Abstract
Initiation of protein synthesis on picornavirus RNA requires an internal ribosome entry site (IRES). Typically, picornavirus IRES elements contain about 450 nucleotides (nt) and use most of the cellular translation initiation factors. However, it is now shown that just 280 nt of the porcine teschovirus type 1 Talfan (PTV-1) 5′ untranslated region direct the efficient internal initiation of translation in vitro and within cells. In toeprinting assays, assembly of 48S preinitiation complexes from purified components on the PTV-1 IRES was achieved with just 40S ribosomal subunits plus eIF2 and Met-tRNAiMet. Indeed, a binary complex between 40S subunits and the PTV-1 IRES is formed. Thus, the PTV-1 IRES has properties that are entirely different from other picornavirus IRES elements but highly reminiscent of the hepatitis C virus (HCV) IRES. Comparison between the PTV-1 IRES and HCV IRES elements revealed islands of high sequence identity that occur in regions critical for the interactions of the HCV IRES with the 40S ribosomal subunit and eIF3. Thus, there is significant functional and structural similarity between the IRES elements from the picornavirus PTV-1 and HCV, a flavivirus.
For the vast majority of eukaryotic cell mRNAs the initiation of protein synthesis is achieved after the recognition of the cap structure (m7GpppN), located at the 5′ terminus of all cytoplasmic mRNAs, by the cap-binding complex eIF4F (reviewed in reference 12). This translation initiation factor is a heterotrimer comprising eIF4E (which binds to the cap), eIF4A (which has RNA helicase activity), and eIF4G (a scaffold protein). The eIF4F complex acts as a bridge between the mRNA and the small ribosomal subunit. The eIF4G subunit has binding sites for a number of proteins, as well as eIF4E and eIF4A (two independent sites); these proteins include eIF3 (associated with the 40S ribosome subunit), poly(A)-binding protein, and Mnk-1 (an eIF4E-kinase). The importance of eIF4G is demonstrated by the observation that cleavage of this protein occurs within cells infected by many different picornaviruses (e.g., poliovirus and foot-and-mouth disease virus [FMDV]) and results in the inhibition of cellular protein synthesis (for a review, see reference 2). The translation of picornavirus RNA is not inhibited under these conditions (except for hepatitis A virus [HAV] RNA) (3).
Picornavirus RNAs are uncapped and the initiation of translation on picornavirus RNA occurs up to 1,300 nucleotides (nt) away from the 5′ terminus. This process is directed by an internal ribosome entry site (IRES). The picornavirus IRES elements are generally about 450 nt in length (2). Two major classes of picornavirus IRES element have been defined. The entero- and rhinovirus IRES elements constitute one class, whereas the cardio- and aphthovirus IRESs (including the encephalomyocarditis virus [EMCV] and FMDV elements) make up a second group of elements. The HAV IRES (see reference 6) has different characteristics from these two major groups; for example, it requires the intact eIF4F complex (1, 3). We have recently characterized the IRES element from the porcine teschovirus type 1 (PTV-1) Talfan (16). This virus causes polioencephalomyelitis in swine and has been classified recently as the prototype member of the teschovirus genus within the picornavirus family (32). It was shown that the PTV-1 IRES is present within a sequence of 405 nt (16), but the boundaries of this element were not defined.
All picornavirus IRES elements are predicted to contain extensive secondary structure but these structures are completely different between the distinct classes of IRES element. Except for the PTV-1 IRES (16), they all have a polypyrimidine tract about 20 nt upstream of an AUG codon near the 3′ terminus of the IRES (for reviews, see references 2 and 23). There is no significant sequence similarity between the different classes of picornavirus IRES element.
The cardio- and aphthovirus IRES elements and the teschovirus IRES each function efficiently in the rabbit reticulocyte lysate (RRL) system and within mammalian cells in the presence or absence of the enterovirus 2A protease (16, 35), which induces the cleavage of eIF4G. In contrast, the entero- and rhinovirus IRES elements function rather poorly in the RRL system unless it is supplemented with HeLa cell proteins (5, 8). Within BHK cells, the activity of these IRES elements is rather low but is strongly stimulated by the coexpression of an enterovirus 2A protease (4, 35, 36).
IRES elements have also been identified and characterized from within the genomes of hepaciviruses (HCV) (33, 41) and pestiviruses (e.g., classical swine fever virus [CSFV] (10, 28) which are members of the flavivirus family. These IRES elements are about 300 to 350 nt in length and are unrelated to the well-characterized picornavirus IRES elements. They also contain extensive secondary structure, including a pseudoknot, which is important for activity, just upstream of the initiation codon (10, 28, 42). Some evidence suggests that the coding sequence, downstream of the IRES, is also important for the activity of the HCV IRES, but at least in certain circumstances this is not the case (33, 34).
In recent years, it has become possible to monitor the reconstitution of the initiation step of protein synthesis by using toeprinting assays with purified initiation factors, ribosomal subunits and RNA transcripts (see, for example, references 26, 27, and 31). These studies have shown that the assembly of 48S preinitiation complexes on FMDV and EMCV IRES elements functions most efficiently with the initiation factors eIF2, eIF3, eIF4A, eIF4B, and eIF4F, together with some auxiliary RNA-binding proteins, e.g., the polypyrimidine tract-binding protein (PTB). In contrast, the assembly of these complexes on HCV or CSFV IRES elements functions efficiently with only the factors eIF2 and eIF3 (28). Indeed, a binary complex simply between 40S subunits and the HCV IRES can be observed. The initiation factor eIF4E is not required by any of the viral IRES elements except for HAV (1, 3). Similarly, the factors eIF1 and eIF1A are only necessary for translation initiation when ribosomes have to scan along the RNA to reach the initiation codon (29). There are currently no reports of reconstitution of translation initiation with purified components on any enterovirus or rhinovirus IRES element. It is known, however, that these elements continue to function when eIF4G is cleaved. Furthermore, the ability of the dominant-negative mutants of eIF4A to block poliovirus IRES function (25, 40) suggests that the residual C-terminal fragment of eIF4G, together with eIF4A, is required for translation initiation.
We have now defined the boundaries of the PTV-1 IRES and identified its requirements for the formation of 48S preinitiation complexes. The results indicate that the properties of the PTV-1 IRES are entirely different from all other known picornavirus IRES elements.
MATERIALS AND METHODS
Plasmid construction.
All DNA manipulations were performed by using standard methods (37).
