Abstract
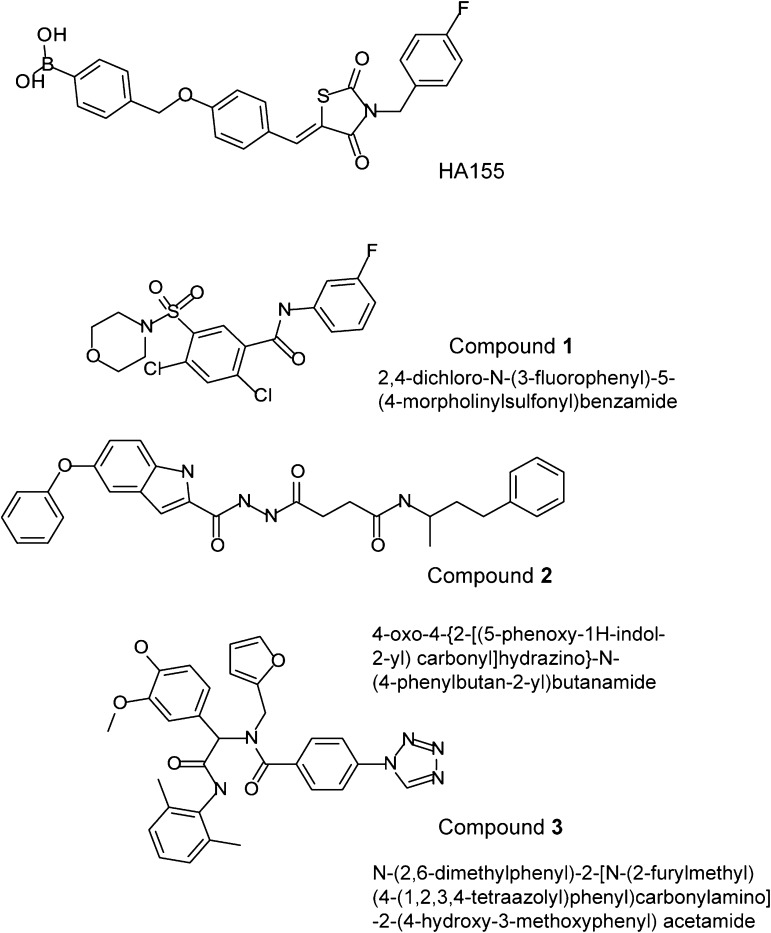
Autotaxin (ATX), a lysophospholipase D, plays an important role in cancer invasion, metastasis, tumor progression, tumorigenesis, neuropathic pain, fibrotic diseases, cholestatic pruritus, lymphocyte homing, and thrombotic diseases by producing the lipid mediator lysophosphatidic acid (LPA). A high-throughput screen of ATX inhibition using the lysophosphatidylcholine-like substrate fluorogenic substrate 3 (FS-3) and ∼10,000 compounds from the University of Cincinnati Drug Discovery Center identified several small-molecule inhibitors with IC50 vales ranging from nanomolar to low micromolar. The pharmacology of the three most potent compounds: 918013 (1; 2,4-dichloro-N-(3-fluorophenyl)-5-(4-morpholinylsulfonyl) benzamide), 931126 (2; 4-oxo-4-{2-[(5-phenoxy-1H-indol-2-yl)carbonyl]hydrazino}-N-(4-phenylbutan-2-yl)butanamide), and 966791 (3; N-(2,6-dimethylphenyl)-2-[N-(2-furylmethyl)(4-(1,2,3,4-tetraazolyl)phenyl)carbonylamino]-2-(4-hydroxy-3-methoxyphenyl) acetamide), were further characterized in enzyme, cellular, and whole animal models. Compounds 1 and 2 were competitive inhibitors of ATX-mediated hydrolysis of the lysophospholipase substrate FS-3. In contrast, compound 3 was a competitive inhibitor of both FS-3 and the phosphodiesterase substrate p-nitrophenyl thymidine 5′-monophosphate. Computational docking and mutagenesis suggested that compounds 1 and 2 target the hydrophobic pocket, thereby blocking access to the active site of ATX. The potencies of compounds 1–3 were comparable to each other in each of the assays. All of these compounds significantly reduced invasion of A2058 human melanoma cells in vitro and the colonization of lung metastases by B16-F10 murine melanoma cells in C57BL/6 mice. The compounds had no agonist or antagonist effects on select LPA or sphingosine 1-phosphate receptors, nor did they inhibit nucleotide pyrophosphatase/phosphodiesterase (NPP) enzymes NPP6 and NPP7. These results identify the molecular surface of the hydrophobic pocket of ATX as a target-binding site for inhibitors of enzymatic activity.
Introduction
Autotaxin (ATX, NPP2) is member of the nucleotide pyrophosphatase/phosphodiesterase (NPP) family of enzymes that was originally discovered as a secreted factor that promoted the invasion and motility of melanoma cells (Stracke et al., 1992). The dual lysophospholipase (LPL) and phosphodiesterase (PDE) activity of ATX is catalyzed by the same active site (Gijsbers et al., 2003). While hydrolysis of nucleotide pyrophosphates does not appear to be physiologically relevant in vivo (Clair et al., 2003; Koh et al., 2003; Baker et al., 2006), hydrolysis of lysophosphatidylcholine (LPC), which is considered to be the physiologically relevant substrate of ATX, generates the bioactive lipid lysophosphatidic acid (LPA). A variety of biologic processes are mediated by LPA via activation of multiple G protein–coupled receptors (GPCRs) (Tigyi, 2010; Yanagida et al., 2013). Some of these responses—including angiogenesis, chemotaxis, cell invasion, migration, proliferation, and suppression of apoptosis—are particularly important in tumor biology because they affect the growth, progression, and metastasis of many types of cancers (Mills and Moolenaar, 2003; Houben and Moolenaar, 2011; Brindley et al., 2013). Upregulated ATX expression has been reported in numerous cancers, including thyroid, prostate, breast, and ovarian cancers, as well as melanoma (Yang et al., 1999, 2002; Kehlen et al., 2004; Nouh et al., 2009; David et al., 2010; Wu et al., 2010). In addition to malignant diseases, ATX has been implicated in neuropathic pain, fibrotic diseases, cholestatic pruritus, lymphocyte homing, chronic inflammatory conditions, and thrombotic diseases (Ikeda and Yatomi, 2012; Kremer et al., 2012; Nikitopoulou et al., 2012; Tager, 2012; Moolenaar et al., 2013; Ueda et al., 2013). Because of the established role of LPA in these malignancies, inhibition of ATX represents a therapeutically attractive target for the disruption of the ATX-LPA-LPA receptor signaling axis (Albers and Ovaa, 2012).
At the present time there is no approved ATX inhibitor available for therapy; however, development of ATX inhibitors has gained increasing interest with several small-molecule ATX inhibitors having been described (Ferry et al., 2008; Saunders et al., 2008; Albers et al., 2010; Gierse et al., 2010; Hoeglund et al., 2010; Mize et al., 2011; Gotoh et al., 2012; Kawaguchi et al., 2013; St-Cœur et al., 2013). A considerable amount of effort has been devoted to the characterization of inhibitors that interact with the active site of ATX. Recent crystal structures of ATX, including a cocrystal with the irreversible inhibitor (Z)-4-((4-((3-(4-fluorobenzyl)-2,4-dioxo-1,3-thiazolan-5-yliden)-methyl)phenoxy)methyl)benzeneboronic acid ATX inhibitor IV (HA155) (Hausmann et al., 2011) and the active-site inhibitor BoA set of compounds (Kawaguchi et al., 2013) have provided new insight into the catalytic site of this enzyme and also shed light on the access of the lipid substrate to the active site (Hausmann et al., 2011; Nishimasu et al., 2011). The structure reported by Nishimasu et al. (2011) shows LPA in a hydrophobic channel that suggests an exit route for the product connecting the catalytic site to the enzyme surface.
Our objective in the present study was to apply a new high-throughput screening (HTS) strategy with the synthetic LPC-like substrate fluorogenic substrate 3 (FS-3) for novel ATX inhibitors using 10,000 compounds representative of the chemical diversity space of the ∼360,000 compounds contained in the University of Cincinnati Drug Discovery Center (UC-DDC) chemical library. Previous HTS strategies targeting ATX (Ferry et al., 2008; Albers et al., 2010) used the nucleotide-like PDE substrate p-nitrophenyl thymidine 5′-monophosphate (pNP-TMP). In our strategy, hits that inhibited ATX activity by 50% at a concentration of 10 µM were selected for further characterization using pNP-TMP and another LPC analog, 3-acyl-7-dimethylaminonaphtyl–1-LPC (ADMAN-LPC). Using these LPL and PDE substrates our hits fell into two categories (Fig. 1). The first category of compounds, exemplified by 2,4-dichloro-N-(3-fluorophenyl)-5-(4-morpholinylsulfonyl) benzamide (UC-DDC number 918013, designated as compound 1) and 4-oxo-4-{2-[(5-phenoxy-1H-indol-2-yl)carbonyl]hydrazino}-N-(4-phenylbutan-2-yl)butanamide (UC-DDC number 931126, designated as compound 2) are competitive inhibitors of the hydrolysis of LPC-type substrates only (termed herein as single inhibitors).
