Abstract
The interaction between the gp120 and gp41 subunits of the human immunodeficiency virus envelope glycoprotein serves to stabilize the virion form of the complex and to transmit receptor-induced conformational changes in gp120 to trigger the membrane fusion activity of gp41. In this study, we used site-directed mutagenesis to identify amino acid residues in the central ectodomain of gp41 that contribute to the stability of the gp120-gp41 association. We identified alanine mutations at six positions, including four tryptophan residues, which result in mutant envelope glycoprotein complexes that fail to retain gp120 on the cell surface. These envelope glycoproteins readily shed their gp120 and are unable to mediate cell-cell fusion. These findings suggest an important role for the conserved bulky hydrophobic residues in stabilizing the gp120-gp41 complex.
The envelope glycoprotein of human immunodeficiency virus (HIV) type 1 promotes the sequential processes of binding to host cell receptors and fusion of the viral and cellular membranes to mediate viral entry. On the virion surface, the envelope glycoprotein complex exists as a trimeric spike comprising the surface and receptor-binding subunit gp120, which is bound through a noncovalent association with the transmembrane (TM) subunit gp41 (reviewed in reference 24). In this complex, the gp41 subunit is thought to exist in a metastable state that is stabilized by association with gp120. As a result of the sequential binding to CD4 and coreceptor (reviewed in references 2 and 40), conformational changes in gp120 trigger a major structural reorganization of the gp41 glycoprotein (6). Numerous studies lead to a model in which the N- and C-terminal heptad repeat regions of gp41 refold to form α-helices that are ultimately packed to produce the fusion-active six-helix bundle (reviewed in references 10, 31, and 35 and references therein). The formation of this highly stable core in gp41 is thought to contribute to overcoming the energy barrier to membrane fusion (11, 16, 36).
The atomic structure of the native gp120-gp41 complex is unknown, in part because of the intrinsic lability of the gp120-gp41 association (3). This instability is reflected in the facile shedding of gp120 from the cell and virion surface (26). Stabilization of this association through genetic means is thought to represent a promising approach to the production of envelope glycoprotein immunogens (3, 41), and several laboratories have sought to define amino acid residues involved in maintaining the native structure.
Broad outlines of the gp120-gp41 interface in the native envelope glycoprotein complex have emerged. In early studies, Sodroski and colleagues applied mutagenesis to the gp120 molecule to identify the N-terminal (C1) and C-terminal (C5) regions as important for the association with gp41 (14, 39). Within gp41, key determinants of the association with gp120 have been demonstrated in the central ectodomain region (5, 25). This region is highly conserved among HIV isolates (19) and spans the region between the N- and C-terminal heptad repeats. Central in this region is a disulfide-bonded loop that may facilitate gp41 chain reversal in the folding of the fusion-active six-helix bundle. In type C retroviruses, this central region of the TM glycoprotein is joined to the C-terminal region of the surface glycoprotein by a disulfide bridge (30). In HIV, scanning cysteine mutagenesis of the C1 and C5 regions of gp120 and of the disulfide-bonded loop region of gp41 has identified multiple sites at which paired cysteine side chains are able to bridge the gp120-gp41 interface (3). One such mutant, SOS gp140 (3), is significantly stabilized in its native oligomeric form. The recent finding that this complex can be indirectly stabilized through specific mutation in the N-terminal helix of gp41 (SOSIP gp140) emphasizes the dynamic nature of the gp120-gp41 interface (29).
Interactions at the gp120-gp41 interface are therefore critical in maintaining the structure of the envelope glycoprotein complex and in modulating its function in viral entry. In order to further probe the structure and function of this interface, we applied scanning mutagenesis within the central ectodomain of gp41.
Results and discussion.
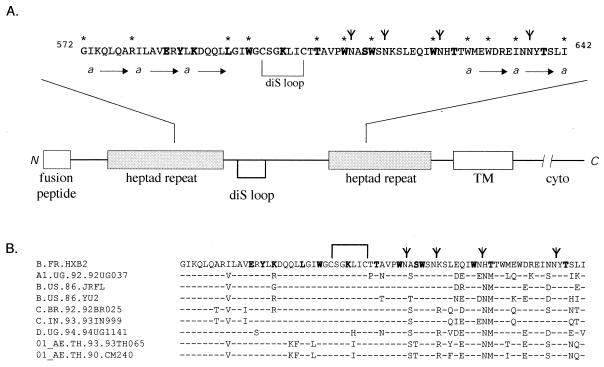
The central ectodomain of gp41 shows remarkable conservation among diverse isolates of HIV within the disulfide-bonded loop and at hydrophobic residues and a significantly lesser degree of identity at hydrophilic amino acids and near the glycosylation sites clustered in the C-terminal region (Fig. 1). In our studies, we focused on conserved hydrophobic residues and included additional positions within the central ectodomain region. The present mutations bridge regions previously examined in our studies of the N- and C-terminal heptad-repeat regions (23, 34).