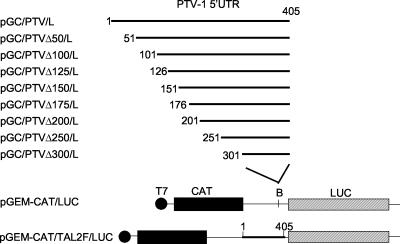
The reporter plasmid pGEM-CAT/LUC has been described previously (35), it expresses from a T7 promoter a dicistronic mRNA encoding chloramphenicol acetyltransferase (CAT) and luciferase (LUC) (see Fig. 1), each cistron has its own initiation and termination codons. A heminested set of cDNA fragments corresponding to regions of the PTV-1 Talfan 5′-untranslated region (5′UTR) were generated by PCR with one of a series of forward primers plus the reverse primer TAL2R (Table 1) by using the pGEM-IB plasmid (see Fig. 1) (16) as a template. The fragments obtained were purified and digested with BamHI, and the products were ligated into BamHI-linearized and phosphatase-treated pGEM-CAT/LUC to generate the pGC/PTV/L series of plasmids shown in Fig. 1. The orientation and veracity of the PTV-1 cDNA was confirmed by DNA sequencing.
FIG. 1.
Structure of plasmids used to determine the boundaries of the PTV-1 IRES element. The cDNA fragments indicated were generated by using the PCR as described in Materials and Methods and inserted into the BamHI (B)-digested vector pGEM-CAT/LUC (35). The plasmid pGEM-CAT/TAL2F/LUC was described previously (16) and is shown for completeness.
TABLE 1.
PCR primers
| Primer | Sequence (5′-3′)a | Location (nt) on PTV RNA |
|---|---|---|
| TAL2R | CCCGGATCCTTTCAACTGACTATACAAAGT | 405-385 |
| BT1F | AGCGGATCCTCTGGACTTGTAACTGGTAA | 1-20 |
| BT51F | AGCGGATCCACGCTAGTTTTGGATTATCT | 51-70 |
| BT101F | AGCGGATCCTGTGTTTAAACACAGAAATC | 101-120 |
| BT126F | AGCGGATCCACTTGGTTATGAATTCATTG | 126-145 |
| BT151F | AGCGGATCCACCCCTCTGAAAGACCTGCT | 151-170 |
| BT176F | AGCGGATCCCGAGCTAAAGCGCAATTGTC | 176-195 |
| BT201F | AGCGGATCCGGTATTGCACCAATGGTGGC | 201-220 |
| BT251F | AGCGGATCCCTGGGTAATGGGACTGCATT | 251-270 |
| BT301F | AGCGGATCCGTGCTGCCTGACAGGGTCGC | 301-320 |
All of the primers include a BamHI site (indicated in italics).
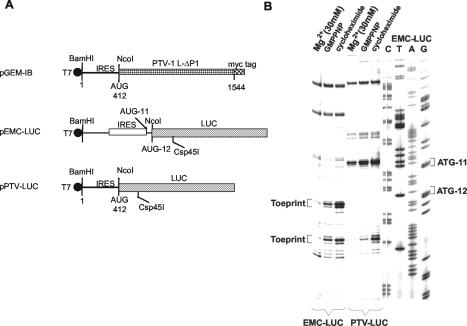
Plasmid pGEM3Z/J1 expresses the 1D-2A region from an enterovirus, swine vesicular disease virus, under the control of a T7 promoter and has been described previously (36). Plasmid pEMCV-LUC was constructed by inserting the NcoI-PstI fragment, including the LUC sequence from pGL3R (Promega), between the NcoI and PstI sites of plasmid pTE1 (9) that harbors the EMCV IRES. The plasmid pPTV-LUC was prepared by removing the IRES from pEMCV-LUC by using BamHI and NcoI and replacing it with the BamHI-NcoI fragment, including the PTV-1 IRES cDNA from pGEM-IB (16).
Expression assays.
In vitro transcription-translation (TNT) reactions were performed within RRL by using the Promega TNT T7 kit with [35S]methionine essentially as described by the manufacturer. The T7 LUC control DNA (T7 LUC; Promega) was included as a marker. Aliquots (3 μl) of the reactions were removed after 120 min of incubation, mixed with SDS plus DTT sample buffer, boiled, and then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (20) and autoradiography.
Transient-expression assays within cells infected with the recombinant vaccinia virus vTF7-3 (11), which expresses the T7 RNA polymerase, were performed as described previously (35). Briefly, BHK cells were infected with vTF7-3 and transfected by using Lipofectin (8 μg; Life Technologies) with plasmid DNA (2.5 μg). In some of these assays, cotransfection of the reporter plasmids (pGEM-CAT/LUC derivative, 2 μg) with pGEM3Z/J1 (0.5 μg) was performed. In each case, the cell extracts were prepared after 20 h and analyzed by SDS-PAGE, followed by Western blotting with antibodies specific for CAT (5prime-3prime, Inc.) and LUC (Promega) as required. Detection was achieved by using peroxidase-labeled anti-species antibodies (Amersham) and chemiluminescence reagents (Amersham). LUC expression was also quantified by using a LUC assay kit (Promega) and a luminometer (Bio-Orbit).
In vitro transcription reactions.
The plasmid pPTV-LUC was linearized within the LUC coding sequence by using Csp45I, runoff RNA transcripts of ca. 530 nt were prepared by using T7 RNA polymerase and purified by phenol extraction and ethanol precipitation. The size and integrity of transcripts was checked by gel analysis.
Preparation of factors, ribosomal subunits, and Met-tRNAiMet.
40S ribosomal subunits, Met-tRNAiMet, and the native translation initiation factors eIF2, eIF3, eIF4A, and eIF4B were prepared as previously described (24); the preparations of eIF2 and eIF3 were then further purified by using fast-performance liquid chromatography, and eIF4F was purified as described previously (13). Recombinant eIF1 and eIF1A were prepared as described previously (29).
Assembly and toeprinting of 48S and 80S complexes in nuclease-treated RRL.
To assemble the translation initiation complexes, nuclease-treated RRL (Promega) was used with the addition of either guanylimidodiphosphate (GMPPNP, 0.4 mM) or cycloheximide (0.5 mg/ml) as described previously (7). Briefly, the mixtures were incubated for 5 min at 30°C, and the RNA transcripts were added and incubated for another 5 min. The 5′-32P-labeled toeprint primer (5′-GCAATTGTTCCAGGAACC-3′), RT-Mix (including deoxynucleotide triphosphates), and avian myeloblastosis virus reverse transcriptase (Roche) were then added, and the samples were incubated for 15 min at 30°C. The mixtures were analyzed by using a 6% sequencing gel. The cDNA products generated in the toeprint assay were compared to a dideoxynucleotide sequence ladder obtained by using the same primer with the pPTV-LUC or pEMCV-LUC plasmid. These two plasmids contain identical sequences downstream of the LUC initiation codon.
Reconstitution of 48S translation initiation complexes from purified components.