Fig. 1.
Chemical structures of ATX inhibitors identified in the present study and HA155.
A second category of compounds, represented by the compound N-(2,6-dimethylphenyl)-2-[N-(2-furylmethyl)(4-(1,2,3,4-tetraazolyl)phenyl)carbonylamino]-2-(4-hydroxy-3-methoxyphenyl) acetamide (UC-DDC number 966791, designated as compound 3), competitively inhibited the hydrolysis of both types of substrates via a competitive mechanism (termed herein as dual-inhibitor). Computational docking of compounds 1 and 2 into the ATX crystal structure suggests that they interact with a surface in the hydrophobic pocket of ATX. In contrast, compound 3 interacts with a surface at the catalytic site. Alanine replacement of residue Phe275 in the hydrophobic inhibitor-interacting surface reduced the inhibition of FS-3 hydrolysis by compound 1, whereas it had no effect on the action of the dual-inhibitor compound 3. The hydrolysis of pNP-TMP by the F275A mutant was also blocked by compound 3. All three compounds dose-dependently reduced the ATX-dependent invasion of A2058 melanoma cells in vitro. The single inhibitor compound 1 and the dual inhibitor compound 3, when administered to C57BL/6 mice inoculated with the B16 melanoma cells, reduced the number of metastatic lung nodules. These results identify the hydrophobic channel surface as a novel inhibitory site in the ATX structure amenable to drug discovery.
Materials and Methods
Reagents.
An ∼10,000-member diversity library was provided by the UC-DDC. Compounds 1 and 3 were also purchased from TimTec (Newark, DE) and ChemBridge (San Diego, CA) for the animal studies. The compounds were diluted in dimethyl sulfoxide (DMSO) at a 10 mM stock concentration at −80°C. PNP-TMP was purchased from Sigma-Aldrich (St. Louis, MO), FS-3 was purchased from Echelon (Salt Lake City, UT), and ADMAN-LPC was synthesized as previously described (Baker et al., 2006).
Expression, Purification, and Activity of ATX.
The expression, purification, and enzymatic assay conditions of human ATX were described previously (Gupte et al., 2011).
Primary HTS Assay Conditions.
Before the start of the assay, 20 µl of 4 nM ATX/well were dispensed into 384-well plates. The assay was started immediately by dispensing 10 µM of the test compounds in DMSO (final DMSO concentration 0.093%), DMSO alone (0% inhibition control), or LPA (final concentration 10 µM, 100% inhibition control) to the appropriate wells. Next, 10 µl of 4 µM FS-3 (1 µM final) in assay buffer [50 mM Tris, 1 mM MgCl2, 1 mM CaCl2, 3 mM KCl, 140 mM NaCl, and 15 µM bovine serum albumin (BSA)] was added to all wells. Plates were briefly centrifuged for 35 seconds at 337g. An initial fluorescence was read by fluorescence excitation at 475 nm and detection of emission at 535 nm using a Envision Plate reader (PerkinElmer, Waltham, MA). The plates were then incubated for 3 hours at 37°C. Autotaxin activity was determined after 3 hours by the change in fluorescence. Dose-response curves were recorded to determine IC50 values for compounds that caused > 50% inhibition of ATX enzyme activity.
Secondary Assay Conditions for Inhibition of PDE Activity Using pNP-TMP Substrate.
The assay contained overall concentrations of 4 nM ATX, 1 mM pNP-TMP, and 10 µM of compound in assay buffer. Absorbance was monitored at 485 nm after 4 hours of incubation to measure enzyme activity.
Tertiary Screen in the Presence of 0.01% Triton X-100.
A detergent-dependent screen was adapted from the previously described enzyme assay, for the identification of promiscuous inhibitors. Triton X-100 was added to assay buffer at a concentration of 0.01% to test interference with FS-3 and pNP-TMP hydrolysis assays.
Amplex Red Choline Release Assay.
Inhibition of ATX-mediated hydrolysis of various chain-length LPC substrates was determined via Amplex Red choline release assay as described previously (Hoeglund et al., 2010; North et al., 2010). Briefly, 60-μl triplicate reaction volumes were loaded into 96-well, half-area plates (Corning Inc., Corning, NY) in assay buffer consisting of 50 mM Tris, 5 mM CaCl2, and 30 μM fatty acid–free BSA (pH 7.4). Final concentrations of reaction constituents were 0.1 U/ml choline oxidase (MP Biomedicals, Solon, OH), 1 U/ml horseradish peroxidase (Thermo Fisher Scientific, Waltham, MA), 10 μM Amplex Red (Life Technologies, Carlsbad, CA), 10 nM ATX, 100 μM LPC (14:0, 16:0, 18:0, or 18:1 chain lengths from Avanti Polar Lipids, Alabaster, AL), and 10 μM inhibitor. Fluorescence was measured at excitation and emission wavelengths of 560 and 590 nm, respectively, initially and after 2-hour incubation at 37°C. From the background-corrected endpoint data, percent ATX inhibition versus vehicle control was calculated for each triplicate data set and reported ± standard deviation.
Quaternary Screen Using ADMAN-LPC.
ADMAN-LPC at a final concentration of 30 µM was resuspended in assay buffer containing 2 mg/ml BSA with or without 10 µM inhibitor compound and 30 nM ATX. The reaction was incubated at 37°C for 4 hours. Lipids were extracted using a modification of the Bligh and Dyer procedure by adding 3.5 volumes of citrate phosphate buffer, pH 4, before addition of chloroform and methanol (2:1 v/v). Lipids were dried and resuspended in 30 µl chloroform/methanol (1:1) and separated on silica gel 60 thin layer chromatography plates (Merck, Billerica, MA) using CHCl3/MeOH/NH4OH (60:35:8 v/v/v). Fluorescent LPC and LPA species were visualized by ultraviolet transillumination and quantified via densitometric analysis in ImageJ software (NIH, Bethesda, MD) (Schneider et al., 2012) to compare LPA production and ATX inhibition.
Determination of the Mechanism of Autotaxin Inhibition.
The mechanism of ATX inhibition was determined using an activity assay with inhibitor concentrations of 0, 0.5, and 2 times the experimentally determined IC50 value and FS-3 substrate concentrations ranging from 0.3–20 µM. The initial reaction rate was determined by plotting background-corrected fluorescence at emission wavelength 485 nm as a function of time using 528-nm excitation of the fluorochrome. Data were then transformed using a carboxyfluorescein standard curve, which is analogous to the fluorescent FS-3 hydrolytic product. Reaction rate was then plotted as a function of FS-3 concentration and simultaneously fitted via nonlinear regression using the Michaelis-Menten equations for competitive, noncompetitive, uncompetitive, and mixed-mode inhibition as described in our previous publication (Gupte et al., 2011), using GraphPad Prism v5 (GraphPad Software, Inc., La Jolla, CA). The model providing the highest global nonlinear fit (R2) value, taking into account GraphPad’s interpretation rules for the calculated α value, was selected as the mechanism of inhibition. Ki (compound affinity for enzyme) was subsequently calculated based on the corresponding regression analysis.
Testing of Off-Target Effects of Select ATX Inhibitors.
Compounds 1 and 3 were submitted to Psychoactive Drug Screening Program (PDSP) of the National Institute for Mental Health for analysis of effect at 10 µM on 32 various human transporters and receptors.
Molecular Docking of the Inhibitors to the ATX Crystal Structure.
All of the molecular docking studies were carried out using Autodock Vina (Trott and Olson, 2010). The crystal structure of ATX (PDB ID 2XRG) was chosen as the structure of reference protein (Hausmann et al., 2011). All water molecules, sugars, iodoacetamide, and nonpolar hydrogen atoms were removed; zinc heteroatoms records were retained. The binding site was defined by centering the docking box around the catalytic domain containing the cocrystallized inhibitor HA155. Both the enzyme and ligands were prepared using the Raccoon software (The Scripps Research Institute, La Jolla, CA). Docking simulations were run using the default settings of this program.
Site-Directed Mutagenesis and Transfection.