FIG. 1.
Central ectodomain of HIV gp41. (A) Schematic diagram of gp41 and the central ectodomain. The gp41 glycoprotein is drawn, and the following key features are shown: the N-terminal fusion peptide, the N- and C-terminal heptad-repeat regions, the disulfide-bonded loop (diS loop), and the TM and cytoplasmic (cyto) domains. The heptad-repeat motifs are designated by the “a” position and arrows, and glycosylation sites (N-X-T/S) are designated by a modified “Y.” Amino acid residues changed in these studies are in boldface. Amino acid positions that are important for the gp120-gp41 association (this study and others [5, 23, 25, 34]) are indicated by asterisks and in the text. (B) Comparison of central gp41 ectodomain amino acid sequences (19). The Epilign program available at the Los Alamos National Laboratory HIV database (http://hiv-web.lanl.gov) was used to prepare the alignment, and representative HIV isolates are shown.
In choosing sites for mutagenesis, we based certain choices on the known structure of the fusion-active six-helix bundle (7, 33, 36) and the larger trimer-of-hairpins gp41 ectodomain structure (4, 43). Although it is unlikely that many of the structural elements in the fusion-active core pre-exist in the native gp120-gp41 complex (see references 10 and 35), we wanted to examine whether this known structure might provide insight toward probing of the native complex. In this context, we chose to mutate amino acids that lie buried within the N-terminal coiled coil (E584 and Y586), as well as those that are exposed on its surface (K588, L593, I595, and W596) (see references 4, 25, and 43). To assess the role of these residues in the maintenance of the native gp120-gp41 association, we changed each amino acid to alanine, a small helix-inducing residue that contributes little to protein-protein interactions (9). Similarly, within the C-terminal region, T606, W610, S613, W614, W623, and T639 were changed to alanine and T626 was altered to methionine, a common polymorphism found in HIV isolates (Fig. 1B). The S613A, T626M, and T639A mutations eliminate glycosylation signals, and the tryptophan residues were targeted on the basis of their conservation. The cysteine residues that define the disulfide-bonded loop of gp41 are essential for envelope glycoprotein biosynthesis (32) and were not altered. The effect of a charge at K588 and the highly conserved residue K601 was examined by mutation to glutamate or glutamine. In our studies, certain previously examined mutations were included for completeness, i.e., E584A, T626M, and T639A (5) and L593A and K601E (25).
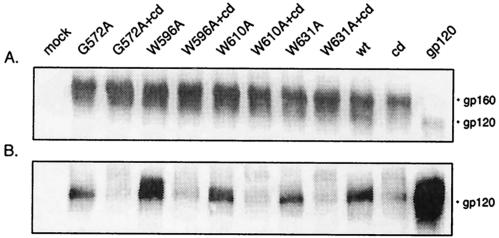
We assessed the stability of the mutant gp120-gp41 complex by determining the relative retention of gp120 on the cell surface. This determination required a careful analysis of envelope glycoprotein biosynthesis in order to exclude ancillary defects in expression or proteolytic maturation. In our analyses, wild-type and mutant envelope glycoproteins were expressed by transient transfection in COS-7 cells. Overall expression levels were found to be similar for all of the envelope glycoproteins examined (Fig. 2A), and in all cases gp120 was shed into the cell culture medium (Fig. 2B). Expression was quantitated from Western blots by chemifluorescence, using a Fuji FLA-3000G imager and Image Gauge software (Fuji), and the analysis of four independent studies is shown in Table 1. The overall expression levels of the mutants relative to the wild-type envelope glycoprotein ranged from 0.84 to 1.13, with an average of 1.03 (± 0.07 [standard deviation]). No significant differences in the relative degree of gp120 shedding were discerned among the mutant envelope glycoproteins (range, 0.93 to 1.83; average, 1.36 ± 0.28). Shedding was, however, reduced to background levels when proteolytic maturation was blocked by a mutation at the furin-like cleavage site in gp120 (REKR to REKT).