48S complexes were assembled from purified components as previously described (7, 26, 30). Briefly, the 48S complexes were assembled by incubating the RNA transcripts for 10 min at 30°C in the reconstitution buffer (20 mM Tris-HCl [pH 7.5], 100 mM potassium acetate, 2.5 mM magnesium acetate, 0.1 mM EDTA, 1 mM dithiothreitol) with 1 mM ATP, 0.4 mM GMPPNP, 0.25 mM spermidine-HCl, Met-tRNAAiMet (1.5 pmol), eIF1 (0.5 μg), eIF1A (0.5 μg), eIF2 (2 μg), eIF3 (3 μg), eIF4A (0.5 μg), eIF4B (0.5 μg), eIF4F (0.5 μg), PTB (0.6 μg), and 40S ribosomal subunits (0.15 A260). The mixture was incubated for 10 min at 30°C, and the primer extension assay was performed as described above. The cDNAs generated were analyzed by using 6% polyacrylamide sequencing gels. To assess the requirement for specific factors, individual components of the reconstitution mixture were omitted as indicated in the figure legends. In certain experiments, sucrose gradient centrifugation analysis of the assembled initiation complexes was performed as described previously (26).
Analysis of toeprint data.
Typically the yield of 48S translation initiation complexes is estimated from toeprint data by using the ratio of the integral intensity of toeprint bands to that of the full-length product of reverse transcription. This method is readily applicable to relatively short and unstructured 5′UTRs like that of β-globin mRNA. However, for long and highly structured transcripts (e.g., IRESs) the reverse transcriptase stops at many points before the 5′ end of mRNA, and these stops are often variable in intensity. Thus, it is preferable to compare gel lanes that have similar incorporation of the labeled primer above and including the toeprint signal. Thus, the toeprints that are presented in each figure contain similar amounts of 32P-labeled cDNAs, as determined with a phosphorimager, in different lanes.
RESULTS
Delimitation of the PTV-1 (Talfan) IRES element.
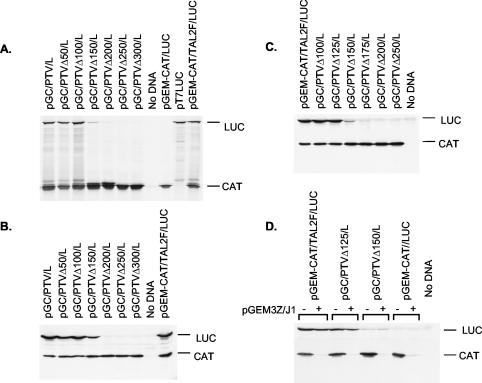
In our earlier studies (16), it was demonstrated that the initiation of protein synthesis occurs at an AUG codon (nt 412 to 414) on the known PTV-1 (Talfan) RNA sequence (15) and that an IRES element was contained within nt 1 to 405 (n.b., the 5′ terminus of the genomic PTV-1 RNA has not been cloned or sequenced). Deletion from nt 1 to 273 (or to 363), not surprisingly, abolished the IRES function (16). To define the limits of the PTV-1 IRES more precisely, a series of 5′ deletions were made, and the residual sequences were inserted into the pGEM-CAT/LUC vector (35) that expresses, from a T7 promoter, a dicistronic mRNA. The resulting plasmids (Fig. 1) were assayed by using in vitro-coupled TNT assays in RRL (Fig. 2A) and also by using transient-expression assays within BHK cells (Fig. 2B). The expression of LUC, as judged by [35S]methionine incorporation in the TNT assay or by immunoblotting of cell extracts, indicates the level of IRES activity. Loss of 50 or 100 nt from the 5′ end of the PTV-1 sequence had no effect on IRES function in either system but the loss of nt 1 to 150 significantly decreased the expression of LUC observed to ca. 20% of that detected from the complete IRES. Deletion of ≥200 nt completely inactivated the IRES (note that LUC activity assays on BHK cell extracts [data not shown] were entirely consistent with the LUC expression assays shown in Fig. 2B). Expression of the upstream cistron, CAT, was similar for each plasmid both within the RRL and as judged by immunoblotting of cell extracts (Fig. 2A and B).
FIG. 2.
Functional analysis of the boundaries of the PTV-1 IRES. (A) The indicated plasmids were used to program TNT reactions containing [35S]methionine. The products were analyzed by SDS-PAGE and autoradiography. (B and C) The plasmids indicated were used to transfect vTF7-3-infected BHK cells. After 20 h, cell extracts were prepared and analyzed for the expression of CAT and LUC by immunoblotting. (D) The indicated plasmids were assayed as for panels B and C either alone (−) or cotransfected with the plasmid pGEM3Z/J1 (36) that expresses an enterovirus 2A protease (+).
To refine further the boundaries of the IRES element, additional deletions were made that removed 125 or 175 nt from the 5′ terminus of the PTV-1 sequence (Fig. 1), and these constructs were assayed in the same way. Within BHK cells, it was apparent that the loss of nt 1 to 125 had no significant effect on IRES activity, but the removal of nt 1 to 175 completely inhibited IRES function (Fig. 2C). Similarly, in RRL, it was found that the loss of nt 1 to 125 had no effect on IRES activity (data not shown). From these results it was concluded that nt 126 to 405 of the PTV-1 (Talfan) RNA, i.e., 280 nt, are sufficient to direct efficient internal initiation of protein synthesis.
To determine whether this minimal element (nt 126 to 405) retained all of the characteristics of the IRES sequence contained within nt 1 to 405, the ability of these elements to direct internal initiation of protein synthesis in the presence of an enterovirus 2A protease was assayed (Fig. 2D). The expression of the enterovirus 2A protease induces eIF4G cleavage (36) and the inhibition of cap-dependent protein synthesis as indicated by the decrease in CAT expression. Consistent with earlier results (16), the IRES-directed LUC expression was maintained from the transcripts containing the PTV-1 nt 1 to 405 and, furthermore, the truncated 5′UTR sequence with just nt 126 to 405 also continued to direct efficient internal initiation of translation under these conditions (Fig. 2D). Thus, this 280-nt region from the PTV-1 5′UTR constitutes a fully functional IRES element.
48S preinitiation complex formation on the PTV-1 (Talfan) IRES.