Amino-terminal FLAG epitope-tagged ATX constructs were subcloned into pcDNA3.1 vector (Invitrogen, Grand Island, NY). Constructs were mutated using the QuikChange II XL site-directed mutagenesis kit (Stratagene, Santa Clara, CA). TOP10 competent cells (Invitrogen) were transformed with the mutant constructs, and clones were verified by complete sequencing of the inserts. Human embryonic kidney cell line 293T was cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal calf serum and 2 mM L-glutamine in a 10-cm dish. The cells were grown overnight at 37°C and 5% CO2 up to 80% confluence before transfection. Transfection was done in the presence of Effectene transfection reagent from Qiagen (Valencia, CA) according to the manufacturer's protocol. Twenty hours post-transfection, the culture medium was changed to serum-free DMEM. Forty-eight hours post-transfection, the conditioned serum-free medium containing secreted ATX mutants was collected and concentrated in ATX assay buffer using 10,000 molecular-weight cutoff filters (Millipore, Billerica, MA) by centrifugation at 3000g.
Inhibition of ATX-Dependent A2058 Melanoma Cell Invasion.
A2058 melanoma cells (gift from Dr. Timothy Clair, National Cancer Institute, National Institutes of Health) were cultured in DMEM containing 10% (v/v) FBS, 2 mM L-glutamine, 100 U ml−1 penicillin, and 100 µg ml−1 streptomycin. Cell invasion across a matrigel-coated membrane was performed using the 24-well BD BioCoat tumor invasion system, 8-µm pore size (BD Biosciences, San Jose, CA). 5 × 104 cells in serum-free DMEM supplemented with 0.1% BSA were added to the upper chamber. The chemoattractant (recombinant ATX plus 1 µM of 18:1 LPC) was added in 0.75 ml of serum-free DMEM/0.1% BSA to the bottom chamber. Where indicated, the various compounds were preincubated in serum-free DMEM/0.1% BSA with recombinant ATX for 30 minutes at 37°C, prior to the addition of 1 µM LPC and placement in the bottom chamber. Cells were left to invade the matrigel for 16 hours at 37°C. After incubation, the medium in the upper chamber was removed and the insert was transferred into a new 24-well plate containing 4 µg/ml of calcein AM (Molecular Probes, Invitrogen, Grand Island, NY) in Hanks’ balanced salt solution. The plates were incubated for 1 hour at 37°C. The fluorescence of invaded cells was measured with a FLEXStation II plate reader (Molecular Devices, Sunnyvale, CA) at excitation and emission wavelengths of 485 and 530 nm, respectively.
Melanoma Lung Colonization Model.
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Tennessee. Eight- to twelve-week-old female C57Bl/6 mice (Charles River, Wilmington, MA) were injected with 7.5 × 104 B16F10 cells in 100-µl conditioned medium via tail veins. The groups then received either compound 1, compound 3, or 4-pentadecylbenzylphosphonic acid, an ATX inhibitor we developed and validated earlier (Gupte et al., 2011) at 30 µg/mouse via intraperitoneal injection starting one day before or after the B16-F10 injection, and daily for an additional 10 days. Control mice were injected with phosphate-buffered saline, using a 10% polyethylene-glycol vehicle. Subsequently, animals were monitored for another 10 days without treatment. On day 21, all mice were sacrificed; the lungs were harvested, inflated, and fixed with 10% formalin. The number of metastatic nodules on the lung surface was counted. The number of lung nodules was compared against the vehicle-treated control group by Student’s t test, and P < 0.05 was considered significant.
Results
The primary objective of the present study was to experimentally identify a new and diverse set of nonlipid compounds that modulate the LPL activity of human recombinant ATX against the FS-3 substrate. To achieve this we developed an HTS method of ATX in 40-µl assay volume (Supplemental Fig. 1). In this assay a 10-µM concentration of LPA, which acts as a feedback inhibitor of ATX, caused an average 85% inhibition of the catalytic activity after a 3-hour incubation. The Z′ value was 0.847 (range 0.554–0.924), with a mean signal-to-background ratio of 4.19 (range 3.14–4.70), indicating the robustness of the assay. To maximize diversity, a chemical library of 9,652 compounds from the UC DDC library of 360,000 compounds was assembled and screened at 10 µM to identify novel compounds that inhibited ATX activation. A compound was defined as a hit if it showed >50% inhibition of FS-3 hydrolysis. From the primary screening, 198 inhibitory compounds (Supplemental Table 1) were selected (2% hit rate). Hits were further reduced by applying a series of filters, including a 10-point dose-response for each confirmed hit. The average Z′ value for the dose-response assay was 0.75, ranging from 0.57 to 0.83. Signal-to-background ratios [(+) ATX to (−) ATX] averaged 3.38 and ranged from 2.28 to 4.14. A compound was selected for further characterization if it had an IC50 ≤ 1 µM. Evaluation of the initial 198 compounds led to 26 compounds (Supplemental Table 2) that were chosen for further screening.
Secondary, Tertiary, and Quaternary Screening of the Primary Hits.
The 26 compounds were subsequently screened against the PDE substrate pNP-TMP (Figs. 2B and 3B). This screen revealed two groups of ATX inhibitors: 14 compounds that inhibited FS-3 hydrolysis only, and 12 compounds that inhibited both pNP-TMP and FS-3 hydrolysis. The 26 compounds were next counter-screened in the presence of 0.01% Triton X-100, discarding 11 promiscuous aggregators (data not shown). Finally, the ability of the compounds at 10 µM to inhibit ADMAN-LPC hydrolysis with a hit cutoff at 50% was used as the concluding criterion (Figs. 2B and 3C). Only compound 3 in the dual-inhibitor group and compounds 1 and 2 in the single-inhibitor group met these criteria and were further characterized in cell-based assays and in vivo. These three compounds were tested for the inhibition of the ATX-mediated hydrolysis of LPC 14:0, LPC 16:0, LPC 18:0, and LPC 18:1 (Table 1) and found to inhibit the cleavage of all four molecular species of the substrate with similar efficacy when applied at 10 µM.
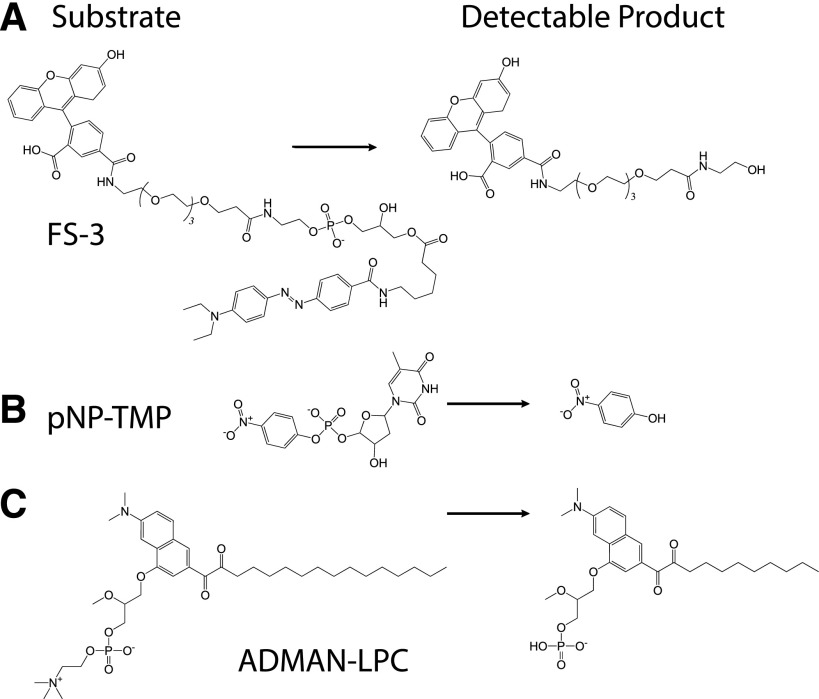
Fig. 2.
Schematic of enzyme reactions used for measuring ATX activity. (A) FS-3 substrate used for high-throughput screening and in the counter-screen in the presence of 0.01% Triton X-100. (B) pNP-TMP substrate used for nucleotide hydrolysis by ATX. (C) ADMAN-LPC, a synthetic, fluorescent LPC analog used to monitor ATX-catalyzed hydrolysis of LPC.
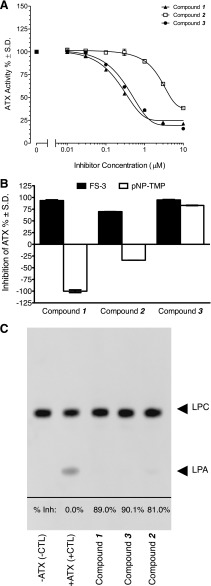
Fig. 3.
Identification and characterization of ATX inhibitors by high-throughput screening. (A) Dose-response curve of the three most potent hits from the high-throughput screening. (B) Comparison of substrate selectivity against FS-3 (filled bars) and pNP-TMP (open bars) for the three inhibitors applied at 10 µM. The inhibition was normalized to cleavage of the substrate in the absence of the inhibitors designated as 100%. (C) Inhibition of ATX-mediated hydrolysis of ADMAN-LPC by inhibitors. A representative thin layer chromatography image shows that ATX inhibitors applied at 10 µM inhibited the hydrolysis of 30 μM ADMAN-LPC by 30 nM ATX.
