Summary
Formins are actin assembly factors that act in a variety of actin-based processes. The conserved formin homology 2 (FH2) domain promotes filament nucleation and influences elongation via interaction with the barbed end. FMNL3 is a formin that induces assembly of filopodia but whose FH2 domain is a poor nucleator. The 3.4 Å structure of an FMNL3 FH2 dimer in complex with tetramethylrhodamine-actin uncovers details of formin-regulated actin elongation. We observe distinct FH2-actin binding regions; interactions in the knob and coiled-coil subdomains are necessary for actin binding while those in the lasso/post interface are important for the stepping mechanism. Biochemical and cellular experiments test the importance of individual residues for function. This structure provides details for FH2 mediated filament elongation via processive capping and supports a model in which C-terminal non-FH2 residues of FMNL3 are required to stabilize the filament nucleus.
Introduction
The dynamic nature of the actin cytoskeleton is essential for a variety of cellular processes and is controlled by a large number of actin binding proteins1. The formin family of actin assembly factors, influencing both actin nucleation and elongation, is central to the regulation of many actin-based processes. Formins are characterized by the presence of formin homology 1 (FH1) and formin homology 2 (FH2) domains generally found toward the carboxy-terminus. The FH1 contains multiple poly-proline repeats that interact with profilin bound actin monomers2 The adjacent FH2 domain functions as a homodimer3.
Crystal structures of FH2 domains from Bni1p4,5, mDia16, and DAAM17,8 have been solved. All are highly conserved alpha helical structures, which combine to create a ring-shaped head-to-tail dimer mediated by two equivalent “lasso/post” interactions. A flexible linker of varying length allows considerable flexibility in orientation of the subunits4,8. Both FH2 subunits interact with the barbed end of actin and are able to bind and release from the terminal actin subunits as polymerization occurs, allowing processive movement with the elongating filament4. A mechanism for FH2 mediated capping has been proposed in which the FH2 domain moves as a Brownian ratchet during filament elongation5, with additional insights on the mechanism of processivity coming from biochemical and modeling studies9. The structure of the FH2 domain of Bni1p bound to actin (Bni1p FH2-actin) provided the basis for the ratchet model by capturing a step in the mechanism during which the FH2 domains were interacting with three actin subunits5,10,11. Although biochemical studies suggest that Bni1p FH2 is an extremely stable dimer4, in the Bni1p FH2-actin structure the FH2 is not dimeric, but rather forms a helical concatenation of several Bni1p monomers in head-to-tail fashion that is unlikely to be formed under physiological conditions.
Formin FH2 domains vary dramatically in both their nucleation and elongation activities12-15, and this range of activities provides the potential for wide range control of cell morphology. Some formins, including FMNL3, require not only the FH1 and FH2, but also the C-terminus for potent nucleation15-17 suggesting even broader functionality across the protein family.
To investigate the mechanism by which formins nucleate actin filaments and promote elongation, we determined the high resolution structure of an actin-formin complex. Our 3.4 Å crystal structure of the FH2 domain of FMNL3 bound to tetramethylrhodamine (TMR)-labeled actin represents the first mammalian formin to be crystallized in the presence of actin and visualizes an additional step in processive elongation. Furthermore, it suggests an explanation for the poor nucleation ability of FMNL3’s FH2 domain and allows us to propose a model for FMNL3 nucleation that requires actin monomer binding independently by both the FH2 domain and the C-terminus.
Structural Overview
We solved the X-ray crystal structure of the FH2 domain of FMNL3 (amino acids 555 – 954) in complex with tetramethylrhodamine-actin (TMR-actin) to 3.4 Å (Table 1, Fig 1). The asymmetric unit of the crystal contains two heterotetramers packed together in a head-to-head manner (Supplementary Fig. 1), with the presumed biological unit being composed of two actin monomers and two FH2 domains (Supplementary Fig. 1). Within each actin-FH2 tetramer, the two FH2 domains interact in a head-to-tail orientation encircling the barbed end of two actin monomers (Fig. 1). When the FMNL3/actin complex used for crystallization was analyzed by sedimentation equilibrium analytical ultracentrifugation, we observed a single species with a molecular weight of 166 kDa +/- 15 kDa (Supplementary Fig. 1), corresponding to the 2:2 complex of FH2:actin observed in the crystal structure. The head-to-head packing of tetramers is quite different from the structure of Bni1p bound to TMR-actin5, in which the actin subunits form a flat actin polymer with FH2 domains spiraling around it (Supplementary Fig. 1). Due to the helical arrangement of subunits in the Bni1p FH2-actin structure, symmetric dimerization was not observed between FH2 domains (Supplementary Fig. 1) and each copy of the asymmetric unit contained a single FH2 bound actin subunit. In contrast, the FMNL3/actin complex shows two-fold (non-crystallographic) symmetry (Fig. 1).
Table 1.
Data collection and refinement statistics
| FMNL3 FH2/TMR-actin | |
|---|---|
| Data collection | |
| Space group | P 1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 126.2, 126.3, 129.9 |
| α, β, γ (°) | 90.0, 93.1, 90.0 |
| Resolution (Å) | 19.87-3.40(3.50-3.40) a |
| Rmeas | 10.3(60.1) |
| I/σ | 10.3(2.6) |
| Completeness (%) | 99.2(99.9) |
| Redundancy | 3.8(3.8) |
| Refinement | |
| Resolution (Å) | 19.82-3.40 |
| No. reflections | 55,408 |
| Rwork/ Rfree | 23.0/27.7 |
| No. atoms | |
| Protein | 22688 |
| Ligand/ion | 188 |
| Average B-factors | 105.4 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.448 |
Highest resolution shell is shown in parenthesis.
Figure 1. Structure of FMNL3 FH2 domain bound to TMR-actin.
