Abstract
Two types of vaccine-derived polioviruses have been recently designated to emphasize the different origins of the evolved viruses: circulating vaccine-derived polioviruses (cVDPV) associated with outbreaks of paralytic disease and strains isolated from chronically infected immunodeficient individuals (iVDPV). We describe here a type 3 VDPV (PV3/EST/02/E252; later E252) isolated from sewage collected in Tallinn, Estonia, in October 2002. Due to aberrant properties in subtyping, the virus was subjected to detailed characterization. Partial genomic sequencing suggested that the closest relative was the oral vaccine strain PV3/Sabin, but the two virus strains shared only 86.7% of the 900 nucleotides (nt) coding for the capsid protein VP1. Phylogenetic analysis of the nearly complete genome [nt 19 to poly(A)] revealed multiple nucleotide substitutions throughout the genome and a possible Sabin 3/Sabin 1-recombination junction site in the 2C coding region. A calculation based on the estimated mutation frequency of the P1 region of polioviruses suggested that the E252 virus might have replicated in one or more individuals for approximately 10 years. No persons chronically excreting poliovirus are known in Estonia. Amino acid substitutions were seen in all known antigenic sites, which was consistent with the observed aberrant antigenic properties of the virus demonstrated by both monoclonal antibodies and human sera from vaccinated children. In spite of the apparent transmission potential, no evidence was obtained for circulation of the virus in the Estonian population.
Polioviruses, members of the Enterovirus genus in the Picornaviridae family, are important human pathogens causing the acute paralytic disease poliomyelitis. The worldwide program for eradication of wild-type poliovirus is coordinated by the World Health Organization (WHO). The program includes two simultaneous approaches: intensive immunizations mainly with the trivalent oral poliovirus vaccine (OPV) (35) and systematic surveillance of the remaining cases. The surveillance includes, most importantly, epidemiological and virological investigation of acute flaccid paralytic (AFP) cases. The AFP surveillance is supplemented in some countries by analysis of poliovirus circulation in human populations through investigation of wastewater specimens contaminated with human fecal materials (environmental surveillance) and/or by scrutinizing the results of routine virus diagnostics (enterovirus surveillance).
The evolution rate of polioviruses is very high, partly due to the high error frequency in RNA synthesis: roughly 10−4 per base pair per replication cycle. For wild polioviruses circulating in human populations, nucleotide substitutions accumulate at a rate of approximately 1% per year and consist primarily of changes at synonymous codon positions thus not leading to amino acid substitutions (22). The genetic diversity of poliovirus strains is exploited in molecular epidemiology, a key component of poliovirus surveillance (33), currently based on sequence analysis of the VP1 coding region of the genome. The replication of the attenuated poliovirus strains (Sabin vaccine strains) in the human gut also leads to genetic variation and may result in reversions of the vaccine strains to pathogenic phenotypes reminiscent of wild polioviruses.
In addition to the high mutation frequency, the divergence of poliovirus strains is increased by recombination. Intertypic recombination is a frequent phenomenon in poliovirus vaccinees, and strains with a recombinant genome have been isolated from both healthy vaccinees and from patients with vaccine-associated poliomyelitis (5, 15-17, 24). In natural intertypic recombinant poliovirus strains, the recombination junctions are usually located in the genomic region encoding the nonstructural proteins. However, in some strains, the recombination site has been shown to reside in the capsid protein VP1 coding region (2, 27).
OPV recipients are known to excrete OPV-derived polioviruses for various periods but usually not more than for a few months. During extended replication in immunodeficient individuals, the virus may accumulate point mutations and the modified virus, referred to as immunodeficiency-associated vaccine-derived poliovirus (iVDPV), has occasionally caused paralytic disease in the vaccinee (4, 18, 23, 26). VDPVs may also be generated through circulation of OPV-derived virus strains in human populations with deficient herd immunity (circulating VDPV [cVDPV]). Some VDPVs characterized in the literature cannot be classified in these two main categories, and their origin remains open to question (10, 11, 37). Recently, cVDPVs have been associated with four outbreaks of poliomyelitis. Type 2 VDPV circulated in Egypt for over 10 years (1983 to 1993) and was isolated from 30 patients (7, 41). An outbreak of poliomyelitis on the island of Hispaniola was associated with type 1 cVDPV (21). In the Philippines, type 1 cVDPV was involved in three poliomyelitis cases in 2001 (6, 8), and in Madagascar in 2002, type 2 cVDPV was the causative agent of four paralytic cases (9, 34). In each of these outbreaks, the cVDPV strains were recombinants, originating from either a type 1 or 2 OPV strain and in which most of the genomic region encoding the nonstructural proteins was suggested to be derived from an unknown non-polio enterovirus.
