Abstract
Norwalk virus is the prototype strain for members of the genus Norovirus in the family Caliciviridae, which are associated with epidemic gastroenteritis in humans. The nonstructural protein encoded in the N-terminal region of the first open reading frame (ORF1) of the Norwalk virus genome is analogous in gene order to proteins 2A and 2B of the picornaviruses; the latter is known for its membrane-associated activities. Confocal microscopy imaging of cells transfected with a vector plasmid that provided expression of the entire Norwalk virus N-terminal protein (amino acids 1 to 398 of the ORF1 polyprotein) showed colocalization of this protein with cellular proteins of the Golgi apparatus. Furthermore, this colocalization was characteristically associated with a visible disassembly of the Golgi complex into discrete aggregates. Deletion of a predicted hydrophobic region (amino acids 360 to 379) in a potential 2B-like (2BL) region (amino acids 301 to 398) near the C terminus of the Norwalk virus N-terminal protein reduced Golgi colocalization and disassembly. Confocal imaging was conducted to examine the expression characteristics of fusion proteins in which the 2BL region from the N-terminal protein of Norwalk virus (a genogroup I norovirus) or MD145 (a genogroup II norovirus) was fused to the C terminus of enhanced green fluorescent protein. Expression of each fusion protein in cells showed evidence for its colocalization with the Golgi apparatus. These data indicate that the N-terminal protein of Norwalk virus interacts with the Golgi apparatus and may play a 2BL role in the induction of intracellular membrane rearrangements associated with positive-strand RNA virus replication in cells.
Norwalk virus is the prototype strain for the genus Norovirus in the family Caliciviridae. Viruses in this genus have characteristically been associated with acute epidemic gastroenteritis in humans, but their host range extends to other species such as swine, cattle, and mice (12, 23). The noroviruses are now recognized as the most important agents of nonbacterial gastroenteritis outbreaks (10, 26, 30, 35), with as many as 23 million estimated cases of norovirus-associated illness occurring in the United States each year (31).
The RNA genome of Norwalk virus is 7,654 nucleotides (nt) in length and is polyadenylated at the 3′ end (20). By analogy with better-characterized caliciviruses (4), the 5′ end of Norwalk virus is likely covalently linked to a VPg protein. Direct demonstration of this linkage has been difficult in the absence of a cell culture system (12). The genome encodes three open reading frames (ORFs): ORF1 is the largest and encodes a polyprotein precursor of approximately 200 kDa that is cleaved by a viral proteinase into the mature nonstructural proteins required for virus replication; ORF2 encodes the major structural capsid protein VP1; and ORF3 encodes the minor structural protein VP2 (9, 19, 28).
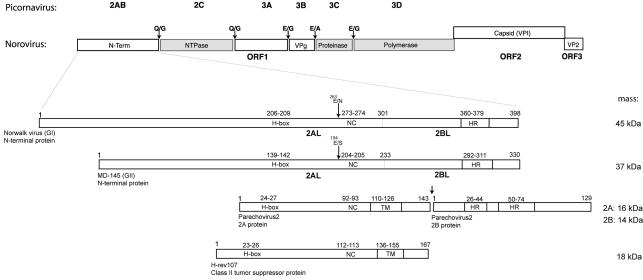
The caliciviruses belong to the picornavirus-like supergroup of the positive-strand RNA viruses, which is characterized by the colinearity and conservation of three major enzymes: a nucleoside triphosphatase (NTPase) (2C in picornaviruses), a proteinase with a chymotrypsin-like fold (3C), and an RNA-dependent RNA polymerase (3D) (see Fig. 1) (10a, 25, 36). Upon comparison of the caliciviruses and picornaviruses, this colinearity extends to a number of products derived from the regions flanking the 2C protein (the 2C-like [2CL] protein in caliciviruses) that may not be universally conserved. One of these proteins, which occupies a polyprotein position equivalent to that of the picornavirus 3B protein (the 3B-like [3BL] protein), has been implicated in a conserved function as the protein covalently linked to the 5′ end of the viral RNA (VPg) (5). A parallel between picornaviruses and caliciviruses was also drawn for proteins from the 3A (3A-like) region (24, 43).
FIG. 1.
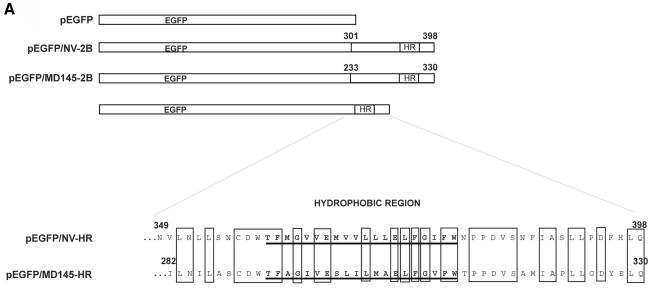
Schematic diagram showing the organization of the Norwalk virus genome and a comparison of the N-terminal protein of Norwalk virus (a GI norovirus) with that of MD145 (a GII norovirus), human parechovirus 2 proteins 2A and 2B, and the cellular class II tumor suppressor H-rev107. The genomes of viruses in the genus Norovirus are organized into three ORFs. ORF1 encodes a large polyprotein that undergoes cleavage by the virus-encoded 3CL proteinase to release the mature nonstructural proteins required for virus replication. The corresponding nonstructural proteins of the picornaviruses are shown, and proteins of the noroviruses with known similar functions are shaded; these include a 2CL NTPase, a 3CL proteinase, and a 3D-like RNA-dependent RNA polymerase. The dipeptide cleavage sites that are recognized by the norovirus 3CL proteinase and are conserved in GI and GII members of the genus Norovirus are indicated. The putative 3CL cleavage sites at E263/N and E194/S, recently proposed for the GI and GII norovirus N-terminal proteins, respectively (40), are indicated by arrows. ORF2 encodes the major capsid protein VP1, and ORF3 encodes a minor structural protein of unknown function, VP2. The norovirus N-terminal protein shares conserved motifs (H-box and NC) with a group of cellular proteins involved in the control of cell proliferation, represented by H-rev107 (17), and with certain picornavirus “H-NC” 2A proteins, represented here by human parechovirus 2. This region is labeled 2AL below the enlarged diagrams of the Norwalk virus and MD145 N-terminal proteins. A C-terminal TM region identified in the parechoviruses and a predicted membrane-associated alpha-helix region (labeled TM) of H-rev107 are shown. The computer algorithm TMPred identified potential TM regions in the Norwalk virus and MD145 N-terminal proteins as well as in the human parechovirus 2 2B protein, which we designate here as a hydrophobic region (HR). The regions of the Norwalk virus (aa 301 to 398) and MD145 virus (aa 233 to 330) with possible 2BL homology are indicated. The calculated molecular masses of the norovirus N-terminal proteins, human parechovirus 2 proteins 2A and 2B, and H-rev107 are given on the right. The GenBank or EMBL accession numbers for the sequences used in this analysis are M87661 (Norwalk virus), AY032605 (MD145), AAC79756 (human parechovirus 2), and X92814 (H-rev107). The predicted borders of human parechovirus 2 proteins 2A and 2B were adapted from the work of Johansson et al. (21) and correspond to aa 782 to 924 and 925 to 1053 of the parechovirus polyprotein, respectively. Numbering of amino acids on the diagram corresponds to that of the individual 2A or 2B protein.