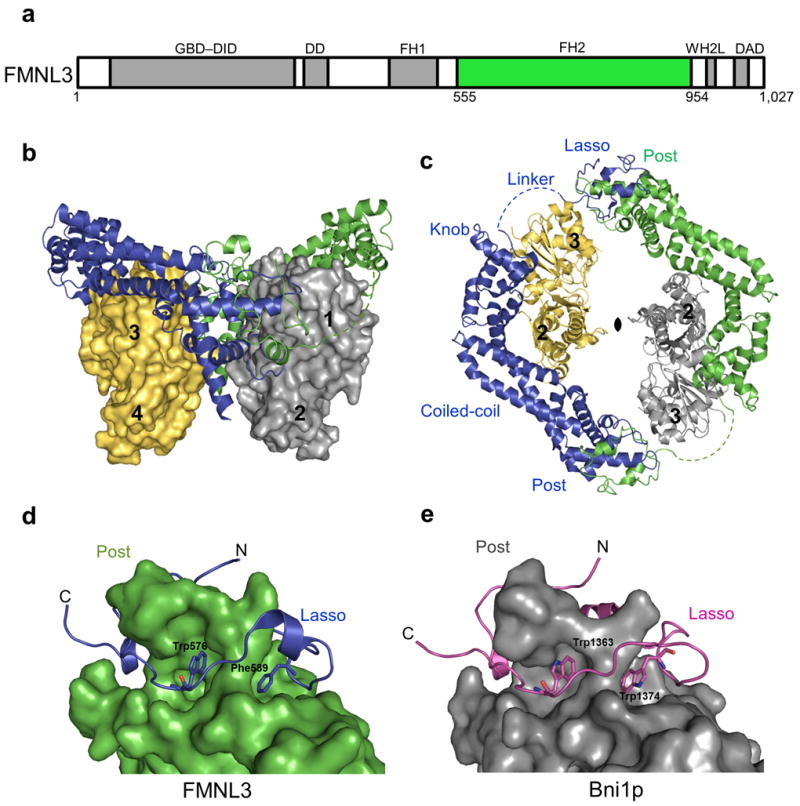
(a) Domain organization of FMNL3 (1027 amino acids) with the GTPase binding domain (GBD), diaphanous inhibitory domain (DID), dimerization domain (DD), formin homology 1 (FH1), WASP-homology 2-like (WH2L) motif, and diaphanous autoinhibitory domain (DAD) colored in grey and the formin homology 2 (FH2) domain (555-954) in green. (b-c) Overall structure of the FMNL3 FH2-actin biological unit with an FH2 dimer (blue and green) bound to two actin subunits (yellow and grey). Actin subdomains are numbered. (c) Top view of the complex with the 2-fold non-crystallographic symmetry axis indicated. The FH2 subdomains are labeled with the disordered linker represented as a dashed line. (d-e) FH2 domain dimerization through the lasso and post subdomains of FMNL3 and Bni1p (1Y64) respectively. Conserved hydrobphobic residues in the lasso of FMNL3 (W576, F589) and Bni1p (W1363, W1374) aid in dimerization.
The overall structural features of the FMNL3 FH2 monomers are similar to other existing formin FH2 domain structures, each having five subdomains: lasso, linker, knob, coiled-coil, and post (Fig. 1)4-8. The structure of the FH2 domain core is conserved among formins for which crystal structures are available, and the head-to-tail dimer of FMNL3 FH2, in which the lasso of each monomer wraps around the post of its partner, has been observed in the structures of the yeast Bni1p and human DAAM14,5,7,8. Two aromatic residues in the lasso, Trp576 and Phe589 in FMNL3, are highly conserved in formins (Supplementary Fig. 2) and presumably help to stabilize dimer formation by interacting with the post (Fig. 1). The FMNL3 knob subdomain (Lys635-Ala755) is clearly similar to that of Bni1p (Lys1417-Ser1537), with r.m.s.d of 2.96 Å between analogous Cα atoms. The coiled-coil and post subdomains have an r.m.s.d. of 1.58 Å between analogous Cα atoms, however differences between the alignments of the knob subdomains to the critical actin binding helix D result in a comparative shift in the relative orientation of the coiled-coil and post subdomains (Supplementary Fig. 3).
The TMR-actin in our structure contains bound ATP (Supplementary Fig. 4), and its overall conformation is similar to those of actin in the the Bni1p/TMR-actin complex5 as well as the TMR-actin-AMPPNP structure19(Supplementary Fig. 4). A top-down view (as in Fig. 1), shows there are no interactions between the two actin subunits, and at their closest point the side chains of adjacent Glu270 residues in the loops between subdomains 3 and 4 are 6.5 Å apart (Fig. 2). In contrast, actin subunits in the Bni1p/TMR-actin structure are in contact, burying a surface of 196 Å2 (Fig. 2)20. The separation of actin subunits in the FMNL3/actin structure shows that each actin monomer can be held in place only through interactions with FH2 subunits, suggesting that flexibility in the FH2 linker region could allow the actin subunits to remain in contact with the FH2 domains while sliding past one another during elongation (see Discussion). While the ability of the FH2 domain to form a symmetric complex with TMR-actin may be specific to FMNL3, our structure suggests this mode of dimerization and of actin binding that we have visualized the first time may be broadly applicable to other formins.
Figure 2. Distinct actin-actin interactions in the FMNL3 FH2-actin complex compared to the Bni1p-actin complex.
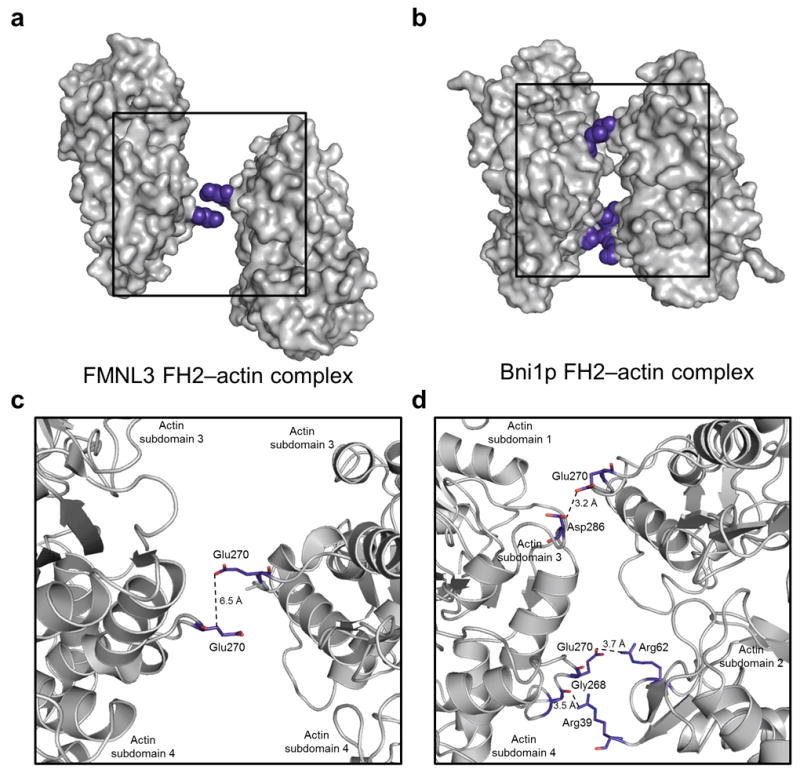
(a-b) Orientation of two actin subunits (gray surface) from the FMNL3 complex (panel a) and the Bni1p complex (panel b). Actin-actin closest contacts are highlighted in blue spheres and boxed with enlargement in panels c and d. The distances between the closest sidechain atoms are indicated by dotted lines to illustrate the difference in actin organization within the FMNL3 and Bni1p FH2-actin complexes.
Formin/actin binding interface
Interactions between FMNL3 and actin cluster to three regions on the inner surface of the FH2 domain and involve the knob, coiled-coil, and lasso/post subdomains (Fig. 3). Several solvent exposed residues in the knob and post subdomains that are highly conserved among formins and are crucial for formin-actin binding4,5 are in close proximity in the FMNL3 FH2-actin structure. For example, in the knob subdomain, helix D of both Bni1p and FMNL3 lie in a groove between subdomains 1 and 3 on the barbed end of actin (Fig. 3 and Supplementary Fig. 3). One residue in the D helix is Ile649 of FMNL3, which is conserved among all formins (Supplementary Fig. 2) and interacts with actin subdomain 1 (Tyr143, Gly146). Two additional residues in helix D, Lys645 and Asn646, make close contacts with actin and are important for activity (see functional data below). The FMNL3 Ile649 residue interacts with actin in the same location as Ile1431 in Bni1p, but the trajectory of helix D differs between the two proteins, resulting in rotation of the FMNL3 FH2 core with respect to actin (Supplementary Fig. 3).
