Abstract
Despite considerable interest in the enediyne family of antitumor antibiotics, assembly of their polyketide core structures in nature remains poorly understood. Discriminating methods to access enzyme-bound intermediates are critical for elucidating unresolved polyketide and non-ribosomal peptide biosynthetic pathways. Here, we describe the development of broadly applicable techniques for the mild chemical release and analysis of intermediates bound to carrier proteins (CPs), providing access to these species even in sensitive systems. These techniques were applied to CalE8, the polyketide synthase (PKS) involved in calicheamicin biosynthesis, facilitating the unambiguous identification of enzyme-bound polyketides on an enediyne PKS. Moreover, these methods enabled the preparation of fully unloaded CalE8, providing a "clean slate" for reconstituted activity and allowing us to demonstrate the preferential accumulation of a PKS-bound octaketide with evidence of programmed processing control by CalE8. This intermediate, which has the expected chain length for enediyne core construction, could only be indirectly inferred previously. These studies prove that this polyketide is an authentic product of CalE8 and may be a key precursor to the enediyne core of calicheamicin, as it is the only programmed, enzyme-bound species observed for any enediyne system to date. Our experimental advances into a generally inaccessible system illustrate the utility of these techniques for investigating CP-based biosynthetic pathways.
INTRODUCTION
Iterative catalysis is defined by the orchestrated use and reuse of a variable set of enzyme domains each functioning in the catalytic cycle a fixed number of times. Typically a carrier protein (CP) delivers the growing intermediate to client domains in a “programmed” and highly processive fashion to synthesize and release the final product. Among natural products, fatty acids and most polyketide metabolites are assembled in a stepwise manner that can involve 50, or more, individual reactions, but no free intermediates. These intermediates remain covalently bound to the CP domain via a thioester linkage throughout repeated chain extension and processing cycles, confounding efforts to understand the intrinsic programming of these enzymes. In this paper, we introduce methods for the mild yet efficient chemical release of CP-bound intermediates, providing access to products sequestered by carrier proteins, even if labile. Furthermore, this approach avoids harsh conditions and organic solvents, allowing for recovery of active but fully unloaded enzyme that can be exploited in downstream experiments.
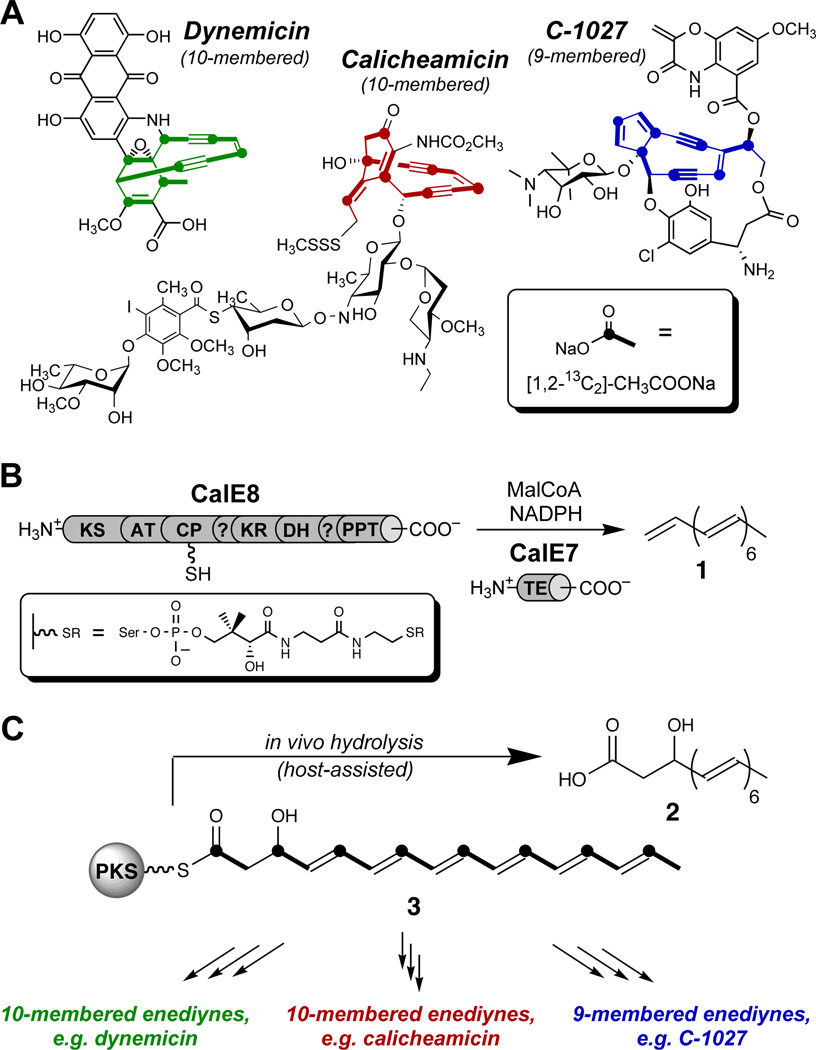
The techniques described here can, in principle, be applied to any CP-based system, but their development was motivated by our studies on the biosynthesis of the enediynes, a natural product family with clinical relevance as targeted anticancer therapeutics.1 Stable isotope-labeling studies show that these antitumor antibiotics, which are often categorized as 9-membered or 10-membered, actually fall into three distinct structural subclasses (Figure 1).2–4 Enediyne core biosynthesis is initiated by an unusual but highly conserved family of iterative type I polyketide synthase (PKS) enzymes; a discrete thioesterase (TE) is also conserved among enediyne biosynthetic clusters.5–8 These enzymes have been the subject of a number of biochemical and structural studies,9–13 yet the isolable polyketides reported thus far provide only indirect evidence toward establishing the true, biosynthetically relevant product of each enediyne PKS.
Figure 1. Enediyne antibiotics and related octaketides.
(A) Representative structures of enediyne natural products from each of three structural families, showing incorporation patterns of isotopically labeled acetate (see inset) for the core structures. (B) In vitro production of heptaene 1 by the calicheamicin PKS CalE8 and TE CalE7. KS, β-ketoacyl synthase; AT, acyl transferase; CP, carrier protein; KR, ketoreductase; DH, dehydratase; PPT, phosphopantetheinyl transferase. Inset: 4'-phosphopantetheine tether installed post-translationally on the CP domain. (C) A biosynthetic proposal for divergence inspired by production of octaketide 2 during heterologous expression of CalE8 alone.