The assembly of 48S preinitiation complexes and 80S initiation complexes on RNA transcripts containing the PTV-1 IRES was studied initially within RRL as described previously for the EMCV IRES element (7). Analysis of the 48S and 80S initiation complex formation in RRL, which contains all components necessary for translation initiation on the PTV-1 IRES (as described above), is an important prerequisite to the analysis of the formation of these complexes by using purified components. This assay determines whether the formation of 48S or 80S complexes with this IRES is efficient and whether the complexes are stable enough to be revealed by toeprinting. Otherwise, the potential absence of a toeprint signal in the reconstitution assays (see below) may not be interpreted unambiguously since it may result from either the absence of an essential component in the purified system or insufficient sensitivity of the assay. The assembly of the 48S and 80S initiation complexes was assayed by using toeprinting assays on a monocistronic RNA transcript (from pPTV-LUC, Fig. 3A) containing nt 1 to 414 of the PTV-1 sequence with a primer complementary to the LUC reporter sequence. For comparison, an analogous transcript containing the EMCV IRES was analyzed in parallel (as used previously [7]). In the presence of GMPPNP, a nonhydrolyzeable analog of GTP, stable assembly of 48S complexes on the PTV-1 RNA was observed as indicated by a toeprint at 16 to 18 nt downstream of the initiation codon (Fig. 3B). The location of the toeprint on the PTV-1 RNA is in the same position relative to the primer as the toeprint resulting from 48S complex formation at AUG-12 of the EMCV transcript. This result is consistent with the structure of the transcripts shown in Fig. 3A if initiation occurs at the PTV-1 AUG codon at nt 412 to 414 (as demonstrated previously [16]). The assembly of 48S preinitiation complexes on EMCV RNA occurs at both AUG-11 and AUG-12 in this assay (as seen previously [7, 26]). The assembly of 80S complexes on both the PTV-1 and EMCV RNA transcripts was assayed in a similar way but in the presence of GTP plus the inhibitor cycloheximide (to block translocation). Under these conditions a stronger toeprint signal was observed on both RNAs at the same positions (Fig. 3B). In the presence of a high concentration of Mg2+ (30 mM), the loss of all of the specific toeprints was observed, a result consistent with previous observations using the EMCV IRES (7). It should be noted that the formation of the 48S complexes in the presence of GMPPNP and the formation of 80S complexes in the presence of cycloheximide on the PTV-LUC mRNA was confirmed by sucrose gradient sedimentation (data not shown).
FIG. 3.
Assembly of initiation complexes on the PTV-1 IRES in rabbit reticulocyte lysate. (A) Monocistronic RNA transcripts containing the PTV-1 IRES (nt 1 to 412) were prepared by using T7 RNA polymerase from Csp45I linearized pPTV-LUC and pEMCV-LUC. The plasmid pPTV-LUC was constructed by introducing the BamHI-NcoI fragment from pGEM-IB (16) into similarly digested pEMCV-LUC (9). (B) The RNA transcripts were incubated in RRL in the presence of GMPPNP or with GTP plus cycloheximide or with 30 mM Mg2+ (as a negative control). The formation of the initiation complexes, as detected by the toeprints at 16 to 18 nt downstream of the AUG initiation codon, was assayed as described in Materials and Methods. The products were analyzed by gel electrophoresis and autoradiography in parallel with dideoxynucleotide DNA sequencing reactions.
Reconstitution of 48S preinitiation complex formation on PTV-1 RNA with purified components.
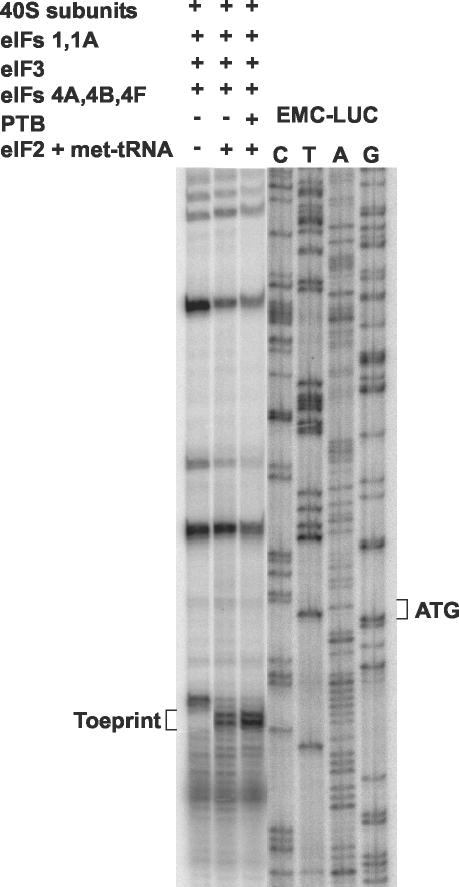
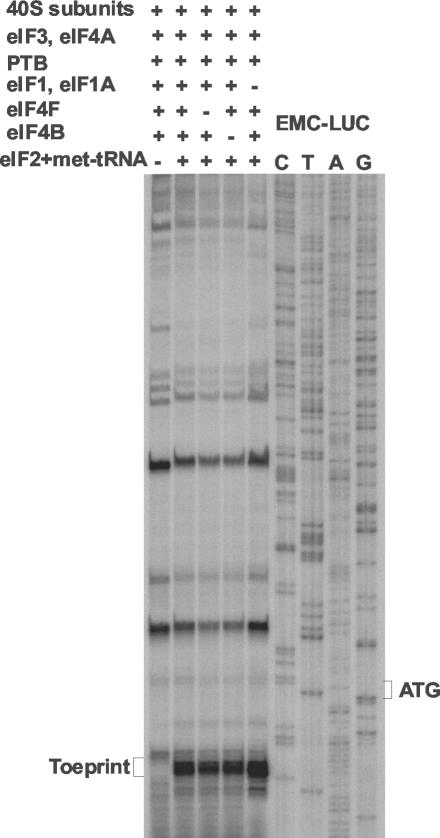
The assembly of 48S preinitiation complexes on the PTV-1 IRES by using purified components was assayed in the presence of each of the following initiation factors: eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, and eIF4F. In the presence of all of these factors together a toeprint indicative of 48S complex formation was observed (Fig. 4). The addition of PTB, which is needed for efficient 48S complex formation on certain picornavirus IRESs (2), only slightly increased the intensity of the toeprint signal on the PTV-1 RNA. This increase in complex formation was much less than that observed when the EMCV IRES was used in similar experiments (A. Pisarev and I. N. Shatsky, unpublished results). In the absence of eIF2 and Met-tRNAiMet, no toeprint at the +16 to +18 position was observed, but a weaker signal was detected closer to the AUG codon.
FIG. 4.
Reconstitution of 48S preinitiation complex formation on the PTV-1 IRES by using purified components. The formation of 48S preinitiation complexes was assayed on the PTV-LUC RNA transcript as shown in Fig. 3 but with purified components. Reactions contained the purified initiation factors plus PTB (0.6 μg) as indicated and a control reaction was performed in which eIF2 plus Met-tRNAAiMet were omitted. The position of the toeprint at 16 to 18 nt downstream of the initiation codon is indicative of 48S preinitiation complex formation. The sequence of the pEMC-LUC plasmid was used as a marker, as in Fig. 3 (note that the sequences of this plasmid and of pPTV-LUC are identical downstream of the LUC initiation codon).