Figure 3. FH2-actin binding interfaces.
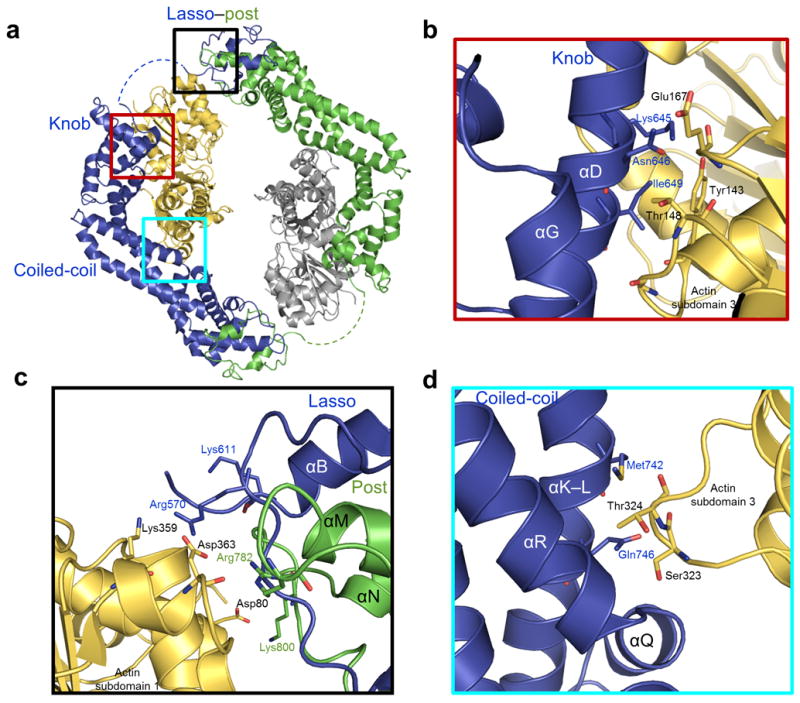
(a) Location of the post, knob, and coiled-coil actin binding regions are boxed in black, red, and cyan respectively. (b-d) Regions boxed in a highlight interacting residues in the FH2 domain binding interface within 3.5 Å of actin for the knob (panel b), lasso/post (panel c), and coiled-coil (panel d) subdomains of FMNL3. Coloring is the same as in Figure 1.
At the lasso/post-actin interface (Fig. 3) there are interactions from both the post and the lasso that participate in actin binding. Within the post, a conserved lysine in the loop between helix N and O (Lys1601 in Bni1p, Lys800 in FMNL3) has been shown to be important for FH2 function in many formins5-7. In FMNL3, this loop makes contacts with subdomain 1 of actin, as is observed in the Bni1p/actin structure. However, the specific contacts are different due to the distinct actin positions in the two structures. In the FMNL3 complex, the actins are positioned side-by-side, while in the Bni1p complex they are offset by a 30 Å translation along the filament. As a result, the lysines in Bni1p and FMNL3 interact with distinct helices in subdomain 1 of actin (Supplementary Fig. 5). There are also two arginine residues in this interface, Arg570 in the lasso and Arg782 in the post, that help to stabilize post-actin binding via interactions with the actin C-terminus (Fig. 3).
In addition to the conserved binding regions in the knob and post, the coiled-coil subdomain of FMNL3 also interacts with actin. Amino acids Met742 and Gln746 in helix K/L contact the Ser323 containing loop in subdomain 3 of actin (Fig. 3). This loop in actin also interacts with the FH2 domain of Bni1p, however; the primary contacts are with side chains in helix R of the FH2 domain.
The primary FH2-actin binding contacts observed previously in the knob subdomain are conserved in FMNL3; however, the coiled-coil contacts with actin in FMNL3 are distinct from those in the Bni1 FH2-actin complex. We also observe a lasso/post binding interface with actin that is similar for both FMNL3 and Bni1p, but interacting with an alternate region of actin. It is unclear if the FMNL3 FH2 interface with actin in the crystal is physiological; however, given the flexibility of the linker tethering each half of the FH2-actin complex. It is likely that contacts with the lasso/post FH2 subdomain are a result of crystal packing and may vary under physiological conditions.
Mutational analysis of FH2-actin binding interface
Together, the knob, coiled-coil, and post regions make up an actin/formin binding interface with a buried surface of 1,638 Å2 (compared to 1,724 Å2 for Bni1p)20. To determine the role of each FMNL3 FH2 subdomain in actin assembly, we used site-directed mutational analysis coupled with biochemical assays for actin binding and filament elongation. Analysis of residues in the binding interface allowed us to assign functional roles to FH2 subdomains that mediate various activities. For actin binding, we used sedimentation velocity analytical ultracentrifugation. The FH2 domain of wild-type (WT) FMNL3 forms a single stable complex with TMR-actin with a sedimentation coefficient of 7.3 S. In contrast, TMR-actin alone is monomeric, 3.4 S species (Fig. 4). The ability of FMNL3 to form a mono-dispersed complex with TMR-actin in solution is specific to FMNL3 out of the formins tested, as constructs containing the FH1-FH2 domains of mDia1 and mDia2 are unable to form stable complexes with TMR-actin (Supplementary Fig. 6).
Figure 4. Effects of FMNL3 mutations on actin filament elongation.