FIG. 2.
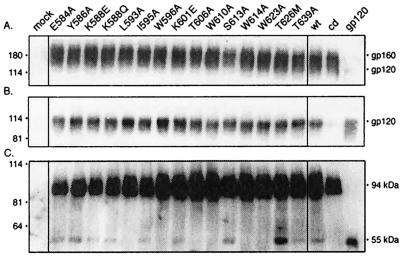
Expression of mutant envelope glycoproteins and gp120 shedding. Mutations were introduced into the HXB2 envelope glycoprotein expression plasmid by QuikChange (Stratagene, Inc.) mutagenesis (23, 34). Mutant and wild-type (wt) envelope glycoproteins were transiently expressed in COS-7 cells upon transfection with FuGene-6 reagent (Roche Molecular Biochemicals). Standards for gp160 and gp120 were obtained by expression of the cleavage-defective (cd) envelope glycoprotein and soluble gp120. (A) Cell lysates were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, and the envelope glycoprotein was visualized by Western blot analysis with anti-gp120 monoclonal antibody Chessie B13 (1) and ECL-Plus detection (Amersham Pharmacia Biotech) (23, 34). A residuum of cell-associated gp120 is seen in cells expressing soluble gp120. (B) Cell culture supernatants were collected, and gp120 was immunoprecipitated with the anti-HIV immunoglobulin HIVIG (28). The cleavage-defective envelope glycoprotein served as a control for gp120 shedding and was subtracted for quantitation. In general, the mutant glycoproteins shed slightly more gp120 than the wild type (1.36 ± 0.28), but significant differences among the mutants were not discerned. (C) Cells were biotinylated with the membrane-impermeant agent NHS-LC-biotin (Pierce Chemical) (23, 34). Cell surface proteins were isolated with NeutrAvidin-agarose (Pierce Chemical) and deglycosylated by treatment with peptide-N-glycosidase F (New England Biolabs). The resulting polypeptides were detected by Western blot analysis with the gp120-specific monoclonal antibody Chessie 12 (1) and ECL-Plus imaging (23, 34). All mutant envelope glycoproteins were transported to the cell surface comparably to the wild type (1.20 ± 0.27). A dark image is used to display the deglycosylated gp120s. HXB2 gp160 and gp120 were deglycosylated to provide size markers (94 and 55 kDa, respectively).
TABLE 1.
Expression and stability of gp120 in envelope glycoproteinsa
| Mutant(s) | Avg (SD)
|
|||
|---|---|---|---|---|
| Envelope glycoproteinb | gp120 sheddingc | Cell surface gp160d | Cell surface gp120d | |
| E584A | 1.00 (0.17) | 0.93 (0.78) | 0.97 (0.27) | 0.49 (0.15) |
| Y586A | 1.05 (0.06) | 1.20 (0.87) | 0.94 (0.21) | 0.66 (0.28) |
| K588E | 1.13 (0.14) | 1.11 (0.69) | 1.13 (0.36) | 0.75 (0.30) |
| K588Q | 1.09 (0.04) | 1.13 (0.78) | 1.01 (0.38) | 0.73 (0.25) |
| L593A | 1.09 (0.10) | 1.75 (0.85) | 1.16 (0.22) | 0.02 (0.05) |
| I595A | 1.10 (0.09) | 1.54 (0.77) | 1.20 (0.42) | 0.57 (0.09) |
| W596A | 1.04 (0.11) | 1.83 (0.74) | 1.31 (0.37) | 0.13 (0.10) |
| K601E | 1.03 (0.22) | 1.65 (0.79) | 1.06 (0.08) | 0.44 (0.40) |
| T606A | 1.00 (0.08) | 1.27 (0.56) | 1.21 (0.11) | 0.00 (0.11) |
| W610A | 1.06 (0.15) | 1.04 (0.56) | 1.43 (0.24) | 0.00 (0.11) |
| S613A | 0.84 (0.15) | 1.25 (0.92) | 0.80 (0.08) | 0.78 (0.24) |
| W614A | 1.03 (0.10) | 1.23 (0.60) | 1.76 (0.79) | −0.01 (0.11) |
| W623A | 0.99 (0.10) | 1.40 (0.69) | 1.73 (0.65) | 0.06 (0.08) |
| T626M | 0.97 (0.14) | 1.30 (0.56) | 1.24 (0.22) | 2.19 (1.18) |
| T639A | 1.00 (0.10) | 1.74 (0.41) | 1.10 (0.13) | 0.59 (0.25) |
| Wild type | 1.00 | 1.00 | 1.00 | 1.00 |
| Cleavage-defective mutant | 0.91 (0.13) | 0.00 | 0.79 (0.20) | 0.00 |
| All | 1.03 (0.07) | 1.36 (0.28) | 1.20 (0.27) | 0.49 (0.56) |
Imaging and Fuji Image Gauge quantitation (Measure Profile tool) of four independent and complete expression studies. The cleavage-defective mutant was used to determine the background in gp120 measurements, and all averages are ratios relative to the wild type. Italicized averages are constrained by definition. Mutants judged deficient in gp120-gp41 association are in boldface. Average values for all gp41 mutants are shown at the bottom.