The 2AB region upstream of picornavirus 2C and its genetic equivalent in the caliciviruses are highly divergent within and between families. In members of the genera Vesivirus and Lagovirus, two proteins, designated p5.6 and p32 for feline calicivirus (FCV) and p16 and p23 for rabbit hemorrhagic disease virus (RHDV), are derived from this region by 3C-like (3CL)-mediated cleavage (43, 47). In contrast, a single protein (designated here as the N-terminal protein) of approximately 45 or 37 kDa is derived from this region in Norovirus genogroup I (GI) or genogroup II (GII), respectively (3, 29), and marked sequence variation of this protein has been reported (8, 37). An indication for further proteolytic processing of the norovirus N-terminal protein was recently obtained in coexpression studies of the Camberwell virus (GII) N-terminal protein and 3CL proteinase in mammalian cells (40). The norovirus 37- or 45-kDa N-terminal protein was shown to be distantly related to 2A proteins of a diverse set of picornaviruses (including parechovirus, kobivirus, avian encephalomyelitis virus, and Ljungan virus) and a number of cellular proteins, including H-rev107, a protein implicated in the control of cellular proliferation as a class II tumor suppressor (17, 21).
In this study, we extended comparative sequence analysis of the Norwalk virus N-terminal protein to identify a 2B-like (2BL) region, implicating the norovirus protein in the membrane-mediated activities characteristic of picornavirus protein 2B (38, 46). Confocal microscopic analysis showed that the Norwalk virus N-terminal protein expressed in transfected cells was colocalized with markers of the Golgi apparatus and caused an apparent disassembly of the Golgi complex into discrete aggregates. In addition, the interaction of the N-terminal protein in the Golgi apparatus was facilitated by the presence of a predicted C-terminal hydrophobic region (HR). The observed phenotype of the Norwalk virus N-terminal protein expression was consistent with a possible 2BL function for at least the part of the protein that involves interaction with host cell membranes. This phenotype further strengthens a molecular parallel between picornaviruses and caliciviruses.
MATERIALS AND METHODS
Cells.
Crandell-Rees feline kidney (CRFK) cells and HeLa cells were maintained in Eagle's minimum essential medium containing amphotericin B (2.5 μg/ml), chlortetracycline (25 μg/ml), penicillin (250 U/ml), streptomycin (250 μg/ml), and 10% heat-inactivated fetal bovine serum. Norovirus strains Norwalk virus (Hu/NV/Norwalk virus/1968/US) (22) and MD145-12 (Hu/NV/MD145-12/1987/US) (11) have been described previously.
Plasmids.
A full-length cDNA clone of the Norwalk virus genome was engineered into the pSPORT1 plasmid (Invitrogen, Carlsbad, Calif.) by using a cloning strategy similar to that described for the generation of an infectious cDNA clone of the FCV genome (42). Briefly, overlapping cDNA fragments were generated from purified Norwalk virus RNA by reverse transcription-PCR with SuperScript II reverse transcriptase and Elongase DNA polymerase (Invitrogen) and then cloned into the pSPORT plasmid. A unique BsmBI restriction enzyme site followed by a bacteriophage T7 RNA polymerase promoter was engineered immediately upstream of the 5′ end of the viral genome. A unique MluI restriction enzyme site was engineered downstream of a poly(A30) tract at the 3′ end of the genome. The assembled full-length cDNA clone was designated NV FL101.
A gene fragment corresponding to nt 5 to 1198 of the Norwalk virus genome, which encodes the N-terminal protein, was amplified by PCR (Elongase amplification system; Invitrogen) using NV FL101 as the template. The NV forward primer (5′ AGGCCTACGCGTCTCGAGAAGATGATGATG GCGTCAAAAGACGTCGTT CCTACTGCTGCTAGCAGTGAAAATGC 3′) was designed to introduce an XhoI restriction site (underlined) upstream of the first AUG codon (boldfaced). The NV reverse primer (5′ ATATAAATAATATTCTAGATTACTGTAGATGGAAATCTGGCAGGAGTGAAGCTATAAAGTTGG 3′) was designed to introduce a stop codon (boldfaced) and an XbaI site (underlined). The XhoI-XbaI fragment encoding the N-terminal protein was ligated into the corresponding restriction enzyme sites of the eukaryotic expression plasmid pCI (Promega, Madison, Wis.) with T4 DNA ligase (New England Biolabs, Beverly, Mass.). The recombinant plasmid was transformed into Max-efficiency Escherichia coli DH5α cells (Invitrogen), and clones were selected in the presence of carbenicillin (50 μg/ml). The resulting plasmid was designated pC/NV-N wild type (wt). For construction of the Norwalk virus C-terminal deletion constructs, the NV forward primer containing the XhoI restriction site described above was used to amplify DNA fragments from plasmid pC/NV-N wt as a template with the following individual reverse primers: NV 3′ Δ10/XbaI (5′ ATATAAATAATATTCTAGATTATATAAAGTTGGAAACATCAGGTGGG 3′), NV 3′ Δ20/XbaI (5′ ATATAAATAATATTCTAGATTAAAAG ATTCCAAAGAGTTCTAAAAGG 3′), NV 3′ Δ30/XbaI (5′ ATATAAATAATATTCTAGATTACACCATCTCCACGACCCCCATGAACG 3′), NV 3′ Δ40/XbaI (5′ ATATAAATAATATTCTAGATTAATCACAGTTTGAAAGTAAGTTAAGC 3′), and NV 3′Δ50/XbaI (5′ ATATAAATAATATTCTAGATTACTGTAGATGGAAATCTGG CAGGAGTGAAGCTATAAAGTTGGG 3′). Each of these reverse primers included an XbaI site (underlined) and an engineered stop codon (boldfaced).
A similar strategy was used to PCR amplify nt 5 to 990 of the MD145 genome from a cDNA clone (cORF1) of MD145 ORF1 (3) by using two primers: MD145forward primer (5′ CCGCTCGAGAATGAAGATGGCGTCTAACGACGCTTC 3′), which was designed to introduce an XhoI restriction site (underlined) upstream of the first AUG (boldfaced), and MD145 reverse primer (5′ GCTCTAGATCATTGTAACTCGTAATCACCTAGCAAG 3′), designed to intro-duce a stop codon (boldfaced) and an XbaI site (underlined). The selected plasmid was designated pC/MD145-N wt.
The H-rev107 gene (18) was amplified, by using a Titan reverse transcription-PCR kit (Roche, Indianapolis, Ind.), from total RNA isolated from approximately 5 × 106 to 1 × 107 HeLa cells with the RNeasy minikit (Qiagen, Valencia, Calif.). The H-rev107 forward primer was 5′ AGGCCTACGCGTCTCGAGAAGATGCGTGCGCCCATTCCAGAGCCTAAGCCTGGAGACC 3′ and contained an XhoI site (underlined), and the H-rev107 reverse primer was 5′ ATATAAATAATATTCTAGATTATTGCTTTTGTCGCTTGTTTCTTGAGAACATGACTCC 3′, with an engineered XbaI site (underlined). The XhoI/XbaI-cut DNA fragment was cloned into the compatible sites of the pCI vector as described above, and the selected plasmid was designated pC/H-rev107. Our cloned H-rev107 gene contained two nucleotide differences (CG to GC) from H-rev107-1 (EMBL accession no. X92814), which resulted in a single amino acid change at residue 90 (Thr to Ser) of the deduced protein.
Plasmid p47/331, encoding amino acids (aa) 47 to 331 of the FCV ORF1 polyprotein (corresponding to the FCV p32 protein) in the pCI vector, and an antiserum specific for this region (designated anti-B4) have been described previously (43).