The requirement of 48S complex assembly on the PTV-1 IRES for the initiation factors eIF1 and eIF1A (required for ribosome scanning), eIF4B (which stimulates helicase activity of eIF4A and eIF4F), and eIF4F was determined by the omission of these factors in separate reactions (Fig. 5). Efficient 48S assembly was observed in the presence of all of the initiation factors potentially required for 48S assembly together with PTB. The omission of eIF1 with eIF1A had no adverse effect on 48S assembly. Similarly, omission of eIF4B had no significant effect on 48S complex formation. Thus, these factors are not required for translation initiation directed by the PTV-1 IRES. Each of these results is similar to those obtained with the EMCV IRES (26, 27). However, in contrast to the properties of the EMCV IRES, when eIF4F was omitted the assembly of the 48S complex, as indicated by the formation of the toeprint, was still observed with similar efficiency (Fig. 5). As in Fig. 4, when eIF2 plus Met-tRNAiMet were omitted the normal toeprint was not observed, but a weaker toeprint closer to the AUG was again detected.
FIG. 5.
48S preinitiation complex formation on the PTV-1 IRES in the absence of eIF4F. The formation of 48S preinitiation complexes was assayed by using purified components on the PTV-LUC RNA transcript as in Fig. 4. The negative control reaction lacked eIF2 and met-tRNA, whereas the positive control reaction included each of the purified initiation factors (as in Fig. 4) plus PTB. The other reactions omitted eIF4F or eIF4B or eIF1 plus eIF1A as indicated. The position of the toeprint is shown.
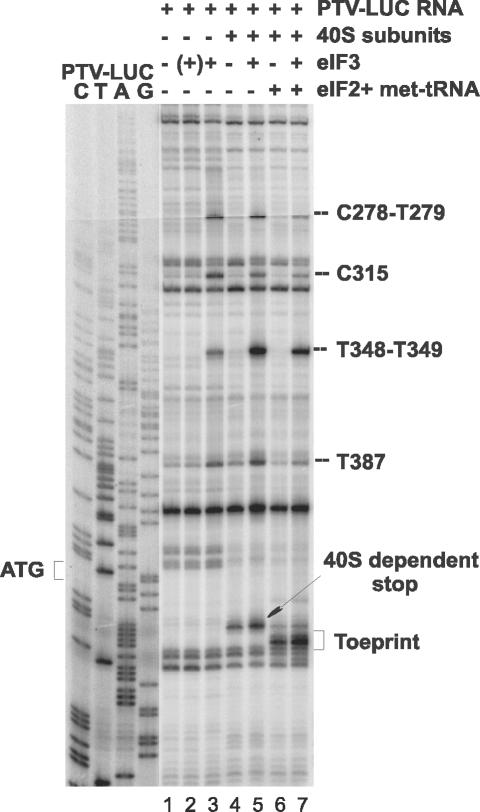
Determination of the minimal requirements for 48S complex formation on the PTV-1 IRES.
The minimal requirements for 48S preinitiation complex formation on the PTV-1 IRES were determined by toeprint analysis with purified 40S ribosome subunits alone or with the addition of eIF2 plus Met-tRNAiMet and eIF3 (Fig. 6). No toeprint indicative of 48S complex formation was observed in the presence of eIF3 and 40S ribosome subunits but in the absence of eIF2 plus Met-tRNAiMet. However, 48S complexes were generated in the presence of the RNA plus 40S subunits with just eIF2 and Met-tRNAi. The presence of eIF3 enhanced the formation of the 48S specific toeprint. A weaker toeprint (just upstream of the 48S complex signal) was also observed in the presence of 40S subunits alone, as observed in Fig. 4 and 5. The addition of eIF3 into the assays also generated novel toeprints at nt 387, 348 and 349, 315, and 278 and 279 (Fig. 6, lanes 3, 5, and 7); these were induced by the presence of eIF3 alone (Fig. 6, lane 3). To demonstrate that the incubation with eIF3 did not induce RNA breakage (and hence the appearance of “apparent” toeprints), the PTV RNA was incubated with eIF3, deproteinized, and then assayed in the toeprinting assay. No more bands were generated on this eIF3-treated RNA than on the naked RNA (see Fig. 6, lanes 1 and 2). Thus, we concluded that eIF3 must interact directly with the PTV-1 IRES to produce the novel toeprints. The toeprint assays can reveal interaction sites upstream of the 48S preinitiation site complex since not all RNA molecules will carry this complex.
FIG. 6.
Determination of the minimal components necessary for 48S preinitiation complex formation on the PTV-1 IRES. The formation of 48S preinitiation complexes was assayed by using purified components on the PTV-LUC RNA transcript as in Fig. 4. The toeprint analysis was performed on the RNA after incubation alone (lane 1), in the presence of 40S subunits (lanes 4 to 7), or with the purified initiation factors as indicated. For lane 2, the PTV RNA was incubated with eIF3, deproteinized, and then used in the toeprint assay in the absence of other factors or 40S subunits (hence the “(+)” eIF3 annotation). Products were analyzed by gel electrophoresis in parallel with sequencing reactions. The positions of the 48S toeprint, the 40S-dependent stop, and products generated by the interaction of eIF3 with the RNA are indicated.
Binary complex formation between the PTV-1 IRES and 40S ribosomal subunits.
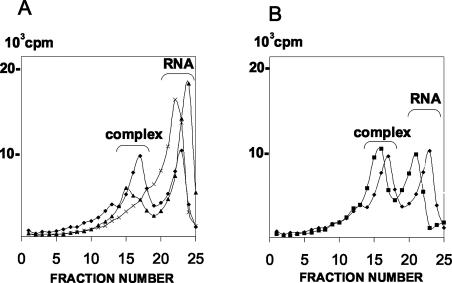
A weaker toeprint, just upstream of that generated by the 48S complex, was observed on the PTV-1 RNA in the presence of 40S subunits alone or when eIF2 and met-tRNA were omitted (see Fig. 4 to 6). This suggested that free 40S subunits may be able to form a binary complex with the PTV-1 IRES, as observed for the HCV and pestivirus IRESs (17, 28). It should be noted, however, that the intensities of bands above a toeprint simply reflect the ease with which reverse transcriptase consecutively overcomes elements of the secondary structure of the RNA, whether it is stabilized or not by interaction with the 40S subunit. They do not allow conclusions about the overall affinity of the RNA for 40S subunits to be made since this probably involves multiple contacts. To determine whether a direct interaction between 40S ribosome subunits and the PTV-1 IRES could be observed, 32P-labeled RNA transcripts corresponding to the PTV-1 IRES were prepared and incubated with 40S ribosomal subunits alone or also in the presence of eIF3. The reactions were analyzed by sucrose gradient centrifugation (26). For comparison, the HCV IRES and EMCV IRES RNAs were analyzed in parallel by using the same molar ratios of RNA and 40S subunits. The distribution of the labeled RNA transcripts in the gradient was determined and is shown in Fig. 7. As expected, no binary complex formation was observed on the EMCV IRES RNA but, in contrast, a clear peak corresponding to this complex was observed on the HCV IRES (Fig. 7A). Similarly, a binary complex was observed on the PTV-1 IRES with 40S subunits alone (Fig. 7A). A more rapidly migrating ternary complex was observed on the PTV-1 RNA in the presence of 40S subunits plus eIF3 (Fig. 7B). Binary complex formation between the PTV-1 IRES and 40S subunits seems to occur at least as efficiently as with the HCV IRES. The formation of this binary complex presumably accounts for the appearance of the 40S-dependent toeprint that is observed in the absence of eIF2, which is closer to the AUG than the 48S preinitiation complex-dependent signal (see above).