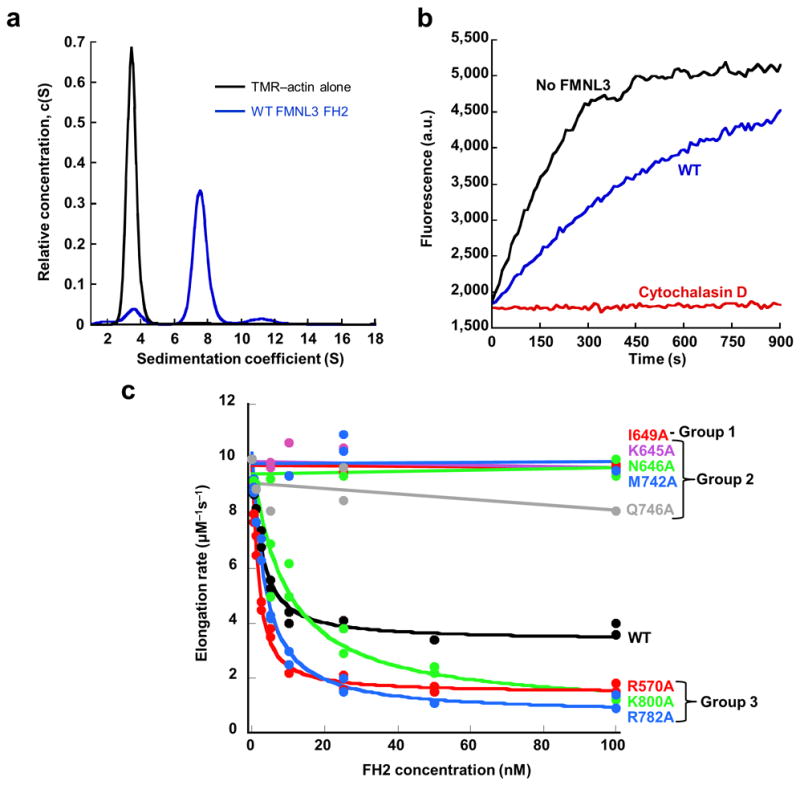
(a) Velocity analytical ultracentrifugation sedimentation profiles for 4.7 μM TMR-actin alone (black curve) and in the presence of 9.7 μM wild-type FMNL3 FH2 (WT) (blue curve) monitored at 557 nm, the peak absorbance wavelength for TMR-actin. TMR-actin alone sediments with a sedimentation coefficient of 3.4 S while a peak shift to 7.4 S is observed in the presence of the WT FH2 domain that form a stable complex with TMR-actin in solution. (b) Pyrene-actin elongation assay performed with 0.5 μM actin monomers (25% pyrene) from filament seeds stabilized with phalloidin. Actin elongation (black curve) is partially inhibited by 100 nM WT FMNL3 FH2 (blue curve) and completely inhibited b 40 nM cytochalasin D (red curve). (c) Concentration dependence of filament elongation inhibition by FMNL3 mutants. FH2 mutant actin binding (data found in Table 2) and elongation data were used to separate FH2 mutants into Group 1 (no TMR-actin binding, no elongation inhibition), Group 2 (binds TMR-actin, no elongation inhibition), or Group 3 (binds TMR-actin, enhanced elongation inhibition). Elongation rates are calculated from the slope of the first 10% of the time course and plotted versus concentration of FH2 domain.
To observe actin filament elongation in vitro, we monitored filament growth as pyrene-labeled actin monomers added to pre-formed filament seeds in the presence and absence of FMNL3 FH2. With WT FMNL3 FH2, actin filaments polymerize at a rate slower than actin alone16 (Fig. 4). We made alanine substitutions for selected residues in all three of the actin-interacting subdomains. In addition to the highly conserved Ile649 and Lys800, mutations were made in the knob and post subdomains, as well as in the coiled-coil. We categorized the mutants based on their abilities to influence TMR-actin binding and filament elongation.
The mutations segregate into three distinct functional groups. Group 1 mutants are unable to bind actin and do not inhibit filament elongation. Group 2 mutants retain the ability to form a complex with TMR-actin, but their capacity to inhibit actin filament elongation is eliminated. Group 3 mutants can bind to actin, however they slow filament elongation to a greater extent than the WT FH2 domain.
The I649A substitution is the only mutant belonging to Group 1. This construct does not cause a peak shift in TMR-actin sedimentation profile, suggesting weak or eliminated binding to TMR-actin (Table 2). Furthermore, it does not slow actin elongation even at a concentration over 50-fold higher than the IC50 for WT FH2 (1.7 nM, Table 2 and Fig. 4), indicating that it is unable to interact with barbed ends. This same mutation in FH1-FH2 containing constructs of FMNL3 also abolishes actin elongation effects and formation of filopodia16,21. Together, these data suggest that, similar to other formins4,7,13,22, the knob subdomain of the FMNL3 FH2 is essential for actin binding via specific interactions involving Ile649.
Table 2.
Actin binding and elongation activity of FMNL3 mutants
| FMNL3 construct | aIC50 (nM) | bImax (%) | cFraction of actin bound | Capping Protein Protection time (sec)d | |
|---|---|---|---|---|---|
| FH2 | FH1-FH2 | ||||
| WT | 1.7 | 65 | 0.91 | 413 | * |
| Group 1 | |||||
| I649A | --- | --- | 0.08 | 150 | 145 |
| Group 2 | |||||
| K645A | --- | --- | 0.98 | * | * |
| N646A | --- | --- | 0.56 | 208 | * |
| M742A | --- | --- | 0.98 | 295 | * |
| Q746A | --- | --- | 0.95 | 350 | * |
| Group 3 | |||||
| R570A | 0.9 | 84 | 0.98 | Nd | Nd |
| R782A | 2.5 | 92 | 0.70 | Nd | Nd |
| K800A | 8.3 | 91 | 0.88 | Nd | Nd |
The concentration of FH2 required for 50% inhibition of filament elongation determined from the fit of the concentration dependent elongation rates in Figure 4c.
The maximum percent inhibition determined from the fit of the concentration dependent elongation rates in Figure 4c.
The fraction of actin bound to FMNL3 is the amount of bound actin divided by the sum of the amount free actin and bound actin where free actin and bound actin are the areas of the velocity sedimentation profile peaks between 2 and 5 S and between 5 and 10 S, respectively.
Time required for capping protein to terminate elongation. For actin alone, the time is 147 sec.
capping protein did not terminate elongation prior to the full utilization of actin monomer.
Two mutants in the knob (K645A, N646A) and two in the coiled-coil (M742A, Q746A) fall into Group 2, retaining TMR-actin binding (Table 2) but unable to alter the elongation rate (Fig. 4). Additional mutations to residues in both the knob and coiled-coil do not affect FMNL3’s ability to bind actin or alter filament elongation (Supplementary Fig. 6).
These data suggest that Group 2 mutants either do not interact with the barbed end or do not change barbed end elongation rate appreciably. To test these possibilities, we performed elongation assays in which heterodimeric capping protein was added 50 sec after elongation was initiated. For filaments in the absence of formin, capping protein terminates all elongation within 150 sec (Table 2 and Supplementary Figure 7). For WT FMNL3 FH2, the time before elongation termination is considerably longer (>400 sec). As expected, the I649A mutant is ineffective at capping protein protection, with termination in 145 sec. Group 2 constructs vary in their ability to protect from capping protein, with K645A effectively protecting ends andN646A, M742A and Q746A providing intermediate protection. These results suggest that, while Group 2 mutants have altered processivity properties, they still interact with barbed ends. Interestingly, when a region containing the FH1 domain is added to these constructs, the protection ability of the WT and Group 2 constructs increases significantly (Table 2 and Supplementary Figure 7), suggesting the FH1 domain contributes to processivity. In contrast, the FH1-FH2 construct of the I649A mutant still does not protect against capping protein, suggesting it has extremely low barbed end affinity.