The pEGFP-C1 protein fusion plasmid (BD Biosciences Clontech, Palo Alto, Calif.) was used to engineer proteins in which the enhanced green fluorescent protein (EGFP) was fused at its C terminus to selected regions of the norovirus (Norwalk virus or MD145) N-terminal protein or H-rev107. Primer pairs containing an engineered XhoI restriction enzyme site (underlined) in the forward (For) primer and a SacII site (underlined) in the reverse primer (Rev) were used in PCRs to produce DNA fragments for cloning. For construction of pEGFP/NV-2B (containing aa 301 to 398 of the N-terminal protein fused to EGFP), primers NV2B/GFPFor (5′ GGACTCAGATCTCGAGGACACCCGACCCAAGATTGGTCCCGAGACACTCCAG) and NV/GFPRev (5′ TAATATTATCCGCGGTTTACTGTAGATGGAAATCTGGCAGGAGTGAAG 3′) were used. For construction of pEGFP/NV-HR (containing aa 349 to 398 fused to EGFP), primers NV HR/GFPFor (5′ GGACTCAGATCTCGAGGAAACGTGCTTAACTTACTTTCAAAC 3′) and NV/GFPRev were used. Plasmid pEGFP/MD145-2B (containing aa 233 to 330 fused to EGFP) was constructed by using primers MD2B/GFPFor (5′ GGACTCAGATCTCGAGGAAGACCCTATCAAGACTGGAATAGGAAACCCCTC 3′) and MD/GFPRev (5′ TAATATTATCCGCGGTTTATTGTAACTCGTAATCACCTAGCAAGG 3′), and plasmid pEFGP/MD145-HR (containing aa 282 to 330 fused to EGFP) was constructed by using MD HR/GFPFor (5′ GGACTCAGATCTCGAGG AATCCTCAACATCTTGGCCTCATGT 3′) and MD/GFPRev. In addition, the region encoding the H-rev107 membrane-associated alpha-helix (aa 136 to 155) described by Husmann et al. (18) was amplified from pC/H-rev107 with primers 5′ GGACTCAGATCTCGAGGAAGTGACCAGGTCAG AGATGTCATCATC 3′ and 5′ TAATATTATCCGCGGTTTATTGCTTTTGTCG CTTGTTTCTTGAG 3′. The PCR fragments were cleaved with XhoI and SacII and ligated into the compatible sites of the pEGFP-C1 plasmid. Plasmids were then transformed into bacteria as described above and selected on a medium containing kanamycin (30 μg/ml). All plasmid constructions were sequenced to verify that no mutations were introduced during the PCR step.
Site-directed mutagenesis.
Site-directed mutagenesis was performed using the QuikChange kit (Stratagene, La Jolla, Calif.). The cysteine residue at position 274 (in the conserved NC domain) of the Norwalk virus N-terminal protein was changed to alanine by mutagenesis of TG (nt 821 and 822) to GC. Primers used for the mutagenesis were 5′ GCAGTCACGAACAACGCCTTCGAATTTTGTTGCCAGGTC 3′ (forward) and 5′ GACCTGGCAACAAAATTCGAAGGCGTTGTTCGTGACTGC 3′ (reverse). The selected plasmid (designated pC/NV-NC/mut) was sequenced to verify that no other mutations were introduced into the N-terminal protein gene during the mutagenesis procedure.
Sequence analysis.
Nucleotide sequences were determined by using an ABI Prism DNA sequencing kit and an ABI Prism 310 genetic analyzer (both from Perkin-Elmer Applied Biosystems, Foster City, Calif.). The computer algorithm in TMPred (16) was used for the prediction of membrane-spanning regions in protein sequences. In the absence of experimental data that defined the precise nature of the interactions of the identified regions with membranes, we designated each predicted transmembrane (TM) region an HR in this study.
Development of region-specific antisera to the Norwalk virus N-terminal protein.
A Norwalk virus PCR amplicon (containing the coding sequence from aa 1 to 223 of the N-terminal protein) was generated by using primer 5′ AACTCAGATCGGGATCCGATGATGATGGCGTCAAAAGACGTCGTTCCTACTGCTGC 3′, which corresponds to nt 5 to 42 of the Norwalk virus genome and incorporates a BamHI restriction site (underlined), and primer 5′ CTTCCCCTGAGATGCGGCCGCAGGGGAGTGTACACCCACCGTCTTGCC 3′, which is complementary to nt 644 to 670 of the Norwalk virus genome and includes a NotI restriction site (underlined). The PCR product was ligated into the pCR2.1 plasmid by TA cloning (Invitrogen), and selected clones were verified by sequence analysis. The Norwalk virus-specific insert was digested with BamHI and NotI and ligated into the compatible sites of the pET-28(b) plasmid, which generated an N-terminal His6 tag. For protein production, the recombinant plasmid was transformed into BL21(DE3) competent bacterial cells (Novagen, Madison, Wis.). Expression of protein in bacterial cells was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the His-tagged N-terminal fragment was purified by nickel-nitrilotriacetic acid chromatography according to the manufacturer's (Qiagen) protocol. Antisera were generated in guinea pigs as described previously (44). Attempts to express the entire Norwalk virus and MD145 N-terminal proteins in bacteria failed.
Coupled in vitro transcription and translation.
Proteins were expressed in vitro from the pCI-based plasmids by using the TNT T7 Reticulocyte Lysate Coupled Transcription & Translation kit (Promega) in the presence of [35S]methionine (>1,000 μCi/mmol) (Amersham Biosciences, Piscataway, N.J.). The TNT reaction mixtures were incubated at 30°C for 1.5 h. Proteins were resolved in 10-to-20% gradient polyacrylamide gels (Invitrogen-Novex) by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by autoradiography.
Cell transfection.
Plasmid DNA (1.0 μg) was transfected into CRFK or HeLa cells by using Lipofectamine Plus or 2000 reagent (Invitrogen) as described by the manufacturer. Transfected cells were allowed to express the protein for 24 to 48 h before observation.
IF and confocal microscopy.
CRFK and HeLa cells grown in six-well tissue culture plates were transfected with plasmid DNA and fixed with cold 100% methanol or 4% paraformaldehyde (PFA) at 24 or 48 h posttransfection for immunofluorescent (IF) staining. When PFA was used as the fixative, cells were permeabilized in 0.1% Triton X-100 for 10 min prior to the addition of antibodies. Following washing with phosphate-buffered saline (PBS), pH 7.4, an antiserum specific for the Norwalk virus N-terminal protein (1:100) was added and incubated at room temperature for 3 or 12 h at 4°C. The wells were washed three times with PBS, and fluorescein isothiocyanate (FITC)-conjugated, affinity-purified goat antibodies (1:20) to guinea pig immunoglobulin G (IgG) (ICN Biomedicals, Aurora, Ohio) were added to detect binding of the primary antibody.