Total envelope glycoprotein (gp120 and gp160) in cell lysates (Fig. 2A).
gp120 in cell culture medium (Fig. 2B). Absolute values of mutants and the wild-type reference varied among experiments, but overall averages revealed little variation among mutants.
Peptide-N-glycosidase F deglycosylated gp160 (94 kDa) and gp120 (55 kDa) on cell surface (Fig. 2C).
The inability to detect differences in gp120 shedding through our measurements of gp120 in the cell culture medium may reflect, in part, the multiple kinetic components that impinge on gp120 accumulation. To specifically examine the stability of the gp120-gp41 complex on the cell surface, we labeled expressing cell cultures with a membrane-impermeant biotinylation reagent and cell surface gp120 was further resolved from gp160 by enzymatic deglycosylation to yield the discrete 55- and 94-kDa polypeptides (from gp120 and gp160, respectively) (Fig. 2C). The predominant form in all cases was the unprocessed gp160 precursor, presumably reflecting saturation of the proteolytic capacity of the cellular furin-like proteases through overexpression (3, 23, 34). Nonetheless, the 55-kDa gp120 polypeptide was readily discerned in the majority of mutant envelope glycoproteins (E584A, Y586A, K588Q, K588E, I595A, K601E, S613A, T626M, and T639A). Interestingly, little or no cell surface gp120 was detected in the L593A, W596A, T606A, W610A, W614A, and W623A mutants. Quantitative analysis of all of the mutants (Table 1) showed a general reduction in the amount of gp120 relative to the wild-type envelope glycoprotein, with a broad range of 0.00 to 2.19 (average, 0.49 ± 0.56). Among those mutants with the notable lesser amounts of cell surface gp120 were L593A, W596A, T606A, W610A, W614A, and W623A. Only the T626M glycoprotein retained more gp120 than did the wild type.
The deficiency in cell surface gp120, in conjunction with the finding that gp120 was shed by these mutants, was taken as presumptive evidence that the gp120-gp41 complex is unstable. Nonetheless, it has been difficult to rigorously exclude the alternative possibility that these mutations reduce the amount of gp120 on the cell surface by affecting proteolytic cleavage (see also references 5, 23, 25, and 34). In order to resolve this question, and to define the observed defect as one in gp120-gp41 association, we examined the effect of a known cleavage-defective mutation (REKR to REKT) (12) on the amount of gp120 that accumulated in the culture medium. We reasoned that if the gp120 was not generated by specific proteolytic cleavage in the gp41 mutants, then the observed shedding might reflect nonspecific proteolysis of a misfolded gp160 molecule. In this case, the addition of a second mutation at the specific gp160 cleavage site would not be expected to affect the level of gp120 accumulation. By contrast, if furin-like cleavage of gp160 was largely retained in the initial gp41 mutants, then gp120 shedding should be markedly reduced by the introduction of an authentic cleavage defect.
The cleavage-defective mutation was introduced into plasmids encoding the W596A and W610A mutants, as well as into two previously described mutants displaying a similar phenotype (G572A [23] and W631A [34]), and the extent of gp120 shedding by the double mutants was determined (Fig. 3). In all cases, gp120 shedding by the original gp41 mutant glycoproteins was significantly reduced in the cleavage-defective double mutants. These findings argue strongly that gp120 shedding by these gp41 mutants is not the result of nonspecific proteolysis of misfolded gp160. If specific proteolytic cleavage is partially compromised in these mutants, this is not reflected in the extensive degree of gp120 shedding. These findings support our initial conclusion that the absence of gp120 on the surface of cells expressing W596A, W610A, G572A, and W631A (and, by extension, the other association mutants) reflects a marked instability of the gp120-gp41 association.