Group 3 mutations cluster in the lasso/post region and include two post mutations (R782A and K800A) and one lasso mutation (R570A). The ability of K800A to bind actin is surprising based on its position in the crystal structure, high conservation among formins (Supplementary Fig. 2), and defects in actin interaction for other formins altered at this residue4,22. FMNL3FH2 K800A forms a stable complex with actin monomers (Table 2) and inhibits elongation to a greater extent than WT, with a maximal inhibition (Imax) of 91% compared to 65% for WT (Fig. 4c, Table 2). One explanation for this difference is that other residues in the N/O loop are more important for actin interaction. To assess this possibility, we made alanine substitutions for all of the residues in this loop proximate to actin (Supplementary Fig. 5). Single alanine substitutions of amino acids 801 to 804 have no effect on binding or elongation in our assays (Supplementary Fig. 6). Furthermore, the double mutation of R804A in a K800A background does not alter elongation inhibition more than the K800A mutation alone, suggesting this loop is not vital for actin binding in FMNL3 (Supplementary Fig. 6).
Substitution of the two arginine residues in the post-actin binding interface also slows filament polymerization. Both R570A and R782A form a complex with TMR-actin (Table 2) and have similar elongation IC50 values to WT (approximately 2 nM); however, they are capable of inhibiting elongation more completely that WT (Imax values of 84% and 92% respectively) (Table 2). The ability of Group 3 mutants to alter actin elongation rates suggests the lasso/post region is critical in mediating the stepping activity of FMNL3 during processive elongation.
Assembly of filopodia by FMNL3 FH2 domain mutants
Transfection of an FH1-FH2 domain construct of FMNL3 is sufficient to induce filopodia when over-expressed in multiple cell lines21. To assess the effects of FH2 domain mutants on assembly of filopodia, we expressed FMNL3 mutants as GFP-FH1-FH2 constructs in Jurkat cells. As observed previously, the WT construct converts the cell surface from a ruffled morphology to one displaying abundant filopodia, while the Group 1 mutant (I649A) does not (Fig. 5a). Surprisingly, both the Group 2 and Group 3 mutants induce filopodia (Fig. 5a) with similar numbers and lengths as WT FH1-FH2 (Supplementary Fig. 8). However, two Group 2 mutants (N646A and M742A) display a striking difference to WT in localization of GFP-FH1-FH2 in the filopodium. While filopodia induced by WT display highly enriched GFP signal at their distal tips, those produced by the N646A and M742A mutants do not (Fig. 5). Quantification of GFP intensity along the filopodium shows a significant decrease in tip enrichment for N646A and M742A (Fig. 5). These results suggest that the N646A and M742A mutants are deficient in barbed end binding affinity, in agreement with their reduced abilities to protect from capping protein (Supplementary Fig. 7). In contrast, the K645A and Q742A group 2 mutants display similar tip intensities as WT, consistent with their increased abilities to protect from capping protein.
Figure 5. Effects of FMNL3 mutants on formation of filopodia in Jurket cells.
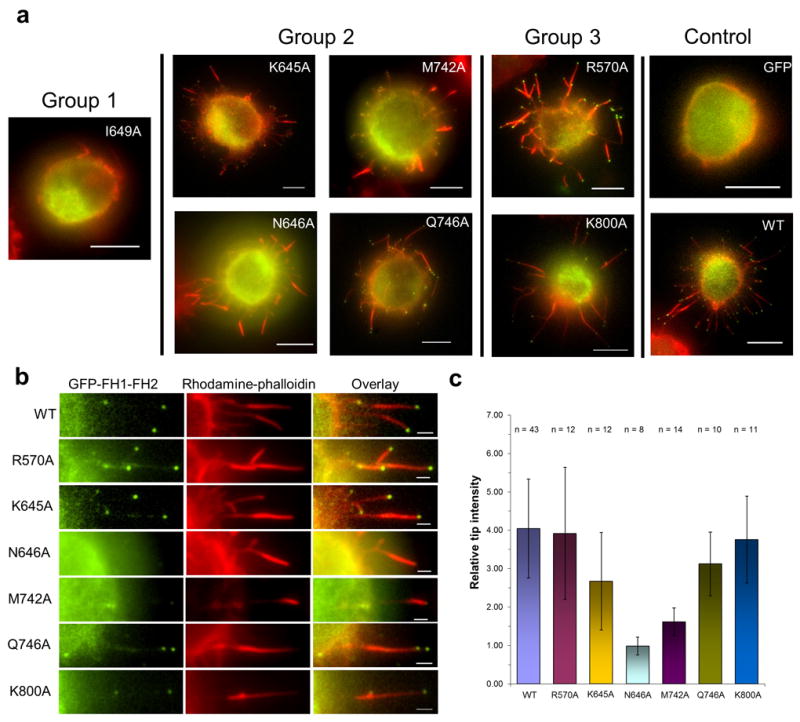
Expression of GFP alone or FH1-FH2 GFP-fusion constructs of FMNL3 (WT or Group 1, 2, or 3 mutants) in Jurkat cells for 6 hours followed by formaldehyde fixation and actin filament staining with rhodamine phalloidin. (a) Surface formation of filopodia was observed for all FMNL3 mutant constructs except for I649A (Scale bar is 5 μm). (b) Enlarged image of filopodia for WT and each of the mutant constructs. N646A and M742A do not display the characteristic GFP localization at the tips of the filopodia as seen for WT. Scale bar is 1 μm. (c) Quantification GFP intensity at tips of individual filopodia shows that intensity is reduced significantly for N646A and M742A. Error bars represent standard deviation (n > 10 from two experiments).
Discussion
Molecular mechanism for FH2 mediated actin filament elongation
A nucleating ratchet model for formins has been proposed5 in which the FH2 domain exists in two conformations. In the accessible, open state, the dimer binds two actin subunits with a free binding site available, suggesting FH2 stepping occurs prior to actin addition. In the blocked, closed state, the FH2 dimer interacts with three actin subunits (Fig. 6). The FH2 dimer acts as a Brownian ratchet, gated by translocation of the FH2 domain as it tracks with the end of the actin filament5. More recent studies of Bni1p processivity suggests a “stepping second” model, in which advancement of the FH2 dimer follows monomer addition9,23 The structure of FMNL3 FH2-actin suggests a distinct step in the processive model in which all actin binding sites of the FH2 dimer are occupied, interacting with only two actin subunits (Fig. 6). While this open state, capable of adding a actin subunit, can be incorporated into either the nucleating ratchet 5 or the stepping second model9,23,24, we use the latter as the basis for our model.
Figure 6. Model for FMNL3 mediated actin assembly.