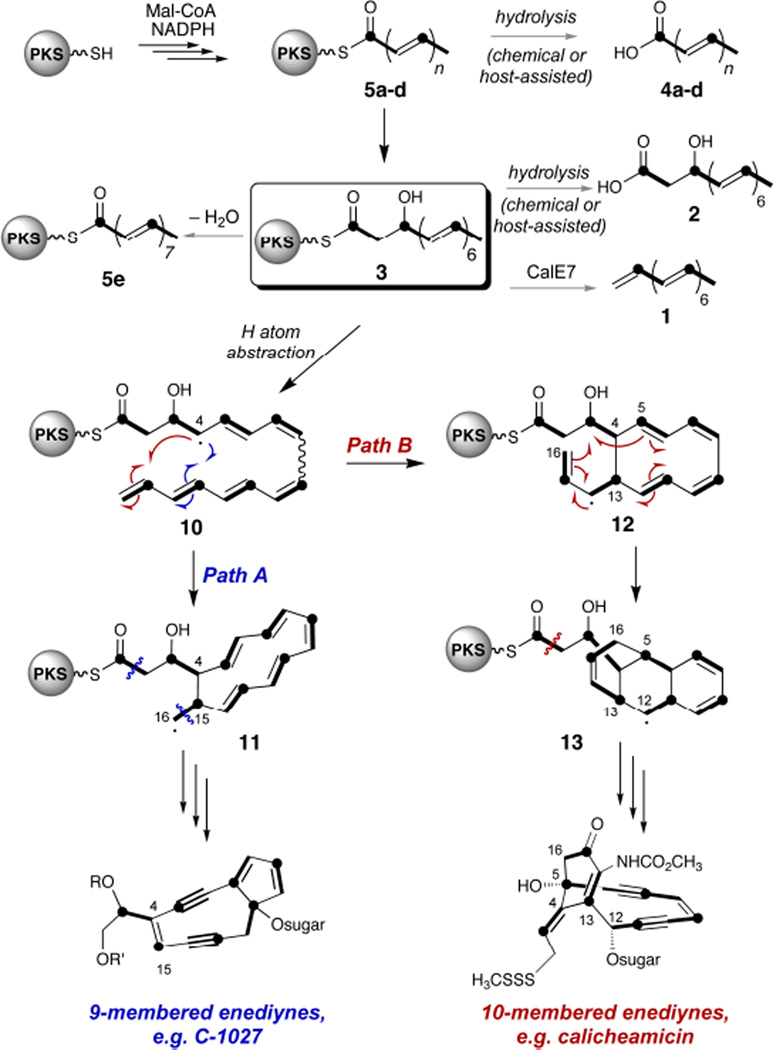
We previously isolated heptaene 1 as a major product of reconstitution reactions with CalE8 and CalE7, the PKS and TE involved in the biosynthesis of the 10-membered enediyne calicheamicin (Figure 1B).14 Recent evidence suggests that 1 is not incorporated into the final enediyne natural products, although it is a common product of enediyne PKS/TE pairs from all subfamilies.15,16 Accordingly, our attention shifted toward elucidating the chemistry of CalE8 without its cognate thioesterase. We recently reported the isolation and characterization of β-hydroxy acid 2 as a major fermentation product of Escherichia coli cultures heterologously expressing CalE8, along with the surprising observation that cultures must be protected from ambient light to ensure appreciable yields.17 The in vivo production of this octaketide, presumably by hydrolysis of hypothetical β-hydroxy thioester 3, implies programmed processing control by CalE8 during the last round of polyketide chain extension. Octaketide 2 and/or its PKS-bound analog 3 have been envisioned as precursors to heptaene formation,14,15 however we imagine an additional, more central role for 3 as the last common intermediate in enediyne biosynthesis. We have offered a mechanistic proposal to account for its divergence to the different enediyne core structures, directed by accessory enzymes specific to each subfamily (Figure. 1C).17
Validating the programmed production of CalE8-bound octaketide 3 would rule out any possibility that the observed fermentation product 2 is an artifact of modification by endogenous E. coli factors following polyketide release. Recombinant enediyne PKSs carry covalently-bound polyketide intermediates produced in vivo, as evidenced, for example, by the yellow color of the purified proteins.14,18 We sought to determine whether CP-bound 3 contributes to the notable yellow color of recombinant CalE8 and, if not, to identify the enzyme-bound polyketide(s) responsible. Protein mass spectrometry, which has been a useful tool for detecting CP-bound species in other systems,19,20 confirmed installation of the 4'-phosphopantetheine tether for another (also yellow) enediyne PKS; however, enzyme-bound polyketides were not identified from those experiments.15 Non-hydrolyzable malonyl-CoA analogs have been employed to offload intermediates from type III21 and linear22,23 PKS systems but have not found the same success with highly processive iterative type I PKSs.24 Furthermore, these analogs rely on a chain extension step to catalyze transfer, so their capacity to release products that are the full, programmed chain length (such as 3) may be limited. Accordingly, we favored a strategy involving the direct chemical cleavage of CP-bound intermediates for isolation and analysis.
The chemical release and characterization of intermediates from CalE8 using traditional methods25–29 have been fraught with major technical barriers. Many researchers interested in enediyne biosynthesis have reported similar experimental intentions and roadblocks,15,18 hinting at the unusual difficulty in probing this particular system. We were thus driven to develop new methods for the chemical release of CP-bound species. The techniques we describe here provide access to these intermediates, resulting in the first characterization of enzyme-bound polyketides from enediyne systems and allowing us to establish the profile of polyketides that are synthesized in vivo and remain bound to CalE8.
These methods have been further exploited to interrogate CalE8 in vitro, separated from native E. coli enzymes and from its cognate TE. A comprehensive picture of reconstituted CalE8 activity in the absence of CalE7 was previously inaccessible owing to low turnover. The protocol we describe provides the means to assay PKS-bound products synthesized exclusively in vitro, enabling a direct measure of CalE8 activity and a deeper understanding of the chemical capabilities of this multifunctional enzyme. Our approach has facilitated rigorous control over experimental factors, allowing us to elucidate how particular variables, including light exposure and substrate availability, affect polyketide assembly by CalE8. We now demonstrate the preferential accumulation of β-hydroxy thioester 3 in vitro, establishing this programmed octaketide as the favored product of the enediyne PKS CalE8 alone and underscoring its likely role as a true biosyn-thetic intermediate of calicheamicin. In addition to revealing key insights into the chemical propensities of enediyne PKSs, these experiments illustrate the general utility of these technical advances for studying CP-based biosynthetic regimes. Furthermore, these tools can be used to probe tailoring enzymes that require CP-bound substrates, increasing the experimental accessibility of this growing category.30
EXPERIMENTAL SECTION
Reagents
Media components and antibiotics were purchased from BD Biosciences (San Jose, CA) and Gold-Bio (St. Louis, MO), respectively. Organic solvents, HPLC solvents, and chemical reagents were acquired from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. LCMS-grade formic acid was obtained from Fluka (a subsidiary of Sigma-Aldrich) for use on the Shimadzu LC-IT-TOF.
Expression and Purification of CalE8 and CalE8-C211A
The expression and purification of hexahistidine-tagged CalE8 and CalE8-C211A have been described previously.14 Briefly, E. coli Rosetta2(DE3) cells harboring pE-CalE8His (or pECalE8H-C211A) were grown in LB medium supplemented with kanamycin (25 µg/mL), chloramphenicol (25 µg/mL) and glycerol (10%, unless noted) at 37 °C to OD600 = 0.6 and cooled for 10 min at 4 °C. Protein expression was induced with IPTG at a final concentration of 1 mM. For light-exposed induction, no special precautions were taken. For cultures grown in the dark, flasks were covered completely with foil or incubated in a closed-door shaker with the door window covered in both aluminum foil and a black fleece blanket. After a specified induction time at 19 °C, cells were pelleted (4000 x g) and resuspended in lysis buffer (300 mM sodium chloride, 50 mM potassium phosphate pH 7.6, 10 mM imidazole, 10% glycerol) with 1 mg/mL lysozyme. Following sonication, the lysate was clarified by centrifugation (25,000 x g) and protein was purified using Ni-NTA resin (Qiagen) according to the manufacturer’s instructions. Protein concentrations were determined by Bradford assay (Bio-Rad, Hercules, CA) using bovine serum albumin as a standard.
HPLC analysis of free or derivatized polyketides produced by CalE8
Routine HPLC separation was carried out on an Agilent 1200 series system (Santa Clara, CA) and analyzed using a diode array detector. Unless noted, samples were injected onto a 4.6 mm × 250 mm Prodigy ODS3 100 Å, 5 µm column (Phenomenex, Torrence, CA) and, unless noted, separated using a gradient of 20% acetonitrile in water with 0.1% (v/v) formic acid each to 80% acetonitrile in water with 0.1% (v/v) formic acid each over 30 min with a flow rate of 1 mL/min.
LCMS analysis of free or derivatized polyketides produced by CalE8
High-resolution LCMS was performed on a Shimadzu LC-IT-TOF (Columbia, Maryland) equipped with a single-wavelength UV/vis detector. Samples were injected onto a Phenomenex Luna C18(2) column (2.0 × 150 mm, 3 µm). The mobile phase and gradient were the same as for HPLC, with a flow rate of 0.2 mL/min. UV/vis peaks in the HPLC traces were correlated to corresponding masses based on their extracted ion chromatograms (EIC). All reported compounds were verified to be absent from CalE8-C211A control reactions based on HPLC chromatogram overlays, EIC overlays, and direct comparison of mass spectra.