CRFK and HeLa cells used for confocal microscopy experiments were grown on coverslips in 35-mm-diameter dishes, transfected with plasmid DNA, and fixed with either methanol or PFA as described above. After a wash with PBS, the cells were incubated with either preimmune serum (pre) or an antiserum raised against the Norwalk virus N-terminal protein (post) together with a mouse monoclonal antibody against human golgin 97 (1:200) (Molecular Probes, Eugene, Oreg.) or against the Golgi 58-kDa protein (p58) (1:100) (Sigma, St. Louis, Mo.) for 2 h at room temperature. For detection of primary antibody binding, slides were incubated in the dark for 1 h with FITC-labeled, affinity purified goat antibodies (1:20) to guinea pig IgG and Alexa Fluor-labeled goat antibodies to mouse IgG (1:200) (Molecular Probes). In addition, 4′,6-diamidino-2′-phenylindole dihydrochloride hydrate (DAPI) (Roche) was used for staining of the nuclei. The slides were washed twice with PBS and then with water. For visualization of EGFP expression, cells were fixed with PFA, followed by staining with anti-human golgin 97 and DAPI. One drop of antifade reagent-mounting mixture prepared by using the reagents in the ProLong Antifade Kit (Molecular Probes) was placed on the center of each slide after staining, a coverslip was added, and the mounting agent was allowed to dry. After sealing, the slides were stored at −20°C. Images were collected by using a 63× oil immersion objective, zoom 1 or 2, on a TCS-NT/SP confocal microscope (Leica Microsystems, Exton, Pa.) by using argon-krypton laser excitation at 488 nm for FITC (green) and 568 nm for Alexa-Fluor (red). The UV fluorescence for DAPI (blue) was excited by using an argon laser at 364 nm. The collected images were analyzed in Adobe Photoshop (Adobe Systems Incorporated, San Jose, Calif.).
Immunoprecipitation assay.
Immunoprecipitation assays were performed as described previously (44). A 5-μl aliquot of the TNT reaction product was incubated with 5 μl of either preimmune serum or an antiserum specific for the Norwalk virus N-terminal protein at 4°C for 1 h. An equal volume of protein G beads-Gamma Bind Sepharose (Pharmacia Biotech AB, Uppsala, Sweden) was added, and the samples were gently mixed at 4°C for 12 h. Each sample was washed, and precipitated proteins were analyzed by SDS-PAGE followed by autoradiography.
RESULTS
The N-terminal protein of Norwalk virus may include three different domains.
The N-terminal protein that is encoded at the beginning of ORF1 of the GI Norwalk virus genome includes 398 aa residues and has a calculated mass of 45 kDa (15) (Fig. 1). It shares 36% amino acid identity with the N-terminal protein of the GII virus MD145, which includes 330 aa residues and has a calculated mass of approximately 37 kDa (3). Apparently, GI viruses (relative to GII viruses) have extra residues in the least conserved part close to the extreme N terminus of the N-terminal protein (8, 37). In a comparably positioned region of the N-terminal proteins of FCV and RHDV, a 3CL-mediated cleavage was observed (43, 47). Cleavages at E263/N and E194/S have recently been proposed in the GI and GII norovirus N-terminal proteins, respectively (40), but these are located further downstream of this variable region.
Approximately 30 aa downstream of this variable region between the GI and GII noroviruses, a norovirus-specific “2A-like” (2AL) domain is located. It is distantly related to the 2A proteins of some picornaviruses, such as parechovirus 2, and a group of cellular proteins that includes the class II tumor suppressor H-rev107 (17) (Fig. 1). All these proteins include two conserved regions: an “H-box” with a histidine (H) residue and an “NC” motif consisting of an asparagine (N) and cysteine (C) dipeptide sequence. The picornavirus 2A and cellular 2AL proteins may be anchored to membranes through putative TM alpha-helices that are located approximately 20 aa downstream of the NC motif (16, 17). The norovirus N-terminal proteins have an HR located further downstream at a distance of approximately 80 aa from the NC motif. Our comparative sequence analysis showed this structure to be part of the approximately 100 aa domain, corresponding to aa 301 to 398 of Norwalk virus and aa 233 to 330 of the MD145 virus, that is conserved at the C terminus of the N-terminal protein in all caliciviruses. We designated this sequence the “2B-like” (2BL) domain because of its polyprotein position between the 2AL and 2CL proteins (domains), its small size, and its conserved hydrophobic profile, all of which properties distinguish picornavirus 2B proteins (38, 46).
Specificity of antibodies developed against the N-terminal part of the Norwalk virus N-terminal protein.
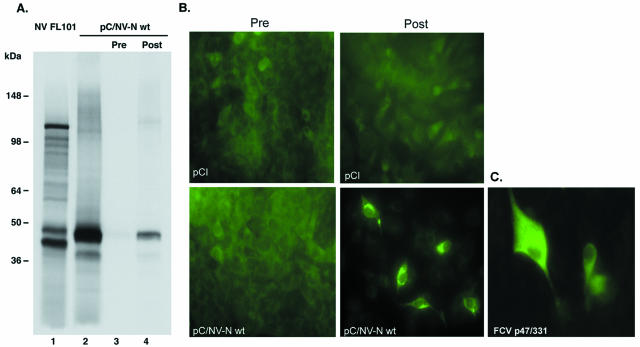
To characterize the Norwalk virus N-terminal protein, aa 1 to 223 fused with a His6 tag were expressed in bacteria, purified, and used to raise antibodies in guinea pigs. These antibodies were tested against the cognate protein that was expressed from pC/NV-N wt under the control of the cytomegalovirus promoter in transfected cells and under the control of the T7 promoter in the in vitro TNT system. A [35S]methionine-labeled 45-kDa protein was precipitated from the TNT reaction mixture programmed with pC/NV-N wt by using the postimmunization antiserum (Fig. 2A, lane 4) but not the preimmunization serum (Fig. 2A, lane 3). The precipitated 45 kDa protein comigrated with the dominant product of the pC/NV-N wt TNT reaction (Fig. 2A, lane 2) and with a product from a TNT reaction that was directed by a full-length Norwalk virus cDNA clone designated NV FL101 (Fig. 2A, lane 1). The specificity of the antiserum was then confirmed on pC/NV-N wt-transfected feline kidney (CRFK) cells, where only the post-N-terminal antiserum showed a positive reactivity (Fig. 2B). This antiserum consistently failed to react with mock-transfected cells (Fig. 2B). It was concluded from these tests that the antiserum developed was specific for the N-terminal protein of Norwalk virus.
FIG. 2.
Specificity of the antiserum raised against the N-terminal protein of Norwalk virus. (A) SDS-PAGE analysis of radiolabeled TNT products derived from either the full-length Norwalk virus cDNA clone NV FL101 (lane 1) or pC/NV-N wt (lane 2). pC/NV-N wt TNT products were analyzed by immunoprecipitation with either preimmune serum (Pre) (lane 3) or an N-terminal protein-specific antiserum (Post) (lane 4). (B) Reactivities of preimmune serum and the N-terminal protein-specific antiserum with the Norwalk virus N-terminal protein expressed in CRFK cells, as visualized by IF. (C) Distribution of the FCV p32 protein expressed in CRFK cells from pCI-based plasmid p47/331, visualized with a p32-specific (region B4) antiserum (43).
The expressed Norwalk virus N-terminal protein is colocalized with Golgi markers and may promote Golgi disassembly.
Since it is known that 2B proteins of certain picornaviruses are associated with the Golgi compartment and induce disassembly of the Golgi complex (39), we investigated whether the norovirus N-terminal protein possesses similar properties through its 2BL domain. Examination of IF staining in pC/NV-N wt-transfected CRFK cells revealed two patterns (Fig. 2B). One pattern consisted of marked, discrete aggregates, and the other was a more diffuse distribution of the protein throughout the cytoplasm. A time course examining the temporal occurrence of the aggregates found that they could be observed as early as 5 h posttransfection and that the numbers of cells with aggregates increased over time (data not shown). In contrast, aggregates were not observed in IF staining of CRFK cells transfected with a pCI-based plasmid encoding the p32 protein of the FCV ORF1 polyprotein (Fig. 2C).