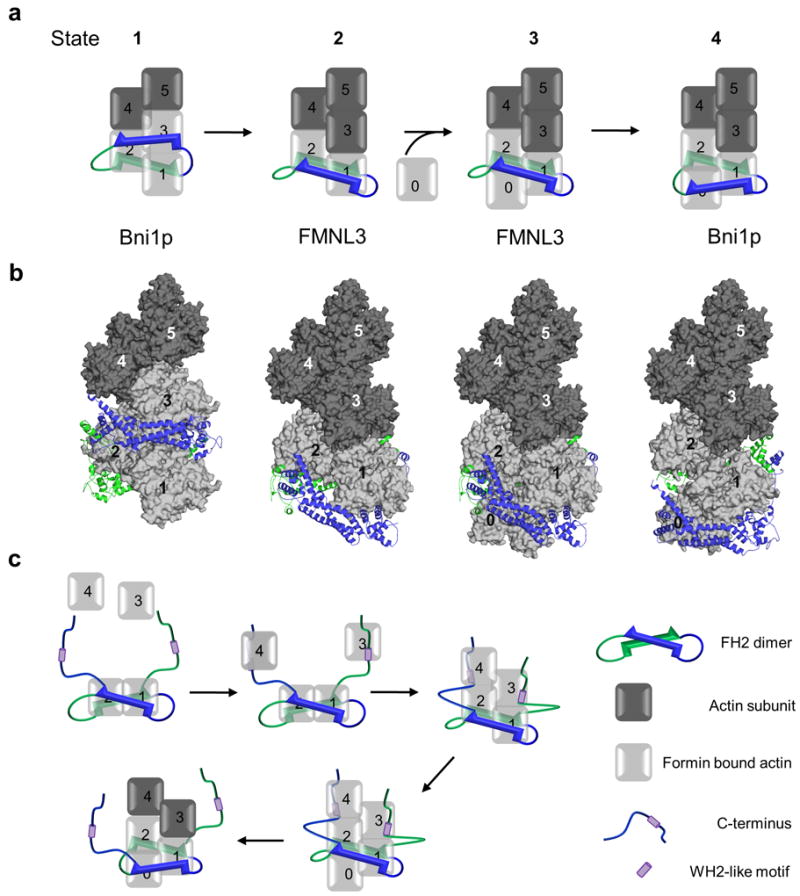
(a,b) Model of a 4-step elongation mechanism in which the FH2 dimer moves processively with the growing actin filament. An FH2 dimer (blue and green) forms a complex with either two (FMNL3 complex) or three (Bni1p complex) actin subunits at the barbed end (light grey surface). Numbered actin subunits aligned in a filament based on the Holmes model are shown in dark gray (surface). The translocation one subunit of the FH2 dimer (blue) is modeled while the second subunit (green) remains stationary. Panel a shows a schematic representation of the structure based filament model in b with the barbed ends of the actin filament oriented down. See Online Methods for a detailed description of the model design. (c) Model describing FMNL3 FH2-C mediated actin filament nucleation activity. The C-terminus alone is capable of binding actin monomers through a WH2-like motif allowing for the formation of an actin filament nucleus where the FH2 domain binds two actin subunits (as seen in the crystal structure) and the WH2-like motifs position additional monomers in close proximity to the FH2 bound actin subunits in a manner that favors nucleation. The WH2-like motifs then quickly release from their bound monomers in order for the FH2-mediated processive elongation to occur.
FH2 subdomains must alternately bind and release actin in order to create an accessible site for subsequent actin subunit addition. When bound to the FH2 dimer, actin subunits are in a strained (i.e. non-filament) orientation. Upon release from the FH2 dimer, they can adopt a favorable filament conformation. It is known that actin filaments can tolerate slight variations in subdomain and subunit position induced by actin binding proteins 25,26, and therefore the deviations from ideal filament contacts27 are not unexpected.
To model States 2 and 3 using the FMNL3 FH2-actin structure, it is necessary to break the two-fold symmetry observed in the crystal structure. Translation of one actin-FH2 domain by 20 Å in the direction of the barbed end of the filament (Supplementary Fig. 9) results in a complex structure with linker lengths similar to those proposed for Bni1p5. Although this translation disrupts interactions at the lasso/post interface, mutational analysis suggests these contacts are not critical. This results in an open conformation in which the FH2 dimer is bound to two actin subunits, allowing for actin subunit addition (Fig. 6 and Supplementary Fig. 9). Addition would then induce transition of the FH2 domain from the penultimate actin to the terminal actin.
Functional significance of the FH2-actin binding regions
Biochemical analysis of each of the actin binding regions of the FH2 domain identifies residues key to the elongation mechanism. Mutants in groups 1 and 2 affect binding and/or elongation activity. In cells, only the Group 1 mutant, I649A, cannot cause assembly of filopodia, while cells transfected with mutants in Group 2 are able to generate filopodia but show varying degrees of FMNL3 tip localization correlating with the abilities to protect filament barbed ends from capping protein (Table 2). Predictably, expression of Group 3 mutants that slow filament elongation do not exhibit defects in the number or length of filopodia, or on the localization on FMNL3 (Fig. 5 and Supplementary Fig. 8).
It is intriguing that all group 2 and 3 mutants maintain interactions with barbed ends, but vary in elongation rate. This suggests alterations in gating, which refers to the percentage of time an FH2 remains in the open and closed states. The decreased gating of group 2 mutants suggests more time spent in the open state, whereas the decreased gating of group 3 mutants suggests more time in the closed state. It will be interesting to compare elongation rates in the presence of profilin, which accelerates elongation and shifts the rate-limiting step to profilin-actin binding to the FH1 domain9.
Of the two often-mutated actin-binding residues in the FMNL3 FH2 domain, only mutation of I649 is sufficient to abolish actin assembly activity. Mutation of K800 merely slows elongation, whereas this mutation disables processive capping in Bni1p5. The increased inhibition of elongation in the K800A mutation suggests an effect on FH2 gating, potentially stalling the processivity through a decrease in anticoopertivity between FH2 subunits5 and slowing filament elongation.
In addition to the knob and post FH2-actin binding interfaces that have been identified previously5, interaction of the coiled-coil subdomain of FMNL3 with actin also contributes to the mechanism, perhaps explaining the ability of FMNL3 FH2 to bind to monomeric actin. The coiled-coil-actin binding interface could also compensate for the weaker actin binding by the lasso/post subdomains and explain why no mutation in this region was capable of disrupting an FH2-actin complex.
Intriguingly, inclusion of the FH1 domain enhances barbed end binding, as suggested by the increased ability to protect barbed ends from capping protein (Table 2). Possible explanations include increased stability of the FH2 dimer during elongation, or contribution to barbed end affinity. FH2 domains of both FMNL115 and FMNL3 are more dynamic dimers than are those of mDia115 and Bni1p19 (HNH, unpublished observations), raising the possibility that sequences N-terminal to the FH2 could stabilize dimers of FMNL formins.
FMNL3 FH2-C actin filament nucleation model
The capacity of FMNL3 FH2 to form a complex with actin monomers could explain why the it is such a poor nucleator, as such a complex may inhibit conversion to a productive actin nucleus. In contrast, filament nucleation readily occurs using a construct containing both the FH2 and the C-terminus16 (Fig. 1, amino acids 955-1027). The C-terminus contains a WH2-like motif that tightly binds actin monomers16. Alignment of the WASP WH2-actin structure28 with the FMNL3 FH2-actin structure shows helix D occupying a very similar binding site to the WH2 (Supplementary Fig. 9), suggesting FH2 and C-terminal domains cannot simultaneously bind to a single actin subunit, leading to a filament nucleation model (Fig. 6). In this model, the FH2 binds two actin subunits while the WH2-like motifs interact independently with additional actin monomers, bringing four subunits together (Supplementary Fig. 9). This nucleation model could be specific to formins that are poor nucleators, whereas the nucleating ratchet model5 would apply to rapid nucleators. However, as the C-terminus enhances FH2-mediated nucleation for a variety of formins independently15,17 or in concert with actin nucleation promoting factors29,30, this model might be more broadly applicable.