We report here the complete genomic sequence of a highly divergent variant of the Sabin strain of type 3 poliovirus (PV3) isolated from the main sewage channel of Tallinn, Estonia, in October 2002. Estonia has been free of poliomyelitis cases since early 1960s (20). The current virus was isolated during routine screening for wild-type poliovirus circulation. It was not linked to any epidemics, and the origin of the virus remains unresolved.
MATERIALS AND METHODS
Reference virus strains and cell lines.
The reference PV3 strain Sabin was kindly provided by J. Martin (National Institute for Biological Standards and Control [NIBSC], Potters Bar, Hertfordshire, United Kingdom). Poliovirus type 3/Leon/USA/1937 (VR-62) was obtained from the American Type Culture Collection, Manassas, Va. The PV3/Saukett H strain was originally from Institute Merieux, Marcy l'Etoile, France. Recombinant mouse L cells expressing human poliovirus receptor (L20B) (30) and human rhabdomyoma (RD) cells were provided by the WHO Polio Labnet. The green monkey kidney (GMK) cell line had been maintained in the laboratory since the 1960s.
Virus isolation.
Wastewater samples for environmental poliovirus surveillance were regularly collected in Tallinn by the “trap principle” essentially as described previously (3). Tightly bound multilayer pads of cotton gauze (about 10 by 10 by 2 cm) were fixed with a string to the sewer wall and immersed into the sewage stream in the inlet sewer entering the main sewage treatment plant or similar places elsewhere in the sewage network. After 6 days of exposure, the pads were removed and transported to the laboratory in a container with 10 ml of cold Hanks' balanced salt solution (pH 7.5). The container was shaken vigorously for 5 min, followed by squeezing the pad several times. All liquid was finally squeezed from the pad and poured into a sterile centrifuge tube. pH was adjusted to 8.0 to 8.4 with 1 M HCl or NaOH, and the sample was clarified by low-speed centrifugation for 20 min. Supernatant was poured into a new sterile tube and extracted with 0.5 ml of chloroform per 10 ml of supernatant. The chloroform-extracted elute was examined for the presence of poliovirus as recommended for stool extracts in the WHO Polio Laboratory Manual (40).
During the intensified sampling period, two 1-liter samples of raw sewage were drawn from the inlet of the sewage treatment plant and transported at 4°C within a few hours to the National Public Health Institute (KTL) in Helsinki, Finland. At KTL, the two-phase separation method was used essentially as described previously (31), except that the entire sample was concentrated for analysis in cell culture.
Serotyping and ITD.
The serotype of the isolate was confirmed at KTL by neutralization with polyclonal antisera obtained from the National Institute of Public Health and the Environment (RIVM), Bilthoven, The Netherlands, according to the protocol recommended by the WHO (40).
Antigenic intratypic differentiation (ITD) was performed with two validated assays. First, an enzyme immuno assay (EIA) was carried out with cross-absorbed antisera for PV3/Sabin and PV3/non-Sabin-like (RIVM) (39). Second, a neutralization assay was carried out with a mixture of Sabin-specific monoclonal antibodies (MAbs) for the three poliovirus serotypes as described by Martin et al. (27).
Reverse transcription-PCR restriction fragment length polymorphism (RT-PCR-RFLP) was used for molecular ITD (1). The sequence of the VP1 capsid protein coding region was determined as previously described (12).
Plaque purification.
The isolate was purified by using a plaque assay in monolayer cultures of GMK cells and passaged twice in GMK cells at 36°C. The same virus stock (from now on referred to as PV3/EST02/E252, or E252) was used in all subsequent studies.
Sequencing and sequence analysis.
Nearly complete genome sequencing of the isolate E252 was carried out as previously described (2) with primers designed by a primer-walking strategy. Electropherograms obtained from cycle sequencing (ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit; Applied Biosystems, Espoo, Finland) in an automated sequencer (model 310; Applied Biosystems) were analyzed by Sequencing Analysis, v3.1, and Sequence Navigator, v.1 (Applied Biosystems). At least two corresponding electropherograms were performed and analyzed for each genomic region. Multiple sequence alignments were made using Clustal X (1.8). Distance matrices were estimated using the DNADIST and PROTDIST programs (PHYLIP, version 3.572c) (13) with the maximum-likelihood model of nucleotide substitution. Dendrograms were drawn with NEIGHBOR (PHYLIP) using the neighbor-joining option and visualized with NJPlot. Bootstrap analysis was performed using SEQBOOT (PHYLIP) with 1,000 replicates. Similarity analysis of the complete genome alignments was performed with the SimPlot software package (version 2.5) (25). The complete genome sequences of PV3/Sabin and PV1/Sabin used in comparisons were from GenBank (accession no. X00925 and V01150, respectively). The illustrations of the molecular structure of E252 were generated by the RasWin Molecular Visualization program (version 2.6; Glaxo Wellcome Research and Development, Stevenage, Herefordshire, United Kingdom) (36). The PV3/Sabin coordinates were from the Protein Data Bank, under identification no. 1PVC (14).
