAGCATGTGACAATCAACAACTTTGTATACTT-3′) and LGH4233 (5′-GATCAAGTATACAAAGTTGTTGATTGTCACATGCTTC-3′) were annealed to form the p21P-3 probe. LGH4268 (5′-GATCGATTGTGACTATTTGTGAAACAATAATGATTAAAGGGGGTGGTATTTCC-3′) and LGH4269 (5′-GATCGGAAATACCACCCCCTTTAATCATTATTGTTTCACAAATAGTCACAATC-3′) were annealed to form the RAP-P (RRE) probe. LGH4272 (5′-GATCCTTCCAAAAATGGGTGGCTAACCTGTCCAAAATATGGGAAC-3′) andLGH4273 (5′-GATCGTTCCCATATTTTGGACAGGTTAGCCACCCATTTTTGGAAG-3′) were annealed to form the PAN-P (PAN-1) probe. The preparation of 32P-labeled oligonucleotides and other procedures involved in the EMSA analysis were done as previously described (10). Quantitation of binding was measured by using an Instant Imager.
Cross-linking assays.
The in vitro-translated or cotranslated 35S-labeled proteins (4 μl) were mixed with 9 μl of cross-linking buffer (10 mM potassium phosphate buffer[pH 8.0], 10 mM DTT) and 1 μl of freshly diluted 0.1% glutaraldehyde (48). After incubation for 1 h at 20°C, the cross-linking mixture was boiled in 2× SDS gel loading buffer and fractionated by electrophoresis on SDS-8% polyacrylamide gels. The gels were fixed and dried, and [35S]Met-labeled protein bands were detected by autoradiography after overnight exposure on Kodak X-ray film.
GST affinity assays.
Recombinant GST and GST-C/EBPα were purified from plasmid DNA-transformed Escherichia coli (strain BL21) as described elsewhere (38). GST or GST-C/EBPα immobilized on beads was pretreated with 0.2 U of DNase I and 0.2 μg of RNase A per μl for 30 min at 20°C in pretreating buffer (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 2.5 mM CaCl2, 100 mM NaCl, 5% glycerol, 1 mM DTT). The beads were washed twice with binding buffer B (20 mM Tris-Cl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM DTT) and blocked in the same buffer containing 10 mg of bovine serum albumin/ml for 30 min at 4°C. After blocking, the bead-immobilized GST fusion proteins were resuspended in binding buffer B containing 1 mg of bovine serum albumin/ml, 10 μl of in vitro-translated [35S]Met-labeled wild-type ZTA or mutant proteins made with the TNT quick coupled transcription-translation system was added to each sample mixture, and the mixture was incubated for 1 h at 4°C. The beads were washed three times with binding buffer B at 20-min intervals. The beads were resuspended in 15 μl of 2× SDS gel loading buffer and boiled for 5 min before being loaded onto gels for SDS-PAGE. After electrophoresis, the gels were fixed in 50% methanol-40% H2O-10% acetic acid for 30 min and dried for X-ray autoradiography.
ChIP assays.
After treatment of 20 ml of EBV-positive Akata cells (5 × 106) with IgG (50 μg/ml; Cappel) for 40 h, 2 ml of formaldehyde solution (11% formaldehyde, 0.1 M NaCl, 1 mM EDTA, 50 mM HEPES [pH 8]) was added to the culture, and the mixture was incubated at 37°C for 30 min. The cross-linking reaction was stopped by the addition of 4 ml of 1 M glycine (final concentration, 0.125 M) to the cell culture. After centrifugation, the cell pellet was washed once with 5 ml of cold wash buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 0.5% NP-40, 1 mM PMSF, 1 μg of aprotinin/ml, 1 μg of pepstatin/ml). The cell pellet was resuspended in 500 μl of RT sonication buffer (1% SDS, 10 mM EDTA, 50 mM Tris-Cl [pH 8.0], 1 mM PMSF) in a 1.5-ml microtube and sonicated for 30 s at the minimal setting to shear genomic DNA to ∼400-bp fragments. The 500-μl sonicated mixture was diluted with 5 ml of cold dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-Cl [pH 8.0], 167 mM NaCl, 1 mM PMSF) and precleared with 120 μl of 50% protein A or G-Sepharose beads for at least 8 h at 4°C. After centrifugation to pellet the Sepharose beads, the supernatant was divided into 500-μl aliquots, which were stored at −70°C.
For each immunoprecipitation assay, 1 μg of anti-ZTA MAb, anti-C/EBPα PAb (71), or anti-KSHV RAP PAb (70) or 1 μl of PBS (negative control) was added to each separate 500-μl supernatant aliquot, and the mixture was incubated at 4°C for 2 h with constant mixing. Next, 30 μl of 50% protein A or G-Sepharose beads was added to each mixture, and incubation was done at 4°C for 4 h with constant mixing. After centrifugation, the Sepharose beads were washed once with 1,000 μl of dilution buffer at room temperature for 3 min, twice with 1,000 μl of dilution buffer at 4°C for 20 min each time, and three times with 1,000 μl of dilution buffer at room temperature for 3 min each time. After the removal of dilution buffer, the beads were resuspended in 100 μl of Tris-EDTA (pH 8.0) (TE). RNase A (50 μg/ml) was added to the TE suspension, and the mixture was incubated at 37°C for 30 min; 5 μl of 10% SDS and 50 μg of proteinase K (500 μg/ml) were added, and the mixture was incubated for 4 h at 37°C. To reverse the cross-linked DNA-protein complex, the mixture was incubated at 65°C overnight. After centrifugation, the supernatant was transferred to a new microtube and diluted with 100 μl of fresh TE. The solution was extracted with an equal volume of phenol and then with an equal volume of chloroform. DNA in the solution was precipitated with 1/10 volume of 3 M sodium acetate (pH 5.0) and 2 volumes of ice-cold 100% ethanol at −20°C for 4 h. After centrifugation at maximum speed and 4°C, the DNA pellet was rinsed with 500 μl of 70% ethanol, vacuum dried, and resuspended in 20 μl of TE. For PCR detection, 2 μl from each DNA-TE solution was used as a template. Primers LGH3207 and LGH3208 were used for detection of the Zp. The conditions for PCR (final volume, 50 μl) were 94°C for 5 min for 1 cycle; 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 30 cycles; and 72°C for 10 min for 1 cycle. The PCR products were analyzed on 2.5% agarose gels.
RESULTS
Evidence against a heterodimeric interaction between ZTA and C/EBPα subunits.
Because ZTA interacts strongly with C/EBPα both in vivo and in vitro (71) and both are bZIP family proteins, there appeared to be a strong possibility that their monomer subunits might be able to form ZTA-C/EBPα heterodimers. Chang et al. previously showed that both ZTA and c-FOS-ZTA fusion protein subunits or c-JUN and c-FOS subunits form heterodimers at high efficiencies when cotranslated together, as assayed by the formation of appropriate intermediate-size gel-shifted bands in EMSA experiments (8). However, cotransfection of ZTA with C/EBPα inhibits the ability of the complex to recognize and gel shift a consensus C/EBPα binding site (71). This result could be explained easily if the two formed classic bZIP family heterodimers, which would presumably not recognize either C/EBPα or ZTA binding sites. We reasoned that evidence for or against the formation of closely folded ZTA-C/EBPα heterodimers could still be obtained by cross-linking of the native proteins after cotranslation, a process that was previously carried out successfully by Chang et al. (8) with a c-FOS-ZTA fusion protein.
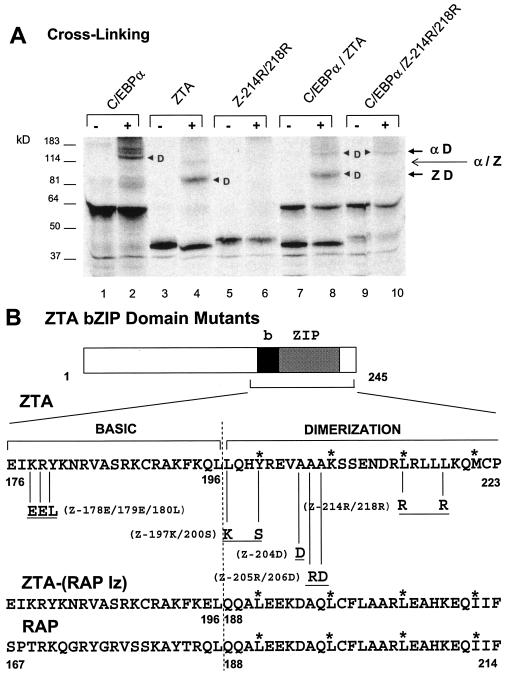
Therefore, in vitro-cotranslated ZTA and C/EBPα were cross-linked with 0.1% glutaraldehyde and subjected to SDS-PAGE (Fig. 1A, lane 8) for comparison with parallel singly in vitro-translated and cross-linked C/EBPα or ZTA protein samples (lanes 2 and 4). The results showed that after cross-linking, the cotranslated ZTA-C/EBPα sample displayed just two rather than three dimeric forms. The higher-molecular-weight dimeric form exactly matched the migration of C/EBPα homodimers, and the lower-molecular-weight form exactly matched the migration of ZTA homodimers; however, no additional intermediate-size forms were found to migrate in between the two homodimeric forms (Fig. 1A, lane 8). A negative control experiment with a ZTA mutant that was expected to be unable to homodimerize (Z214R/218R) was performed in parallel (Fig. 1A, lane 6). Cross-linking of cotranslated C/EBPα and Z214R/218R confirmed that, in addition to the inability of Z214R/218R to homodimerize with itself, no heterodimers were formed when it was cotranslated with C/EBPα (Fig. 1A, lane 10). Furthermore, a similar cross-linking experiment in which individually translated C/EBPα was mixed with the wild-type ZTA protein also yielded only homodimeric bands (data not shown).