Methods
Cloning, expression, and purification of recombinant proteins
The FH2 domain of mouse FMNL3 (amino acids 555-954) was cloned by PCR into the pGEX-KT vector as described previously16 with the additon of a 6X Histidine tag followed by a tobacco etch virus (TEV) protease cleavage site added via the BamH1 site within the multiple cloning region31. The QuikChange Site Directed Mutagenisis protocol (Stratagene) was performed using enzymes from New Enlgand Biolabs to create FH2 domain mutants with alanine substitutions for R570, K645, N646, I649, R652, K653, M742, Q746, R782, K800, S801, T802, D803, R804, S840, N843, D847 and for the double mutant K800/R804. The FMNL3 FH1-FH2 domain containing construct (amino acids 504-954) in the eGFP-C1 vector (Clonetech) used for mammalian cell culture was constructed as previously described21.
The same protein expression scheme was used to generate GST-fusion WT and the mutant FMNL3 FH2 domains using a modified protocol described previously32. Protein expression constructs were transformed into Rosetta 2, non-DE3 E. coli (Novagen) and grown at 37°C to an OD600 of 0.8 – 1.0. The cultures were then transferred to 16°C and protein expression was induced overnight with the addition of 0.5 mM isopropyl-1-thio-β-galactopyranoside.
Following protein expression, all purification steps were performed at 4°C or on ice. The bacterial cells were pelleted at 1,500xg, resuspended in EB (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 5 mM EDTA, 1 mM DTT, 1 pill/50 mL of Complete protease inhibitors (Roche)), and lysed by sonication. The cell debris was removed by ultracentrifugation at 125,000xg and the supernatant was applied to a glutathione-Sepharose 4B (GE Bioscience) column equilibrated in WB (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 5 mM EDTA, 1 mM DTT, 0.05% thesit (Sigma)). The GST-fusion FMNL3 bound beads were thoroughly washed in WB followed by WB without thesit to remove the detergent and prepare the column for digestion with TEV protease 31. The protease was added at a 1:20 ratio of TEV to fusion protein to 50% slurry of beads and the suspension was mixed at 4°C for 16 hours. Following digestion, the cleaved, untagged FMNL3 was washed from the column in WB without thesit. The eluted protein was concentrated with a 30,000 molecular weight cutoff Amicon Ultra-15 (Millipore) to remove the remaining TEV protease and to prepare the FMNL3 for further purification using gel filtration chromatography. Concentrated FMNL3 protein was loaded onto a Superdex 200 16/60 column (GE Bioscience) and eluted over one column volume in the final buffer (5 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, and 1 mM DTT) using the ÄKTAexplorer system (GE Bioscience) to separate the untagged protein from any remaining GST-fusion protein. Fractions containing only untagged FMNL3 protein were pooled and concentrated to 7-10 mg/mL for crystallization.
The plasmid to express capping protein (human α1β2) was a gift from John Cooper (Washington University St. Louis). After bacterial expression, protein was purified by Ni-NTA affinity chromatography (Qiagen) and Superdex200 gel filtration (GE Biosciences) and stored frozen in 10 mM imidazole pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT.
Actin purification and labeling
Rabbit skeletal muscle actin was purified from acetone powder (Spudich and Watt, 1971). Tetramethylrhodamine-5’-maleimide (TMR) (Molecular Probes) was used to label monomeric actin as described previously19.
Crystallization and structure determination
To form the protein complex used for crystallization, FMNL3 FH2 and TMR-actin were mixed at a 2:1 molar ratio of formin to actin. The mixture was gel filtered over a Superose 6 10/300 column (GE Bioscience) in 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, and 10 mM imidazole pH 7.0. The complex eluted as a single species and the peak fractions were pooled and concentrated in an Amicon Ultra-15 (Millipore). The concentration of the complex used for crystallization was not determined due to the interference from the TMR probe when measuring protein concentration at 280 nm. Therefore, absorbance measurements were taken throughout concentration of the complex at 557 nm, the peak absorbance for TMR-actin, until an A557 of 3.5 was obtained for use in crystallization.
Crystals of FMNL3 FH2 bound to TMR-actin were grown at 20°C under hanging drop vapor diffuse conditions. Optimal crystals formed when the protein complex was mixed with the reservoir solution containing 11% PEG 10000, 0.2 M magnesium acetate, 0.05 M MES pH 6.5, and Silver Bullet #33 and #70 (Hampton Research). A typical drop consisted of 2 μl FMNL3 FH2/TMR-actin complex, 1 μl reservoir solution, 0.5 μl Silver Bullet #33, and 0.5 μl Silver Bullet #70. Crystal appeared in 2-3 days and continued to grow for an additionally 5-6 days. The crystals grew to be 120×120×40 μm and were flash-frozen in liquid nitrogen with 30% glycerol for a cryoprotectant.
Diffraction data was collected using the GM/CA CAT 23-ID-B beamline at the Advanced Photon Source, Argonne National Laboratory. A 3.4 Å resolution data set was processed using the program XDS34 (Table 1). The X-ray structure of the FMNL3 FH2-actin complex was solved by molecular replacement with AutoMR followed Autobuild in PHENIX35 using a threaded model generated by the Phyre server36 of FMNL3 FH2 (based on the DAAM1 FH2 domain structure 2J1D7) and a TMR-actin structure5 as search models. The structure was refined through an iterative process of model building using COOT37 and refinement in which NCS restraints were applied using phenix.refine 38.
Analytical ultracentrifugation
Analytical ultracentrifugation was performed using a Beckman Proteomelab XL-A ultracentrifuge and an AN-60 rotor with protein suspended in buffer containing 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, and 10 mM Imidazole pH 7.0. Velocity sedimentation experiments were conducted using WT FMNL3 FH2 as well as mutant constructs of the same domain. WT and mutant FMNL3 at a concentration of 9.4 μM was centrifuged at 35,000 rpm at 20°C in the presence of 4.7 μM TMR-actin. The absorbance was monitored at 557 nm every minute using a continuous scan method with a radial step size of 0.003 cm. Scans 1-200 were analyzed using Sedfit 8739 and the buffer density and viscosity and the partial specific volume were calculated using the Sednterp program (developed by David Hayes & Tom Laue). Sedimentation equilibrium analytical ultracentrifugation was performed and analyzed as described previously32. The absorbance was monitored at 557 nm for pelleting speeds of 7,000, 10,000, and 14,000 rpm with TMR-actin concentrations of 4.7, 2.35, and 1.18 μM and excess FMNL3 FH2 at concentrations of 60, 30, and 15 μM respectively to prevent dissociation of the complex. The data was fit to a single species with a predicted molecular weight of 165 kDa.