FIG. 7.
Binary complex formation on the PTV-1 IRES. (A) 32P-labeled RNA transcripts containing the PTV-1 IRES (♦), the HCV IRES (▴), or the EMCV IRES (×) were incubated with 40S subunits alone. The products were analyzed by sedimentation on sucrose gradients that were then fractionated. The labeled RNA in each fraction was quantified by scintillation counting. The position of the unbound RNAs and the binary complexes of RNA plus 40S subunit are indicated. Sedimentation is from right to left across the gradient. (B) The PTV-1 IRES was incubated with 40S subunits alone (♦) or with 40S subunits plus eIF3 (▪) and analyzed as described for panel A.
Sequence and structural similarity between the PTV-1 IRES and the HCV IRES.
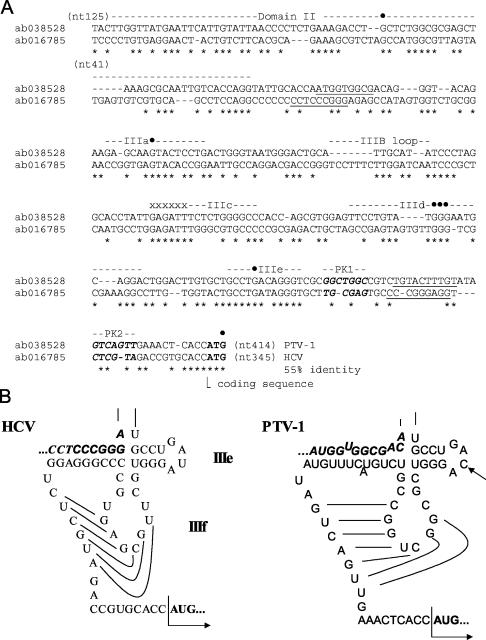
The functional properties of the PTV-1 IRES described above closely resemble those determined previously for the HCV and pestivirus IRES elements (28). The HCV IRES has been extensively studied and is believed to contain two major domains, termed domain II and domain III; the latter is a complex structure comprising multiple subdomains (IIIa to IIIf), including a pseudoknot (see references 14 and 38). To examine the relatedness of the PTV-1 IRES and the HCV IRES elements, an alignment of their nucleotide sequences was performed by using CLUSTAL W (Fig. 8A). It is apparent that the two elements share a significant level of sequence identity, particularly within certain regions. This alignment should be regarded as preliminary, however, since the secondary structure of the PTV-1 IRES has yet to be analyzed by using structure-probing assays. Despite this caveat, the colinearity of multiple highly conserved sequence motifs is striking. The alignments between these conserved motifs are clearly more tentative. However, on the basis of this alignment, within the entire PTV-1 IRES element up to the initiation codon (nt 125 to 414), there is a 55% sequence identity with the HCV IRES. For comparison, the level of sequence identity between the EMCV and FMDV IRES elements is ca. 50%.
FIG. 8.
Sequence and structural similarity between the PTV-1 IRES and the HCV IRES. (A) The cDNA sequences of the PTV-1 IRES and HCV IRES were aligned by using CLUSTAL W and manually edited. The regions of the HCV IRES corresponding to domain II and the subdomains IIIa to IIIf are indicated (14, 38). Individual nucleotides within the HCV IRES that are protected from modification by the interaction with the 40S subunit (17, 22) are indicated (•). A highly conserved sequence within the stem of domain IIIb that is critical for interaction of the HCV IRES with eIF3 (17) is marked by x's. The initiation codons are shown in boldface, and sequences involved in base pairing to form the pseudoknot (HCV domain IIIf) are indicated in boldface italics and marked PK1 and PK2, whereas sequences required for the stem of the pseudoknot are underlined. (B) A secondary structure model for the subdomains IIIe and IIIf (the pseudoknot) of the HCV IRES element is shown (this model is based on previously published studies [14, 38]). The corresponding region of the PTV-1 IRES is also shown in a similar conformation. The single-nucleotide difference between the HCV IRES domain IIIe and the equivalent region of PTV-1 is indicated by an arrow. The initiation codon is indicated in boldface, and sequences downstream of domain II (underlined in panel A) that are involved in the formation of the pseudoknot are indicated in boldface italics.
Of particular interest is the very high degree of sequence identity (11 of 12 nt identical) within the region of the HCV IRES that corresponds to the entire subdomain IIIe (Fig. 8A and B). Thus, it is very likely that this region will exist as a near-identical stem-loop structure in both IRES elements with a single substitution in the loop region. The domain III of the HCV IRES is known to play a critical role in the formation of the complex with the 40S ribosome. Taking domain IIIe of the PTV IRES as a starting point, it was possible to create a model for part of the secondary structure of the PTV-1 IRES (Fig. 8B) that looks very similar to this portion of the secondary structure of the HCV IRES. This model includes, in addition to the IIIe domain, the functionally important pseudoknot element with a long stem structure that locks the fold and stabilizes the pseudoknot. The sequences (labeled PK1 and PK2, Fig. 8A) within the HCV IRES that base pair to form the pseudoknot are not well conserved in the PTV-1 IRES sequence. However, the sequences in equivalent positions within the PTV-1 IRES are complementary to each other, and thus a pseudoknot structure can be predicted to form in the same region (Fig. 8B).
One other important region for the interaction of the HCV IRES with the 40S subunit, domain IIId, is positioned just upstream of domain IIIe in the sequence (Fig. 8A). This region of the PTV sequence is not included in the current secondary structure model. However, the formation of an HCV-like domain IIId structure of the PTV IRES appears to be highly probable. In the aligned sequences, the motif UGGG is present in both sequences at a similar position. This motif is located in the loop of the HCV domain IIId, and its three consecutive G residues are highly accessible to nuclease and chemical attack in the HCV IRES. These nucleotides are protected from modification by interaction with 40S ribosomal subunits (17, 22). Preliminary T1 nuclease probing analysis of the PTV IRES (data not shown) revealed that these three G residues are highly accessible in this RNA as well, whereas flanking G residues are not.