Cysteamine-promoted cleavage of CalE8-bound polyketides
A 1 M solution of cysteamine-HCl (Fluka/Sigma-Aldrich, St. Louis, MO) was prepared in nickel elution buffer at neutral pH. Freshly purified solutions of CalE8 in nickel elution buffer were supplemented with the buffered cysteamine solution to a final concentration of 0.2 M cysteamine, capped, and wrapped in aluminum foil. Cysteamine treatment proceeded for 16 h at 4 °C. Samples were then extracted twice with ethyl acetate and concentrated by rotary evaporator. The residue was immediately resuspended in an appropriate solvent, filtered, and analyzed by HPLC/LCMS.
Slow release of CalE8-bound polyketides (solvolysis)
Protein solutions were kept in nickel elution buffer (300 mM sodium chloride, 50 mM potassium phosphate pH 7.6, 250 mM imidazole, 10% glycerol) and capped and wrapped in foil to protect from light. Following a 48 h incubation at room temperature (22 °C), solutions were extracted twice with ethyl acetate and concentrated by rotary evaporator. Samples were redissolved immediately in an appropriate solvent, filtered, and analyzed by HPLC/LCMS.
Ammonium sulfate precipitation of CalE8
CalE8, either freshly purified in nickel elution buffer or from an in vitro reaction time point, was supplemented with cold (4 °C) saturated ammonium sulfate to give a 35% saturated solution. Samples were incubated on ice and protected from light, but inverted periodically to ensure good mixing. After 30 min, samples were spun down at 15,000 rpm at 4 °C. Supernatant was discarded. The ammonium sulfate pellets could be stored at −80 °C or treated immediately.
Neutral hydrolysis of CalE8-bound polyketides
Ammonium sulfate pellets of CalE8 (or CalE8-C211A) were resuspended buffer (or water) without glycerol. Following a 48 h incubation at room temperature (and in the dark), samples were extracted and prepared for analysis as described above.
Cleavage of CalE8-bound polyketides with small molecules
Ammonium sulfate pellets of CalE8 (or CalE8-C211A) were resuspended in various aqueous solutions of nucleophiles: 0.2 M free hydroxylamine (freshly prepared and pH-adjusted to 7), 50 mM n-butylamine, or 50 mM benzylamine. Samples were incubated for 16 h at 4 °C and protected from light exposure. Hydroxylamine-treated samples were extracted directly as described above. Basic amine-treated solutions were acidified to pH <3 with HCl and extracted immediately with EtOAc to minimize acid exposure of labile polyenes. Extracts were prepared for analysis as described above.
Alkaline hydrolysis of CalE8-bound polyketides
Ammonium sulfate pellets of CalE8 (or CalE8-C211A) were resuspended in 0.2 M sodium hydroxide and incubated in the dark for 16 h at 4 °C. Solutions were acidified to pH <3 and extracted immediately with EtOAc to minimize acid exposure of labile polyenes. Extracts were prepared for analysis as described above.
Preparation of fully unloaded CalE8 for in vitro reactions
CalE8 was expressed and purified as above, and nickel elution fractions containing the PKS were supplemented with neutral cysteamine to a final concentration of 0.2 M. Following a 16 h incubation at 4 °C, protein samples were dialyzed against 50 mM potassium phosphate pH 7.0 with 10% glycerol and 2 mM DTT. Protein concentrations were determined by Bradford assay (Bio-Rad, Hercules, CA) using bovine serum albumin as a standard. Enzymes were flash frozen and stored at −80 °C until use.
In vitro reactions of CalE8
Untreated or cysteamine-treated CalE8 (1 mg/mL), malonyl-CoA (0.5 mM or 2 mM), and NADPH (0.5 mM or 2 mM) were incubated at 24 °C in 50 mM potassium phosphate buffer (pH 7.0) containing 10% glycerol. Control reactions contained CalE8-C211A (1 mg/mL) in place of CalE8, and MalSNAc was often substituted for MalCoA. Reaction aliquots were precipitated with ammonium sulfate to allow for analysis of CalE8-bound intermediates by alkaline hydrolysis as described above. Alternatively, reaction aliquots were extracted directly to analyze free polyketides released from the PKS.
Enzymatic production and HPLC purification of MalSNAc
MalSNAc was synthesized enzymatically using the malonyl-CoA synthetase MatB from Rhizobium leguminosarum.31 The original assay conditions32 were modified to utilize N-acetyl cysteamine as a malonate acceptor in place of expensive Coenzyme A and optimized for bulk production. Recombinant MatB was expressed with a hexahistidine tag in E. coli Rosetta2(DE3) cells and purified using nickel affinity chromatography as previously described.24 For a 40 mL reaction, 242 mg ATP (Amresco, Solon, OH) was weighed into a 50 mL Falcon tube and dissolved in 20 mL buffer (50 mM potassium phosphate, pH 7.0). The resulting acidic solution was adjusted to pH 7 using 1 N sodium hydroxide, and phosphate buffer was added to bring the volume to 37.4 mL. To this solution was added 1 mL of a sodium malonate stock solution (2.0 M, pH 7), 280 µL of a MgCl2 stock solution (1 M), 42.5 µL neat N-acetylcysteamine (Aldrich, St. Louis, MO), and 25 mg purified, recombinant MatB (1mL of a 25 mg/mL solution). This final reaction mixture was filtered through a 0.2 µm filter and incubated at ambient temperature for 4–7 days for optimum yield. Protein was removed using an Amicon Ultra filtration device, and the bulk mixture was lyophilized. The bulk solids were re-suspended in 10% ACN/H2O + 0.1% HCOOH for injection onto the HPLC. Purification was achieved on a preparative C18 column with an isocratic method of 15% ACN/H2O + 0.1% HCOOH and a flow rate of 6.5 mL/min. Lyophilized, purified MalSNAc was stored as a powder at −80°C or as a stock solution at −20°C.
RESULTS
Cysteamine-promoted cleavage of PKS-bound intermediates
In pursuit of the chemical release of PKS-bound intermediates, we purified recombinant hexahistidine-tagged CalE8 and subjected this yellow protein to a battery of different conditions, including acid, base, hydroxylamine, amines, thiols, heat, denaturants, and combinations thereof. Despite the fact that many of these same approaches have been used successfully for other systems,25–29 applying these methods to CalE8 failed to produce clear results in our hands. The lack of progress with established methods compelled us to find alternative procedures for accessing CP-bound intermediates.