Barbed end elongation assays
Barbed end elongation assays were performed as previously described16. Fluorescence (365/410 nm) was monitored in a 96-well fluorescence plate reader (Tecan Infinite M1000, Mannedorf Switzerland) for 2700 s. Slopes of pyrene fluorescence from elongation time courses were taken at 10% completion using Kaleidagraph (Synergy Software, Reading, PA) and were converted to elongation rates (assuming 10 μM-1 s-1 for actin alone40). For capping protein competition assays, the formin construct (100 nM( was pre-mixed with filament seeds (1.5 μM) for 50 sec, then actin monomers were added (0.5 mM, 20% pyrene) and elongation was measured for 50 sec. Capping protein (20 nM) was then added and elongation measurement continued.
Cell culture and fluorescence microscopy
Jurkat T-cell (human) cells (American Type Culture Collection) were transfected as previously described21. Images were acquired using Nikon TE-2000E microscope with 100× 1.4 NA objective, a Roper Cool Snap camera, and Nikon Elements software version 3.22. Cells were measured for number of filopodia per cell and filopdium length using Elements software. Relative tip intensity was calculated by taking a line scan perpendicular at the filopodium tip and divided by the line scan perpendicular at the halfway toward the cell body along the filopodium.
Supplementary Material
Acknowledgments
We wish to thank our phenomenal undergraduates Anne Kelley for mutagenesis and cloning and Ming Lee for crystallization trials and screening. We thank Dr. James Moseley for thoughtful comments and careful reading of the manuscript. We also thank Stephen Corcoran and the staff at the GM/CA CAT 23-ID-D beamline at Argonne National Laboratories Advance Photon Source and Drs. Christopher Bahl and Dean Madden for help with the structural determination and characterization. This work was supported by National Institutes of Health grant R01 GM069818 (to H.N.H) and F31 GM089149 (to E.G.H) as well as Howard Hughes Medical Institute predoctoral fellowship 52006921 and National Science Foundation GK-12 fellowship 0947790 (to M.E.T).
Footnotes
Author Contributions
M.E.T. purified, crystalized, and characterized the FH2-actin complex and also collected data on the crystals and determined the structure. E.G.H. performed the pyrene actin assays. T.J.G. carried out the cellular characterization and analysis of the mutant protein constructs. The experimental design and data analysis was carried out by M.E.T., H.N.H., and F.J.K. The manuscript was prepared by M.E.T., H.N.H., and F.J.K.
Accession Codes
Atomic coordinates and structure factor files for the FMNL3 FH2-TMR-actin complex have been deposited in the Protein Data Bank under ID code 4EAH.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Pollard TD, Cooper Ja. Actin, a central player in cell shape and movement. Science. 2009;326:1208–12. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang F, Drubin D, Nurse P. Cdc12P, a Protein Required for Cytokinesis in Fission Yeast, Is a Component of the Cell Division Ring and Interacts With Profilin. The Journal of cell biology. 1997;137:169–82. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annual review of biochemistry. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–23. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 5.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–94. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 6.Shimada A, et al. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Molecular cell. 2004;13:511–22. doi: 10.1016/s1097-2765(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, et al. Structure of the FH2 domain of Daam1: implications for formin regulation of actin assembly. Journal of molecular biology. 2007;369:1258–69. doi: 10.1016/j.jmb.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita M, et al. Crystal structure of human DAAM1 formin homology 2 domain. Genes to cells : devoted to molecular & cellular mechanisms. 2007;12:1255–65. doi: 10.1111/j.1365-2443.2007.01132.x. [DOI] [PubMed] [Google Scholar]
- 9.Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell motility and the cytoskeleton. 2009;66:606–17. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shemesh T, Otomo T, Rosen MK, Bershadsky AD, Kozlov MM. A novel mechanism of actin filament processive capping by formin: solution of the rotation paradox. The Journal of cell biology. 2005;170:889–93. doi: 10.1083/jcb.200504156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno H, et al. Rotational Movement of the Formin mDia1 Along the Double Helical Strand of an Actin Filament. Science. 2011;331:80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- 12.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–35. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 13.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. The Journal of biological chemistry. 2006;281:14383–92. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 14.Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. The Journal of biological chemistry. 2005;280:28023–33. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- 15.Gould CJ, et al. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Current biology. 2011;21:384–90. doi: 10.1016/j.cub.2011.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heimsath EG, Higgs HN. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. The Journal of biological chemistry. 2012;287:3087–98. doi: 10.1074/jbc.M111.312207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vizcarra CL, et al. Structure and function of the interacting domains of Spire and Fmn-family formins. :1–6. doi: 10.1073/pnas.1105703108/-/DCSupplemental. www.pnas.org/cgi/doi/10.1073/pnas.1105703108. [DOI] [PMC free article] [PubMed]
- 18.Moseley J, Sagot I, Manning A. A conserved mechanism for Bni1-and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Molecular biology of. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graceffa P, Dominguez R. Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. The Journal of biological chemistry. 2003;278:34172–80. doi: 10.1074/jbc.M303689200. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski Ra. PDBsum new things. Nucleic acids research. 2009;37:D355–9. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris ES, Gauvin TJ, Heimsath EG, Higgs HN. Assembly of filopodia by the formin FRL2 (FMNL3) Cytoskeleton (Hoboken, N J) 2010;772:755–772. doi: 10.1002/cm.20485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott BJ, Neidt EM, Kovar DR. The functionally distinct fission yeast formins have specific actin-assembly properties. Molecular biology of the cell. 2011;22:3826–39. doi: 10.1091/mbc.E11-06-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Current biology : CB. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul AS, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. The Journal of biological chemistry. 2009;284:12533–40. doi: 10.1074/jbc.M808587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E. Remodeling of actin filaments by ADF / cofilin proteins. 2011 doi: 10.1073/pnas.1110109108/-/DCSupplemental. www.pnas.org/cgi/doi/10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed]
- 26.Galkin VE, Orlova A, Schröder GF, Egelman EH. Structural polymorphism in F-actin. Nature structural & molecular biology. 2010;17:1318–23. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes KC, Angert I, Kull FJ, Jahn W. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003:423–427. doi: 10.1038/nature01927.1. [DOI] [PubMed] [Google Scholar]
- 28.Chereau D, et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16644–9. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graziano B, et al. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Biology of the Cell. 2011;22:4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breitsprecher D, et al. Rocket Launcher Mechanism of Collaborative Actin Assembly Defined by Single-Molecule Imaging. Science. 2012;336:1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapust RB, et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein engineering. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 32.Harris ES, Li F, Higgs HN. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. The Journal of biological chemistry. 2004;279:20076–87. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- 33.Spudich Ja, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. The Journal of biological chemistry. 1971;246:4866–71. [PubMed] [Google Scholar]
- 34.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of applied crystallography. 1993;26:795–800. [Google Scholar]
- 35.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley La, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Afonine PV, et al. Joint X-ray and neutron refinement with phenix.refine. Acta crystallographica. Section D, Biological crystallography. 2010;66:1153–63. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophysical journal. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophysical journal. 2005;88:1387–402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