Assay for temperature sensitivity.
Temperature sensitivity of the virus E252 was assayed by comparison of plaque formation in GMK and RD cells at 36 and 39.5°C. Serial 10-fold dilutions were prepared from stock viruses PV3/Sabin, E252, and PV3/Leon. Monolayer cultures of both cell lines on six-well plates were inoculated with 100 μl of virus dilutions. After adsorption for 30 min at either 36 or 39.5°C, the unabsorbed virus inoculums were removed. Cell monolayers were washed twice with Hanks' balanced salt solution and overlayed with 2 ml of 0.5% carboxymethyl cellulose in minimal essential medium supplemented with 1% fetal calf serum. The plates were incubated at 36 or 39.5°C and stained with crystal violet at 1 (RD) or 2 (GMK) days postinfection.
Microneutralization assay.
A standard microneutralization assay was used to analyze neutralizing antibodies against the isolate E252 in human sera. Briefly, 50 μl of fourfold serum dilutions beginning at 1:4 was mixed with a standard amount of pretitrated virus (30 to 300 50% tissue culture infective doses [TCID50]/50 μl) in 96-well microtiter plates and incubated for 1 h at 36°C. After a further overnight incubation at 4°C, 2 × 104 GMK cells were added to each well, and the incubation was continued for 6 days at 36°C. The wells were stained with crystal violet, and the virus-induced cytopathic effect (CPE) was determined by photometry.
Human sera.
The first study group included 15 OPV-vaccinated Estonian children ages 3 to 7 years. One serum specimen was drawn from each child between November 2002 and February 2003. The second study group consisted of 23 Finnish children vaccinated with three doses of inactivated polio vaccine (IPV) at ages 4, 6, and 12 months. Serum samples for antibody analysis were drawn at the age of 18 months (29).
Neutralizing MAbs.
PV3-specific MAbs 204, 472, 495, 520, 838, 875, 879, 881, and 882, corresponding to antigenic sites 1 to 3, were kindly provided by Philip D. Minor (NIBSC).
Nucleotide sequence accession number.
The nucleotide sequence of the complete poliovirus genome determined in this study has been submitted to GenBank under accession no. AY421739.
RESULTS
Isolation of an aberrantly reacting PV3.
Estonian polio surveillance includes AFP, enterovirus, and environmental surveillance. No cases of poliomyelitis caused by wild-type virus have been detected in the country since the early 1960s. In 2002, altogether 217 fecal specimens from patients suffering from various neurological or gastrointestinal symptoms were examined in cell cultures susceptible to polioviruses. One poliovirus (PV3/Sabin) was isolated, while 40 specimens were found to contain non-polio enteroviruses. Eighteen specimens from 2002, kept at the Tallinn laboratory, were reexamined at KTL. Non-polio enteroviruses (coxsackievirus B3, echovirus 6, and echovirus 13) were isolated from 11 specimens. All of the reexamined specimens were negative for polioviruses.
Environmental surveillance comprised monthly sampling of two locations in Tallinn. Five of the 13 samples examined during the second half of 2002 revealed echovirus 13, and 1 of them (22 October 2002), in addition, a PV3 labeled PV3/EST/02/E252 (E252), is characterized in detail below. As a response to this observation, four other locations in Tallinn were sampled in December 2002. One PV3 was isolated from the sample collected on 16 December and was shown to be typically Sabin-like. Furthermore, two 1-liter samples of raw sewage collected on 18 December 2002 and 22 January 2003 were concentrated at KTL in Helsinki. The latter sample revealed Sabin-like PV3. Samples collected and analyzed in Tallinn in early 2003 have revealed two isolates of PV2 with Sabin-like characteristics.
Preliminary characterization of the isolate.
Serotype 3 of poliovirus E252 was confirmed by complete neutralization with polyclonal antisera specific for type 3. RT-PCR-RFLP revealed an atypical restriction pattern with one of the three enzymes (Hpa), while the other two (Dde and Hae) gave a result typical of PV3/Sabin. In EIA with cross-absorbed rabbit antisera to Sabin-like and non-Sabin-like PV3, the isolate E252 was not recognized by either antiserum. However, retesting under more sensitive assay conditions suggested the double-reactive nature of the isolate. In the second antigenic ITD assay based on MAbs (27), E252 was not neutralized with Sabin-specific MAbs. The nucleotide sequence of the VP1 capsid protein was 86.7% identical to the PV3/Sabin sequence (GenBank accession no. X00925).