FIG. 1.
ZTA does not heterodimerize with C/EBPα. (A) Cross-linking of in vitro-translated 35S-labeled proteins with glutaraldehyde showing that C/EBPα and ZTA fail to form any novel heterodimer bands migrating at an intermediate position compared to the homodimer bands detected in control samples with each translated alone. Lanes: 1 and 2, C/EBPα from pYNC172a alone; 3 and 4, wild-type ZTA from pYNC100 alone; 5 and 6, mutant Z214R/218R from pFYW49 alone; 7 and 8, cotranslated C/EBPα and ZTA; 9 and 10, cotranslated C/EBPα and Z214R/218R. −, samples without cross-linking; +, samples with cross-linking. Arrowheads indicate dimers; αD, C/EBPα homodimers; ZD, ZTA homodimers; α/Z, expected position of C/EBPα-ZTA heterodimers. (B) Schematic diagram showing the specific amino acid changes in the relevant bZIP domain regions of the five ZTA mutants used in this study (upper sequence) and a comparison of the relevant amino acid sequences of the bZIP domain regions of wild-type ZTA, the ZTP-(RAP lz) fusion protein, and wild-type RAP. Asterisks indicate the expected leucine zipper positions.
These results imply that monomeric ZTA and C/EBPα subunits do not form closely folded heterodimers even after cotranslation and that the interaction presumably is a higher-order oligomeric type of interaction, probably involving native dimers, which evidently did not produce subunit cross-linking under these conditions. Because supershifted C/EBPα-ZTA or C/EBPα-RAP complexes also were not detected in EMSA experiments, it was suggested that at least the DNA-bound forms of the presumed dimer-dimer complexes may need some additional cellular scaffold factor, such as the HCF protein, involved in herpes simplex virus VP16-OCT1 complexes (71, 72). However, another alternative explanation may be that the complexes form much larger oligomers.
Both the basic domain of ZTA and the ability of ZTA to homodimerize are required for the interaction with C/EBPα.
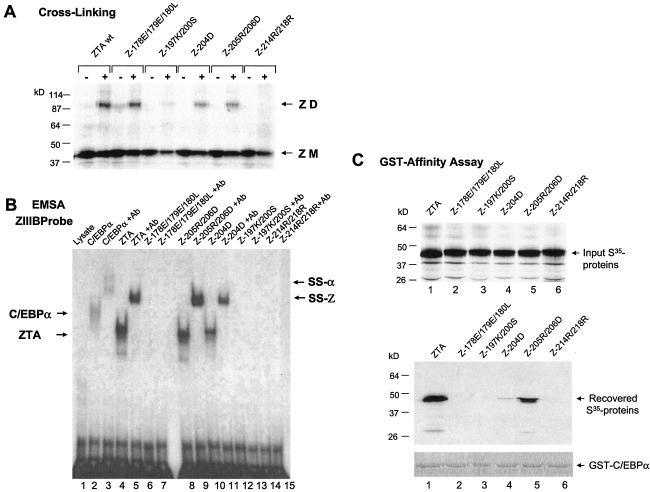
Five ZTA mutants harboring various point mutations in the bZIP domain (Fig. 1B) were used to examine the domain requirements for ZTA-C/EBPα interactions in GST affinity assays (Fig. 2C). Initially, the abilities of each of these mutants to homodimerize and to bind to a ZRE DNA binding site were tested by performing both in vitro cross-linking assays (Fig. 2A) and EMSA experiments to evaluate binding to a ZIIIB ZRE site oligonucleotide probe derived from the Zp (Fig. 2B). As expected, Z178E/179E/180L, which harbors mutations only within its basic domain, retained its ability to homodimerize but could not bind to the ZRE DNA probe (Fig. 2A and B). In contrast, Z197K/200S and Z214R/218R failed to bind to the ZRE DNA probe and were also either completely or partially impaired in their ability to homodimerize. On the other hand, Z204D and Z205R/206D, with evidently noncritical mutations within the leucine zipper domain, retained their abilities both to homodimerize and to bind ZIIIB DNA (although Z204D did show twofold-reduced gel shift efficiency). These results confirmed the interpretation of Flemington and Speck (22) that residues 200 and 214 are critical for leucine zipper formation as well as other evidence that ZTA homodimerization is required for DNA binding (8, 30, 31).
FIG. 2.
Both the ZTA basic domain and the ability of ZTA to homodimerize are critical for the interaction with C/EBPα. (A) Cross-linking analysis of 35S-labeled in vitro-translated ZTA bZIP region point mutant proteins to determine their abilities to homodimerize in comparison with that of the wild-type (wt) ZTA protein. Only Z214R/218R from pFYW48 failed to homodimerize, and Z197K/200S from pFYW46 homodimerized at a lower efficiency. ZD, dimers; ZM, monomers. (B) EMSA showing the relative abilities of the five in vitro-translated ZTA bZIP region mutant proteins to bind to a 32P-labeled ZRE DNA probe (ZIIIB). Z178E/179E/180L (pFYW44), Z197K/200S (pFYW46), and Z214R/218R (pFYW49) all failed to bind to the ZIIIB site from the Zp. Positive controls included wild-type C/EBPα from pYNC172a (lanes 2 and 3) and wild-type ZTA from pYNC100 (lanes 4 and 5). Appropriate antibody (Ab) supershifts were carried out in the second lane of each pair to confirm that the shifted bands contained the expected protein. Arrows designate the positions of directly shifted bands (C/EBPα and ZTA) and antibody-supershifted complexes (SS-α and SS-Z, respectively). (C) GST affinity binding assays conducted with GST-C/EBPα to measure in vitro interaction affinities with the wild-type ZTA protein (lane 1) and each of the five ZTA bZIP region mutant proteins (lanes 2 to 6). (Upper panel) Input 35S-labeled in vitro-translated proteins. (Lower panels) 35S-labeled proteins recovered after binding to and elution from GST-C/EBPα beads (pSEW-C05). Only 35S-labeled wild-type ZTA from pYNC100 and 35S-labeled Z205R/206D from pFYW48 bound strongly to GST-C/EBPα; 35S-labeled Z204D bound at a lower efficiency. These results suggest that the basic domain as well as the ability of ZTA to homodimerize are both required for its interaction with C/EBPα.
In GST affinity assays, the intact GST-C/EBPα fusion construct was found to interact only with wild-type ZTA and the Z204D and Z205R/206D mutants (Fig. 2C, lanes 1, 4, and 5), which retained both their ability to homodimerize and to bind to DNA. However, the interaction of Z204D with GST-C/EBPα was much weaker than that of Z205R/206D, and Z204D had a DNA binding affinity slightly lower than that of Z205R/206R (Fig. 2B), perhaps because the mutation is closer to the critical basic domain. The inability of Z178E/179E/180L to interact with GST-C/EBPα (Fig. 2C, lane 2), despite its strong ability to homodimerize, implies that the basic domain of the ZTA protein is critical for the C/EBPα-ZTA dimer-dimer interaction. However, the inability of Z197K/200S and Z214R/218R to interact with GST-C/EBPα (Fig. 2C, lanes 3 and 6) either may be related to their inability to homodimerize, which potentially precludes the formation of native tetrameric C/EBPα-ZTA complexes, or implies that the specific protein contacts extend into the leucine zipper region (Fig. 1B). Although we previously alluded to preliminary data suggesting that the Z187E/179E/180L mutant could still stably interact with C/EBPα (71), the results shown here clearly prove that notion to be incorrect.
Replacement of the ZTA leucine zipper domain with the KSHV RAP leucine zipper domain does not abolish its interaction with C/EBPα.
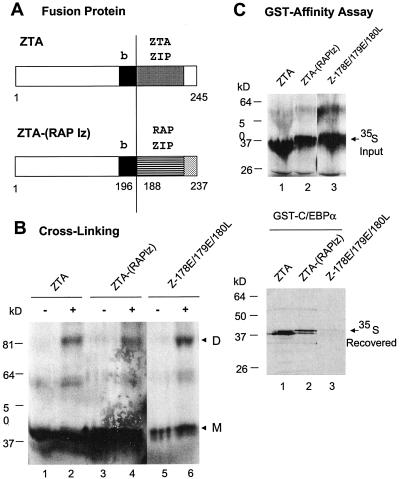
Because the ability of ZTA to homodimerize seemed essential for its interaction with C/EBPα, we examined whether specific amino acid sequences within the ZTA leucine zipper domain might be dispensable for such an interaction. Therefore, we replaced the ZTA leucine zipper domain with the positionally equivalent leucine zipper domain from KSHV RAP (Fig. 3A). Although the resulting hybrid ZTA-(RAP lz) fusion protein did not bind to either AP1 or ZRE DNA probes (data not shown), we confirmed that it could still homodimerize just as efficiently as could both the wild-type ZTA protein (Fig. 3B, lane 2) and the Z187E/179E/180L mutant protein (lane 6) in a subunit cross-linking assay (lane 4). Finally, the results of a GST affinity assay revealed that the ZTA-(RAP lz) fusion protein still interacted very efficiently with C/EBPα (Fig. 3C, lower panel, lane 2), suggesting that specific amino acid sequences of the ZTA leucine zipper domain which differ extensively between ZTA and RAP (Fig. 1B) are not required for its ability to interact with C/EBPα. As expected, in the positive and negative control lanes, GST-C/EBPα interacted strongly with wild-type ZTA (Fig. 3C, lane 1) but again did not interact with the basic region mutant Z187E/179E/180L (lane 3). Therefore, physical contacts between ZTA and C/EBPα seem to involve the basic DNA recognition motif segment of ZTA.
FIG. 3.