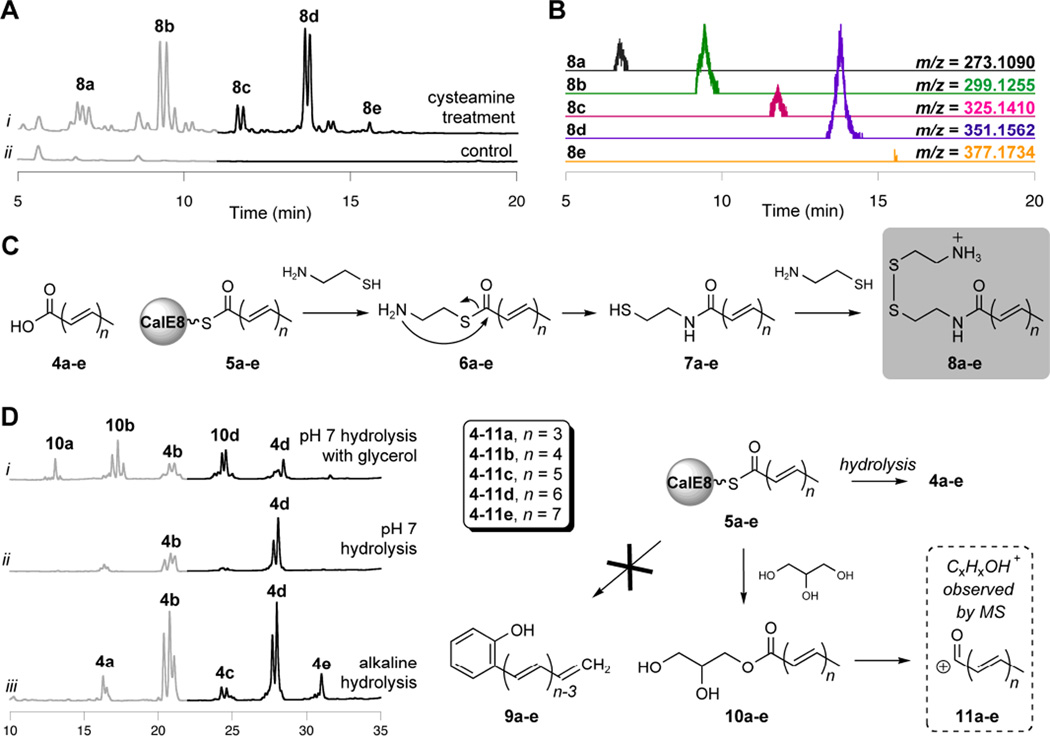
We speculated that cysteamine, which resembles the 4'-phosphopantetheine tether of the CP-domain, could intercept polyketide intermediates. We further anticipated that, following transthioesterification, an intramolecular rearrangement would result in an amide derivative with improved stability.33,34 Indeed, incubation of CalE8 with free cysteamine generated a small but reproducible array of extractable products when compared to an inactive PKS mutant control (CalE8-C211A14) that had been prepared and treated in parallel (Figure 2A). The UV/visible spectra of these adducts resemble the free polyene acids (4a–e) that were isolated as fermentation products,17 suggesting that the PKS is loaded with carbonyl-conjugated polyenes 5a–e in vivo and carries them through the purification process. Rather than the expected cysteamine adducts, however, LCMS and fragmentation analysis indicated derivatization of the acyl-polyene series with cystamine, the oxidized disulfide of cysteamine, yielding mixed disulfides 8a–e (Figure 2B, Table S1, and Figure S1). Direct nucleophilic attack by the nitrogen of cysteamine is not the likely reaction pathway for this derivatization, as simple primary alkylamines did not release CalE8-bound intermediates efficiently during preliminary experiments. Instead, we believe that, following transthioesterification and the expected intramolecular rearrangement, the resulting free thiols of the polyketide adducts are oxidized to the mixed disulfides 8a–e with excess free cysteamine (Figure 2C). This reaction cascade introduces a free amine, providing a serendipitous and dramatic increase in MS analytical sensitivity.
Figure 2. Chemical release of CalE8-bound polyketides.
(A) Derivatized polyketides released upon cysteamine treatment of purified CalE8. HPLC traces (λ = 325 nm, gray; λ = 375 nm, black) of adducts extracted following treatment of CalE8 with 0.2M cysteamine are shown. (B) Extracted ion chromatograms, showing observed masses for 8a–e (ESI+). (C) Reaction cascade leading to cystamine adducts 8a–e. (D) Solvolysis and hydrolysis of polyketides bound to CalE8. HPLC traces (λ = 325 nm, gray; λ = 375 nm, black) of released products are shown; labelled peaks were confirmed by LCMS analysis. Within each panel, chromatogram intensity scales are identical to allow for a direct comparison.
These results constitute an efficient nonenzymatic release of polyketides from an enediyne PKS. Mechanistically, these adducts must derive from activated acid precursors (not occluded, random materials), proving the covalent nature of the observed PKS-bound intermediates. Despite the promise of this mild approach to releasing bound polyketides, no evidence was found for PKS-bound intermediates with alternative processing levels, such as the precursor to β-hydroxy acid 2. Although cysteamine release represents a substantial technical advance in our studies of the PKS-bound products of CalE8, it relies on the use of a sulfur-based nucleophile in a setting where Michael acceptors are abundant, so competitive pathways involving polyene degradation are likely also at work. Thus, we continued our search for improved methods to release CalE8-bound polyketides.
Hydrolytic release of CalE8-bound intermediates
Subtle changes in the UV/visible absorption spectrum of purified CalE8 with time suggested the slow, spontaneous hydrolysis of polyketides from the PKS. Following a 48-hour incubation period, released polyketides can be extracted in trace amounts from freshly-purified CalE8 (Figure S2). In addition to members of the familiar polyene-conjugated carboxylic acid series (4a–e), LCMS analysis revealed a second set of polyketides that gave the general molecular formula CxHxO, suggesting an intramolecular release from CalE8 to give phenolic polyenes 9a–e. To control for involvement from buffer components, aliquots of freshly-purified CalE8 were precipitated with ammonium sulfate and resuspended in water or phosphate buffer with various additives. We thus discovered that glycerol, which is present in nickel elution buffer at 10%, promotes formation of the observed CxHxO series at the expense of the free acid series (Figure 2D, traces i and ii). Returning to the LCMS data, we identified exact masses consistent with glycerol oxyesters that matched the extracted ion chromatograms for several corresponding CxHxO profiles. This observation suggests that the CxHxO series represents in-source fragmentation of glycerol adducts 10a–e to the corresponding acylium ions (11a–e) and that solvolysis competes with simple hydrolysis.
The discovery of competing glycerolysis of PKS-bound intermediates encouraged us to revisit earlier cleavage strategies. Removal of glycerol by ammonium sulfate precipitation provided the next technical jump forward – hydroxylamines, alkylamines, and benzylamines were found to be capable of intercepting CalE8-bound intermediates to yield the corresponding hydroxamic acid and amide derivatives, although competing hydrolysis persists (Figure S3). Treatment of CalE8 with sodium hydroxide following ammonium sulfate precipitation resulted in efficient hydrolysis of bound polyketides and enabled detection of the less abundant members of the polyene-conjugated acid series (Figure 2D, trace iii). Based on the relative intensities of extracted products, this is the most effective technique to date for assaying CalE8-bound intermediates.