TABLE 1.
Inhibition of ATX-mediated hydrolysis of different LPC species
ATX inhibition by the selected compounds was assessed via Amplex Red choline release assay. Fluorescence was read initially and after 2-hour incubation at 37°C at Ex/Em λ of 560/590 nm. Data (relative fluorescence) were then recorded as a mean value of the triplicates for each sample and reported as % inhibition of ATX-mediated LPC hydrolysis.
| % Inhibition (± S.D.) |
||||
|---|---|---|---|---|
| Compound | LPC 14:0 | LPC 16:0 | LPC 18:0 | LPC 18:1 |
| 918013 (Compound 1) | 41.0 ± 5.1 | 55.4 ± 1.7 | 43.6 ± 2.6 | 51.8 ± 2.4 |
| 931126 (Compound 2) | 55.9 ± 0.5 | 69.0 ± 1.2 | 57.6 ± 0.9 | 61.9 ± 2.0 |
| 966791 (Compound 3) | 70.3 ± 1.8 | 80.3 ± 1.0 | 78.2 ± 1.1 | 78.8 ± 0.7 |
Mechanism of Inhibition of the Hit Compounds.
The mechanism of inhibition of ATX activity by the three compounds was determined by measuring the Ki, and/or Ki′ values against ATX-mediated hydrolysis of FS-3 and pNP-TMP (Table 2). These experiments indicated that all three compounds inhibit the hydrolysis of the LPL substrate FS-3 by a competitive mechanism. In addition, competition assays were conducted for compound 3 against pNP-TMP hydrolysis in which the compound was also found to act via competitive mechanisms.
TABLE 2.
Characterization of the hit compounds
| Compound | Partition Coefficient LogP | Assays and Result Type |
|||||
|---|---|---|---|---|---|---|---|
| FS-3 |
pNP-TMP |
||||||
| IC50 | Mechanism of Inhibition | Ki | IC50 | Mechanism of Inhibition | Ki | ||
| nM | nM | nM | nM | ||||
| 1 | 2.99 | 31.42 | Competitive | 12.98 | N/A | N/A | N/A |
| 2 | 3.54 | 6.50 | Competitive | 19.64 | N/A | N/A | N/A |
| 3 | 3.98 | 53.05 | Competitive | 7.14 | 314 | Competitive | 933 |
IC50, concentration that inhibits the cleavage of LPC by 50%; Ki, inhibitory constant; N/A, not applicable due to lack of inhibition of pNP-TMP hydrolysis.
Molecular Docking of the Inhibitors into the ATX Structure.
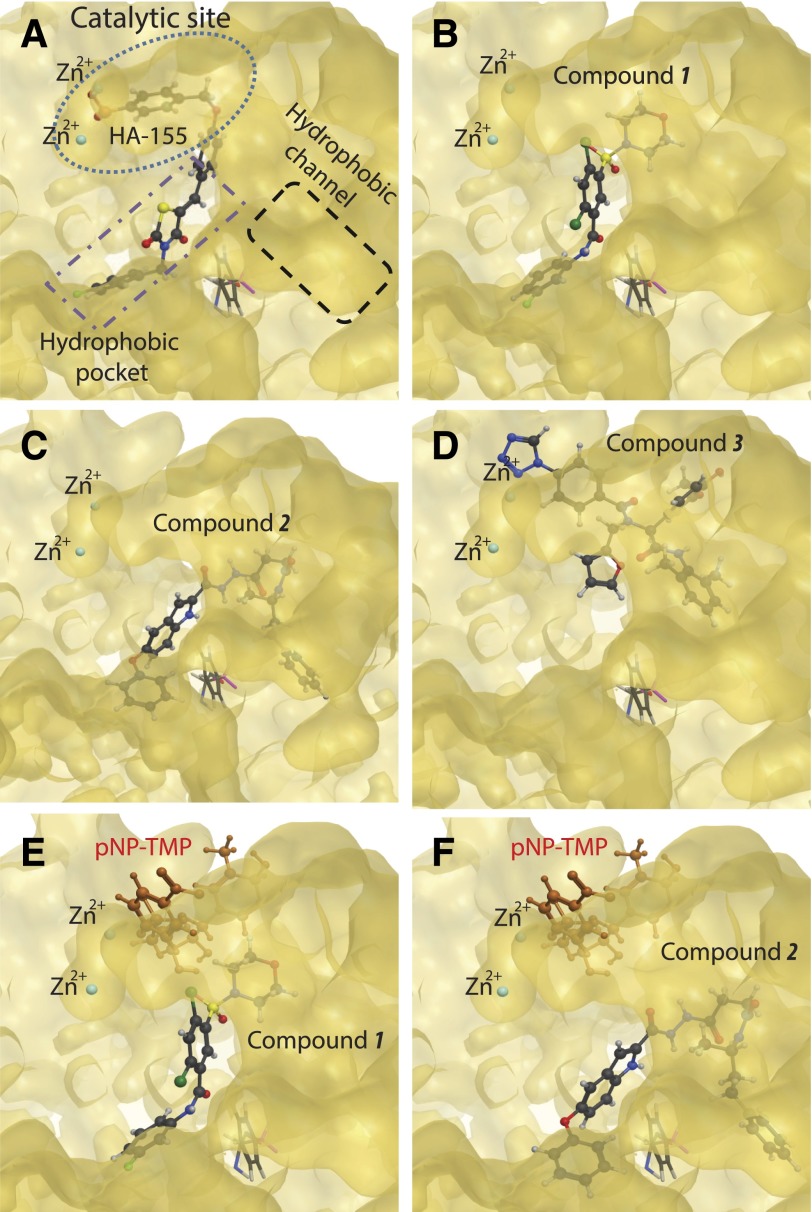
The differences in substrate-specific inhibition by the two groups of compounds led us to hypothesize that they might interact with different surfaces of ATX, as has been suggested for other ATX inhibitors showing substrate-selective effects (Hoeglund et al., 2010). Computational docking studies were performed using one of the recently solved crystal structures of ATX (2XR9) (Fig. 4). The crystallographic studies identified three hot spots in the ATX structure (Fig. 4A): 1) the catalytic site with Thr210 and the two Zn atoms; 2) a hydrophobic pocket, which accommodates the hydrocarbon tail of the lipid substrate and product; and 3) a hydrophobic channel that is hypothesized to provide entry to LPC and exit to LPA. The best docking poses obtained for compounds 1 and 2 show that these molecules are oriented similarly within the hydrophobic pocket (Fig. 4, B and C). The aromatic moieties of these compounds were inserted deeply into the pocket. However, these compounds do not reach the catalytic site. In contrast, compound 3 fits near the Zn2+ atoms at the catalytic site, almost in complete overlap with the position occupied by the covalently bound inhibitor HA155. The computed placement of compound 3 is consistent with the disruption of the catalysis of both LPL and PDE substrates, just as shown experimentally (Fig. 4D).
Fig. 4.
Molecular surfaces involved in catalysis, substrate binding, and substrate/product access in ATX. (A) The three binding surfaces in the cocrystal of HA155 in ATX. The catalytic center surface with the two Zn2+ ions is marked by the blue dotted line. The hydrophobic channel proposed to shuttle the substrate and the product in and out from the catalytic site is marked with a black dashed line. The hydrophobic pocket surface is delineated in the purple dotted-dashed line. (B–D) The energy minimized docked positions calculated by Molecular Operating Environment, Version 2009.10 (Chemical Computing Group Inc., 2012) for compounds 1, 2, and 3, respectively. The orientation of the enzyme is the same in all panels. (E and F) pNP-TMP docked to the complexes of ATX with compound 1 and compound 2, respectively. Note the complete separation of this docked position of pNP-TMP and the two inhibitors. These models explain the lack of inhibition of pNP-TMP hydrolysis by compounds 1 and 2.