FIG. 3.
Effect of a secondary cleavage-defective mutation on gp120 shedding. Four mutant envelope glycoproteins that shed gp120 yet display only gp160 on the cell surface (W596A, W610A, and G572A [23] and W631A [34]) were further mutated to specifically eliminate furin-like proteolytic cleavage of gp160 (12). The effect on gp120 shedding of this second mutation (cleavage-defective [cd] mutant) was assessed by immunoprecipitation, as described in the legend to Fig. 2. wt, wild type.
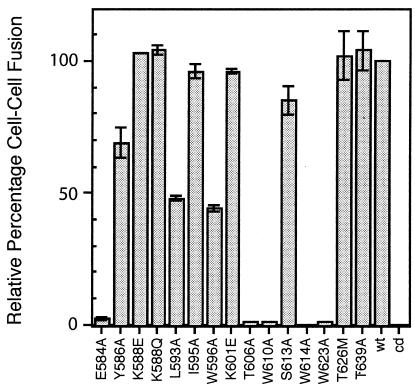
The fusogenic potential of the mutant envelope glycoproteins bearing single mutations in the central gp41 ectodomain was determined by coculturing transfected COS-7 cells with a fusion partner, U87 cells expressing CD4 and CXCR4 coreceptor (15, 21). In these studies, cells expressing the mutant envelope glycoproteins were immunochemically stained to determine the proportion involved in syncytia (Fig. 4). Most mutants that behaved as the wild type did in biochemical analyses were able to mediate cell-cell fusion (viz., K588Q, K588E, I595A, K601E, S613A, T626M, and T639A). Only the Y586A mutant showed a minor deficiency in fusion. Elimination of the glycosylation sites in S613A, T626M, or T639A did not affect the fusogenicity of the envelope glycoprotein (see also references 17 and 27), nor did the reversal of the amino acid charge at positions K588 and K601. By contrast, an alanine mutation at E584 eliminated fusogenicity without an apparent effect on envelope glycoprotein biosynthesis. This finding is consistent with the role of E584 in salt bridge formation in the fusion-active core (25).
FIG. 4.
Syncytium formation by mutant envelope glycoproteins. COS-7 cells expressing the wild-type (wt) or mutant envelope glycoproteins were cocultured with U87 cells expressing CD4 and CXCR4 (15) for 5 h as previously described (21, 23, 34). Cultures were fixed with cold methanol-acetone (1:1) and immunochemically stained with HIVIG to reveal envelope glycoprotein-expressing cells and syncytia. The percentage, relative to the wild type, of envelope glycoprotein-expressing cells involved in syncytia containing three or more nuclei is indicated (± 1 standard deviation). cd, cleavage-defective mutant.
Importantly, mutations that disrupted the maintenance of the gp120-gp41 association (viz., L593A, W596A, T606A, W610A, W614A, and W623A) were, for the most part, devoid of fusion activity. The defect in fusogenicity appeared to be in part related to the extent of residual gp120 on the cell surface, in that only mutants L593A and W596A (2 and 13% residual gp120, respectively) retained some ability to mediate cell-cell fusion. This finding is consistent with our previous experience with association-defective mutants G572A, R579A, W628A, W631A, and I635A (23, 34). The defect in fusion caused by the G572A or R579A mutation is so profound that HIV NL4-3 virions bearing these are entirely noninfectious and revertants could not be rescued by blind passage in cell culture (K. E. Follis and J. H. Nunberg, unpublished data).
In summary, the pattern of envelope glycoprotein phenotypes uncovered in our mutagenesis studies reveals further evidence of the important role of the central ectodomain of gp41 in the association between HIV envelope glycoprotein subunits. Mutations in this region of gp41 give rise to envelope glycoproteins that are specifically deficient in maintaining the gp120-gp41 association on the cell surface. The examples described here add to those catalogued by us (23, 34) and by others (5, 25). Mutations elsewhere in gp41 do not typically generate this specific defect (4, 5, 23, 25, 34, 37, 38). The phenotypes identified in the present study are concordant with those of identical mutations previously described (e.g., L593A and K601E [25] and E584A [5]), with the exception of our wild-type phenotype at T626M and T639A (compare to reference 5). The severe defects reported in reference 5 may be secondary to the intended mutation. Among the positions associated with gp120-gp41 instability in our study, several have previously been identified through other amino acid substitutions (e.g., L593V; W596F, -H, or -L; and W610F or -H) (25).