Effect of culture conditions on the profile of PKS-bound intermediates
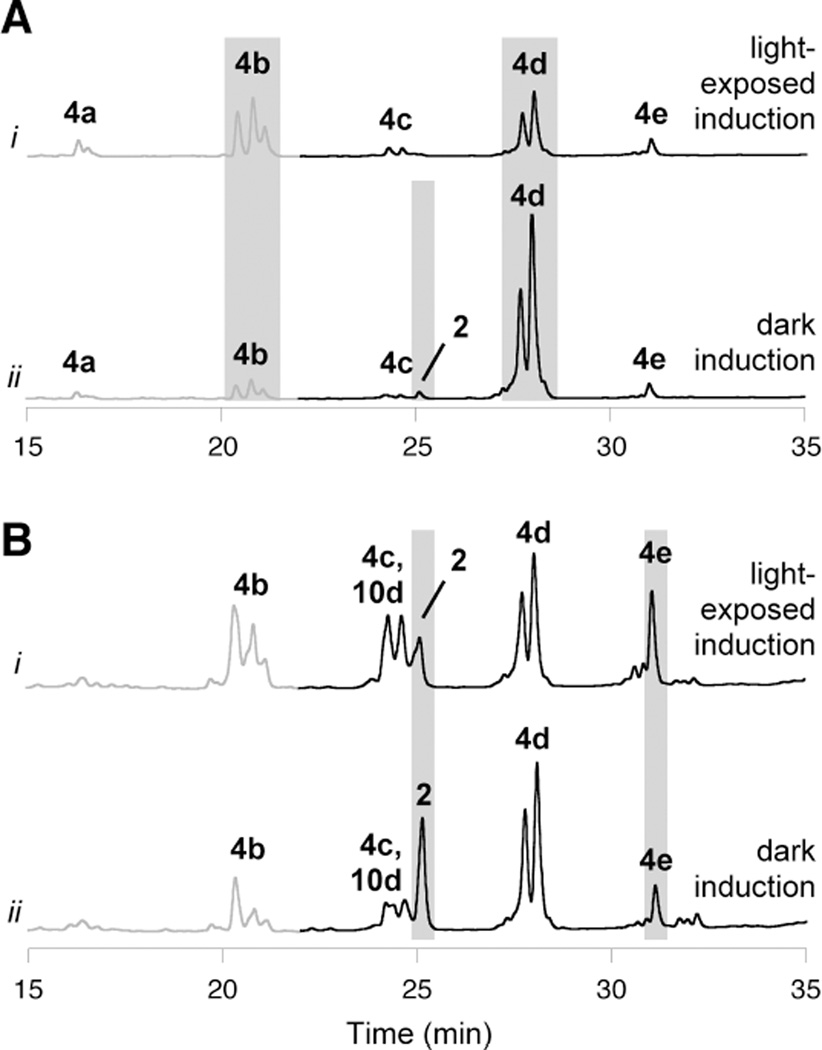
Evidence for programmed control of polyketide processing, as in 3, was not observed during development of cysteamine-based or alkaline hydrolysis protocols; however, those efforts used recombinant CalE8 from light-exposed cultures. Applying these techniques, we profiled the enzyme-bound intermediates synthesized by CalE8 in vivo under different culture conditions, including medium composition, induction time, and light exposure. We recently reported a striking increase in the levels of β-hydroxy acid 2 as a free metabolite when cultures were protected from ambient light,17 so we anticipated that CalE8 from light-protected cultures would harbor some of the corresponding β-hydroxy thioester 3, in addition to acyl-conjugated polyenes such as 5a–e. Accordingly, we cultured parallel batches of E. coli cells expressing CalE8 differing only by light exposure. Upon harvesting the protein, we detected trace amounts of the free acid 2 after base hydrolysis treatment only from CalE8 expressed in the dark (Figure 3A), suggesting that the PKS was indeed loaded with the β-hydroxy thioester 3 under these conditions. Despite the low abundance of β-hydroxy acid 2 in these experiments, the distinctive fine structure indicative of an isolated hexaene moiety could still be observed in its UV/vis spectrum.35 This signature absorbance pattern, in combination with exact mass determination and the enzymatic limitations of CalE8, were essential to assigning the structure of this labile compound.17 The levels of 2 released from purified CalE8 were minute, even at early induction time points, indicating that host-assisted hydrolysis of 3 is efficient.
Figure 3. The effect of light exposure on polyketides synthesized in vivo by CalE8 during heterologous expression.
(A) PKS-bound intermediates that are carried by CalE8 through the purification process and released upon alkaline hydrolysis. Shown are HPLC traces (λ = 325 nm, gray; λ= 375 nm, black) of base hydrolysis products from CalE8 expressed for (i) 18 h under ambient light and (ii) 18 h in the dark. (B) Polyketides found as free metabolites in cultures expressing CalE8. Shown are HPLC traces (λ= 325 nm, gray; λ= 375 nm, black) of compounds extracted from clarified cell lysate from cultures expressing CalE8 for (i) 18 h under ambient light and (ii) 18 h in the dark. Gray boxes highlight notable effects of light exposure. Within each panel, chromatogram intensity scales are identical to allow for a direct comparison.
These experiments also uncovered a surprising correlation between light exposure and chain length of CalE8-bound acyl polyene intermediates (5a–e). Pentaketide 4b was a prominent base-hydrolysis product of CalE8 from light-exposed cultures, and heptaketide 4d dominated in the light-protected batch (Figure 3A). The population of enzyme-bound polyketides from cultures grown in the dark remained steady over a range of induction times. In light-exposed cultures, however, the shift to shorter chain length intermediates was more pronounced with longer induction times (Figure S4). For a comparison to PKS-bound intermediates, polyketides found in vivo as free metabolites were also evaluated under light and dark induction conditions (Figure 3B). In cultures protected from light, the enhanced production of free β-hydroxy acid 2 was accompanied by a lower level of the free polyene acid 4e, suggesting a connection between light exposure and the ratio of these two octaketides.
The development of techniques to directly assay PKS-bound products has also enabled us to confirm the role of glycerol, which was originally used as a medium additive to enhance MalCoA pools.36,37 Whole-cell pellet extracts from cultures expressing CalE8 implied this effect, with improved yields of isolable polyketides from cultures supplemented with glycerol. Given our discovery that glycerol also facilitates polyketide release, we were concerned that supplementation prevents in vivo accumulation of an authentic intermediate. In fact, analysis of CalE8-bound polyketides at various induction time points indicated just the opposite: glycerol supplementation at 1% or higher was necessary to sustain PKS-bound acyl polyene intermediate levels at late induction time points. At earlier induction time points, the profiles of CalE8-bound intermediates were almost identical regardless of the presence of glycerol. These experiments suggest that without appropriate glycerol supplementation, cellular MalCoA concentrations are insufficient at late induction times to support the "reloading" of PKS active sites. Furthermore, these studies illustrate the dynamic nature of CalE8-bound intermediates, with rates of polyketide synthesis, release, and photoreactivity/degradation all contributing to the observed profile. Given the difficulty of evaluating obviously important factors such as MalCoA levels and host-assisted release pathways, we sought improved control over experimental variables and developed an in vitro approach to investigate the chemistry of the enediyne PKS CalE8.
In vitro accumulation of CalE8-bound polyketides
Dissecting the isolated catalytic behavior of CalE8 under the rigorous control provided by an in vitro system previously constituted an immense challenge owing to poor turnover in reactions lacking the thioesterase CalE7 and pre-loading of the PKS with polyketides that were synthesized in vivo during heterologous expression. Coexpression with the thioesterase does yield a colorless CalE8; however, this PKS may still be loaded with lower chain length polyketides that do not absorb in the visible range. We exploited our newly-developed cleavage methods to generate fully-unloaded CalE8 by treating with cysteamine directly after nickel-affinity chromatography to strip off any bound thioesters. After dialysis to remove released adducts, this mild approach provided a “clean slate” for de novo polyketide synthesis without compromising enzyme activity. Reconstitution reactions could then be evaluated for PKS-bound intermediates that are synthesized exclusively in vitro by precipitating CalE8 with ammonium sulfate and hydrolyzing these newly-formed polyketides under basic conditions.