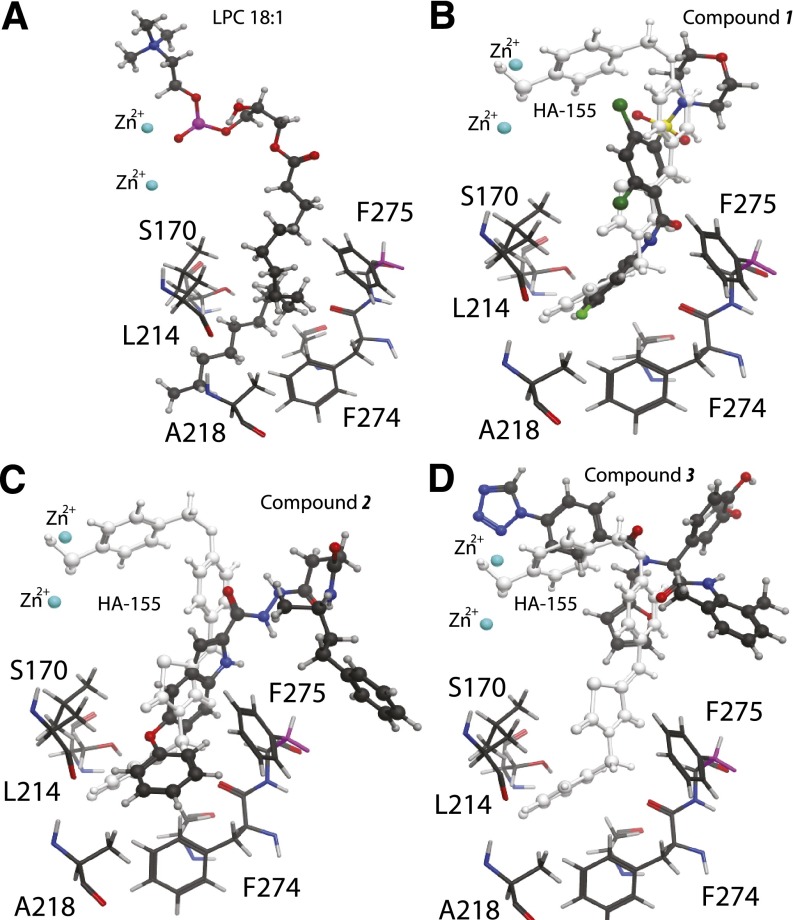
Comparison of the docked positions of compounds 1 and 2 predicted that they are within 4.5 Å of residues Ser170, Phe211, Leu214, Ala218, Phe274, Phe275, Ala305, and Tyr307, which were defined as a hydrophobic pocket that accommodates the lipid tail of the substrate in the crystal structure (Hausmann et al., 2011; Nishimasu et al., 2011). In contrast, compound 3 was anchored within the catalytic binding site indicated by a predicted interaction with residues near the Zn2+ atoms. In silico calculation of energies of interaction of residues lining the hydrophobic pocket with compounds 1 and 2 predict more robust contribution to binding energy than compound 3 (Table 3). Thus, results of our molecular docking suggest that interactions with residues of the hydrophobic pocket play a fundamental role in the ATX inhibitory activity of compounds 1 and 2.
TABLE 3.
Modeled energy of interactions (kJ/mol) and distances (Å) between selected ATX residues and inhibitors
Energy of interaction between individual residues and inhibitors calculated with the Interaction Forces and Energies module of Molecular Operating Environment for the optimized enzyme-inhibitor complexes.
| Residue | Compound 1 Energy Distance | Compound 2 Energy Distance | Compound 3 Energy Distance | |||
|---|---|---|---|---|---|---|
| Tyr307 | −14.51 | 1.78 | −4.65 | 3.00 | −3.12 | 1.49 |
| Phe275 | −6.02 | 1.74 | −8.67 | 1.66 | −4.85 | 3.12 |
| Leu214 | −3.74 | 3.87 | −3.11 | 2.74 | −0.36 | 4.29 |
| Phe274 | −1.17 | 1.70 | −2.17 | 2.88 | NA | 8.31 |
| Thr210 (catalytic site) | NA | 4.90 | NA | 6.79 | −4.77 | 3.45 |
| Phe211 | −1.07 | 2.40 | −3.69 | 1.95 | −30.55 | 2.62 |
| Ala218 | −0.95 | 2.14 | −0.08 | 1.66 | NA | 11.41 |
| Ala305 | −0.92 | 2.82 | −0.94 | 3.80 | NA | 9.74 |
| Ser170 | −0.34 | 2.26 | −0.02 | 4.63 | NA | 7.60 |
NA, not applicable because distance is > 4.5 Å.
Mutagenesis of Hydrophobic Residues Predicted to Interact with the Single Inhibitor Compounds.
Although in silico predictions suggested Tyr307 and Phe211 have the strongest binding energy, when mutating these residues we encountered problems with lack of expression and/or activity of the ATX mutants (Supplemental Fig. 2) as noted previously (Hausmann et al., 2011; Nishimasu et al., 2011). From the dock simulation we hypothesized a possible amino-aromatic interaction of Phe275 with regions of the inhibitors.
Thus, Phe275 was selected as a target hydrophobic residue, while Tyr83 was selected as a target residue in the vicinity, but outside of the hydrophobic channel and the catalytic site (Fig. 5). We hypothesize that these two mutants would have different sensitivity to the inhibitors. Specifically, the Y83A, 10.28 Å away, should maintain wild-type properties when challenged with any of the inhibitors, whereas the single inhibitors should be more affected by the F275A replacement (Fig. 5). Replacement of Phe275 and Tyr83 with alanine proved to be successful by yielding an enzyme that was active, cleaving LPL and PDE substrates (Supplemental Fig. 2). Characterization of the F275A mutant showed a greater loss in inhibitory potency by compound 1 than by compound 3 (Table 4). The inhibitory potency of compound 2 also decreased by 3-fold. Furthermore, characterization of the Y83A mutant supported our hypothesis by revealing that this mutation had no significant impact on the effect of either type of inhibitor. The differential effect of the Phe275 mutation on the inhibitory potency of the single inhibitors lends support to the unique interactions these compounds have with the surface of the hydrophobic pocket.
Fig. 5.
Landmark residues (sticks) lining the hydrophobic pocket based on the ATX crystal structure. Docked substrate and inhibitors are shown in ball and stick models. (A) LPC 18:1 substrate, (B) Compound 1, (C) Compound 2, and (D) Compound 3 docked and energy minimized using the Molecular Operating Environment, Version 2009.10 software (Chemical Computing Group Inc., 2012). In (B) and (C) the HA155 inhibitor-bound crystal structure is overlayed in white. Phe275 is shown for reference in all panels. Note the close proximity of Phe275 to docked compounds 1 and 2 and the relatively large distance to compound 3.
TABLE 4.
Characterization of ATX mutants
Compounds 1 and 2 had no effect on pNP-TMP hydrolysis.
| Compound 1 |
Compound 2 |
Compound 3 |
||||
|---|---|---|---|---|---|---|
| Substrate ATX Mutant | FS-3 IC50 | pNP-TMP IC50 | FS-3 IC50 | pNP-TMP IC50 | FS-3 IC50 | pNP-TMP IC50 |
| µM | ||||||
| WT | 3.0 ± 0.01 | NE | 0.13 ± 0.00 | NE | 0.41 ± 0.01 | 0.15 ± 0.01 |
| F275A | > 10.00 | NE | 0.32 ± 0.01 | NE | 2.5 ± 0.06 | 1.58 ± 0.05 |
| Y83A | 0.15 ± 0.01 | NE | 0.06 ± 0.00 | NE | 0.19 ± 0.03 | 0.11 ± 0.01 |
IC50, concentration that inhibits cleavage by 50%; NE, no effect.
Characterization of the ATX Inhibitors on Other Pharmacologically Relevant Targets.
As a member of the NPP family of enzymes, ATX shares homology with NPP6 and NPP7, which are type C phospholipases. NPP6 cleaves phosphocholine from LPC, sphingosylphosphorylcholine, and glycerophosphorylcholine (Sakagami et al., 2005). NPP7 hydrolyzes sphingomyelin to generate ceramide and can also cleave phosphocholine from LPC and platelet-activating factor to generate monoacyl- and alkyl-acetyl glycerols (Duan et al., 2003; Wu et al., 2006). The selectivity of the ATX inhibitors was examined against NPP6 and NPP7. None of the inhibitors had a significant effect on NPP6/7-mediated hydrolysis of the synthetic substrate, p-nitrophenylphosphocholine.
To determine potential off-target effects that the three hit compounds may have we submitted them to the National Institute of Mental Health PDSP (http://pdsp.med.unc.edu/indexR.html). The compounds were tested for modulation of 32 types and subtypes of GPCRs, including seven lysophospholipid receptors—including sphingosine-1-phosphate (S1P)2/3/4/5 and LPA1/2/3—16 orphan receptors, six transporters, and three ion channels that included the HERG2 channel. The results of this extensive screen showed no activating or inhibitory action by compounds 1 and 3 on any of these targets up to 10 µM, the highest concentration tested. We also extended the characterization of the three ATX inhibitors to the non-EDG family LPA receptors LPA4 and LPA5 and found no agonist, antagonist, or inhibitory action up to 10 µM, the highest concentration tested (data not shown).
Effect of the ATX Inhibitors on A2058 Melanoma Cell Invasion.