Other features that are very similar between the PTV and HCV sequences can also be identified. The domain IIIa sequence of the HCV IRES contains a run of 8 of 10 nucleotides that are identical to the corresponding region of the PTV-1 IRES and around the loop region of the IIIb subdomain there is also extensive identity (see Fig. 8A). These regions have a key role in the interaction between the HCV IRES and the 40S ribosomal subunit and eIF3 (17, 22). Furthermore, a contiguous group of 7 nt that are identical between the PTV-1 and HCV sequences, located just upstream of the HCV domain IIIc (see Fig. 8A), have also been identified as critical nucleotides for the interaction of this sequence with eIF3 (17). Thus, it appears that the PTV-1 (Talfan) and HCV RNAs, from different virus families, possess closely related IRES elements.
DISCUSSION
The results presented here clearly show that the PTV-1 IRES element is contained within nt 126 to 405. Initiation of protein synthesis occurs at the AUG codon located at nt 412 to 414 (16). Thus, the PTV-1 IRES, at 280 nt, is much shorter than all known picornavirus IRES elements. Furthermore, as pointed out previously (16), in contrast to all other picornavirus IRES elements, there is no polypyrimidine tract located about 20 nt upstream of an AUG codon at the 3′ end of the PTV-1 IRES. The activity of the PTV-1 IRES is not modified by the cleavage of eIF4G, a key component of the eIF4F cap-binding complex, induced by an enterovirus 2A protease. This result is similar to that obtained with the cardio- and aphthovirus and HCV IRES elements. The activity of the cardio- and aphthovirus IRES elements requires only the C-terminal cleavage product of eIF4G plus eIF4A, whereas, in contrast, the HCV IRES has no requirement for any component of eIF4F (28).
To determine the requirement of the PTV-1 IRES for individual translation initiation factors, the formation of 48S preinitiation complexes was assayed by using toeprinting assays. As with the cardio- and aphthovirus IRES elements, efficient assembly of 48S complexes on the PTV-1 IRES could be detected within RRL (Fig. 3). It was also possible to reconstitute 48S complex assembly on this RNA by using purified components. These studies showed that the initiation factors eIF1, eIF1A, eIF3, eIF4B, and eIF4F were not required for 48S preinitiation complex formation on the PTV-1 IRES. However, eIF2 and Met-tRNA were required (c.f., the cricket paralysis virus intergenic region IRES [43]). The presence of PTB had a modest stimulatory effect on 48S complex formation, although this effect was much less pronounced than that observed with the EMCV IRES (I. N. Shatsky, unpublished results). Taken together, these results contrast strongly with observations for the cardio- and aphthovirus IRES elements (26, 32). These elements are, however, highly reminiscent of the pesti- and hepacivirus IRES elements (28). These IRES elements can directly interact with 40S ribosome subunits to form a binary complex, and this property has also now been demonstrated for the PTV-1 IRES (Fig. 7).
By alignment of the nucleotide sequences of the HCV and PTV-1 IRES elements, it became apparent that these elements are significantly related (ca. 55% sequence identity overall). Within this figure there are distinct islands of very high sequence similarity (e.g., within HCV domains IIIa and IIIe [see Fig. 8A]). These regions correspond to sequences known to be critical for the interaction of the HCV IRES with eIF3 and the 40S subunit (17, 22). There is a single nucleotide difference between the sequence of the HCV domain IIIe and the corresponding region of the PTV-1 RNA. The HCV U297 has been mutated to C297 (as in the PTV-1 sequence), and this modification resulted in a 55% decrease in IRES activity (22), we are currently exploring the effect of the reverse mutation within the context of the PTV-1 IRES. It is clear, however, that the PTV-1 IRES works very efficiently in RRL and within BHK cells (Fig. 2) (16).
Secondary structure models have been derived for the pestivirus and HCV IRES elements (10, 14, 38, 42); these have two principal domains, including a pseudoknot just upstream of the initiation codon. The pseudoknot is required for activity of these IRES elements (10, 28, 42). Starting from the highly conserved IIIe domain, it was possible to derive a secondary structure model for part of the PTV IRES (Fig. 8B). Apart from the domain IIIe, the sequences forming this structure have a rather low sequence similarity with the respective regions of the HCV IRES. However, they occupy equivalent positions in the IRES sequences. The most striking example for this is the long stem sequence that locks the pseudoknot structure. In the HCV IRES, one of the sequences forming the stem (Fig. 8B) is located in a position between domains II and IIIa far upstream from other sequences forming the pseudoknot. The predicted stem sequence for the PTV pseudoknot is located in an analogous position in the PTV IRES sequence (see Fig. 8). In addition, we have presented some evidence for the existence in the PTV IRES of a stem-loop that is equivalent to domain IIId in the HCV IRES secondary structure. Taken together, these features of the PTV IRES comprise the entire set of structural elements known to be critical for highly specific recognition of the 40S ribosomal subunit by the HCV and pestivirus IRESs (17, 22). In addition, we have presented some data in support of the structural and positional equivalence of their eIF3 binding sites. Thus, the data from our structural analysis are in excellent agreement with the functional tests presented in the present study.
The role of the HCV/CSFV domain II in IRES function is not yet clear; however, it has been shown that the loss of sequences from within the domain II of the HCV or CSFV IRES sequence resulted in an ∼5-fold drop in IRES function (10, 33a). These results closely match the data obtained from deleting 1 to 150 nt from the PTV-1 5′UTR sequence, which removes about 25 nt of the PTV IRES corresponding to part of the HCV domain II. The complete loss of IRES function requires the loss of the entire domain II structure.
It is not yet apparent whether the complete PTV-1 IRES sequence can be modeled onto the current HCV IRES secondary structure prediction, and further structure probing studies are in progress to facilitate this process.
We conclude that the PTV-1 IRES represents a new class of picornavirus IRES with properties that are very different from all other picornavirus IRES elements (as suggested previously [16]), and we have now shown that this element closely resembles the IRES elements from HCV and the pestiviruses. The ability to function in the complete absence of eIF4F marks a clear separation between the PTV-1 IRES and the cardiovirus/aphthovirus IRES elements, although in other respects the biological properties (e.g., activity in RRL and insensitivity to eIF4G cleavage) are shared with this group of IRES elements. It is also clear that the conserved J-K domains present near the 3′ end of the cardio- and aphthovirus IRES elements, which are critical for the interaction with eIF4G (18, 21, 39), are not present within the PTV-1 IRES. However, evidence for direct interaction of the PTV-1 IRES with eIF3 was obtained (Fig. 6). The interaction of the PTV-1 IRES with 40S ribosomal subunits to form a binary complex (Fig. 7) is also currently unique within the picornavirus family. It will be interesting to determine whether other picornaviruses possess similar IRES elements. Possible candidates include porcine enterovirus type 8 and the simian picornavirus type 2, since there is significant sequence similarity within their 5′UTRs to the PTV-1 IRES, although the viruses are quite distinct overall (19, 44).