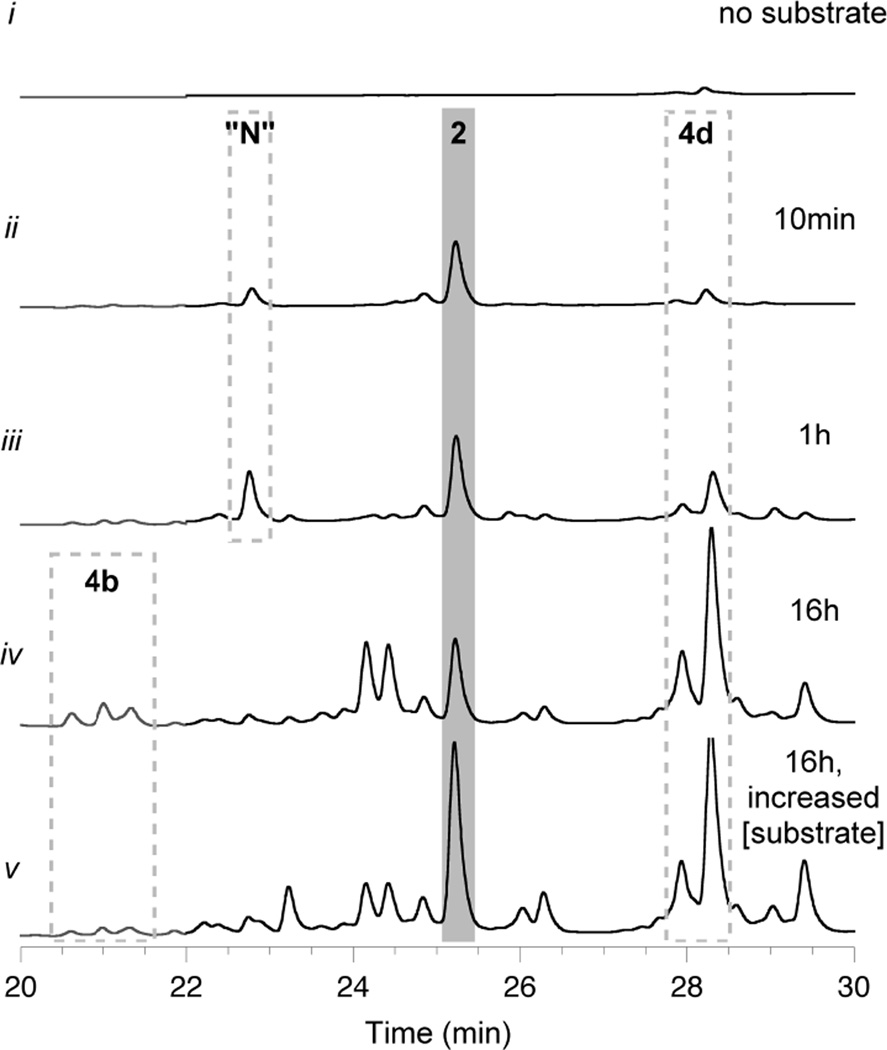
A reaction time course was established for cysteamine-treated CalE8 provided with 0.5 mM each MalCoA and NADPH (Figure 4). At 10 minutes and at 1 hour, β-hydroxy acid 2 was the most prominent component of the base-hydrolyzed polyketides, demonstrating that CalE8 generates β-hydroxy thioester 3 without any interference or assistance from endogenous E. coli proteins. The early and almost exclusive accumulation of octaketide 3 supports the theory that this PKS-bound intermediate is the true programmed product of CalE8.17 Additional minor constituents released from the PKS at these early time points included the heptaketide 4d (one chain extension shorter than the expected length) and a nonaketide with apparent molecular formula C18H22O3. This overextension product may be the result of aberrant PKS activity owing to unnaturally long lifetimes of 3 in the absence of a downstream biosynthetic enzyme and/or with substrate concentrations outside of the physiologically relevant range. The octaketide 4e was barely discernible among hydrolysis products – an important detail, as CalE8 is enzymatically capable of the facile dehydration that would convert 3 to 5e and does not require any cofactors to effect that transformation. This observation provides further evidence that retention of the hydroxyl group in 2/3 is a programmed event.
Figure 4. In vitro synthesis and accumulation of polyketides on CalE8.
PKS-bound intermediates that are assembled on cysteamine-treated CalE8 during reconstitution reactions and released upon alkaline hydrolysis. Shown are HPLC traces (λ = 325 nm, gray; λ = 375 nm, black) of base hydrolysis products from in vitro reactions of CalE8 supplied with no substrate (trace i), 0.5 mM each MalCoA and NADPH (traces ii - iv), or 2 mM each MalCoA and NADPH (trace v). "N" = nonaketide overextension product. Chromatogram intensity scales are identical, allowing for a direct comparison.
At the 16-hour time point, PKS-bound octaketide 3 persisted but was no longer the main constituent. Levels of heptaketide 4d were notably enhanced relative to earlier time points, and polyene acids 4a and 4b were also now observed among hydrolysis products. We suspected that these truncated intermediates reflect a shortage of substrates and/or cofactors at late time points. Indeed, repeating the time course with higher initial concentrations of MalCoA and NADPH revealed both a lag in accumulation of the heptaketide 4d at the 1-hour time point (Figure S5) and almost indiscernible levels of the shorter polyene acids 4a and 4b following an overnight reaction (Figure 4, trace v.). Moreover, in the reaction supplied with higher substrate concentrations, PKS-bound octaketide 3 was maintained at higher levels over the whole time course, as evidenced by the increased levels of 2 following alkaline hydrolysis.
In addition to analyzing CalE8-bound intermediates, a separate reaction aliquot was extracted directly to assay free polyketides. At early time points, low levels of previously reported truncated 2-pyrone release products were observed.14,18 At late reaction time points, solvolysis products of the heptaketide acyl polyene (4d and 10d) were also present, consistent with its accumulation and slow release. The free β-hydroxy acid 2 has never been detected as a spontaneous hydrolysis product by LCMS.
For all of the reconstituted reactions, parallel negative control reactions were set up in which either substrates were withheld (e.g., Figure 4, trace i) or CalE8 was replaced with the inactive point mutant CalE8-C211A.14 We also confirmed that both the PKS-bound and free polyketide profiles of CalE8 are consistent when MalCoA is replaced by malonyl-SNAc (MalSNAc), which offers an economic advantage, as it can be synthesized enzymatically from inexpensive materials.
Effect of light exposure during reconstitution reactions with CalE8
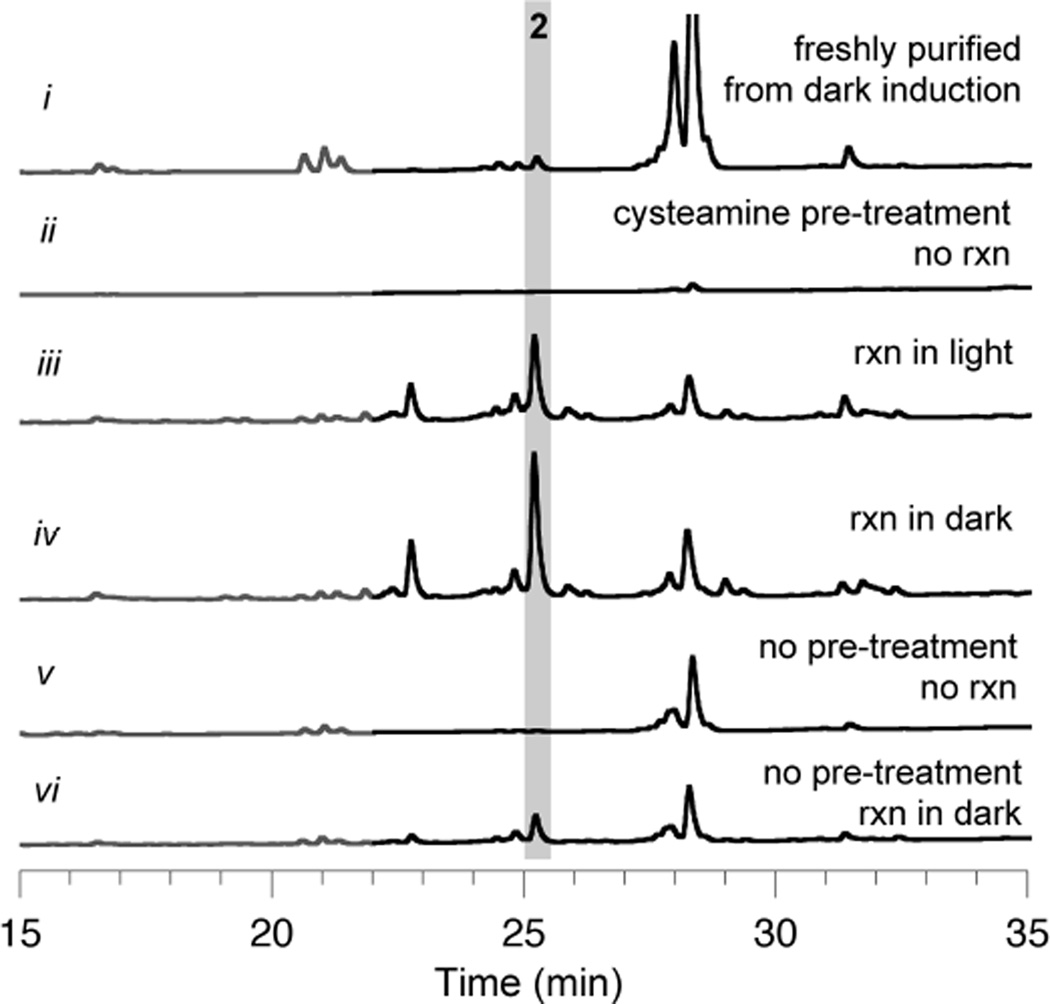
To clarify the effect of light on polyketide synthesis by CalE8, cysteamine-cleared CalE8 was used to set up two parallel reactions that differed only by light exposure. After 1 hour, base-hydrolysis profiles indicated that the levels of PKS-bound octaketide 3 are drastically lower in the light-exposed reaction, implying photodegradation and/or inefficient synthesis of 3 (Figure 5). In contrast to trends observed for in vivo polyketide accumulation on CalE8, it is noteworthy that shorter polyketides were not preferentially amassed on the PKS during in vitro reactions exposed to light.