Complete genomic sequence.
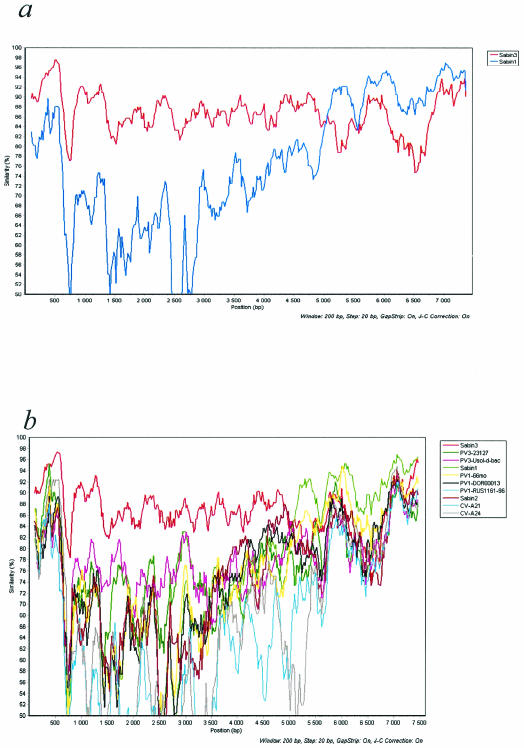
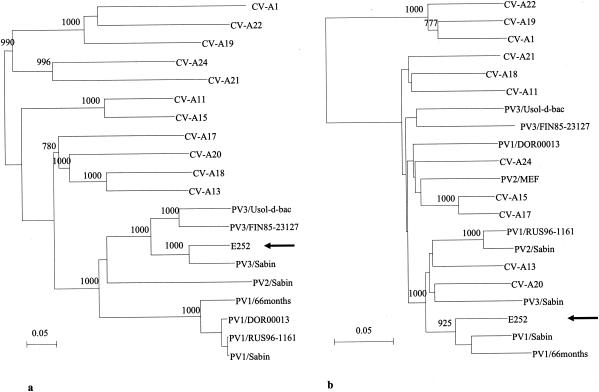
The (almost) complete genomic sequence of isolate E252 was determined from nucleotide (nt) 19 to the end of the coding region and compared to that of PV3/Sabin and other sequence data available in GenBank. According to a similarity analysis with a 200-nt sliding window, the closest relative of E252 from the 5′ noncoding region (NCR) close to the 3′ end of the 2C coding region was PV3/Sabin, the rest of the genome being more closely related to PV1/Sabin (Fig. 1). The most divergent part of the genome of E252 was the approximately 100-nt-long hypervariable region in the 5′ NCR. This region included two deletions (3 and 8 nt, respectively) compared to PV3/Sabin, and the identity to PV3/Sabin was only approximately 50%. Nearly complete genomic sequencing revealed multiple nucleotide substitutions throughout the genome as compared to PV3/Sabin or PV1/Sabin. The phylogenetic relationships of the E252 with other polioviruses and human enterovirus C (HEV-C) in VP1 and three-dimensional (3D) polymerase coding regions are shown in Fig. 2a and b, respectively.
FIG. 1.
Similarity analysis of complete PV genomes. Analysis was done with a sliding window of 200 nt moving in steps of 20 nt, with all positions with a gap deleted, and the nucleotide similarity was plotted by using the JC model of nucleotide substitution. (a) E252 is compared against PV1/Sabin and PV3/Sabin (b) E252 is compared against 12 different poliovirus or HEV-C genomes. The GenBank accession numbers of the sequences used are as follows: PV3/Sabin, X00925; PV3/23127, X04468; PV3/Usol-d-bac, AJ293918; PV1/Sabin, V01150; PV1/DOR00013, AF405690; PV1/RUS1161-96, AF462419; PV2/Sabin, X00595; CV-A21, D00538; CV-A24, D90457; and PV1/66months, AJ132961.
FIG. 2.
Neighbor-joining tree of the nucleotide sequence relationships of the isolate E252 and other polioviruses and HEV-C in VP1 (a) and 3D polymerase (b) protein coding regions. The location of E252 is indicated by an arrow. Numbers at nodes represent reliability values for the internal branch (= bootstrap values). Values over 70% are shown. The scale bar indicates the evolutionary distance (0.01 = 1%). The GenBank accession numbers for the sequences are as follows: CV-A1, AF081293 and AF499635; CV-A11, AF081301 and AF499636; CV-A13, AF081303 and AF499637; CV-A15, AF081305 and AF499638; CV-A17, AF081306 and AF499639; CV-A18, AF081307 and AF499640; CV-A19, AF081308 and AF499641; CV-A20, AF081309 and AF499642; and CV-A22, AF081310 and AF499643. For information on other sequences, see the legend to Fig. 1.