Taken together, these observations direct attention to a 70-amino-acid region in the central ectodomain, flanking and including the disulfide-bonded loop, as promoting gp120-gp41 interaction. Evidence in support of the physical association between this region of gp41 and the gp120 subunit was previously provided through the identification of cysteine mutations in gp41 that allow the formation of disulfide bonds with engineered cysteine residues in the C1 or C5 region of gp120 (3). In our studies, the alanine mutation at T606, adjacent to one such cysteine mutation (T605C), eliminates gp120-gp41 association and suggests that this position may interact directly with gp120.
The central ectodomain of gp41 includes a large number of tryptophan and other bulky hydrophobic residues (Fig. 1). When the four tryptophan side chains studied here, and two previously examined tryptophan residues (W628 and W631 [34]), were individually mutated to alanine, all gave rise to envelope glycoproteins that were unable to maintain the gp120-gp41 association. Because bulky hydrophobic amino acids, including tryptophan residues, often contribute to the hydrophobic interactions that stabilize protein-protein interfaces (8, 18, 22), we propose that the tryptophan residues identified here may be structured so as to form a hydrophobic face of gp41 in the native gp120-gp41 complex.
Our findings are consistent with a model in which the association between gp120 and gp41 is mediated across a buried hydrophobic interface in the virion envelope glycoprotein complex. Although the details of the gp120-gp41 interaction are unknown, early speculation suggested a “knob-and-socket” structure involving the disulfide-bonded loop of gp41 (30). Mutational analysis of amino acids within the loop reveals relatively minor phenotypes and tends to disfavor this model (K601E [25]). Recent studies by Sodroski and colleagues have identified mutations in a hydrophobic groove of the gp120 core (20) that is important for the interaction with gp41 (42). This groove in the β-sandwich structure of the inner domain of gp120 includes portions of the C1 and C5 regions that have previously been implicated in the gp120-gp41 association (14, 39). We speculate that these two regions, the β-sandwich structure of the gp120 inner domain and the central ectodomain of gp41, associate via specific hydrophobic interactions to contribute to the gp120-gp41 interface. The reported spectroscopic interaction between tryptophan residues in the central ectodomain of gp41 and a synthetic peptide corresponding to the C5 domain of gp120 (13) lends credence to this model. Strategies to stabilize the native gp120-gp41 association may offer promise for the production of envelope glycoprotein vaccine immunogens.
Acknowledgments
This work was supported by National Institutes of Health grant AI054266.
We thank Kathryn Follis, Kimberly Hardwick, Jessica Elsey, and Vanessa Thompson for generating several of the mutations studied and Fred Prince (New York Blood Center) and George Lewis (University of Maryland) for providing antibody reagents. We are grateful to Min Lu (Weill Medical College of Cornell University) for input throughout the study and in the preparation of the manuscript.
REFERENCES
- 1.Abacioglu, Y. H., T. R. Fouts, J. D. Laman, E. Claassen, S. H. Pincus, J. P. Moore, C. A. Roby, R. Kamin-Lewis, and G. K. Lewis. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retrovir. 10:371-381. [DOI] [PubMed] [Google Scholar]
- 2.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 3.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, J., L. Bergeron, E. Helseth, M. Thali, H. Repke, and J. Sodroski. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 67:2747-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 7.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 8.Creighton, T. E. 1993. Proteins: structures and molecular properties, 2nd ed. W. H. Freeman & Co., New York, N.Y.