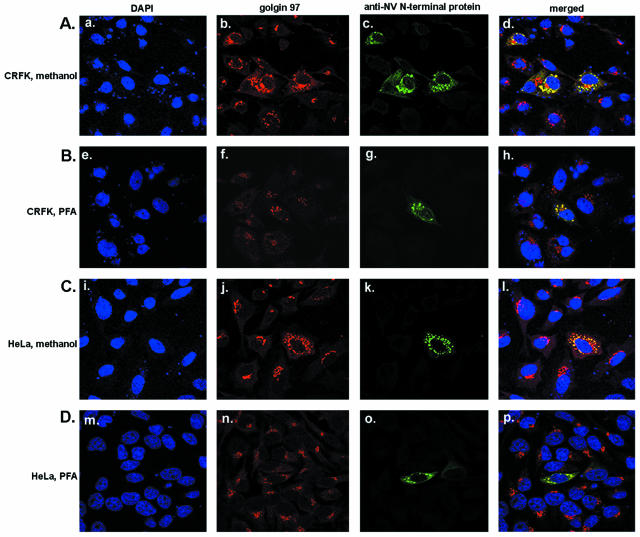
Confocal microscopy was used to determine whether the expression patterns of the Norwalk virus N-terminal protein observed in cells by IF were associated with the Golgi apparatus. Both CRFK cells and human HeLa cells were transfected with the pC/NV-N wt plasmid and then fixed with methanol or PFA after 24 h. The fixed cells were incubated with N-terminal protein-specific antibodies and anti-golgin 97. Anti-golgin 97 is an antibody that recognizes a 97-kDa peripheral membrane protein on the cytoplasmic face of the Golgi apparatus and that characteristically exhibits a perinuclear Golgi staining pattern in nontransfected cells. Cells were then incubated with secondary antibodies and DAPI (for staining of cellular nuclei). The Norwalk virus N-terminal protein was expressed in a fraction of cells, which facilitated comparative analysis of intact and transfected cells. The N-terminal protein was colocalized with the golgin 97 protein in both cell lines, as indicated by the presence of yellow color in the merged images (Fig. 3). This colocalization was observed in cells fixed with either methanol (Fig. 3A and C) or PFA (Fig. 3B and D), although the staining pattern of the golgin 97 antibody alone appeared more diffuse in the PFA-fixed cells (Fig. 3B, panel f, and D, panel n). Evidence for the colocalization of the Norwalk virus N-terminal protein with the Golgi apparatus was also obtained with a p58-specific monoclonal antibody that recognizes an epitope located on a 58-kDa microtubule-binding peripheral Golgi membrane protein (data not shown).
FIG. 3.
Expression of the Norwalk virus N-terminal protein in CRFK and HeLa cells and observation by confocal microscopy. CRFK (A and B) and HeLa (C and D) cells were grown on coverslips and transfected with a plasmid (pC/NV-N wt) encoding the Norwalk virus N-terminal protein. At 24 h posttransfection, cells were fixed with either methanol (A and C) or PFA (B and D) and were incubated with a guinea pig antiserum against the Norwalk virus N-terminal protein together with a mouse monoclonal antibody directed against human golgin 97. Secondary antibodies were fluorescein-conjugated, affinity-purified goat antibodies against guinea pig IgG, used to detect expression of the N-terminal protein (green), and Alexa red-conjugated goat antibodies raised against mouse IgG, used for detection of golgin 97 (red). Nuclei were stained with DAPI (blue). Yellow color in merged images indicate colocalization.
The dispersed aggregates, like those observed in the initial IF assays (Fig. 2B), were stained with both the Golgi- and N-terminal protein-specific antibodies in the two cell lines (Fig. 3) and were not evident in cells negative for the N-terminal protein. This result suggested that expression of the Norwalk virus N-terminal protein in cells was associated with disassembly of the Golgi apparatus. However, this experiment did not rule out a nonspecific effect on the Golgi apparatus due to a possible “overexpression” of the N-terminal protein in cells.
Deletions in the HR of the 2BL domain affect the colocalization of the N-terminal protein with the Golgi apparatus.
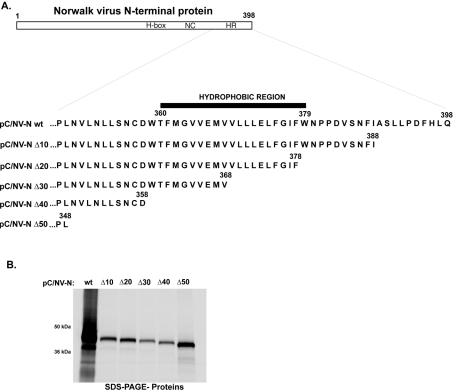
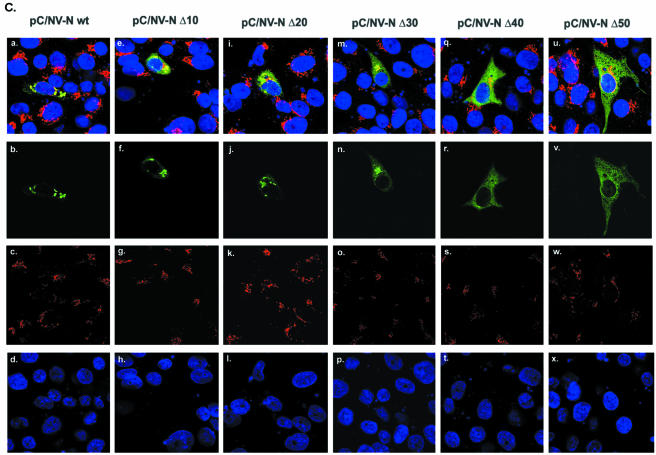
To gain insight into the determinants responsible for the phenotype induced by the Norwalk virus N-terminal protein, we constructed a series of plasmids encoding mutant forms of the protein. First, we investigated a predicted involvement of the 2BL domain in this phenotype by testing C-terminally truncated mutants that lacked a part of the HR (aa 360 to 379). We engineered cDNA constructs encoding sequential deletions of 10 to 50 aa from the C terminus of the Norwalk virus N-terminal protein encoded in pC/NV-N wt (Fig. 4A). SDS- PAGE analysis of the protein products derived by TNT from the five plasmids (designated pC/NV-N Δ10, pC/NV-N Δ20, pC/NV-N Δ30, pC/NV-N Δ40, or pC/NV-N Δ50) showed the expected differences in the observed molecular masses of the encoded proteins (Fig. 4B). Analysis of the transfected cells by confocal microscopy with the N-terminal protein-specific antibodies and anti-golgin 97 showed that the gradual increase in the amount of the N-terminal protein that was deleted correlated with a reduced colocalization of the expressed protein with the Golgi apparatus relative to that of the wt protein (Fig. 4C, panels a, e, i, m, q, and u). We consistently observed distinct colocalization and accumulation of the truncated N-terminal protein with the Golgi marker in cells transfected with pC/NV-N Δ10, pC/NV-N Δ20, or pC/NV-N Δ30 (Fig. 4C, panels e, i, and m, respectively). In contrast, the two variants of the N-terminal protein with larger truncations, expressed from plasmids pC/NV-N Δ40 and pC/NV-N Δ50, were more diffuse throughout the cells, and colocalization with the Golgi marker was reduced (Fig. 4C, panels q and u, respectively). Of interest, the presence of dispersed aggregates that were costained with both anti-N-terminal protein and golgin 97 antibodies decreased markedly as the HR was sequentially deleted (compare pC/NV-N wt [Fig. 4C, panel a] with pC/NV-NV Δ50 [panel u]). The detection of varying phenotypes among mutant forms of the N-terminal protein expressed under similar conditions supported the association of the wt N-terminal protein with disassembly of the Golgi apparatus. In addition, these data were compatible with a role for the HR in the interaction of the N-terminal protein with the Golgi apparatus.