Amino acid substitutions.
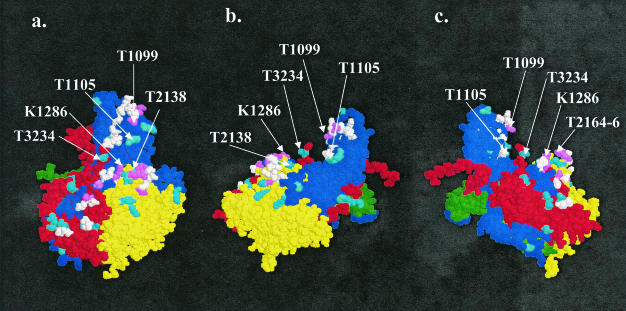
While most of the nucleotide substitutions in the coding region of the genome were synonymous, in the capsid protein coding part they resulted in as many as 40 amino acid substitutions (4.1% of all positions) relatively evenly distributed through the three major proteins VP1, VP2, and VP3. No substitutions were seen in VP4. The E252 had differences in all antigenic sites when compared to PV3/Sabin (Table 1). In the 3D atomic model of a protomer, based on the X-ray crystallographic analysis of PV3/Sabin (14), most of the substituted positions were located close to the outer surface within or near the designated neutralization antigenic sites (Nags; Fig. 3). Yet major parts of the Nag site motifs were left unaffected, although we cannot tell whether the conformation of the nonsubstituted part of the peptide chain remained conserved. Substitutions also occurred elsewhere, including the beta barrels, but these substitutions were usually conservative by nature, whereas those observed in the inter-beta chain loops or terminal strings of the proteins had less limitation as regards the category of amino acid.
TABLE 1.
Amino acid sequences of different antigenic sites of PV3
| Virus strain | Amino acid sequence
|
|||
|---|---|---|---|---|
| Site 1a (VP1, aa 89-100)b | Site 2B (VP2, aa 164-172) | Site 3A (VP1, aa 286-290) | Site 3B (VP3, aa 58-60, 70-71, 77, 79) | |
| Sabin 3 | E V D N E Q P T T R A Q | N A V T S P K R E | R N N L D | E S T V T D S |
| Saukett G | ----- E------ | T-------- | K D-- N | - N----- |
| Saukett H | ----- E------ | T-------- | K D-- N | - N----- |
| 23127FIN | ------- A- N V- | T-------- | K D G- A | - N-- R N- |
| E252 | -------- A- T- | T T T----- K | K--- E | - R----- |
FIG. 3.
Predicted molecular structure of the protomer E252. A space-fill model of a protomer (14) is shown at three different angles. The picture was generated with the RasWin Molecular Visualization Program (version 2.6) (36). (a) Front view. (b) Left-side view. (c) Right-side view. VP1, blue; VP2, yellow; VP3, red; VP4, green. A changed amino acid is shown by cyan, an antigenic site is in white, and a changed amino acid in an antigenic site is in violet. Identification of amino acids by the one-letter code: T, threonine; K, lysine. The first number after the one-letter code indicates the capsid protein: 1, VP1; 2, VP2; and 3, VP3. The three-number code indicates the position of the amino acid in a given capsid protein.
Relatively fewer amino acid substitutions, compared to the parental strains, were seen in the nonstructural proteins (2% of all positions). There were differences in 56 positions either between the PV3/Sabin and PV1/Sabin strains or between either of them and the E252 virus. The E252 virus was parent-like in 26 cases, other serotype-like in 9 cases, and unlike either of the parents in 21 cases. Five of the nine other serotype-like substitutions were in 3D—all conservative by nature.
Antigenic properties of the isolate.
Isolate E252 was neutralized completely with polyclonal PV3-specific antisera, but not with a mixture of Sabin-specific MAbs used in the ITD assay. To further characterize the antigenic properties of the isolate, neutralizing activities of nine antigenic site 1- to 3-specific MAbs were determined with a standard microneutralization assay. The isolate E252 was not neutralized with any of the tested PV3-specific MAbs. In contrast, PV3/Sabin was neutralized with eight and wild-type PV3/Saukett was neutralized with six of the nine tested MAbs (Table 2).
TABLE 2.
Reactivity of isolate E252 with MAbs specific for different antigenic sites (1, 2, and 3) of PV3
| Virus strain | Reactivity with MAb toa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Site 1
|
Site 2
|
Site 3 | |||||||
| 204 | 472 | 495 | 520 | 881 | 838 | 875 | 882 | 879 | |
| PV3/Sabin | + | + | + | + | + | − | + | + | + |
| E252 | − | − | − | − | − | − | − | − | − |
| PV3/Saukett H | + | + | + | + | − | + | − | + | − |
+, antibody titer of ≥4; −, antibody titer of <4.