The fact that viruses from two different virus families share a common type of IRES element is remarkable. It either suggests that genetic exchange of this element between virus families has occurred or that there are a very limited number of ways in which an exclusively cytoplasmic RNA can directly recruit ribosomes in a cap-independent manner.
Acknowledgments
This study was supported in part by a grant from INTAS (project number 011-0293) to G.J.B. and I.N.S. and by grant 02-04-48798 from the Russian Foundation for Basic Research to I.N.S. L.S.C. acknowledges receipt of a studentship from BBSRC.
REFERENCES
- 1.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75:7854-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Borman, A. M., and K. M. Kean. 1997. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 237:129-136. [DOI] [PubMed] [Google Scholar]
- 4.Borman, A. M., P. Le Mercier, M. Girard, and K. M. Kean. 1997. Comparison of picornaviral IRES-driven internal initiation in cultured cells of different origins. Nucleic Acids Res. 25:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, B., and E. Ehrenfeld. 1979. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology 97:396-405. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. A., A. J. Zajac, and S. M. Lemon. 1994. In vitro characterization of an internal ribosome entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dmitriev, S. E., A. V. Pisarev, M. P. Rubtsova, Y. E. Dunaevsky, and I. N. Shatsky. 2003. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 533:99-104. [DOI] [PubMed] [Google Scholar]
- 8.Dorner, A. J., B. L. Semler, R. J. Jackson, R. Hanecak, E. Duprey, and E. Wimmer. 1984. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 50:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evstafieva, A. G., T. Y. Ugarova, B. K. Chernov, and I. N. Shatsky. 1991. A complex RNA sequence determines the internal initiation of encephalomyocarditis virus RNA translation. Nucleic Acids Res. 19:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher, S. P., and R. J. Jackson. 2002. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5′ untranslated region important for IRES function. J. Virol. 76:5024-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 13.Grifo, J. A., S. M. Tahara, M. A. Morgan, A. J. Shatkin, and W. C. Merrick. 1983. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 258:5804-5810. [PubMed] [Google Scholar]
- 14.Honda, M., M. R. Beard, L.-H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaku, Y., A. Sarai, and Y. Murakami. 2001. Genetic reclassification of porcine enteroviruses. J. Gen. Virol. 82:417-424. [DOI] [PubMed] [Google Scholar]
- 16.Kaku, Y., L. S. Chard, T. Inoue, and G. J. Belsham. 2002. Unique characteristics of a picornavirus internal ribosome entry site from the porcine teschovirus-1 Talfan. J. Virol. 76:11721-11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieft, J. S., K. Zhou, R. Jubin, and J. A. Doudna. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7:194-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolupaeva, V. G., T. V. Pestova, C. U. T. Hellen, and I. N. Shatsky. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273:18599-18604. [DOI] [PubMed] [Google Scholar]
- 19.Krumbholz, A., M. Dauber, A. Henke, E. Birch-Hirschfeld, N. J. Knowles, A. Stelzner, and R. Zell. 2002. Sequencing of porcine enterovirus groups II and III reveals unique features of both virus groups. J. Virol. 76:5813-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.López deq Uinto, S., and E. Martínez-Salas. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6:1380-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukavsky, P. J., G. A. Otto, A. M. Lancaster, P. Sarnow, and J. D. Puglisi. 2000. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 7:1105-1110. [DOI] [PubMed] [Google Scholar]
- 23.Meerovitch, K., and N. Sonenberg. 1993. Internal initiation of picornavirus RNA translation. Semin. Virol. 4:217-227. [Google Scholar]
- 24.Merrick, W. C. 1979. Assays for eukaryotic protein synthesis. Methods Enzymol. 60:101-108. [DOI] [PubMed] [Google Scholar]
- 25.Pause, A., N. Methot, Y. V. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestova, T. V., C. U. T. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestova, T. V., I. N. Shatsky, and C. U. T. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. T. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova, T. V., S. I. Borukhov, and C. U. T. Hellen. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394:854-859. [DOI] [PubMed] [Google Scholar]
- 30.Pestova, T. V., I. B. Lomakin, J. H. Lee, S. K. Choi, T. E. Dever, and C. U. T. Hellen. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403:332-335. [DOI] [PubMed] [Google Scholar]
- 31.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. T. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 32.Pringle, C. R. 1999. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch. Virol. 144:2065-2070. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis-C virus-RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Reynolds, J. E., A. Kaminski, A. R. Carroll, B. E. Clarke, D. J. Rowlands, and R. J. Jackson. 1996. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA 2:867-878. [PMC free article] [PubMed] [Google Scholar]
- 34.Rijnbrand, R., P. J. Bredenbeek, P. C. Haasnoot, J. S. Kieft, W. J. M. Spaan, and S. M. Lemon. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts, L. O., R. A. Seamons, and G. J. Belsham. 1998. Recognition of picornavirus internal ribosome entry sites within cells: influence of cellular and viral proteins. RNA 4:520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakoda, Y., N. Ross-Smith, T. Inoue, and G. J. Belsham. 2001. An attenuating mutation in the 2A protease of swine vesicular disease virus, a picornavirus, regulates cap- and internal ribosome entry site-dependent protein synthesis. J. Virol. 75:10643-10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, N.Y.
- 38.Sarnow, P. 2003. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J. Virol. 77:2801-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stassinopoulos, I. A., and G. J. Belsham. 2001. A novel protein-RNA binding assay: functional interactions of the foot-and-mouth disease virus internal ribosome entry site with cellular proteins. RNA 7:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svitkin, Y. V., A. Pause, A. Haghighat, S. Pyronnet, G. Witherell, G. J. Belsham, and N. Sonenberg. 2001. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is directly proportional to the degree of mRNA 5′ secondary structure. RNA 7:382-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, C., S. Y. Le., N. Ali, and A. Siddiqui. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526-537. [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, J. E., T. V. Pestova, C. U. T. Hellen, and P. Sarnow. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511-520. [DOI] [PubMed] [Google Scholar]
- 44.Zell, R., M. Dauber, A. Krumbholz, A. Henke, E. Birch-Hirschfeld, A. Stelzner, D. Prager, and R. Wurm. 2001. Porcine teschoviruses comprise at least eleven distinct serotypes: molecular and evolutionary aspects. J. Virol. 75:1620-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]