A ZTA-(RAP lz) fusion protein still interacts with C/EBPα. (A) Schematic diagram showing the structures of wild-type ZTA and the ZTA-(RAP lz) fusion protein. b, ZTA basic domain; ZTA ZIP and RAP ZIP, C-terminal leucine zipper domains of ZTA and RAP, respectively. (B) Cross-linking of 35S-labeled in vitro-translated wild-type ZTA from pYNC100 (lanes 1 and 2), ZTA-(RAP lz) fusion fusion from pFYW04 (lanes 3 and 4), and the basic domain mutant Z178E/179E/180L from pFYW44 (lanes 5 and 6) demonstrating that the fusion protein retains the ability to homodimerize efficiently. −, samples without cross-linking; +, samples with cross-linking. D, dimers; M, monomers. (C) GST affinity assay showing that the ZTA-(RAP lz) fusion protein still binds efficiently to C/EBPα. (Upper panel) Input 35S-labeled in vitro-translated proteins. (Lower panel) Recovered bound 35S-labeled proteins eluted from GST-C/EBPα beads (pSEW-C05). Lanes: 1, wild-type ZTA (codons 1 to 245) from pYNC100; 2, ZTA-(RAP lz) fusion protein from pFYW04; 3, basic domain mutant ZTA178E/179E/180L from pFYW44.
The Zp contains three C/EBPα binding sites.
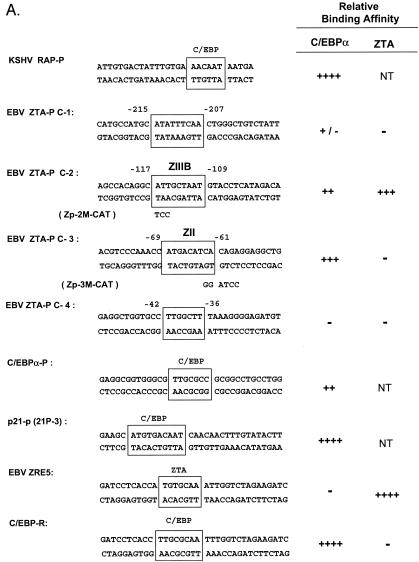
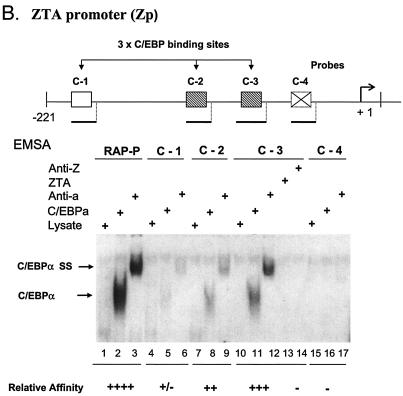
Because ZTA is known to augment transcription from the C/EBPα promoter (71), we examined whether C/EBPα may also have a reciprocal role in activating the Zp. First, we investigated whether C/EBP DNA binding sites occur within the Zp. Although C/EBP binding sites are notoriously difficult to recognize by visual inspection, we considered four putative C/EBPα binding motifs (Fig. 4A) from positions −215 to −207 (C-1), −117 to −109 (C-2), −69 to −61 (C-3), and −42 to −36 (C-4). The abilities of these sites to bind to C/EBPα were tested by EMSAs with in vitro-translated C/EBPα and 32P-labeled double-stranded oligonucleotide DNA probes. Two of these four putative Zp sites bound relatively strongly to C/EBPα, with C-2 (−117 to −109) binding less efficiently than C-3 (−69 to −61) (Fig. 4B, lanes 8 and 11) or the RAP-P positive control (lane 1), whereas C-1 (−215 to −207) was an extremely weak C/EBP binding site (lane 5) and C-4 failed to bind at all (lane 16). The specificity of the EMSA DNA-bound band was confirmed by supershift experiments with an added anti-C/EBPα PAb (Fig. 4B, lanes 3, 6, 9, and 12). These two sites correspond to the C/EBP-protected sites identified previously by Kouzarides et al. (31) by DNase I footprinting analyses encompassing positions −121 to −107 (contains the C-2 site) and positions −75 to −57 (contains the C-3 site).
FIG. 4.
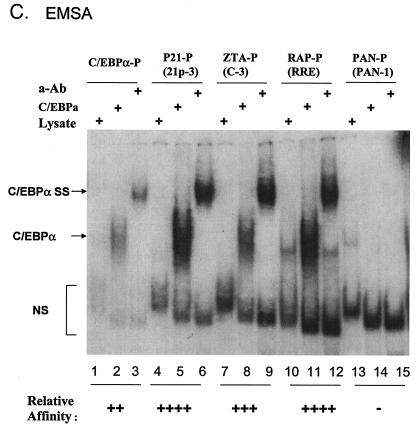
Identification of two strong C/EBP binding sites in the EBV Zp. (A) DNA sequences of double-stranded oligonucleotide probes used in EMSA experiments and semiquantitive summary of their relative affinities for C/EBPα: +, ++, +++, and ++++, approximate values of one, two-, four-, and eightfold, respectively; NT, not tested. Known or projected C/EBPα recognition motifs are boxed. Several probes containing C/EBP sites that were evaluated previously (8) or that were recently identified in various cellular or viral promoters are also included (71, 72). (B) EMSA experiment with Zp-derived oligonucleotide probes showing that in vitro-translated C/EBPα from pYNC172a binds more strongly to the C-3 site (lanes 11 and 12) than to the C-2 site (lanes 8 and 9), only weakly to the more distal C-1 site (lanes 5 and 6), and not at all to the C-4 site (lanes 16 and 17). Wild-type ZTA from pYNC100 also failed to bind to the C-3 site (lanes 13 and 14). The relative positions of the four C site probes between positions −221 and +1 in the proximal Zp are indicated in the upper diagram. Anti-Z, added anti-ZTA PAb; Anti-α, added anti-C/EBPα PAb; SS, supershift. (C) EMSA experiment comparing the binding affinities of the Zp C-3 site and selected oligonucleotide probes containing various other known C/EBP sites. C/EBPα-P (lanes 1 to 3) is the C/EBP binding site from the C/EBPα promoter; P21-P (21p-3; lanes 4 to 6) is the strongest C/EBP binding site from the p21CIP-1 promoter (71); ZTA-P (C-3; lanes 7 to 9) is the C/EBP C-3 site from the Zp; RAP-P (RRE; lanes 10 to 12) is the C/EBP-II site from the KSHV RAP promoter (66); and PAN-P (PAN-1; lanes 13 to 15) is a negative control sequence taken from the KSHV PAN promoter, which contains no C/EBP sites. In each group, the first lane contains an unprogrammed reticulocyte lysate, the second lane contains in vitro-translated C/EBPα (pYNC172a), and the third lane contains both C/EBPα and added anti-C/EBPα PAb. NS, nonspecific binding.
Interestingly, the C-3 C/EBP binding site overlapped the previously defined ZII motif of the Zp (Fig. 4A), which reportedly functions as a recognition site for CREB, ATF-1, ATF-2, and c-JUN, but had no consensus ZRE motif and did not bind to ZTA in our EMSA experiments (Fig. 4B, lanes 13 and 14). The sequence of the C-3 C/EBP binding motif (ATGACATCA) differs significantly from all other C/EBP consensus sequences known to us. Therefore, to compare the affinity of the C-3 site in the Zp to those of several other known or consensus C/EBP binding sites (Fig. 4A), we also performed EMSA experiments with three different oligonucleotide probes encompassing proven functional C/EBP binding sites from the human C/EBPα promoter, the human p21 promoter (p21-3), and the KSHV RAP promoter (Fig. 4C). An oligonucleotide probe from the KSHV PAN promoter, which does not contain a C/EBP recognition motif, was used as a negative control (Fig. 4C, lanes 14 and 15). For semiquantitive results, all probes were prepared at the same specific activities (counts per minute per microgram), and equal input DNA levels were used. In comparison to these other C/EBP binding sites, the C-3 site in the Zp (Fig. 4C, lanes 8 and 9) bound to C/EBPα with a twofold-higher affinity than did the C/EBPα promoter autoregulation site (lanes 2 and 3) but with a twofold-lower affinity than did the C/EBP binding sites found in either the KSHV RAP promoter (lanes 11 and 12) or the p21CIP-1 promoter (lanes 5 and 6) (66, 71).
Comparison of the ability of C/EBPα and ZTA homodimers and of C/EBPα-ZTA complexes to bind to single or overlapping ZRE and C/EBP binding motifs.
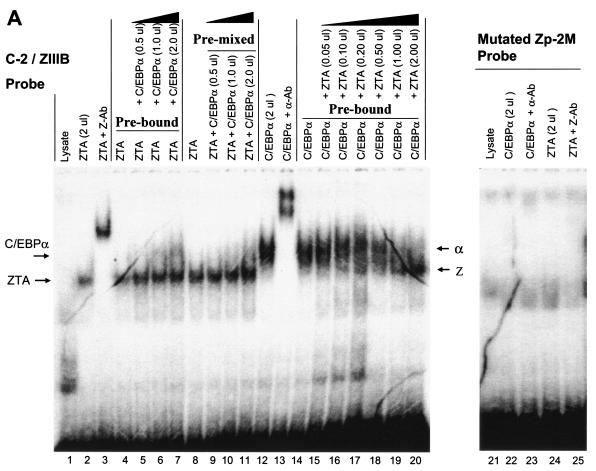
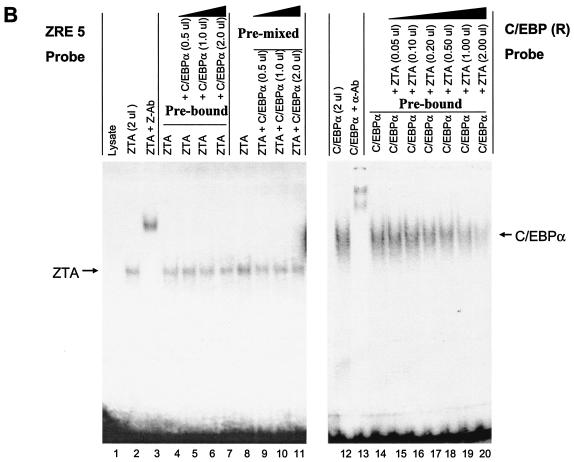
The other Zp C/EBPα binding site (C-2) overlaps completely with the previously defined ZIIIB motif (Fig. 4A), which binds to ZTA strongly (21, 31), even though the sequence of the ZRE motif used here (TTAGCAA) deviates slightly from the consensus sequences identified originally by Lieberman et al. (39, 40) and Chang et al. (8). To confirm that the 32P-labeled combined C-2/ZIIIB motif probe can be bound by either C/EBPα or ZTA, EMSAs were performed with in vitro-translated C/EBPα or ZTA. The results showed that both C/EBPα and ZTA each bound efficiently and independently to the 30-bp C-2/ZIIIB motif probe (Fig. 5A, lanes 2 and 12) and could be supershifted with the appropriate specific antibody (lanes 3 and 13).