Figure 5. The effect of light exposure and pre-treatment with cysteamine on in vitrosynthesis and accumulation of polyketides on CalE8.
PKS-bound intermediates that are assembled on CalE8 during reconstitution reactions and released upon alkaline hydrolysis. Shown are HPLC traces (λ= 325 nm, gray; λ= 375 nm, black) of base hydrolysis products from in vitro reactions of CalE8 supplied with 0.5 mM each MalSNAc and NAPDH. Chromatogram intensity scales are identical, allowing for a direct comparison.
To verify that photo-oxidation events do not cause irreversible damage to the protein itself, two batches of CalE8 were prepared: one exposed to light during induction and the other kept in the dark. Both were treated with cysteamine to clear polyketides synthesized during protein expression, then used to set up in vitro reactions. The reconstituted activities of these two batches of CalE8 were found to be identical (Figure S6), verifying that light exposure primarily affects the synthesis, degradation, and release of early enediyne precursors, not CalE8 itself.
Finally, it should be noted that cysteamine pre-treatment was critical to establishing the prominence of CalE8-bound octaketide 3. Levels of the analogous β-hydroxy acid 2 in base-hydrolysis extracts were greatly diminished when the PKS was not “unloaded” pre-reaction (Figure 5, trace vi). Enzyme-bound intermediates synthesized in vivo during the expression of CalE8 apparently interfere with subsequent in vitro polyketide synthesis. Some new polyketide assembly was observed with untreated CalE8, so complete inactivation did not occur. Regardless, the protocol developed for PKS pre-treatment with cysteamine was essential for separation of in vitro polyketide synthesis from that which happened in vivo prior to protein purification.
DISCUSSION
The protocols we have developed for assaying CP-bound intermediates allow us to delineate a comprehensive picture of CalE8 chemistry in the absence of the thioesterase CalE7, both in a heterologous host and in reconstitution reactions. Until now, this particular family of highly-reducing iterative PKSs has been resistant to traditional methods of evaluating CP-bound intermediates. By exploiting the unique structural and chemical properties of cysteamine, we have demonstrated the first controlled, nonenzymatic release of intermediates bound to enediyne PKSs. Moreover, the mild nature of cysteamine cleavage makes it possible to reconstitute activity following treatment, a feature that has been unexpectedly critical to downstream studies. We also recognized glycerol interference as a major impediment to the practical application of other cleavage approaches. Ammonium sulfate precipitation of the protein can remove glycerol without degrading the acid-sensitive polyenes produced by this system, facilitating efficient alkaline hydrolysis of the remaining enzyme-bound intermediates. These methodological advances have enabled the analysis of polyketides found "on" and "off" of the calicheamicin PKS CalE8, from both in vivo and in vitro experimental settings.
The in vitro behavior of CalE8 substantiates our previous assertion17 that the enzyme-bound octaketide 3 is a native intermediate in the biosynthesis of calicheamicin. The corresponding β-hydroxy acid 2 is the most prevalent base hydrolysis product of CalE8 at early reaction time points under all variations of substrate concentration and light exposure (if the PKS was "unloaded" by cysteamine prior to reaction). Moreover, octaketide 3 persists on CalE8 even following overnight reactions, without offloading by hydrolysis or pyrone formation. CalE8 from light-exposed reactions harbors lower levels of 3 but does not specifically accumulate (or release) an alternative polyketide. This detail favors a model in which the observed effects of ambient light are not part of the natural biosynthetic pathway, but instead are related to degradation of photosensitive intermediates.
In the context of heterologous expression, octaketide 3 does not accumulate appreciably on CalE8, rather the free β-hydroxy acid 2 is present as a free metabolite. Notably, the spontaneous hydrolysis of 3 is not observed in vitro. Together, these observations are indicative of host-assisted (rather than programmed) release of this enzyme-bound intermediate in vivo, and are, therefore, compatible with our hypothesis that PKS-bound 3 is a key intermediate in enediyne core assembly.
During fermentation, a strong correlation between light exposure and the chain length of PKS-bound truncation products was observed, with shorter polyketides favored during light-exposed induction. Our in vitro studies were at odds with this result, as light exposure did not promote accumulation of shorter polyketides during reactions. This apparent inconsistency can be reconciled by considering the effects of light on the background processes of E. coli, which has one known photoreceptor.38 BluF (formerly YcgF) initiates a complex signal transduction pathway upon blue-light illumination; known molecular outputs of this cascade include induction of genes related to stress response and biofilm maturation.39,40 Other downstream consequences of BluF activation have yet to be fully characterized, but it is likely that the E. coli populations from cultures grown in the dark exist in a different metabolic and/or oxidative state than those in which BluF has been activated.40 Because polyketide synthesis during fermentation relies on access to primary metabolic pathways and redox sensitive cofactors, comparative data from in vivo experiments must be interpreted with caution. Our methodological advances enabled a more rigorous in vitro approach to studying the effects of light exposure and substrate availability on CalE8 activity. Indeed, our reconstitution experiments indicate that the accumulation of PKS-bound truncated intermediates correlates with limited substrate availability, not irradiation, suggesting that the in vivo effects of light exposure on chain length are achieved indirectly.
Collectively, our data support a biosynthetic model that favors the β-hydroxy thioester 3 as a key intermediate on the path to calicheamicin. Furthermore, the universal production of heptaene 1 by all enediyne PKS/TE combinations evaluated to date16 indicates that 3, the presumed precursor to 1, is produced efficiently by enediyne PKSs from all three subclasses. CalE8 cannot restore antibiotic production to enediyne PKS knockout strains from the 9-membered enediyne family, although (a) CalE8 should be able to interact with the host TE to produce 1 and (b) enediyne PKSs from other 9-membered products could complement these knockouts to restore antibiotic production.15,16 Accordingly, we believe that the PKS-bound β-hydroxy acid 3 represents the last common intermediate in enediyne biosynthesis and the branchpoint for divergence to the different subclasses (Scheme 1). We previously proposed a model for enediyne biosynthesis in which tailoring enzymes specific to each subclass interact directly with (then-hypothetical) 3 to direct this divergence.17 We have now validated the programmed production of 3 by CalE8. Moreover, the protocols we have created can now be exploited to query tailoring enzymes suspected of using PKS-bound 3 as a substrate.
Scheme 1.