- 9.Cunningham, B. C., and J. A. Wells. 1989. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244:1081-1085. [DOI] [PubMed] [Google Scholar]
- 10.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 11.Follis, K. E., S. J. Larson, M. Lu, and J. H. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition to the fusion-active state of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 76:7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O., D. J. Myers, and R. Risser. 1989. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J. Virol. 63:4670-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guilhaudis, L., A. Jacobs, and M. Caffrey. 2002. Solution structure of the HIV gp120 C5 domain. Eur. J. Biochem. 269:4860-4867. [DOI] [PubMed] [Google Scholar]
- 14.Helseth, E., U. Olshevsky, C. Furman, and J. Sodroski. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65:2119-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill, C. M., H. Deng, D. Unutmaz, V. N. Kewalramani, L. Bastiani, M. K. Gorny, S. Zolla-Pazner, and D. R. Littman. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 71:6296-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jelesarov, I., and M. Lu. 2001. Thermodynamics of trimer-of-hairpins formation by the SIV gp41 envelope protein. J. Mol. Biol. 307:637-656. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, W. E., J. M. Sauvron, and R. C. Desrosiers. 2001. Conserved, N-linked carbohydrates of human immunodeficiency virus type 1 gp41 are largely dispensable for viral replication. J. Virol. 75:11426-11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, S., and J. M. Thornton. 1996. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. W. Mellors, J. Mullins, S. Wolinsky, and B. Korber. 2000. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 20.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaCasse, R. A., K. E. Follis, T. Moudgil, M. Trahey, J. M. Binley, V. Planelles, S. Zolla-Pazner, and J. H. Nunberg. 1998. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J. Virol. 72:2491-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen, T. A., A. J. Olson, and D. S. Goodsell. 1998. Morphology of protein-protein interfaces. Structure 6:421-427. [DOI] [PubMed] [Google Scholar]
- 23.Lu, M., M. O. Stoller, S. Wang, J. Liu, M. B. Fagan, and J. H. Nunberg. 2001. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J. Virol. 75:11146-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luciw, P. A. 1996. Human immunodeficiency viruses and their replication, p. 1881-1952. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melinick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 25.Maerz, A. L., H. E. Drummer, K. A. Wilson, and P. Poumbourios. 2001. Functional analysis of the disulfide-bonded loop/chain reversal region of human immunodeficiency virus type 1 gp41 reveals a critical role in gp120-gp41 association. J. Virol. 75:6635-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKeating, J. A., A. McKnight, and J. P. Moore. 1991. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J. Virol. 65:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrin, C., E. Fenouillet, and I. M. Jones. 1998. Role of gp41 glycosylation sites in the biological activity of human immunodeficiency virus type 1 envelope glycoprotein. Virology 242:338-345. [DOI] [PubMed] [Google Scholar]
- 28.Prince, A. M., H. Reesink, D. Pascual, B. Horowitz, I. Hewlett, K. K. Murthy, K. E. Cobb, and J. W. Eichberg. 1991. Prevention of HIV infection by passive immunization with HIV immunoglobulin. AIDS Res. Hum. Retrovir. 7:971-973. [DOI] [PubMed] [Google Scholar]
- 29.Sanders, R. W., M. Vesanen, N. Schuelke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz, T. F., B. A. Jameson, L. Lopalco, A. G. Siccardi, R. A. Weiss, and J. P. Moore. 1992. Conserved structural features in the interaction between retroviral surface and transmembrane glycoproteins? AIDS Res. Hum. Retrovir. 8:1571-1580. [DOI] [PubMed] [Google Scholar]
- 31.Sodroski, J. G. 1999. HIV-1 entry inhibitors in the side pocket. Cell 99:243-246. [DOI] [PubMed] [Google Scholar]
- 32.Syu, W. J., W. R. Lee, B. Du, Q. C. Yu, M. Essex, and T. H. Lee. 1991. Role of conserved gp41 cysteine residues in the processing of human immunodeficiency virus envelope precursor and viral infectivity. J. Virol. 65:6349-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan, K., J. Liu, J. Wang, D. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, S., J. York, W. Shu, M. O. Stoller, J. H. Nunberg, and M. Lu. 2002. Helical packing interactions in the HIV-1 gp41 core: implications for the activation of membrane fusion. Biochemistry 41:7283-7292. [DOI] [PubMed] [Google Scholar]
- 35.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 36.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 37.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weng, Y., Z. Yang, and C. D. Weiss. 2000. Structure-function studies of the self-assembly domain of the human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 74:5368-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyatt, R., E. Desjardin, U. Olshevsky, C. Nixon, J. Binley, V. Olshevsky, and J. Sodroski. 1997. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J. Virol. 71:9722-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 41.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, X., E. Mahoney, G. H. Holm, A. Kassa, and J. Sodroski. 2003. Role of the gp120 inner domain b-sandwich in the interaction between the HIV-1 envelope glycoprotein subunits. Virology 313:117-125. [DOI] [PubMed] [Google Scholar]
- 43.Yang, Z. N., T. C. Mueser, J. Kaufman, S. J. Stahl, P. T. Wingfield, and C. C. Hyde. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J. Struct Biol. 126:131-144. [DOI] [PubMed] [Google Scholar]