FIG. 5.
ZTA interferes with C/EBPα binding to the C/EBP C-2/ZIIIB ZRE site, but C/EBPα has no effect on ZTA binding to the same site. (A) EMSA experiment showing the following results. First, in vitro-translated ZTA (pYNC100) and C/EBPα (pYNC172a) both bind strongly and independently to the 32P-labeled C-2 probe encompassing the ZIIIB ZRE motif (lanes 1, 2, 3, 12, and 13). Second, the addition of C/EBPα at various doses (lanes 4 to 11) does not affect ZTA DNA binding to the C-2 probe, whereas in the reciprocal approach, the addition of ZTA at various doses displaces C/EBPα binding to the C-2 probe (lanes 15 to 20). Third, point mutation of the C-2/ZIIIB site inactivates binding to both proteins (lanes 21 to 25). Z-Ab, anti-ZTA antibody; α-Ab, anti-C/EBPα antibody. (B) EMSA experiment showing the results of similar dose-response mixture binding experiments with in vitro-translated C/EBPα (pYNC172a) and ZTA (pYNC100) and with consensus 32P-labeled ZRE(5) and C/EBP(R) oligonucleotide DNA probes that each bind specifically to one protein but do not cross-react with the other (lanes 1, 2, 3, 12, and 13). Again, the addition of C/EBPα failed to affect the binding of ZTA to the ZRE(5) probe (lanes 4 to 11), but the addition of ZTA gradually displaced C/EBPα binding to the C/EBP(R) probe (lanes 14 to 20).
We were initially interested in the concept that, although C/EBPα-ZTA heterodimers (if they existed) would not be able to recognize either C/EBP or ZTA sites, they might nevertheless be able to recognize the combined C/EBP-ZRE recognition site within the C-2/ZIIIB motif. However, because the bZIP DNA binding domain of ZTA is also critical for the protein-protein interaction with C/EBPα, many additional questions arose about the effect of this interaction on the DNA binding abilities of both proteins. To test whether the interaction with C/EBPα might affect ZTA binding to ZIIIB, we first preincubated the ZTA protein with the 32P-labeled C-2/ZIIIB probe for 30 min and then added C/EBPα at doses increasing by fourfold in a dose-response experiment. Under these conditions, added C/EBPα failed to measurably affect the binding of ZTA to the C-2 probe (Fig. 5A, lanes 4 to 7). To examine whether the prior formation of a ZTA-C/EBPα complex might subsequently inhibit ZTA binding, we also preincubated C/EBPα with ZTA at different ratios for 30 min before the addition of the C-2 DNA probe. However, again, no inhibition of or increase in ZTA DNA binding was observed in the presence of increasing amounts of C/EBPα (Fig. 5A, lanes 8 to 11). The above results imply one or more of the following: (i) the ZTA protein has a higher affinity for the C-2 DNA probe than for C/EBPα; (ii) the DNA-bound form of ZTA is not able to interact with C/EBPα, perhaps because the interactive face of the basic domain of ZTA is no longer exposed; or (iii) even oligomeric C/EBPα-ZTA protein complexes are unable to recognize ZRE motifs.
To carry out the reverse experiments, we preincubated C/EBPα with the C-2/ZIIIB probe and then added the ZTA protein in a dose-response manner beginning at a 1:20 ratio. The results of these experiments revealed that bound C/EBPα was gradually displaced, with ZTA apparently outcompeting C/EBPα for binding to the ZIIIB site when the input protein ratios approached 1:1 (Fig. 5A, lanes 14 to 20). As observed previously, the expected more slowly migrating C/EBPα-ZTA oligomeric complexes were apparently unable to form supershifted bands in these in vitro experiments (71). One could interpret the findings to mean that ZTA simply has a 10-fold-higher affinity for binding to the ZIIIB site than does C/EBPα. However, it seems more likely that once all of the bound or unbound C/EBPα was complexed with ZTA, any added excess ZTA was now free to bind to the C-2/ZIIIB site as ZTA homodimers. When an ATT-to-TCC site-specific mutation was introduced into the C-2/ZIIIB site (Zp-2 M probe), binding both by C/EBPα alone and by ZTA alone was abolished (Fig. 5A, lanes 22 to 24), confirming that the sites within the ATTGCTAAT motif that ZTA and C/EBPα recognize indeed either are exactly superimposed or overlap.
To better understand the interactions observed above with the combined or overlapping C/EBPα and ZTA binding sites in the C-2/ZIIIB DNA probe, we also carried out similar experiments with two additional DNA recognition motif probes, namely, ZRE(5) and C/EBP(R) (Fig. 4A). The ZRE(5) motif, encompassing the sequence ATGTGCAAA, is known to be bound by ZTA but not at all by C/EBPα (40); conversely, the C/EBP(R) motif, encompassing the sequence ATTGCGCAAT, is known to be bound by C/EBPα but not by ZTA (8). These two independent probes were used to evaluate how ZTA and C/EBPα might affect the DNA binding of each other in the absence of competition for the target site. Similar to our results obtained with the C-2/ZIIIB motif, direct ZTA binding to the ZRE(5) motif (Fig. 5B, lanes 2 and 3) proved to be totally unaffected by the addition of C/EBPα (lanes 4 to 11), whereas the direct binding of C/EBPα to the C/EBP(R) motif (lanes 12 and 13) was affected by the addition of ZTA in a dose-responsive manner (lanes 14 to 20), but without ZTA itself replacing C/EBPα. It is clear that, in this experiment, a higher affinity of ZTA than of C/EBPα for the probe cannot account for the displacement of C/EBPα.
Therefore, we conclude (i) that DNA-bound ZTA is evidently unable to interact with C/EBPα and (ii) that the displacement of C/EBPα by ZTA can be explained only by DNA-bound C/EBPα still being capable of interacting with ZTA. These conclusions are consistent with the previous model that ZTA enhances the C/EBPα transactivation of target promoters via complex formation with DNA-bound C/EBPα through a piggyback mechanism (71). However, the reverse interaction evidently does not occur, perhaps because of steric interference effects. Unfortunately, as noted previously, the EMSA experiments do not give direct in vitro evidence for the binding of C/EBPα-ZTA protein complexes to the C/EBPα site probes by the formation of the expected supershifted bands, perhaps because some additional component is necessary to stabilize the DNA-bound forms.
Both C/EBPα and the ZTA protein associate with the Zp during the lytic cycle, as detected by in vivo ChIP assays.
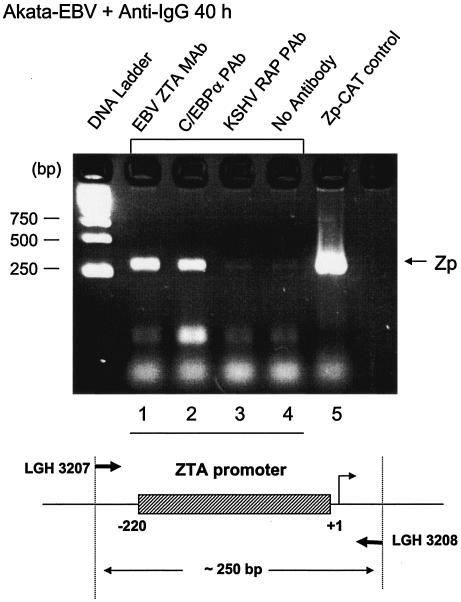
To attempt to verify that C/EBPα and ZTA do both bind to the Zp in vivo, ChIP assays were performed. After induction of the EBV lytic cycle in Akata cells with anti-IgG antiserum for 40 h, C/EBPα and ZTA were each immunoprecipitated from the cross-linked cell lysates by using C/EBPα- or ZTA-specific antibody attached to protein A and G-Sepharose beads. After extensive washing and removal of all proteins, the DNA was purified, and Zp DNA was amplified by PCR with primers (LGH3207 and LGH3208) specific for the 220-bp Zp. The results showed that Zp DNA was detected in both C/EBPα and ZTA immunoprecipitates (Fig. 6, lanes 1 and 2). No Zp DNA (above the basal level) was detected in negative control samples immunoprecipitated with KSHV RAP-specific antibody (Fig. 6, lane 3) or lacking antibody (lane 4). These results confirm that both C/EBPα and ZTA associate with the Zp during the EBV lytic cycle in virus-infected cells. Importantly, we did not detect any such binding to Zp DNA in ChIP assays with uninduced Akata cells (data not shown), confirming either that very little C/EBPα (or ZTA) is present during latency or that latency-associated repressors and the expected closed chromatin structure associated with the Zp during latency prevent the binding of any C/EBPα that is present.
FIG. 6.