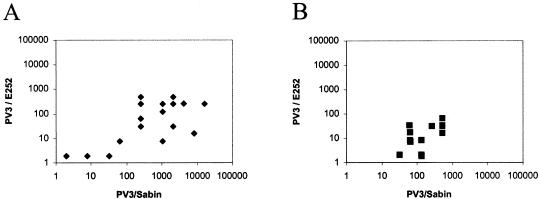
Due to the distinct antigenic properties of the isolate E252, we wanted to know whether antibodies induced by poliovirus vaccinations are capable of neutralizing this isolate. Two sets of human sera were analyzed for neutralizing antibodies against the E252 and PV3/Sabin. All 15 tested sera from OPV-vaccinated Estonian children had neutralizing antibodies to the PV3/Sabin strain, and 12 of them also had neutralizing antibodies to the isolate E252 (Fig. 4B). However, the levels of antibody to the isolate were lower than those to the reference strain. Median antibody titers for PV3/Sabin and E252 were 128 and 16, respectively. Practically similar results were obtained with 23 Finnish children vaccinated with trivalent IPV at the ages of 4, 6, and 12 months (Fig. 4A). In IPV-vaccinated children, median antibody titers to PV3/Sabin and the isolate were 1,024 and 64, respectively.
FIG. 4.
Serum neutralizing antibodies to PV3 strains E252 and Sabin in children vaccinated with IPV (A) or OPV (B). One serum sample from each child was analyzed for neutralizing antibodies by using a standard microneutralization assay with fourfold serum dilutions. The negative sera (titers of <4) were blotted, as they were 1:2.
Temperature sensitivity of the isolate.
The E252 isolate was compared with PV3 strains Sabin and Leon as regards replication capacity at an elevated temperature (39.5°C) in two cell lines routinely used in enterovirus isolation (Table 3). In contrast to PV3/Sabin, isolate E252 had lost its temperature-sensitive phenotype in both GMK and RD(A) cells and replicated at elevated temperature as well as the wild-type PV3/Leon.
TABLE 3.
Temperature sensitivity of E252
| Cell line and PV3 strain | Virus titer (PFU/ml) at growth temp:
|
||
|---|---|---|---|
| 36°C | 39.5°C | Log titer (PFU/ml) reduction at 36°C/39.5°C | |
| GMK cells | |||
| Sabin 3 | 7.65 | 2.30 | 5.35 |
| E252 | 8.06 | 8.27 | −0.21 |
| Leon | 7.07 | 6.58 | 0.49 |
| RD cells | |||
| Sabin 3 | 7.74 | 4.40 | 3.34 |
| E252 | 8.87 | 8.16 | 0.71 |
| Leon | 7.17 | 6.89 | 0.28 |
DISCUSSION
This paper describes the isolation and characterization of the highly evolved and, to the best of our knowledge, most divergent vaccine-derived poliovirus strain that has been found to date. This virus, E252, was isolated from the main sewage collector of Tallinn, the capital of Estonia, during routine screening for wild-type polioviruses.
The initial sequencing of the capsid protein VP1 coding region suggested that the closest relative to the isolate E252 was the oral vaccine strain PV3/Sabin, but the two viruses shared only 86.7% of the VP1 nucleotides. The attenuated Sabin strain and its parental strain PV3/Leon belong to an extinct genotype. Putative reemergence of related wild strains is highly unlikely. Complete genomic sequencing revealed multiple nucleotide substitutions, compared to Sabin 3, throughout the genome. These included all the genetic markers of reverted, neurovirulent PV3/Sabin-derived virus strains. Therefore, we did not test the neurovirulence of the E252 virus in experimental animals. There is plenty of evidence for increased neurovirulence in strains that are genetically much closer to the vaccine strain than E252.
A probable Sabin 3/Sabin 1-recombination junction was found in the 2C coding region. It is noteworthy, however, that the sequences of the P3 regions are not serotype specific and those of HEV-C are not necessarily separable from the corresponding sequences found in polioviruses. Therefore, theoretically it is also possible that the P3 region of the E252 virus was derived from an unidentified genetic cluster C virus.