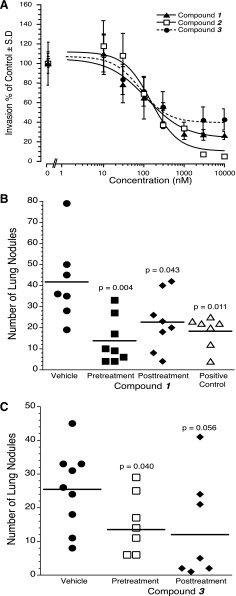
ATX was originally discovered as a proinvasive factor in the culture supernatant of A2058 melanoma cells (Stracke et al., 1992). For this reason, we elected to evaluate the three new inhibitors on the invasion of this human melanoma cell line. We applied recombinant 0.3 nM ATX plus 1 µM of 18:1 LPC with or without increasing concentrations of the inhibitors and quantified the invasion of matrigel-coated BD BioCoat chambers 16 hour later. The dose response-curves generated (Fig. 6A) showed comparable nanomolar IC50 values regardless of the blocking site of the compound in ATX. Specifically, the IC50 values were 118.79 ± 62.9 nM, 153.05 ± 59.5 nM, and 85.35 ± 44.6 nM, for compounds 1, 2, and 3, respectively.
Fig. 6.
Effects of ATX inhibitor hits on cancer cell invasion (A) and lung metastasis compound 1 (B) and compound 3 (C). (A) A2058 human melanoma cells were applied to matrigel-coated BD BioCoat chambers. 0.3 nM ATX plus 1 µM of 18:1 LPC with or without increasing concentrations of the inhibitors was applied, and invasion was quantified 16 hour later. Note that all three compounds showed a dose-dependent inhibition of A2058 melanoma invasion. C57BL/6 mice were inoculated with 7.5 × 104 B16-F10 melanoma cells via the tail vein. Treatment with either compound 1 (B), compound 3 (C), or 4-pentadecylbenzylphosphonic acid (positive control) at 30 µg per mouse via intraperitoneal injection was started one day before (pretreatment) or after (post-treatment) the B16-F10 inoculation, and continued daily for an additional 10 days. Control mice were injected with vehicle phosphate-buffered saline with 10% polyethylene glycol vehicle. The mice were sacrificed on postinoculation day 21 and the lung nodules were counted. P values were calculated using Student’s t test relative to the vehicle group.
Effect of the ATX Inhibitors on B16-F10 Melanoma Cell Metastasis.
After injection into C57BL/6 mice via the tail vein, the syngeneic B16-F10 melanoma cells metastasize primarily to the lungs in an ATX-dependent manner (Baker et al., 2006; Gupte et al., 2011; Gotoh et al., 2012). We showed that in this model administration of an ATX inhibitor up to 48 hours postinoculation of the melanoma cells still causes a significant reduction in the number of metastatic lung nodules, indicating a role for ATX in the colonization and seeding of the lungs with melanoma cells (Gotoh et al., 2012). We tested compounds 1 and 3 by treating mice 1 day prior or 1 day after inoculation of B16-F10 melanoma cells via the tail vein, and continued for an additional 10 days. We also included 4-pentadecyl benzylphosphonic acid, a previously validated ATX inhibitor in the pre- and postinoculation paradigm (Gupte et al., 2011). The single inhibitor compound 1 caused a significant reduction in the number of lung nodules in both treatment paradigms (Fig. 6B). The dual inhibitor compound 3 (Fig. 6C) caused a significant reduction in the metastatic nodules in the pretreatment paradigm but did not reach significance in the post-inoculation paradigm (P = 0.056). 4-Pentadecyl benzylphosphonic acid, which is also a dual-type inhibitor, caused a significant reduction in the number of metastatic nodules in the lungs (Gupte et al., 2010; Gotoh et al., 2012). Altogether, these findings support the hypothesis that inhibition of the ATX hydrophobic pocket can have similar blocking effects of melanoma metastasis in vivo as inhibiting the ATX active site.
Discussion
In this study we identified a surface of the hydrophobic pocket of ATX as a target site of inhibitors of enzymatic activity. The primary objective of the present study was to identify structurally diverse nonlipid inhibitors of ATX. We applied an HTS strategy using purified recombinant human β-ATX with the LPC-like substrate FS-3, which distinguishes our study from two previous HTS screens that used the PDE substrate pNP-TMP to screen a 13,000 member proprietary library with a partially purified mouse β-ATX from 3T3-F442A adipocytes (Ferry et al., 2008) or a 40,000 compound library using human recombinant ATX (Albers et al., 2010). Docking simulations of FS-3 into the ATX structure showed (Supplemental Fig. 3) that the hydrocarbon chain of this substrate extends into the hydrophobic pocket, whereas the fluorochrome occupies a different space distinct from the hydrophobic channel of the enzyme. This model suggests that if one were to use pNP-TMP-like nucleotide substrates for HTS, all compounds that bind to the hydrophobic pocket would be missed. Thus, our results favor the use of LPC-like substrates that engage both the active site and the hydrophobic pocket of ATX. Ferry et al. (2008) noted that in their hands ATX, once purified, precipitated, which led them to apply hexadecyl trimethylammonium bromide or 2-methyl-2,4-pentanediol detergent in their assays. These detergents did have an impact on the IC50 values they detected for their inhibitors. In our case the use of the human recombinant ATX did not present solubility problems, and the assay was linear up to 6 hours, the latest point tested. Nonetheless, we also retested our hits in the presence of 0.01% Triton X-100, not because of the poor stability of the enzyme but rather to eliminate self-aggregating compounds leading to false positives in the ATX inhibition assay (Feng et al., 2005). Our three-tiered assays (Fig. 2; Table 1) using LPC, PDE-, and LPC-like substrates consistently identified several inhibitor hits against the different types of substrates. The ∼10,000 compound diversity set was selected to be representative of the full compound library after application of a range of property and structural filters, affording a set of compounds significantly more drug-like and diverse. Chemoinformatics calculations and comparisons between compounds were performed within Pipeline Pilot (Pipeline Pilot Ver. 8.0.1.600, 2010; Accelrys Software Inc., San Diego, CA). The UC Compound Library was evaluated according to the popular property filters (Lipinski et al., 2001; Veber et al., 2002; Hann and Oprea, 2004) targeting improved oral bioavailability or starting points for optimization toward improved oral bioavailability (i.e., fragment or lead-like), and from this evaluation the diversity set was compiled. The 9,652-compound diversity set we used is adherent to three out of four Lipinski Rules (Lipinski et al., 2001) drug-like property constraints.
Even though the hit rate in the primary screen was only ∼2% using the 10 µM concentration and 50% inhibition criterion, the HTS identified 198 structurally diverse nonlipid inhibitor compounds. These compounds fall into multiple structural categories, and we have already noticed that some structural features, such as presence of an aromatic sulfonamide group, show up in a disproportionately high number of the initial hits. This list of hits is now being used for the development of a structure-based ATX inhibitor pharmacophore and will be discussed in a future report.
Tests conducted using pNP-TMP and the LPC-like substrates distinguished two groups of compounds. The first group inhibited the LPL activity against different naturally occurring LPC species (Table 1) and LPC-like substrates that included FS-3 and ADMAN-LPC (Fig. 3C). In contrast, the second type of compounds inhibited both pNP-TMP and the LPL substrates. The mechanism of inhibition, regardless of the type of substrate, for both types of compounds was consistent with the results of computational docking. Nishimasu and colleagues (Nishimasu et al., 2012) proposed three important molecular interaction surfaces in ATX (Fig. 4A). These included the active site, a hydrophobic pocket harboring the lipid tail, and a channel through which the substrate and the product can shuttle in and out from the active site. Our docking simulations place the dual inhibitor compound 3 in the vicinity of the active site overlapping with the space occupied by the covalent inhibitor HA155 in the crystal structure (Figs. 3D and 4D). The docked position of compound 3 provides a rational explanation for the inhibition of both types of substrates by blocking their interaction with the active site. In contrast, compounds 1 and 2 docked into the hydrophobic pocket sufficiently far away from the active site to allow the hypothesis that these inhibitors selectively block the hydrolysis of lipid substrates (Fig. 5, E and F).
The hydrophobic pocket of ATX is lined by residues Ile168, Phe211, Leu217, Ala218, Leu260, Phe274, Phe275, Trp261, and Met513 (all human ATX residue numbers, [Nishimasu et al., 2011]) (Fig. 6). Based on our docking simulation, Phe275 has an amino-aromatic interaction with compound 1. This is demonstrated by the strong effect of the Phe275 mutant on the potency of compound 1 and the absence of effect on the potency of compound 3. Analysis of the F275A mutant ATX showed that it retained nearly wild-type catalytic activity toward FS-3 and pNP-TMP substrates for compound 3. However, it lost its inhibition by compound 1, and its inhibition by compound 2 was also diminished (Table 4).