ChIP assay showing that C/EBPα binds to Zp DNA in vivo. (Upper panel) ChIP assay results for EBV-positive Akata cells at 40 h after induction by anti-IgG treatment, showing that Zp DNA was recovered from both endogenous ZTA and C/EBPα immunoprecipitates but not from the negative control samples. Lanes contained Zp PCR DNA products obtained with an anti-ZTA MAb (1), an C/EBPα PAb (2), an anti-RAP PAb (3), or no antibody (4). Lane 5, positive control PCR amplification of a Zp-CAT plasmid (pHC41) with the same primers. (Lower panel) Schematic diagram of the Zp region (250 bp) that was targeted for detection by PCR amplification with specific primers LGH3207 and LGH3208.
Because of the presence of independent ZRE sites in the Zp, there was no possibility of being able to confirm here that some ZTA binding was of the indirect piggyback type that is dependent on C/EBPα, although Wu et al. previously used an antibody preclearing approach to successfully demonstrate this point for the C/EBPα promoter (71).
Cotransfected C/EBPα activates the Zp in transient reporter gene assays.
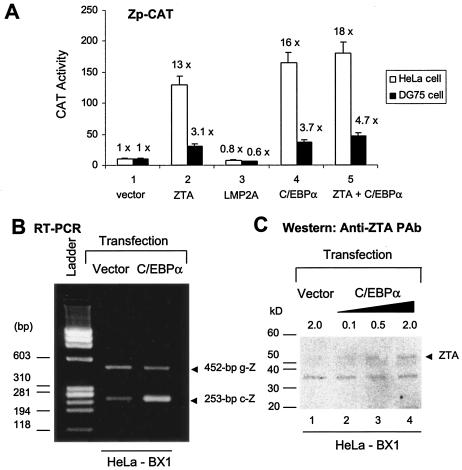
To address the central issue of the role of C/EBPα binding in the functional activity of the Zp, a CAT target reporter gene (Zp-CAT) was cotransfected with C/EBPα expression vector DNA into either HeLa or DG75 cells. Zp-CAT was activated up to 16-fold from the basal level by 1 μg of cotransfected C/EBPα in HeLa cells (Fig. 7A, panel 4, white bars). Cotransfection with a ZTA expression plasmid (0.1 μg) in the same experiment yielded 13-fold activation (Fig. 7A, panel 2), consistent with previous evidence for positive ZTA autoregulation of its own promoter (21, 31). The negative controls used here consisted of an EBV LMP2A expression vector or empty vector plasmid DNA, each of which failed to affect Zp-CAT levels.
FIG. 7.
Exogenous C/EBPα activates both a Zp reporter gene and endogenous ZTA mRNA and protein expression. (A) Transient reporter gene assays showing that C/EBPα activates the Zp-CAT promoter. (White bars) Target Zp (positions −200 to +10)-CAT reporter gene plasmid DNA (pHC41; 0.5 μg) was transfected into HeLa cells either alone or in the presence of cotransfected mammalian expression plasmids encoding empty vector DNA (pcLUC; 1.0 μg) (lane 1), ZTA (pRTS21; 0.1 μg) (lane 2), LMP2A (pLMP2A; 1.0 μg) (lane 3), C/EBPα (pSEW-C01; 1.0 μg) (lane 4), or a combination of ZTA and C/EBPα (lane 5). (Black bars) Assays were carried out with DG75 lymphocytes and the Zp-CAT reporter gene plasmid DNA (5 μg) electroporated either alone or together with expression plasmids encoding empty vector DNA (pcLUC; 10 μg), ZTA (pRTS21; 1 μg), LMP2A (pLMP2A; 10 μg), C/EBPα (pSEW-C01; 10 μg), or a combination of ZTA and C/EBPα. Total DNA was balanced by vector plasmid DNA as a carrier. Fold activation was calculated relative to the basal level of Zp-CAT activity in each cell type (designated 1.0). (B) Exogenous C/EBPα increases the level of ZTA mRNA in cells latently infected with EBV, as shown by the results of RT-PCR performed with mRNA harvested from C/EBPα-transfected HeLa-BX1 cells and primers specific for a spliced segment of the ZTA gene (LGH2617 and LGH2618). Relative to the basal level in HeLa-BX1 cells, the expression of ZTA mRNA (253-bp cDNA product; c-Z) was induced 10-fold in C/EBPα-transfected HeLa-BX1 cells. Some amplification of ZTA genomic DNA (452-bp band; g-Z) indicates equal loading. (C) Exogenous C/EBPα increases the level of ZTA protein in cells latently infected with EBV. Western blotting with an anti-ZTA MAb shows that the expression of C/EBPα from pSEW-C01 in HeLa-BX1 cells increased the total ZTA protein level by up to threefold in comparison to that in cells transfected with the same amount of the empty vector plasmid control.
At higher input doses of ZTA, the Zp response does not increase further and may even decrease because of a negative autoregulatory feature that comes into play, but this scenario does not apply to C/EBPα. Thus, C/EBPα can be considered just as effective as ZTA in activating the Zp. Cotransfection of the two proteins together (Fig. 7A, panel 5) did not produce the same synergistic or additive effects that occur with all other C/EBPα-responsive targets that we have tested, a finding which may be related to the independent abilities of C/EBPα and ZTA to bind to the overlapping C-2 and ZIIIB sites in the Zp or which may be due to either the dose-response effect of ZTA or competition between ZTA and C/EBPα-ZTA complexes. These results were reproducible in DG75 lymphocytes, but because of the much lower DNA transfection efficiency, we could detect only three- to fourfold activation of the Zp by ZTA, C/EBPα, or both (Fig. 7A, black bars).
Expression of exogenous C/EBPα induces endogenous ZTA mRNA and protein expression in HeLa-BX1 cells.
We have shown above by in vitro EMSAs and by cotransfection assays that C/EBPα can bind to and transcriptonally activate the Zp. Furthermore, ChIP studies revealed that C/EBPα does associate with the Zp in vivo after TPA-mediated induction of EBV-infected cells. Therefore, we examined whether exogenously introduced C/EBPα is capable of triggering ZTA expression in latently infected cells. Chen et al. (9) infected HeLa cells with the EBV-BX1(GFP) virus and isolated a cell line latently infected with EBV, HeLa-BX1, which can be easily transfected. HeLa-BX1 cells express most EBV latency genes, such as those for the LMPs and Epstein-Barr nuclear antigens, and can be induced into the lytic cycle by either TPA or exogenous expression of ZTA (9). Total cell mRNA was harvested from HeLa-BX1 cells after transfection with either a mammalian expression plasmid encoding C/EBPα or a control empty expression plasmid. Functional ZTA mRNA is the product of differential splicing and consists of three exons. Two primers that span genomic coordinate positions 102,283 to 102,735 were designed to distinguish between the 452-bp ZTA genomic DNA and the spliced 253-bp ZTA cDNA. RT-PCR was performed to detect changes in the levels of ZTA cDNA. The results showed that the cell culture sample transfected for 48 h with the C/EBPα expression plasmid had a sixfold increase in ZTA mRNA levels compared to the control cell culture sample transfected with just the empty expression plasmid (Fig. 7B, lower band). However, there was no parallel increase in genomic DNA levels (Fig. 7B, upper band).
Induction at the ZTA protein level was also evaluated by Western blotting with anti-ZTA MAb. The results showed that the total amount of ZTA protein present also increased in a dose-responsive manner up to a maximum of threefold in the HeLa-BX1 cell culture sample transfected with the C/EBPα expression plasmid compared to the control cell culture sample transfected with just the empty expression plasmid (Fig. 7C). However, despite this threefold increase in endogenous ZTA protein expression in HeLa-BX1 cells after transfection with C/EBPα, the overall ZTA protein levels were still very low compared to the levels of endogenous ZTA expressed in a parallel cell culture sample of TPA-treated HeLa-BX1 cells (data not shown), suggesting that C/EBPα transfection alone is not highly efficient at inducing endogenous ZTA expression in EBV-infected cells.
Expression of exogenous C/EBPα induces both ZTA and EAD expression in EBV-positive Akata cells.
Wu et al. previously found that the ZTA protein was rarely detectable in untreated Akata B cells latently infected with EBV by IFAs (0.05% of the overall cell population), whereas after anti-IgG treatment, the ZTA protein was induced in greater than 10% of the overall cell population at 40 h, and the majority of these ZTA-positive Akata cells proved also to express C/EBPα and vice versa (71). Therefore, we decided to investigate whether exogenous C/EBPα can induce ZTA expression in latently infected Akata cells, as assayed by a double-label IFA. The cells were transfected with the Flag-C/EBPα expression plasmid or a Flag-empty vector control plasmid by electroporation, and both ZTA expression and C/EBPα expression were monitored by using anti-ZTA PAb and anti-Flag MAb. Although only a very low transfection efficiency (less than 3%) was achieved in this experiment, approximately one-third of the Flag-C/EBPα-positive cells were also induced to express endogenous nuclear ZTA protein (Fig. 8A, lower panel). In comparison, no ZTA protein expression was observed beyond the 0.05% spontaneous basal level in cells transfected with the empty vector control plasmid (Fig. 8A, upper panel).
In a second experiment of this type, we also examined whether the endogenous viral EAD early lytic antigen was induced in Akata cells by exogenous Flag-C/EBPα (Fig. 8B). In this experiment, a majority of the 5% of cells expressing nuclear C/EBPα detected with anti-Flag MAb (red) were also induced to express nuclear EAD detected with anti-EAD (PPF or BMLF1) PAb (green). Some additional cells apparently expressing low levels of either EAD or both EAD and C/EBPα in the cytoplasm were discounted. A small subset of the positive cells appeared to contain viral DNA replication compartments, indicative of the extensive lytic cycle progression that is typically observed when an exogenous ZTA expression vector is introduced. A control antibody (anti-KSHV LANA1 MAb) failed to detect any positive signals with the EAD-positive cells, confirming that no nonspecific effects were occurring (data not shown).