Amino acid substitutions were seen in most of the viral proteins. They were relatively less frequent in the nonstructural proteins than in the major capsid proteins. In the capsid proteins VP1, VP2, and VP3, most of the substitutions were located near the virion surface, where the degree of freedom of polypeptide conformation is greater than in the beta barrels. Several substitutions were seen at the known neutralization antigenic sites. However, we do not know if this was due to immune selection in the host or had occurred by successive random selections associated with putative bottleneck transmission events over the years. The numerous substitutions at the antigenic sites were reflected in the strikingly altered antigenic properties of the virus. None of the nine tested MAbs recognizing one or both of the tested PV3 reference strains was able to neutralize the E252 virus. Furthermore, the capacity of human sera from groups of IPV- or OPV-immunized children to neutralize the virus was significantly reduced compared to that of the corresponding vaccine strains. From this point of view, the virus should have had a potential for transmission in both Estonian and Finnish populations. No evidence for this type of circulation was obtained.
The route of the virus to the sewerage remains unresolved. There have not been any “polio-like” epidemics or even suspected cases of poliomyelitis in Estonia during the time the virus was isolated. All the fecal specimens collected from patients with meningitis or a similar diagnosis during autumn 2002 were subjected to reisolation attempts. In spite of these duplicate analyses of the specimens, no sign of poliovirus circulation was seen before or after sewage isolate E252. The environmental surveillance was intensified by collecting more wastewater samples from distinct areas of Tallinn, but this did not lead to a second isolation of a similar virus. It seems highly unlikely that the numerous substitutions in the genome of E252 would have been generated by widespread circulation of the virus in the local population.
Importation of the evolved virus from abroad cannot be excluded. Wild-type PV3, most probably after importation, was isolated from Tallinn sewage in 1987 and 1988 in the absence of reported cases of poliomyelitis (20). Tallinn is a busy harbor city with numerous visiting ships every week. Occasionally the ships empty their waste containers into the sewers of the City of Tallinn, and one could speculate about this route of introduction of the virus into the city sewers. However, according to the harbor records, no ship emptied its waste containers into the sewerage of Tallinn in October 2002.
The origin of the virus E252 is unknown, but one likely reason for emergence of this virus is a long-term replication in an immunodeficient individual. A Sabin 3/Sabin 1 recombination is typical of most recipients of OPV, whereas the P3 region in discovered cVDPVs has been derived from an unidentified genetic group C virus (including the HEV-C and poliovirus species) (6, 8, 21, 41). There are two problems with this hypothesis, however. First, no poliovirus-excreting immunodeficient patients are known in Tallinn. This does not, of course, exclude their existence, because humoral immunodeficiency sufficient to allow chronic poliovirus infection sometimes can be asymptomatic. Second, the virus was isolated by a trap sampling method. Theoretically, the likelihood of detecting virus excreted by a single person in a sewage system where excreta from a given person are mixed with sewage derived from half a million nonexcretors, is very small (19, 32). The nonselective adsorbent immersed in the sewage is likely to be rapidly saturated with unrelated suspended materials abundant in a city's sewage. We have to assume that part of the E252 virus excreted by the putative immunodeficient individual happened to be in the very location and at the very time where and when the gauze pad was immersed in the sewage.
The AFP surveillance is the “gold standard” in the WHO program aiming at the global eradication of wild-type poliovirus. Environmental surveillance, recognized as a supplementary tool, has shown to be extremely powerful in some situations. For example, a recent survey in Egypt revealed prolonged and extensive circulation of wild-type PV1 at a time when the poliovirus-positive AFP cases were already rare (12). Environmental surveillance has also previously yielded isolation of diverged vaccine-derived strains with a wild-type phenotype without any concurrent reported cases of paralytic poliomyelitis. For example, a PV2 strain recovered from sewage in Israel, in the absence of any polio cases, had 91.7% nucleotide identity with PV2/Sabin in the P1 capsid region and had regained some wild-type characteristics (37).
In conclusion, we have described genetic and phenotypic features of a PV3/PV1 recombinant VDPV that appears to have characteristics of an iVDPV, although the putative chronically infected individual is not known. No evidence for circulation of this virus was obtained, although the observed antigenic changes would help the virus to escape vaccine-induced immunity in human populations. The existence of this kind of viruses in human populations is a serious risk, considering the option of abruptly stopping the use of OPV after the desired and continuing global eradication of wild-type poliovirus-dependent poliomyelitis.
Acknowledgments
Expert assistance from Riina Raud, National Poliovirus Laboratory, Tallinn, Estonia, is gratefully acknowledged. We thank Jaan Ruutman from Tallinn Tervisekaitse for organization of the collection of additional sewage specimens. Galina Lipskaya and Eugene Gavrilin, WHO Euro, are thanked for valuable comments during the analysis. Philip D. Minor, NIBSC, is greatly acknowledged for providing the PV3-specific MAbs. Steven Oberste, Centers for Disease Control and Prevention, Atlanta, Ga., is thanked for sharing the unpublished sequences of HEV-C.
This work was partially supported by WHO TSA I8/181/526 to KTL.