The hits we identified in the DDC library have fulfilled one of our objectives for compounds that are nonlipid with partition coefficients in the range of 2.99–3.98, which makes them more water-soluble than previously identified lipid-like inhibitors with logP values greater than 5 (Albers and Ovaa, 2012). Interestingly, the two single inhibitors had lower logP values than the dual inhibitor compound 3. This observation suggests that compounds with reasonable water solubility can access the hydrophobic binding pocket of ATX.
We had compounds 1 and 3 tested for off-target effects using the Psychoactive Drug Screening Program provided by the National Institute of Mental Health. This extensive screen turned up no activity of these compounds at the 30-plus GPCRs, ion channels, and transporter targets that included the S1P GPCRs S1P2, S1P3, S1P4, and S1P5; the LPA1, LPA2, and LPA3 LPA receptors; and the HERG-2 channel. The lack of agonist or antagonist action of our new ATX inhibitors on LPA and sphingosine-1-phosphate receptors, combined with their lack of inhibitory activity at NPP6 and NPP7 underlines the specificity of these compounds to ATX and paves the way toward their use in cell-based and in vivo disease models.
The solubility, stability, and hence the bioavailability of several earlier ATX inhibitors have presented problems for their further development (Ferry et al., 2008; Albers et al., 2010; Albers and Ovaa, 2012). ATX was originally isolated as a factor that promoted melanoma invasion (Stracke et al., 1992). To assess the utility of the two types of inhibitors we tested their ability to inhibit invasion in a Boyden chamber cell-based assay. Our findings with both types of inhibitors confirmed that blocking the enzyme inhibits the invasion of A2058 melanoma cells. At this early stage of the characterization of these novel ATX inhibitors we deemed it premature to conduct pharmacokinetic studies; however, we tested compounds 1 and 3 in the lung colonization model using B16-F10 syngeneic melanoma cells. These results indicated that the hydrophobic pocket inhibitor compound 1 was effective in significantly reducing the number of lung metastases when applied 1 day prior or 1 day after inoculation with the tumor cells. This observation extends our previous reports that inhibition of ATX reduces the seeding of lung metastases in this animal model. This is the first set of results that shows the antimetastatic utility of a compound that blocks the hydrophobic pocket without interfering with the PDE activity of the enzyme. It is in line with the previous hypothesis that the PDE activity of ATX does not play a role in the prometastatic action of this enzyme in vivo.
The present results taken together establish the utility of the compounds 1 and 2 for in vitro studies. Nonetheless, we view these compounds as primary hits that will have to undergo lead optimization to reach broad applicability in vivo.
Supplementary Material
Acknowledgments
The authors thank the National Institute of Mental Health Psychoactive Drug Screening Program for providing human receptor profiling and Ki data. The NIMH PDSP is directed by B. Roth at the University of North Carolina at Chapel Hill and Project Officer J. Driscol at NIMH, Bethesda, MD, Contract No. NO1MH32004.
Abbreviations
- ADMAN-LPC
analog 3-acyl-7-dimethylaminonaphthyl–1-LPC
- ATX
autotaxin
- BSA
bovine serum albumin
- compound 1
2,4-dichloro-N-(3-fluorophenyl)-5-(4-morpholinylsulfonyl) benzamide
- compound 2
4-oxo-4-{2-[(5-phenoxy-1H-indol-2-yl)carbonyl]hydrazino}-N-(4-phenylbutan-2-yl)butanamide
- compound 3
N-(2,6-dimethylphenyl)-2-[N-(2-furylmethyl)(4-(1,2,3,4-tetraazolyl)phenyl)carbonylamino]-2-(4-hydroxy-3-methoxyphenyl) acetamide
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- FS-3
fluorogenic substrate 3
- GPCR
G protein–coupled receptor
- HA155
(Z)-4-((4-((3-(4-fluorobenzyl)-2,4-dioxo-1,3-thiazolan-5-yliden)-methyl)phenoxy)methyl)benzeneboronic acid ATX inhibitor IV
- HTS
high-throughput screening
- LPA
lysophosphatidic acid
- LPC
lysophosphatidylcholine
- LPL
lysophospholipase
- NPP
nucleotide pyrophosphatase/phosphodiesterase
- PDSP
Psychoactive Drug Screening Program
- PDE
phosphodiesterase
- pNP-TMP
p-nitrophenyl thymidine 5′-monophosphate
- S1P
sphingosine 1-phosphate
- UC-DDC
University of Cincinnati Drug Discovery Center
Authorship Contributions
Participated in research design: Fells, Lee, Fujiwara, Norman, Tsukahara, Patil, Kirby, Nelson, Parrill, Miller, Tigyi.
Conducted experiments: Fells, Lim, Liu, Lee, Fujiwara, Norman, Tsukahara, Patil, Kirby, Nelson.
Contributed new reagents or analytic tools: Fells, Norman, Tsukahara, Patil, Kirby, Nelson, Seibel, Papoian, Bittman, Baker.
Performed data analysis: Fells, Lee, Fujiwara, Lim, Norman, Kirby, Nelson, Seibel, Parrill, Tigyi.
Wrote or contributed to the writing of the manuscript: Fells, Lee, Fujiwara, Norman, Parrill, Baker, Tigyi.
Footnotes
This work was supported by the National Institutes of Health National Cancer Institute [Grant CA092160]; the American Cancer Society [Grant 122059-PF-12-107-01-CDD]; and the Van Vleet Endowment.
 This article has supplemental material available at mol.aspetjournals.org.
This article has supplemental material available at mol.aspetjournals.org.
References
- Albers HM, Dong A, van Meeteren LA, Egan DA, Sunkara M, van Tilburg EW, Schuurman K, van Tellingen O, Morris AJ, Smyth SS,, et al. (2010) Boronic acid-based inhibitor of autotaxin reveals rapid turnover of LPA in the circulation. Proc Natl Acad Sci USA 107:7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers HM, Ovaa H. (2012) Chemical evolution of autotaxin inhibitors. Chem Rev 112:2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DL, Fujiwara Y, Pigg KR, Tsukahara R, Kobayashi S, Murofushi H, Uchiyama A, Murakami-Murofushi K, Koh E, Bandle RW,, et al. (2006) Carba analogs of cyclic phosphatidic acid are selective inhibitors of autotaxin and cancer cell invasion and metastasis. J Biol Chem 281:22786–22793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindley DN, Lin FT, Tigyi GJ. (2013) Role of the autotaxin-lysophosphatidate axis in cancer resistance to chemotherapy and radiotherapy. Biochim Biophys Acta 1831:74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemical Computing Group Inc (2012) Molecular Operating Environment, MOE, Montreal. [Google Scholar]
- Clair T, Aoki J, Koh E, Bandle RW, Nam SW, Ptaszynska MM, Mills GB, Schiffmann E, Liotta LA, Stracke ML. (2003) Autotaxin hydrolyzes sphingosylphosphorylcholine to produce the regulator of migration, sphingosine-1-phosphate. Cancer Res 63:5446–5453 [PubMed] [Google Scholar]
- David M, Wannecq E, Descotes F, Jansen S, Deux B, Ribeiro J, Serre CM, Grès S, Bendriss-Vermare N, Bollen M,, et al. (2010) Cancer cell expression of autotaxin controls bone metastasis formation in mouse through lysophosphatidic acid-dependent activation of osteoclasts. PLoS ONE 5:e9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan RD, Bergman T, Xu N, Wu J, Cheng Y, Duan J, Nelander S, Palmberg C, Nilsson A. (2003) Identification of human intestinal alkaline sphingomyelinase as a novel ecto-enzyme related to the nucleotide phosphodiesterase family. J Biol Chem 278:38528–38536 [DOI] [PubMed] [Google Scholar]
- Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. (2005) High-throughput assays for promiscuous inhibitors. Nat Chem Biol 1:146–148 [DOI] [PubMed] [Google Scholar]
- Ferry G, Moulharat N, Pradère JP, Desos P, Try A, Genton A, Giganti A, Beucher-Gaudin M, Lonchampt M, Bertrand M,, et al. (2008) S32826, a nanomolar inhibitor of autotaxin: discovery, synthesis and applications as a pharmacological tool. J Pharmacol Exp Ther 327:809–819 [DOI] [PubMed] [Google Scholar]
- Gierse J, Thorarensen A, Beltey K, Bradshaw-Pierce E, Cortes-Burgos L, Hall T, Johnston A, Murphy M, Nemirovskiy O, Ogawa S,, et al. (2010) A novel autotaxin inhibitor reduces lysophosphatidic acid levels in plasma and the site of inflammation. J Pharmacol Exp Ther 334:310–317 [DOI] [PubMed] [Google Scholar]