During EBV latency in B cells, the Zp is known to be completely silenced by various cellular repressor factors, such as MEF2D and ZEB (27, 32, 33); therefore, the overexpression of exogenous C/EBPα in a cellular environment that is highly favorable to latency may not always be sufficient to completely overcome endogenous repression of the Zp, resulting in a somewhat lower level of ZTA reactivation than in HeLa-BX1 cells. However, EAD protein was induced at levels two- to threefold higher than those of ZTA, suggesting that C/EBPα may affect other viral promoters as well (such as Rp and BMLF1 itself) and may even be capable of inducing viral DNA replication compartment formation within a small subset of cells.
Mutation of the C/EBP binding sites in the Zp abolishes C/EBPα-mediated transactivation.
To test the functionality of the C/EBP binding sites in terms of the ability of C/EBPα to activate the Zp, we generated three mutant Zp-CAT reporter genes that contained site-specific substitutions at C-2/ZIIIB, C-3/ZII, or both (Fig. 9A). Transient CAT reporter gene assays were performed with cotransfected HeLa cells to evaluate the responsiveness of these mutant Zp-CAT target genes to different effector plasmids expressing C/EBPα, ZTA, or both.
FIG. 9.
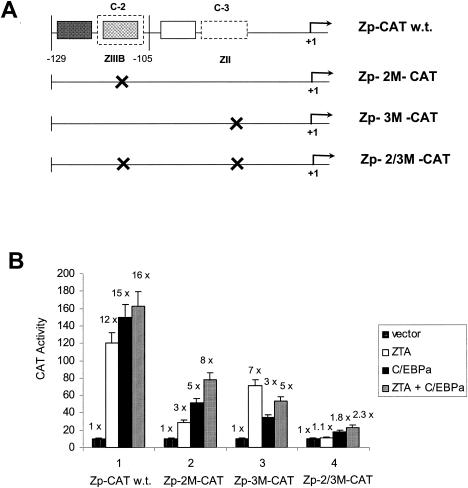
Mutation of the C-3/ZII or C-2/ZIIIB C/EBP binding sites reduces or abolishes C/EBPα and ZTA transactivation of Zp-CAT. (A) Diagram showing the relative locations of the two strong C/EBP binding sites (C-2 and C-3) within the wild-type (w.t.) Zp-CAT target reporter gene and the structures of the three single- or double-point mutant Zp-CAT reporter genes used here. (B) Transient reporter gene assays in which effector plasmids encoding C/EBPα, ZTA, or both were cotransfected into HeLa cells with target Zp-CAT reporter plasmids containing either the intact Zp in Zp-CAT (pHC41) or its mutated derivatives Zp-2 M-CAT (pFYW33), Zp-3 M-CAT (pFYW42), and Zp-2/3 M-CAT (pFYW43). These results demonstrate that both the C-2/ZIIIB and the C-3/ZII sites are essential for high-level C/EBPα-mediated transactivation and that they both contribute to ZTA transactivation.
In comparison to the results obtained with the wild-type Zp (−221 to +39)-CAT reporter (Fig. 9B, panel 1), the Zp-2 M-CAT target, which contains the same mutation in the C-2/ZIIIB site as does the Zp-2 M oligonucleotide used in the EMSA experiments (Fig. 4A and B), showed a significant decrease in C/EBPα-mediated activation, dropping from 15- to 5-fold (Fig. 9B, panel 2). In addition, an expected decrease in activation by ZTA from 12- to 3-fold was observed for Zp-2 M-CAT, presumably because the C-2 site mutation also destroyed the ZRE motif within the ZIIIB domain (Fig. 9B, panel 2). In the presence of cotransfected C/EBPα as well as ZTA, the wild-type Zp again failed to show any additive or synergistic effects (Fig. 9B, panel 1), but an additive (eightfold) activation effect was obtained with the Zp-2 M-CAT target, presumably because both C/EBPα and C/EBPα-ZTA complexes can still bind to the C-3/ZII site in vivo and ZTA can still bind to the ZIIIA domain independently of the mutated C-2/ZIIIB site.
A Zp-3 M-CAT mutant was generated by substituting GGATCC for CACAGA across the part of the defined ZII motif that would be expected to interfere with C/EBPα binding to the C-3 site (Fig. 4A). In this experiment, activation by C/EBPα was reduced from 15- to 3-fold when Zp-3 M-CAT was used as the target (Fig. 9B, panel 3). The more profound reduction in C/EBPα-mediated transactivation by mutation of C-3 rather than by mutation of C-2 probably relates to the C-3/ZII site displaying a higher affinity than the C-2/ZIIIB site for C/EBPα. However, ZTA responsiveness was also impaired somewhat with the C-3/ZII mutation, dropping from 12- to 7-fold (Fig. 9B, panel 3). Although the C-3 site mutation should not directly affect ZTA binding to Zp DNA, it would presumably reduce the binding of ZTA piggyback complexes with endogenous C/EBPα and could also eliminate any ability of DNA-bound C/EBPα or C/EBPα-ZTA complexes to interact cooperatively with ZIIIA or ZIIIB domain-bound ZTA. When both C/EBPα and ZTA were cotransfected (Fig. 9B, panel 3), no additive effect on Zp-3 M-CAT activity was observed (still increased only fivefold). Perhaps because the C-3/ZII site is no longer available to bind C/EBPα, the lack of an additive effect may be caused by competition between ZTA and C/EBPα or ZTA and C/EBPα-ZTA complexes for binding to the single remaining C-2/ZIIIB site, as demonstrated in our EMSA experiment (Fig. 5A).
Finally, double mutation of both the C-2 and the C-3 binding sites in Zp-2/3 M-CAT nearly abolished all responses to both C/EBPα and ZTA (Fig. 9B, panel 4), which were reduced from 15- and 12-fold to 1.8- and 1.1-fold, respectively. Therefore, the integrity of both the C-2 and the C-3 binding sites seems to contribute to both C/EBPα-mediated and ZTA-mediated transactivation of the Zp. Furthermore, there are clearly no other undefined motifs that mediate either C/EBPα or ZTA transactivation effects, and we are left with the notion that ZIIIA may be functionally active only in cooperation with adjacent ZIIIB. Based on our data, we present a revised model for ZTA activation that for the first time includes a role for C/EBPα binding to both ZII and ZIIIB (Fig. 10).
FIG. 10.
Updated summary and model of the known viral and cellular factors that bind to the Zp and play roles in regulating the switch from latency to the lytic cycle. (A) Simplified physical map of known multiple transcription factor and cellular repressor binding sites within the proximal segment of the Zp. (B) Schematic diagram hypothesizing a multistep role for C/EBPα at different stages of the initiation and ZTA-mediated amplification or positive autoregulation of Zp transactivation.
DISCUSSION
In this study, we found that the basic domain of the EBV lytic cycle trigger protein ZTA is critical for the protein-protein interaction with the cellular C/EBPα transcription factor. In addition, in transient CAT reporter gene assays, C/EBPα can activate expression from the Zp mediated by two strong C/EBP binding sites, one of which overlaps the previously defined key ZII motif and the other of which overlaps the ZIIIB ZTA binding site. Although neither ZTA nor C/EBPα was found to associate with the Zp before induction in latently infected Akata cells, after triggering of the lytic cycle by BCR cross-linking with anti-IgG antibody, ZTA and C/EBPα each became strongly associated with the Zp in endogenous ChIP assays. Furthermore, exogenous C/EBPα expression efficiently activated ZTA expression in HeLa-BX1 cells latently infected with EBV, although it was less efficient at doing so in EBV-infected Akata cells; these results suggest that although C/EBPα may play a role, full lytic cycle activation of the Zp in B cells probably also requires the displacement of repressors such as MEF2D at the ZI domain and ZEB at the ZV domain prior to the binding of lytic cycle-associated viral and cellular transcription factors (3).
A strong interaction of two members of the bZIP family of proteins might be expected to occur through classical heterodimerization, as exemplified by c-JUN and c-FOS (12, 52) and by the Marek's disease herpesvirus-encoded MEQ protein and c-JUN (50). However, we were unable to detect the formation of simple heterodimers between ZTA and C/EBPα in cross-linking experiments after in vitro cotranslation. Logically, because both C/EBPα and ZTA each form very strong homodimers, it should be energetically and entropically unfavorable for the two strong homodimers once folded to disassemble and reform heterodimers. In contrast, both c-JUN and c-FOS form only very weak homodimers, a factor which contributes to their heterodimeric interaction being energetically and entropically favorable. Additional evidence against the concept of simple heterodimers was provided when we found that KSHV RAP, which also interacts strongly with C/EBPα, also does not heterodimerize with it (72).
Wu et al. previously proposed that both EBV ZTA and KSHV RAP target the cellular p21 and C/EBPα promoters by piggyback binding to DNA-bound C/EBPα (71, 72). Therefore, our finding that the basic domain of ZTA is critical for the interaction with C/EBPα was intriguing, because the basic domain of ZTA is also required for its specific DNA binding to ZRE sites. In addition, the lack of cooperativity in the responses of the Zp to cotransfection by both ZTA and C/EBPα raised the question of whether or not C/EBPα could also piggyback onto DNA-bound ZTA. Interestingly, we found that the ability of ZTA to bind to either the overlapping ZIIIB site or another specific but nonoverlapping ZRE site in vitro was unaffected by the addition of increasing amounts of C/EBPα. Furthermore, premixing (or cotranslation) of ZTA with C/EBPα prior to binding with ZIIIB yielded similar results. These data suggested that DNA-bound ZTA does not interact with C/EBPα and cannot be displaced by C/EBPα and that ZTA-C/EBPα complexes probably do not recognize ZRE sites. In contrast, ZTA (and RAP) can both displace DNA-bound C/EBPα in EMSA experiments and associate with promoters that contain C/EBPα binding sites in ChIP assays through a C/EBPα-dependent piggyback DNA binding mechanism (71, 72).