FIG.4.
Deletion mutagenesis of the HR of the Norwalk virus N-terminal protein. (A) Schematic diagram of the cDNA plasmids encoding sequential deletions of 10 aa from the C terminus of the Norwalk virus N-terminal protein. The HR of the Norwalk virus N-terminal protein is located between aa 360 and 379. (B) In vitro expression (TNT) of the proteins encoded by the wt and HR deletion plasmids shown in panel A. (C) Confocal microscopy observation of CRFK cells transfected with plasmids encoding wt or mutant forms of the N-terminal protein. Plasmids shown in panel A were transfected into CRFK cells, and the cells were fixed with methanol 24 h posttransfection. N-terminal protein expression (panels b, f, j, n, r, and v), golgin 97 (panels c, g, k, o, s, and w), and cellular nuclei (panels d, h, l, p, t, and x) were detected as described in the legend to Fig. 3. The merged image for each construct is shown in panels a, e, i, m, q, and u.
Expression and cellular localization of EGFP fusion proteins.
The deletion mutagenesis experiments indicated that the HR might be involved in targeting the Norwalk virus N-terminal protein to the Golgi apparatus. We examined whether the HR could confer this ability on an unrelated protein, EGFP. We engineered EGFP-norovirus fusion protein constructs in which selected sequences in the C-terminal regions of the Norwalk virus and MD145 N-terminal proteins were fused in frame with EGFP (Fig. 5A). These sequences included the entire predicted 2BL domain (aa 301 to 398 of Norwalk virus and aa 233 to 330 of the MD145 virus, which were expressed from plasmids pEGFP/NV-2B and pEGFP/MD145-2B, respectively) and an N-terminally truncated 2BL domain (aa 349 to 398 of Norwalk virus and aa 282 to 330 of MD145 virus, expressed from plasmids pEGFP/NV-HR and pEGFP/MD145-HR, respectively) that began close upstream of the HR (Fig. 5A).
FIG. 5.
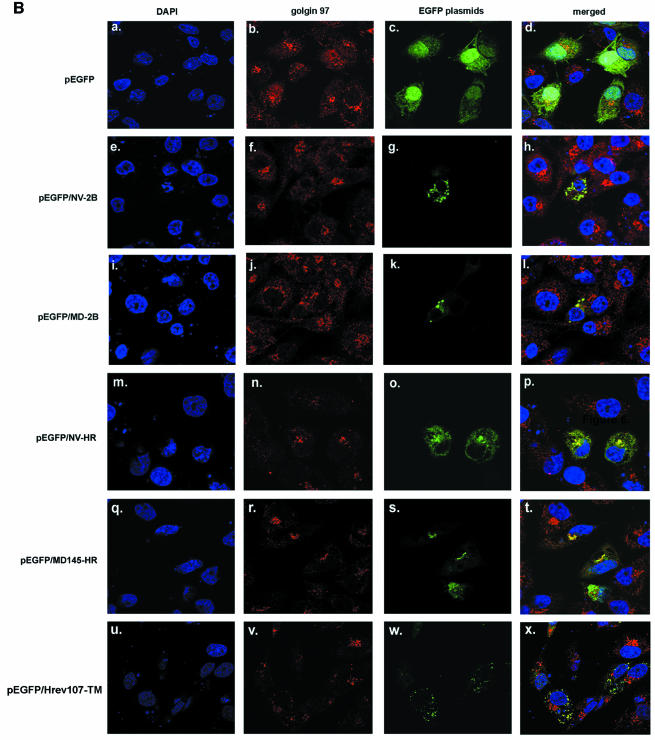
Confocal microscopy of EGFP-TM fusion proteins. (A) Summary of cDNA plasmids encoding EGFP fused with the 2BL region in the C-terminal part of the Norwalk virus (aa 301 to 398) and MD145 (aa 233 to 330) N-terminal proteins. Fusion proteins were also designed to contain the HR (as well as a few amino acid residues immediately upstream of the HR) of Norwalk virus (aa 349 to 398) and MD145 (aa 282 to 330) and a region that included the predicted membrane-associated alpha-helix (TM) of H-rev107 (aa 127 to 162). Identical amino acids in a pairwise comparison of the norovirus proteins in this region are boxed. (B) Confocal microscopy analysis of the expression of control plasmid pEGFP (panels a to d), pEGFP/NV-2B (panels e to h), pEGFP/MD145-2B (panels i to l), pEGFP/NV-HR (panels m to p), pEFPF/MD145-HR (panels q to t), and pEGFP/Hrev107-TM (panels u to x). CRFK cells were grown on coverslips and transfected with EGFP-based plasmids. After 24 h, cells were fixed with 4% PFA and permeabilized in 0.1% Triton X-100. Cells were incubated with anti-human golgin 97, followed by staining with specific secondary antibodies as described in the legend to Fig. 3. Both nonmerged and merged images are shown.
The plasmids were transfected into CRFK cells, and confocal microscopy was conducted by using PFA fixation and direct visualization of EGFP expression. Anti-golgin 97 and a secondary antibody for the detection of golgin 97 binding were used to determine colocalization with the Golgi apparatus. Expression of the pEGFP control showed a diffuse pattern of green fluorescence throughout the cell (Fig. 5B, panel c). The merged image of pEGFP and golgin 97 staining (Fig. 5B, panel d) revealed some yellow color, but in general, EGFP did not appear to accumulate in or disrupt the Golgi apparatus. In contrast, cells transfected with the EGFP-norovirus 2BL fusion proteins characteristically showed the presence of discrete aggregates that appeared to colocalize with golgin 97 (Fig. 5B, panels h and l). In addition, the norovirus 2BL fusion proteins appeared toxic: many of the transfected cells became rounded and detached from the surface of the culture dish (data not shown).
Expression of the Norwalk virus and MD145 EGFP-HR fusion proteins that were engineered to analyze the role of the HR characteristically showed evidence for colocalization of EGFP expression with the Golgi apparatus (Fig. 5B, panels p and t). In the absence of upstream sequences from the N-terminal protein, these fusion proteins did not yield visible aggregates in transfected cells. For comparison, we designed a fusion protein containing the membrane-associated region (TM) of H-rev107 fused to EGFP. In contrast to the findings for the norovirus fusion proteins, the EGFP/H-rev107-TM fusion protein did not accumulate in the Golgi membranes of the perinuclear region (Fig. 5B, panel x). However, the green fluorescence formed a punctate, dispersed pattern throughout the cytoplasm, with some evidence for colocalization with golgin 97. These results suggested that the C-terminal hydrophobic TM region of H-rev107 may interact with membranes in the cells, but through a mechanism likely different from that of the HR of the noroviruses.