- Gijsbers R, Aoki J, Arai H, Bollen M. (2003) The hydrolysis of lysophospholipids and nucleotides by autotaxin (NPP2) involves a single catalytic site. FEBS Lett 538:60–64 [DOI] [PubMed] [Google Scholar]
- Gotoh M, Fujiwara Y, Yue J, Liu J, Lee S, Fells J, Uchiyama A, Murakami-Murofushi K, Kennel S, Wall J,, et al. (2012) Controlling cancer through the autotaxin-lysophosphatidic acid receptor axis. Biochem Soc Trans 40:31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte R, Patil R, Liu J, Wang Y, Lee SC, Fujiwara Y, Fells J, Bolen AL, Emmons-Thompson K, Yates CR,, et al. (2011) Benzyl and naphthalene methylphosphonic acid inhibitors of autotaxin with anti-invasive and anti-metastatic activity. ChemMedChem 6:922–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte R, Siddam A, Lu Y, Li W, Fujiwara Y, Panupinthu N, Pham TC, Baker DL, Parrill AL, Gotoh M,, et al. (2010) Synthesis and pharmacological evaluation of the stereoisomers of 3-carba cyclic-phosphatidic acid. Bioorg Med Chem Lett 20:7525–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann MM, Oprea TI. (2004) Pursuing the leadlikeness concept in pharmaceutical research. Curr Opin Chem Biol 8:255–263 [DOI] [PubMed] [Google Scholar]
- Hausmann J, Kamtekar S, Christodoulou E, Day JE, Wu T, Fulkerson Z, Albers HM, van Meeteren LA, Houben AJ, van Zeijl L,, et al. (2011) Structural basis of substrate discrimination and integrin binding by autotaxin. Nat Struct Mol Biol 18:198–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeglund AB, Bostic HE, Howard AL, Wanjala IW, Best MD, Baker DL, Parrill AL. (2010) Optimization of a pipemidic acid autotaxin inhibitor. J Med Chem 53:1056–1066 [DOI] [PubMed] [Google Scholar]
- Houben AJ, Moolenaar WH. (2011) Autotaxin and LPA receptor signaling in cancer. Cancer Metastasis Rev 30:557–565 [DOI] [PubMed] [Google Scholar]
- Ikeda H, Yatomi Y. (2012) Autotaxin in liver fibrosis. Clin Chim Acta 413:1817–1821 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Okabe T, Okudaira S, Nishimasu H, Ishitani R, Kojima H, Nureki O, Aoki J, Nagano T. (2013) Screening and X-ray crystal structure-based optimization of autotaxin (ENPP2) inhibitors, using a newly developed fluorescence probe. ACS Chem Biol [published ahead of print]. [DOI] [PubMed] [Google Scholar]
- Kehlen A, Englert N, Seifert A, Klonisch T, Dralle H, Langner J, Hoang-Vu C. (2004) Expression, regulation and function of autotaxin in thyroid carcinomas. Int J Cancer 109:833–838 [DOI] [PubMed] [Google Scholar]
- Koh E, Clair T, Woodhouse EC, Schiffmann E, Liotta L, Stracke M. (2003) Site-directed mutations in the tumor-associated cytokine, autotaxin, eliminate nucleotide phosphodiesterase, lysophospholipase D, and motogenic activities. Cancer Res 63:2042–2045 [PubMed] [Google Scholar]
- Kremer AE, van Dijk R, Leckie P, Schaap FG, Kuiper EM, Mettang T, Reiners KS, Raap U, van Buuren HR, van Erpecum KJ,, et al. (2012) Serum autotaxin is increased in pruritus of cholestasis, but not of other origin, and responds to therapeutic interventions. Hepatology 56:1391–1400 [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26 [DOI] [PubMed] [Google Scholar]
- Mills GB, Moolenaar WH. (2003) The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer 3:582–591 [DOI] [PubMed] [Google Scholar]
- Mize CD, Abbott AM, Gacasan SB, Parrill AL, Baker DL. (2011) Ligand-based autotaxin pharmacophore models reflect structure-based docking results. J Mol Graph Model 31:76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenaar WH, Houben AJ, Lee SJ, van Meeteren LA. (2013) Autotaxin in embryonic development. Biochim Biophys Acta 1831:13–19 [DOI] [PubMed] [Google Scholar]
- Nikitopoulou I, Oikonomou N, Karouzakis E, Sevastou I, Nikolaidou-Katsaridou N, Zhao Z, Mersinias V, Armaka M, Xu Y, Masu M,, et al. (2012) Autotaxin expression from synovial fibroblasts is essential for the pathogenesis of modeled arthritis. J Exp Med 209:925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishitani R, Aoki J, Nureki O. (2012) A 3D view of autotaxin. Trends Pharmacol Sci 33:138–145 [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Okudaira S, Hama K, Mihara E, Dohmae N, Inoue A, Ishitani R, Takagi J, Aoki J, Nureki O. (2011) Crystal structure of autotaxin and insight into GPCR activation by lipid mediators. Nat Struct Mol Biol 18:205–212 [DOI] [PubMed] [Google Scholar]
- North EJ, Howard AL, Wanjala IW, Pham TC, Baker DL, Parrill AL. (2010) Pharmacophore development and application toward the identification of novel, small-molecule autotaxin inhibitors. J Med Chem 53:3095–3105 [DOI] [PubMed] [Google Scholar]
- Nouh MA, Wu XX, Okazoe H, Tsunemori H, Haba R, Abou-Zeid AM, Saleem MD, Inui M, Sugimoto M, Aoki J, et al. (2009) Expression of autotaxin and acylglycerol kinase in prostate cancer: association with cancer development and progression. Cancer Sci 100:1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami H, Aoki J, Natori Y, Nishikawa K, Kakehi Y, Natori Y, Arai H. (2005) Biochemical and molecular characterization of a novel choline-specific glycerophosphodiester phosphodiesterase belonging to the nucleotide pyrophosphatase/phosphodiesterase family. J Biol Chem 280:23084–23093 [DOI] [PubMed] [Google Scholar]
- Saunders LP, Ouellette A, Bandle R, Chang WC, Zhou H, Misra RN, De La Cruz EM, Braddock DT. (2008) Identification of small-molecule inhibitors of autotaxin that inhibit melanoma cell migration and invasion. Mol Cancer Ther 7:3352–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Cœur PD, Ferguson D, Morin P, Jr, Touaibia M. (2013) PF-8380 and closely related analogs: synthesis and structure-activity relationship towards autotaxin inhibition and glioma cell viability. Arch Pharm (Weinheim) 346:91–97 [DOI] [PubMed] [Google Scholar]
- Stracke ML, Krutzsch HC, Unsworth EJ, Arestad A, Cioce V, Schiffmann E, Liotta LA. (1992) Identification, purification, and partial sequence analysis of autotaxin, a novel motility-stimulating protein. J Biol Chem 267:2524–2529 [PubMed] [Google Scholar]
- Tager AM. (2012) Autotaxin emerges as a therapeutic target for idiopathic pulmonary fibrosis: limiting fibrosis by limiting lysophosphatidic acid synthesis. Am J Respir Cell Mol Biol 47:563–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigyi G. (2010) Aiming drug discovery at lysophosphatidic acid targets. Br J Pharmacol 161:241–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Matsunaga H, Olaposi OI, Nagai J. (2013) Lysophosphatidic acid: chemical signature of neuropathic pain. Biochim Biophys Acta 1831:61–73 [DOI] [PubMed] [Google Scholar]
- Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623 [DOI] [PubMed] [Google Scholar]
- Wu J, Nilsson A, Jönsson BA, Stenstad H, Agace W, Cheng Y, Duan RD. (2006) Intestinal alkaline sphingomyelinase hydrolyses and inactivates platelet-activating factor by a phospholipase C activity. Biochem J 394:299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JM, Xu Y, Skill NJ, Sheng H, Zhao Z, Yu M, Saxena R, Maluccio MA. (2010) Autotaxin expression and its connection with the TNF-alpha-NF-kappaB axis in human hepatocellular carcinoma. Mol Cancer 9:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida K, Kurikawa Y, Shimizu T, Ishii S. (2013) Current progress in non-Edg family LPA receptor research. Biochim Biophys Acta 1831:33–41 [DOI] [PubMed] [Google Scholar]
- Yang SY, Lee J, Park CG, Kim S, Hong S, Chung HC, Min SK, Han JW, Lee HW, Lee HY. (2002) Expression of autotaxin (NPP-2) is closely linked to invasiveness of breast cancer cells. Clin Exp Metastasis 19:603–608 [DOI] [PubMed] [Google Scholar]
- Yang Y, Mou Lj, Liu N, Tsao MS. (1999) Autotaxin expression in non-small-cell lung cancer. Am J Respir Cell Mol Biol 21:216–222 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.