A mechanistic proposal for the early steps of enediyne biosynthesis and divergence to the different subclasses. A radical pathway is suggested for this process, but cationic and electrocyclic mechanisms can be envisioned as well
CONCLUSION
Enzyme-bound intermediates of polyketide and non-ribosomal peptide synthases can yield details about the mechanism and timing of assembly for members of these two important classes of natural products, facilitating elucidation or engineering of biosynthetic pathways. We have developed methods for the mild chemical release of CP-bound intermediates that can be applied even to labile species, and we have used this approach to catalog polyketide intermediates that are assembled on the calicheamicin enediyne PKS CalE8 during recombinant protein expression and remain bound during subsequent purification steps. We further applied these techniques to probe the intrinsic chemical reactivity of this PKS in complete isolation. CalE8 was not only segregated from its cognate thioesterase but also from any endogenous factors from the heterologous host that could affect its apparent activity and confound the experimental output of in vivo experiments. Application of this technology has clarified the effect of light exposure and substrate availability on polyketide synthesis by CalE8. Furthermore, we have demonstrated the preferential assembly of PKS-bound β-hydroxy thioester 3 as the programmed product of CalE8 and, therefore, a likely key intermediate in enediyne biosynthesis. This work not only provides a breakthrough in understanding the chemical propensities of CalE8, but also sets the stage for assaying potential accessory enzymes involved in enediyne core differentiation. This new set of tools can be broadly applied to both linear and, especially, iterative CP-based biosynthetic regimes to gain insight into these experimentally challenging catalytic systems.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Shimadzu Scientific Instruments, Inc (Columbia, MD) and to the late Professor R. J. Cotter of the Middle Atlantic Mass Spectrometry Laboratory at the Johns Hopkins University School of Medicine for access to a Shimadzu IT-TOF for high resolution LCMS measurements. We also thank Dr. R.F. Li for isolating the matB gene. This work was supported by NIH Grant ES001670.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Supplementary tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Shao RG. Curr. Mol. Pharmacol. 2008;1:50–60. doi: 10.2174/1874467210801010050. [DOI] [PubMed] [Google Scholar]
- 2.Hensens OD, Giner J-L, Goldberg IH. J. Am. Chem. Soc. 1989;111:3295–3299. [Google Scholar]
- 3.Tokiwa Y, Miyoshi-Saitoh M, Kobayashi H, Sunaga R, Konishi M, Oki T, Iwasaki S. J. Am. Chem. Soc. 1992;114:4107–4110. [Google Scholar]
- 4.Lam KS, Veitch JA, Golik J, Krishnan B, Klohr SE, Volk KJ, Forenza S, Doyle TW. J. Am. Chem. Soc. 1993;115:12340–12345. [Google Scholar]
- 5.Liu W, Christenson SD, Standage S, Shen B. Science. 2002;297:1170–1173. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 6.Ahlert J, Shepard E, Lomovskaya N, Zazopoulis E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 7.Zazopoulis E, Huang K, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM. Nat. Biotechnol. 2003;21:187–190. doi: 10.1038/nbt784. [DOI] [PubMed] [Google Scholar]
- 8.Liu W, Ahlert J, Gao Q, Wendt-Pienkowski E, Shen B, Thorson JS. 2003;100:11959–11963. doi: 10.1073/pnas.2034291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du L, Lou L. Nat. Prod. Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 10.Liang ZX. Nat. Prod. Rep. 2010;27:499–528. doi: 10.1039/b908165h. [DOI] [PubMed] [Google Scholar]
- 11.Liew CW, Sharff A, Kotaka M, Kong R, Sun H, Qureshi I, Bricogne G, Liang ZX, Lescar J. J. Mol Biol. 2010;404:291–306. doi: 10.1016/j.jmb.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 12.Liew CW, Nilsson M, Chen MW, Sun H, Cornvik T, Liang ZX, Lescar J. J. Biol. Chem. 2012;287:23203–23215. doi: 10.1074/jbc.M112.362210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim J, Sun H, Fan JS, Hameed IF, Lescar J, Liang ZX, Yang D. Biophys. J. 2012;103:1037–1044. doi: 10.1016/j.bpj.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belecki K, Crawford JM, Townsend C. A. J. Am. Chem. Soc. 2009;131:12564–12566. doi: 10.1021/ja904391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Van Lanen SG, Ju J, Liu W, Dorrestein PC, Li W, Kelleher NL, Shen B. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1460–1465. doi: 10.1073/pnas.0711625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horsman GP, Chen Y, Thorson JS, Shen B. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11331–11335. doi: 10.1073/pnas.1003442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belecki K, Townsend CA. Angew. Chem., Int. Ed. Engl. 2012;51:11316–11319. doi: 10.1002/anie.201206462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong R, Goh LP, Liew CW, Ho QS, Murugan E, Li B, Tang K, Liang ZX. J. Am. Chem. Soc. 2008;130:8142–8143. doi: 10.1021/ja8019643. [DOI] [PubMed] [Google Scholar]
- 19.Dorrestein PC, Kelleher NL. Nat. Prod. Rep. 2006;23:893–918. doi: 10.1039/b511400b. [DOI] [PubMed] [Google Scholar]
- 20.Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tosin M, Spiteller D, Spencer JB. Chembiochem. 2009;10:1714–1723. doi: 10.1002/cbic.200900093. [DOI] [PubMed] [Google Scholar]
- 22.Tosin M, Betancor L, Stephens E, Li WM, Spencer JB, Leadlay PF. Chembiochem. 2010;11:539–549. doi: 10.1002/cbic.200900772. [DOI] [PubMed] [Google Scholar]
- 23.Tosin M, Demydchuk Y, Parascandolo JS, Per CB, Leeper FJ, Leadlay PF. Chem. Commun. (Cambridge, U.K.) 2011;47:3460–3462. doi: 10.1039/c0cc05077f. [DOI] [PubMed] [Google Scholar]
- 24.Vagstad AL, Bumpus SB, Belecki K, Kelleher NL, Townsend C. A.J. Am. Chem. Soc. 2012;134:6865–6877. doi: 10.1021/ja3016389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangan VS, Witkowski A, Smith S. J. Biol. Chem. 1991;266:19180–19185. [PubMed] [Google Scholar]
- 26.Kopka J, Ohlrogge JB, Jaworski JG. Anal. Biochem. 1995;224:51–60. doi: 10.1006/abio.1995.1007. [DOI] [PubMed] [Google Scholar]
- 27.Kopp F, Linne U, Oberthur M, Marahiel M. A. J. Am. Chem. Soc. 2008;130:2656–2666. doi: 10.1021/ja078081n. [DOI] [PubMed] [Google Scholar]
- 28.Awakawa T, Yokota K, Funa N, Doi F, Mori N, Watanabe H, Horinouchi S. Chem. Biol. 2009;16:613–623. doi: 10.1016/j.chembiol.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin S, Huang T, Shen B. Methods Enzymol. 2012;516:321–343. doi: 10.1016/B978-0-12-394291-3.00008-3. [DOI] [PubMed] [Google Scholar]
- 31.An JH, Kim YS. Eur. J. Biochem. 1998;257:395–402. doi: 10.1046/j.1432-1327.1998.2570395.x. [DOI] [PubMed] [Google Scholar]
- 32.Lombo F, Pfeifer B, Leaf T, Ou S, Kim YS, Cane DE, Licari P, Khosla C. Biotechnol. Prog. 2001;17:612–617. doi: 10.1021/bp010045j. [DOI] [PubMed] [Google Scholar]
- 33.Wieland T, Bokelmann E, Bauer L, Lang HU, Lau H. Liebigs. Ann. Chem. 1953;583:129–149. [Google Scholar]
- 34.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Science. 1994;266:776–778. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 35.Christensen RL, Barney EA, Broene RD. Arch. Biochem. Biophys. 2004;430:30–36. doi: 10.1016/j.abb.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 36.Vallari D, Jackowski S, Rock C. O.J. Biol. Chem. 1987;262:2468–2471. [PubMed] [Google Scholar]
- 37.Kealey JT. Proc. Natl. Acad. Sci. U.S.A. 1998;95:505–509. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Horst MA, Key J, Hellingwerf KJ. Trends Microbiol. 2007;15:554–562. doi: 10.1016/j.tim.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Tschowri N, Lindenberg S, Hengge R. Mol. Microbiol. 2012;85:893–906. doi: 10.1111/j.1365-2958.2012.08147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tschowri N, Busse S, Hengge R. Genes Dev. 2009;23:522–534. doi: 10.1101/gad.499409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.