We engineered an N-terminal protein with an amino acid substitution (cysteine 274 to alanine) in the NC motif (17). CRFK cells were transfected with pC/NV-NC/mut and observed by confocal microscopy 24 h after transfection. The confocal microscopy imaging showed that the mutation in the NC motif did not affect colocalization with the Golgi apparatus or the formation of visible aggregates in the cells, indicating that this predicted site may not be involved in the disassembly of the Golgi apparatus by the N-terminal protein (data not shown).
DISCUSSION
Positive-strand RNA viruses have evolved to utilize intracellular membranes for their replication. An understanding of the mechanisms responsible for the action of viral proteins on host cells is relevant to the development of virus control strategies. This study focused on the characterization of the first protein encoded in the Norwalk virus genome, designated here the N-terminal protein. Although Norwalk virus cannot be grown in cell culture, we used computer analysis predictions coupled with a eukaryotic expression system to explore possible functions for this protein. Expression of the Norwalk virus N-terminal protein in feline and human cells resulted in a reproducible pattern of protein distribution in the cytoplasm of transfected cells characterized by colocalization with the Golgi apparatus and the apparent disassembly of the Golgi apparatus into discrete aggregates. The disruption of the Golgi apparatus by the Norwalk virus N-terminal protein is similar to the effect of the poliovirus 2B protein on the Golgi apparatus when the latter is expressed alone in cells (39). The poliovirus 2B protein has recently been described as a viroporin-like molecule that mediates the permeabilization of lipid bilayer membranes (1). Our data suggest that the Norwalk virus N-terminal protein interacts with intracellular membranes and may possess certain 2BL functions during replication. However, the presence of motifs immediately upstream that are conserved with certain picornavirus 2A proteins raises the possibility of 2AL functions for the Norwalk virus N-terminal protein as well. The function of the 2A protein is not established for all members of the family Picornaviridae. The poliovirus 2A protein is a cysteine protease that cleaves itself from the newly synthesized polyprotein at its amino terminus (45). However, other picornavirus 2A proteins, such as those of the human parechoviruses and Aichi virus, apparently lack protease activity and share H-box and NC motifs with certain tumor suppressor proteins (17). Like these picornavirus 2A proteins, the norovirus N-terminal protein has no known protease activity and contains the H-box and NC motifs. It should be noted that the potential 3CL cleavage site in the norovirus N-terminal protein recently proposed by Seah et al. (40) would result in cleavage between the H-box and NC motifs, and the verification of this cleavage site will be important in future studies of the N-terminal protein. Mutagenesis of the cysteine in the Norwalk virus NC motif did not affect the expression phenotype of the Norwalk virus N-terminal protein, and the role of this motif, if any, in Norwalk virus replication cannot yet be determined. It is interesting that both the predicted 2AL and 2BL domains map toward the C-terminal half of the norovirus N-terminal protein. It is possible that the extreme N terminus of this protein (which is highly variable among the noroviruses) may be involved in 2AL or 2BL functions (or both), or it may have a function unique to the caliciviruses.
Our computer analysis of the Norwalk virus N-terminal protein predicted an HR spanning aa 360 to 379 near the C terminus. Deletion of this region from the Norwalk virus N-terminal protein appeared to abolish the ability of the protein to colocalize with, and cause disassembly of, the Golgi apparatus in transfected cells. Targeting of proteins to the Golgi complex is not well understood, but certain localization signals have been described (33). Comparison of the Norwalk virus HR to signals previously identified in cellular proteins directed to the Golgi apparatus (32-34) found no striking amino acid similarity, although phenylalanine residues (characteristic of mammalian Golgi enzyme TM domains) were present. Furthermore, the Norwalk virus HR does not share significant identity with a Golgi targeting signal in the cytoplasmic tails of the coronavirus infectious bronchitis virus E protein and the Uukuniemi bunyavirus G1 protein (2, 6). We explored whether the norovirus HR could function in directing an unrelated protein to the Golgi apparatus by designing EGFP fusion proteins containing the HR with or without upstream sequences from the putative 2BL region of the N-terminal protein. Expression of EGFP fused to the norovirus HR lacking most of the upstream 2BL region showed Golgi colocalization and no evidence for Golgi disassembly. We conclude that the HR plays a role in directing the norovirus N-terminal protein to the Golgi complex but that additional upstream sequences (such as those in the 2BL region) are required for disassembly. It should be noted that interactions of the N-terminal protein with other viral proteins, as well as its levels of expression in an authentic viral replication cycle, could not be determined in this study. However, it is of interest that parechoviruses (with which the noroviruses share 2AL motifs) have recently been associated with disassembly of the Golgi apparatus in infected cells, and it was proposed that parechovirus membranous replication complexes might be derived from membranes carrying a Golgi marker (27).
A recent study of FCV found that nearly all the viral proteins were associated with enzymatically active membranous replication complexes isolated from virus-infected cells (13). Norwalk virus will likely prove similar in its replication strategy. However, there may be important differences in the N-terminal nonstructural proteins among the caliciviruses, just as there are among the picornaviruses. The FCV ORF1 N-terminal proteins (p5.6 and p32) (43) do not share the H-box and NC sequence motifs with H-rev107, although an HR (corresponding to aa 272 to 293 of the FCV ORF1 polyprotein) can be identified by TMPred in the FCV 2B equivalent, p32 (data not shown). However, expression of the FCV p32 protein alone in cells does not show evidence for disassembly of the Golgi apparatus.
The class II tumor suppressor H-rev107 is down-regulated in certain cancerous cells, which has led to the proposal that it controls cellular proliferation as part of its normal function (14, 41). In tumor cells, the gene encoding H-rev107 remains intact, and its expression is apparently regulated at the level of transcription or translation (a key feature of class II tumor suppressors) (41). Although our data showed evidence for a punctate expression pattern of the EGFP/H-rev107-TM fusion protein in cells, with evidence for some colocalization with golgin 97, the perinuclear structure of the Golgi apparatus appeared intact and the fusion protein did not accumulate in this region. A previous study of H-rev107 protein showed that removal of 25 aa from the C terminus of the H-rev107 protein (which would remove nearly all the TM domain) abolished its association with a membranous fraction isolated from transfected cells (41). Of interest, the C-terminal hydrophobic domain of TIG3, an H-rev107-related tumor suppressor protein, has also been associated with its intracellular localization (7). Although H-rev107, the norovirus N-terminal protein, and some picornavirus 2A proteins share relatedness in certain motifs, it remains unclear whether interaction with the Golgi apparatus is a common theme. For viruses, the modification of the cellular protein processing machinery and vesicle transport system could be interpreted as part of the “hostile” takeover of a cell. However, for H-rev107 and other related tumor suppressor proteins, the mechanism responsible for controlling cell growth via an interaction with intracellular membranes is less clear. It will be interesting to compare emerging data on the functions of proteins in this group in an effort to identify common themes in virus and host cell evolution.
Acknowledgments
We extend our appreciation to Owen Schwartz, Head, Biological Imaging Facility of NIAID, NIH, for assistance with the confocal imaging. We thank Albert Kapikian, Head, Epidemiology Section, NIAID, for support of this work.