Various investigators (21, 25, 39, 40) showed previously that TPA-induced ZTA activation is distinct from ZTA-mediated activation of its own promoter. During TPA-induced activation, only the ZI and ZII domains are occupied by proteins, whereas the ZIII domain remains unoccupied until higher levels of ZTA are attained. These data led to the proposal of a two-step pathway for ZTA activation (21). The present study adds additional information about the contribution of C/EBPα to the existing model describing the transcriptional activation of the EBV Zp (Fig. 10).
During the early stages of the lytic cycle after treatment with inducing agents, cellular repressors, such as MEFIID and ZEB, that are bound to the Zp must be displaced or modified prior to any transcriptional activation taking place (3, 33). Subsequently, the Sp1 and Sp3 proteins may bind to the ZI domain (43), and C/EBPα as well as CREB, AP1, or ATF may also bind to the ZII domain to trigger the basal expression of the ZTA protein (although it is not known whether C/EBPα may bind cooperatively or exclusively or even heterodimerize with CREB, AP1, or ATF). Initial low levels of ZTA then upregulate C/EBPα expression by both enhancing C/EBPα-mediated transcription and stabilizing C/EBPα through direct physical interactions (71). The increased level of C/EBPα allows C/EBPα to more efficiently bind to the ZII domain (C-3 site) as well as to the ZIIIB domain (C-2 site) to activate higher levels of ZTA expression. In addition, ZTA may also bind to both ZII and ZIIIB domain-bound C/EBPα to form piggyback complexes. Some degree of cooperativity or even competitive interactions between adjacent C/EBPα-bound ZIIIB and ZTA-bound ZIIIA would also be expected. Finally, increased ZTA expression would result in ZTA filling in both the ZIIIA sites and any unoccupied ZIIIB sites to maximally activate transcription of the Zp (Fig. 10B).
Although C/EBPα also activates the KSHV RAP promoter (66), regulation of the EBV Zp by C/EBPα is evidently very different from that of the KSHV RAP promoter. First, RAP, unlike ZTA, is not known to bind directly to any known ZRE-like or AP1-like sites; therefore, RAP is incapable of binding to its own promoter or directly transactivating expression from its own promoter (or other downstream viral promoters) (65, 72). However, ZTA is able to positively autoregulate itself through direct binding to the two ZRE sites found in the ZIII domain. Further, the positioning of the C/EBP binding sites on the RAP promoter is very different from that on the Zp. The two C/EBP binding sites on the RAP promoter are located 13 bp apart, an arrangement which may generate a palindromic head-to-head ACAAT motif suggestive of cooperative binding. However, only one of these sites (C/EBP-II) is crucial for C/EBPα-mediated transcription, while the other site (C/EBP-I) is auxiliary and dispensable. The C/EBP sites on the Zp are further apart, and one overlaps an important ZRE binding site. Nevertheless, functional analysis revealed that the C-2 and C-3 sites are both indispensable for the overall efficient responses of the Zp to C/EBPα in transient assays, suggesting some form of cooperative interaction between the two sites. In addition, the two C/EBP recognition sequences on the RAP promoter are homologous to each other, with both containing an ACAAT core motif, whereas on the Zp, the two strong C/EBP sites are very different from each other. In fact, the sequence of the C/EBP binding site in the ZII domain does not match the consensus sequences of any previously known C/EBPα binding sites.
There has been some controversy about whether ZTA truly plays a role in activation of the Zp in vivo in latently infected B cells. Despite the identification of two proximal ZTA binding sites, ZIIIA and ZIIIB (40), and evidence for the upregulation of Zp reporter genes by cotransfected ZTA (21, 63), other investigators have argued that transfection of exogenous ZTA into cells latently infected with EBV does not activate endogenous Zp expression (30, 36). However, our ChIP assay data clearly show that endogenous ZTA does indeed bind to the Zp after anti-IgG-mediated cross-linking of the BCR leads to the induction of the lytic cycle in Akata cells. Deng et al. (17) have also described an association of endogenous ZTA with the Zp in ChIP assays with TPA- and ionomycin-treated Raji cells. The stable episomal Zp reporter gene cell lines used by Binne et al. (3) also presumably involve chromatin-associated promoter DNA, even if transient reporter gene assays may not. Our evidence that exogenous C/EBPα can efficiently activate endogenous Zp expression in HeLa-BX1 cells (although less efficiently in Akata cells) suggests that chromatin-bound Zp is accessible directly to activation by some DNA binding transcription factors, even in the presumed absence of histone acetylation effects, which are often associated with chemical inducers and BCR-mediated induction. Furthermore, Wu et al. showed previously that exogenous ZTA alone can readily activate expression from the endogenous C/EBPα promoter, apparently by augmenting C/EBPα-mediated autoregulation (71). All of the experimental data can be reconciled by the interpretation that ZTA alone cannot access the Zp until after the occurrence of some other initiation event that is specific for the Zp but is evidently not relevant to the cellular C/EBPα or p21 promoters. This event seems most likely to be either the removal of the MEF2D repressors at C-1 or interactions with the ZIIIB and ZIIIC factors at C-2, perhaps combined with a specific modification of ZTA itself, such as phosphorylation at S186 (2, 26).
The Zp evidently encompasses cis-responsive elements for many different viral and cellular factors that are concentrated over a compact 220-bp region. Although some studies with transient assays have suggested a role for additional, more distal repressor elements between −221 and −554 upstream from the Zp mRNA start site (46, 47, 56), these would be rather unusual because they would be embedded within the coding region sequences for RTA. However, the effects were relatively weak, and because this far-upstream region proved to be unimportant for BCR-mediated induction in the stable plasmid reporter cell lines used by Binne et al. (3), we chose to omit it from our transient reporter gene assays. A high concentration of cis-responsive elements may be needed to permit interactions between adjacent bound factors or to ensure that ZTA can be activated in different cell types with different repertoires of cellular factors that may augment the functionality of ZTA (or RTA) in activating the Zp. Because low levels of C/EBPα are present before induction and increase greatly in ZTA-positive lytically induced Akata, D98/H1, and HeLa BX1 cells (71), the presence of the C/EBP binding sites may allow the virus to utilize C/EBPα to more fully upregulate ZTA expression in conjunction with direct ZTA-positive autoregulation. However, it is also possible that in certain EBV-permissive cell lines, the expression of C/EBPα occurs at a much lower level, is more tightly regulated, or is counteracted by related cellular factors, such as C/EBPβ or CHOP-10. In these circumstances, other cellular factors, such AP1, CREB, ATF-1/ATF-2, and Smad3/Smad4, which all share the overlapping ZII domain recognition sites with C/EBPα, may be invoked in place of C/EBPα to function cooperatively with ZTA to reach the highest possible level of ZTA transcription within a short period of time.
Acknowledgments
These studies were funded by National Cancer Institute research grants R01 CA73585 and R01 CA81400 to G.S.H. and R01 CA30356 to S.D.H. from the National Institutes of Health. F.Y.W. was partially supported by the Anti-Cancer Drug Development Training Program (grant T32 CA09243).
We thank Gangling Liao, Jian Huang, and Masahiro Fujimuro for valuable technical advice; M. Dan Lane for the rabbit anti-C/EBPα PAb; and Erik Flemington for providing versions of the ZTA bZIP domain point mutants.