REFERENCES
- 1.Agirre, A., A. Barco, L. Carrasco, and J. L. Nieva. 2002. Viroporin-mediated membrane permeabilization: pore formation by non-structural poliovirus 2B protein. J. Biol. Chem. 277:40434-40441. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, A. M., and R. F. Pettersson. 1998. Targeting of a short peptide derived from the cytoplasmic tail of the G1 membrane glycoprotein of Uukuniemi virus (Bunyaviridae) to the Golgi complex. J. Virol. 72:9585-9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belliot, G., S. V. Sosnovtsev, T. Mitra, C. Hammer, M. Garfield, and K. Y. Green. 2003. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 77:10957-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, D. N., J. N. Burroughs, T. J. Harris, and F. Brown. 1978. The structure and replication of calicivirus RNA. Nature 274:614-615. [DOI] [PubMed] [Google Scholar]
- 5.Burroughs, J. N., and F. Brown. 1978. Presence of a covalently linked protein on calicivirus RNA. J. Gen. Virol. 41:443-446. [DOI] [PubMed] [Google Scholar]
- 6.Corse, E., and C. E. Machamer. 2002. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 76:1273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deucher, A., S. Nagpal, R. A. Chandraratna, D. Di Sepio, N. A. Robinson, S. R. Dashti, and R. L. Eckert. 2000. The carboxy-terminal hydrophobic domain of TIG3, a class II tumor suppressor protein, is required for appropriate cellular localization and optimal biological activity. Int. J. Oncol. 17:1195-1203. [DOI] [PubMed] [Google Scholar]
- 8.Dingle, K. E., P. R. Lambden, E. O. Caul, and I. N. Clarke. 1995. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round structured virus. J. Gen. Virol. 76:2349-2355. [DOI] [PubMed] [Google Scholar]
- 9.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181:S254-S261. [DOI] [PubMed] [Google Scholar]
- 10a.Gorbalenya, A. E., and E. V. Koonin. 1993. Comparative analysis of key enzymes of replication and expression of positive-strand RNA viruses. Validity of approach and functional applications. Sov. Sci. Rev. D Physicochem. Biol. 11:1-84. [Google Scholar]
- 11.Green, K. Y., G. Belliot, J. L. Taylor, J. Valdesuso, J. F. Lew, A. Z. Kapikian, and F. Y. Lin. 2002. A predominant role for Norwalk-like viruses as agents of epidemic gastroenteritis in Maryland nursing homes for the elderly. J. Infect. Dis. 185:133-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 13.Green, K. Y., A. Mory, M. H. Fogg, A. Weisberg, G. Belliot, M. Wagner, T. Mitra, E. Ehrenfeld, C. E. Cameron, and S. V. Sosnovtsev. 2002. Isolation of enzymatically active replication complexes from feline calicivirus-infected cells. J. Virol. 76:8582-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajnal, A., R. Klemenz, and R. Schafer. 1994. Subtraction cloning of H-rev107, a gene specifically expressed in H-ras resistant fibroblasts. Oncogene 9:479-490. [PubMed] [Google Scholar]
- 15.Hardy, M., T. Crone, J. Brower, and K. Ettayebi. 2002. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 89:29. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, K., and W. Stoffel. 1993. TMbase: a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 17.Hughes, P. J., and G. Stanway. 2000. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 81:201-207. [DOI] [PubMed] [Google Scholar]
- 18.Husmann, K., C. Sers, E. Fietze, A. Mincheva, P. Lichter, and R. Schafer. 1998. Transcriptional and translational downregulation of H-REV107, a class II tumour suppressor gene located on human chromosome 11q11-12. Oncogene 17:1305-1312. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, X., D. Y. Graham, K. Wang, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 21.Johansson, S., B. Niklasson, J. Maizel, A. E. Gorbalenya, and A. M. Lindberg. 2002. Molecular analysis of three Ljungan virus isolates reveals a new, close-to-root lineage of the Picornaviridae with a cluster of two unrelated 2A proteins. J. Virol. 76:8920-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapikian, A. Z., R. G. Wyatt, R. Dolin, T. S. Thornhill, A. R. Kalica, and R. M. Chanock. 1972. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 10:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. Virgin IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575-1578. [DOI] [PubMed] [Google Scholar]
- 24.Konig, M., H. J. Thiel, and G. Meyers. 1998. Detection of viral proteins after infection of cultured hepatocytes with rabbit hemorrhagic disease virus. J. Virol. 72:4492-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. (Erratum, 28:546.) [DOI] [PubMed] [Google Scholar]
- 26.Koopmans, M., J. Vinje, M. de Wit, I. Leenen, W. van Der Poel, and Y. van Duynhoven. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181:S262-S269. [DOI] [PubMed] [Google Scholar]
- 27.Krogerus, C., D. Egger, O. Samuilova, T. Hyypia, and K. Bienz. 2003. Replication complex of human parechovirus 1. J. Virol. 77:8512-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519. [DOI] [PubMed] [Google Scholar]
- 29.Liu, B., I. N. Clarke, and P. R. Lambden. 1996. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J. Virol. 70:2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopman, B. A., M. H. Reacher, Y. Van Duijnhoven, F. X. Hanon, D. Brown, and M. Koopmans. 2003. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg. Infect. Dis. 9:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munro, S. 1995. A comparison of the transmembrane domains of Golgi and plasma membrane proteins. Biochem. Soc. Trans. 23:527-530. [DOI] [PubMed] [Google Scholar]
- 33.Munro, S. 1998. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 8:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munro, S., and B. J. Nichols. 1999. The GRIP domain: a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 9:377-380. [DOI] [PubMed] [Google Scholar]
- 35.Nakata, S., S. Honma, K. K. Numata, K. Kogawa, S. Ukae, Y. Morita, N. Adachi, and S. Chiba. 2000. Members of the family Caliciviridae (Norwalk virus and Sapporo virus) are the most prevalent cause of gastroenteritis outbreaks among infants in Japan. J. Infect. Dis. 181:2029-2032. [DOI] [PubMed] [Google Scholar]
- 36.Neill, J. D. 1990. Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA polymerase, cysteine protease and 2C polypeptides. Virus Res. 17:145-160. [DOI] [PubMed] [Google Scholar]
- 37.Pletneva, M. A., S. V. Sosnovtsev, and K. Y. Green. 2001. The genome of Hawaii virus and its relationship with other members of the Caliciviridae. Virus Genes 23:5-16. [DOI] [PubMed] [Google Scholar]
- 38.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 39.Sandoval, I. V., and L. Carrasco. 1997. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J. Virol. 71:4679-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seah, E. L., J. A. Marshall, and P. J. Wright. 2003. Trans activity of the norovirus Camberwell proteinase and cleavage of the N-terminal protein encoded by ORF1. J. Virol. 77:7150-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sers, C., U. Emmenegger, K. Husmann, K. Bucher, A. C. Andres, and R. Schafer. 1997. Growth-inhibitory activity and downregulation of the class II tumor-suppressor gene H-rev107 in tumor cell lines and experimental tumors. J. Cell Biol. 136:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VPg for infectivity. Virology 210:383-390. [DOI] [PubMed] [Google Scholar]
- 43.Sosnovtsev, S. V., M. Garfield, and K. Y. Green. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 76:7060-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sosnovtsev, S. V., S. A. Sosnovtseva, and K. Y. Green. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 72:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toyoda, H., M. J. Nicklin, M. G. Murray, C. W. Anderson, J. J. Dunn, F. W. Studier, and E. Wimmer. 1986. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 45:761-770. [DOI] [PubMed] [Google Scholar]
- 46.van Kuppeveld, F. J., J. M. Galama, J. Zoll, P. J. van den Hurk, and W. J. Melchers. 1996. Coxsackie B3 virus protein 2B contains cationic amphipathic helix that is required for viral RNA replication. J. Virol. 70:3876-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirblich, C., H. J. Thiel, and G. Meyers. 1996. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J. Virol. 70:7974-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]