REFERENCES
- 1.Bauer, G., P. Hofler, and H. Zur Hausen. 1982. Epstein-Barr virus induction by a serum factor. I. Induction and cooperation with additional inducers. Virology 121:184-194. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, M., H. Mischak, S. Dammeier, W. Kolch, O. Gires, D. Pich, R. Zeidler, H. J. Delecluse, and W. Hammerschmidt. 1998. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoylphorbol-13-acetate-induced phosphorylation. J. Virol. 72:8105-8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binne, U. K., W. Amon, and P. J. Farrell. 2002. Promoter sequences required for reactivation of Epstein-Barr virus from latency. J. Virol. 76:10282-10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borras, A. M., L. Strominger, and S. H. Speck. 1996. Characterization of the ZI domains in the Epstein-Barr virus BZLF1 gene promoter: role in phorbol ester induction. J. Virol. 70:3894-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calkhoven, C. F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 6.Cayrol, C., and E. Flemington. 1996. G0/G1 growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein-Barr virus transactivator Zta. J. Biol. Chem. 271:31799-31802. [DOI] [PubMed] [Google Scholar]
- 7.Cayrol, C., and E. K. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. N., D. L. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., L. Hutt-Fletcher, L. Cao, and S. D. Hayward. 2003. A positive autoregulatory loop of LMP1 expression and STAT activation in epithelial cells latently infected with Epstein-Barr virus. J. Virol. 77:4139-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H., J. Lee, Y. Wang, D. Huang, R. Ambinder, and S. Hayward. 1999. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and ZTA interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc. Natl. Acad. Sci. USA 96:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu, R., W. J. Boyle, J. Meek, T. Smeal, T. Hunter, and M. Karin. 1988. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell 54:541-552. [DOI] [PubMed] [Google Scholar]
- 13.Christy, R. J., K. H. Kaestner, D. E. Geiman, and M. D. Lane. 1991. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. USA 88:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daibata, M., S. H. Speck, C. Mulder, and T. Sairenji. 1994. Regulation of the BZLF1 promoter of Epstein-Barr virus by second messengers in anti-immunoglobulin-treated B cells. Virology 198:446-454. [DOI] [PubMed] [Google Scholar]
- 16.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273:30057-30060. [DOI] [PubMed] [Google Scholar]
- 17.Deng, Z., C. J. Chen, M. Chamberlin, F. Lu, G. A. Blobel, D. Speicher, L. A. Cirillo, K. S. Zaret, and P. M. Lieberman. 2003. The CBP bromodomain and nucleosome targeting are required for Zta-directed nucleosome acetylation and transcription activation. Mol. Cell. Biol. 23:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell, P. J., D. T. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-Acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemington, E., and S. H. Speck. 1990. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flemington, E., and S. H. Speck. 1990. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc. Natl. Acad. Sci. USA 87:9459-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flemington, E., and S. H. Speck. 1990. Identification of phorbol ester response elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flemington, E. K. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flemington, E. K., J. P. Lytle, C. Cayrol, A. M. Borras, and S. H. Speck. 1994. DNA-binding-defective mutants of the Epstein-Barr virus lytic switch activator ZTA transactivate with altered specificities. Mol. Cell. Biol. 14:3041-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis, A. L., L. Gradoville, and G. Miller. 1997. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J. Virol. 71:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruffat, H., E. Manet, and A. Sergeant. 2002. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. EMBO Rep. 3:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, T. E., J. H. Albrecht, M. Nakanishi, and G. J. Darlington. 2001. CCAAT/enhancer-binding protein-alpha cooperates with p21 to inhibit cyclin-dependent kinase-2 activity and induces growth arrest independent of DNA binding. J. Biol. Chem. 276:29200-29209. [DOI] [PubMed] [Google Scholar]
- 29.Jenkins, P. J., U. K. Binne, and P. J. Farrell. 2000. Histone acetylation and reactivation of Epstein-Barr virus from latency. J. Virol. 74:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolman, J. L., N. Taylor, L. Gradoville, J. Countryman, and G. Miller. 1996. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J. Virol. 70:1493-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kouzarides, T., G. Packham, A. Cook, and P. J. Farrell. 1991. The BZLF1 protein of EBV has a coiled coil dimerisation domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene 6:195-204. [PubMed] [Google Scholar]
- 32.Kraus, R. J., S. J. Mirocha, H. M. Stephany, J. R. Puchalski, and J. E. Mertz. 2001. Identification of a novel element involved in regulation of the lytic switch BZLF1 gene promoter of Epstein-Barr virus. J. Virol. 75:867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraus, R. J., J. G. Perrigoue, and J. E. Mertz. 2003. ZEB negatively regulates the lytic switch BZLF1 gene promoter of Epstein-Barr virus. J. Virol. 77:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 35.Lane, M. D., Q. Q. Tang, and M. S. Jiang. 1999. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem. Biophys. Res. Commun. 266:677-683. [DOI] [PubMed] [Google Scholar]
- 36.Le Roux, F., A. Sergeant, and L. Corbo. 1996. Epstein-Barr virus (EBV) EB1/Zta protein provided in trans and competent for the activation of productive cycle genes does not activate the BZLF1 gene in the EBV genome. J. Gen. Virol. 77:501-509. [DOI] [PubMed] [Google Scholar]
- 37.Liang, C. L., J. L. Chen, Y. P. Hsu, J. T. Ou, and Y. S. Chang. 2002. Epstein-Barr virus BZLF1 gene is activated by transforming growth factor-beta through cooperativity of Smads and c-Jun/c-Fos proteins. J. Biol. Chem. 277:23345-23357. [DOI] [PubMed] [Google Scholar]
- 38.Liao, G., F. Y. Wu, and S. D. Hayward. 2001. Interaction with the Epstein-Barr virus helicase targets Zta to DNA replication compartments. J. Virol. 75:8792-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman, P. M., and A. J. Berk. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin, F. T., O. A. MacDougald, A. M. Diehl, and M. D. Lane. 1993. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. USA 90:9606-9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, P., S. Liu, and S. Speck. 1998. Identification of a negative cis-acting element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene promoter. J. Virol. 72:8230-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, S., A. M. Borras, P. Liu, G. Suske, and S. H. Speck. 1997. Binding of the ubiquitous cellular transcription factors Sp1 and Sp3 to the ZI domains in the Epstein-Barr virus lytic switch BZLF1 promoter. Virology 228:11-18. [DOI] [PubMed] [Google Scholar]
- 44.Liu, S. P., P. Liu, A. M. Borras, T. Chatila, and S. H. Speck. 1997. Cyclosporin A-sensitive induction of the Epstein-Barr virus lytic switch is mediated via a novel pathway involving a MEF2 family member. EMBO J. 16:143-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacCalum, P., L. Karimi, and L. J. Nicholson. 1999. Definition of the transcription factors which bind the differentiation responsive element of the Epstein-Barr virus BZLF1 Z promoter in human epithelial cells. J. Gen. Virol. 6:1501-1512. [DOI] [PubMed] [Google Scholar]
- 46.Montalvo, E. A., M. Cottam, S. Hill, and Y. J. Wang. 1995. YY1 binds to and regulates cis-acting negative elements in the Epstein-Barr virus BZLF1 promoter. J. Virol. 69:4158-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montalvo, E. A., Y. Shi, T. E. Shenk, and A. J. Levine. 1991. Negative regulation of the BZLF1 promoter of Epstein-Barr virus. J. Virol. 65:3647-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakabeppu, Y., K. Ryder, and D. Nathans. 1988. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell 55:907-915. [DOI] [PubMed] [Google Scholar]
- 49.Ossipow, V., P. Descombes, and U. Schibler. 1993. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc. Natl. Acad. Sci. USA 90:8219-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian, Z., P. Brunovskis, F. Rauscher III, L. Lee, and H. J. Kung. 1995. Transactivation activity of Meq, a Marek's disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J. Virol. 69:4037-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rauscher, F. J., III, P. J. Voulalas, B. R. Franza, Jr., and T. Curran. 1988. Fos and Jun bind cooperatively to the AP-1 site: reconstitution in vitro. Genes Dev. 2:1687-1699. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez, A., E. J. Jung, Q. Yin, C. Cayrol, and E. K. Flemington. 2001. Role of c-myc regulation in Zta-mediated induction of the cyclin-dependent kinase inhibitors p21 and p27 and cell growth arrest. Virology 284:159-169. [DOI] [PubMed] [Google Scholar]
- 54.Ruf, I. K., and D. R. Rawlins. 1995. Identification and characterization of ZIIBC, a complex formed by cellular factors and the ZII site of the Epstein-Barr virus BZLF1 promoter. J. Virol. 69:7648-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwarzmann, F., N. Prang, B. Reichelt, B. Rinkes, S. Haist, M. Marschall, and H. Wolf. 1994. Negatively cis-acting elements in the distal part of the promoter of Epstein-Barr virus trans-activator gene BZLF1. J. Gen. Virol. 75:1999-2006. [DOI] [PubMed] [Google Scholar]
- 57.Slomiany, B. A., K. L. D'Arigo, M. M. Kelly, and D. T. Kurtz. 2000. C/EBPα inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol. Cell. Biol. 20:5986-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takada, K., N. Shimizu, S. Sakuma, and Y. Ono. 1986. trans Activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol. 57:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang, Q. Q., and M. D. Lane. 1999. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 13:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Timchenko, N., D. R. Wilson, L. R. Taylor, S. Abdelsayed, M. Wilde, M. Sawadogo, and G. J. Darlington. 1995. Autoregulation of the human C/EBPα gene by stimulation of upstream stimulatory factor binding. Mol. Cell. Biol. 15:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Timchenko, N. A., T. E. Harris, M. Wilde, T. A. Bilyeu, B. L. Burgess-Beusse, M. J. Finegold, and G. J. Darlington. 1997. CCAAT/enhancer binding protein α regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 17:7353-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Timchenko, N. A., M. Wilde, M. Nakanishi, J. R. Smith, and G. J. Darlington. 1996. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 10:804-815. [DOI] [PubMed] [Google Scholar]
- 63.Urier, G., M. Buisson, P. Chambard, and A. Sergeant. 1989. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 8:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, H., P. Iakova, M. Wilde, A. Welm, T. Goode, W. J. Roesler, and N. A. Timchenko. 2001. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell 8:817-828. [DOI] [PubMed] [Google Scholar]
- 65.Wang, S. E., F. Y. Wu, Y. X. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein α is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and, together with the KSHV replication and transcription activator (RTA), cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 77:9590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, S. E., F. Y. Wu, M. Fujimuro, J. C. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of the CCAAT/enhancer-binding protein α in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, X., E. Scott, C. L. Sawyers, and A. D. Friedman. 1999. C/EBPalpha bypasses granulocyte colony-stimulating factor signals to rapidly induce PU.1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32D cl3 myeloblasts. Blood 94:560-571. [PubMed] [Google Scholar]
- 68.Wang, Y. C., J. M. Huang, and E. A. Montalvo. 1997. Characterization of proteins binding to the ZII element in the Epstein-Barr virus BZLF1 promoter: transactivation by ATF1. Virology 227:323-330. [DOI] [PubMed] [Google Scholar]
- 69.Wu, F., Q. Tang, H. Chen, C. ApRhys, C. Farrell, J. Chen, M. Fujimuro, M. Lane, and G. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi's sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. USA 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu, F. Y., H. Chen, S. E. Wang, C. ApRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein α interacts with ZTA and mediates ZTA-induced p21CIP-1 accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, F. Y., S. E. Wang, Q. Q. Tang, M. Fujimuro, C.-J. Chiou, Q. Cheng, H. Chen, S. D. Hayward, M. D. Lane, and G. S. Hayward. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and the posttranslational levels by binding to CCAAT/enhancer-binding protein α and p21CIP−1. J. Virol. 77:8893-8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, D. E., P. Zhang, N. D. Wang, C. J. Hetherington, G. J. Darlington, and D. G. Tenen. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA 94:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.zur Hausen, H., F. O'Neill, and U. Freese. 1978. Persisting oncogenic herpesvirus induced by the tumor promoter TPA. Nature 272:373-375. [DOI] [PubMed] [Google Scholar]