REFERENCES
- 1.Balanant, J., S. Guillot, A. Candrea, F. Delpeyroux, and R. Crainic. 1991. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology 184:645-654. [DOI] [PubMed] [Google Scholar]
- 2.Blomqvist, S., A.-L. Bruu, M. Stenvik, and T. Hovi. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84:573-580. [DOI] [PubMed] [Google Scholar]
- 3.Bottiger, M., and E. Herrstrom. 1992. Isolation of polioviruses from sewage and their characteristics: experience over two decades in Sweden. Scand. J. Infect. Dis. 24:151-155. [DOI] [PubMed] [Google Scholar]
- 4.Buttinelli, G., V. Donati, S. Fiore, J. Marturano, A. Plebani, P. Balestri, A. R. Soresina, R. Vivarelli, F. Delpeyroux, J. Martin, and L. Fiore. 2003. Nucleotide variation in Sabin type 2 poliovirus from an immunodeficient patient with poliomyelitis. J. Gen. Virol. 84:1215-1221. [DOI] [PubMed] [Google Scholar]
- 5.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2001. Acute flaccid paralysis associated with circulating vaccine-derived poliovirus—Philippines, 2001. Morb. Mortal. Wkly. Rep. 50:874-875. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2001. Circulation of a type 2 vaccine-derived poliovirus—Egypt, 1982-1993. Morb. Mortal. Wkly. Rep. 50:41-42, 51. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2002. From the Centers for Disease Control and Prevention. Acute flaccid paralysis associated with circulating vaccine-derived poliovirus—Philippines, 2001. JAMA 287:311. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2002. Public health dispatch: poliomyelitis, Madagascar, 2002. Morb. Mortal. Wkly. Rep. 51:622. [Google Scholar]
- 10.Cherkasova, E., M. Laassri, V. Chizhikov, E. Korotkova, E. Dragunsky, V. I. Agol, and K. Chumakov. 2003. Microarray analysis of evolution of RNA viruses: evidence of circulation of virulent highly divergent vaccine-derived polioviruses. Proc. Natl. Acad. Sci. USA 100:9398-9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherkasova, E. A., E. A. Korotkova, M. L. Yakovenko, O. E. Ivanova, T. P. Eremeeva, K. M. Chumakov, and V. I. Agol. 2002. Long-term circulation of vaccine-derived poliovirus that causes paralytic disease. J. Virol. 76:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Bassioni, L., I. Barakat, E. Nasr, E. M. de Gourville, T. Hovi, S. Blomqvist, C. Burns, M. Stenvik, H. Gary, O. M. Kew, M. A. Pallansch, and M. H. Wahdan. 2003. Prolonged detection of indigenous wild polioviruses in sewage from communities in Egypt. Am. J. Epidemiol. 158:807-815. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1995. PHYLIP (Phylogeny Inference Package), version 3.57c. Department of Genetics, University of Washington, Seattle.
- 14.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 16.Georgescu, M. M., F. Delpeyroux, and R. Crainic. 1995. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J. Gen. Virol. 76:2343-2348. [DOI] [PubMed] [Google Scholar]
- 17.Georgescu, M.-M., F. Delpeyroux, M. Tardy-Panit, J. Balanant, M. Combiescu, A. A. Combiescu, S. Guillot, and R. Crainic. 1994. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 68:8089-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hidalgo, S., M. Garcia Erro, D. Cisterna, and M. C. Freire. 2003. Paralytic poliomyelitis caused by a vaccine-derived polio virus in an antibody-deficient Argentinean child. Pediatr. Infect. Dis. J. 22:570-572. [PubMed] [Google Scholar]
- 19.Hovi, T., M. Stenvik, H. Partanen, and A. Kangas. 2001. Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiol. Infect. 127:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joks, S., and A. Jogiste. 1997. Poliomyelitis in Estonia. Eesti Arst. 2:132-139. [Google Scholar]
- 21.Kew, O., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. Andre, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van Der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. deq Uadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 22.Kew, O., M. Mulders, G. A. Lipskaya, E. Da Silva, and M. Pallansch. 1995. Molecular epidemiology of polioviruses. Semin. Virol. 6:401-414. [Google Scholar]
- 23.Kew, O. M., R. W. Sutter, B. K. Nottay, M. J. McDonough, D. R. Prevots, L. Quick, and M. A. Pallansch. 1998. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J. Clin. Microbiol. 36:2893-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipskaya, G. Y., A. R. Muzychenko, O. K. Kutitova, S. V. Maslova, M. Equestre, S. G. Drozdov, R. P. Bercoff, and V. I. Agol. 1991. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J. Med. Virol. 35:290-296. [DOI] [PubMed] [Google Scholar]
- 25.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, J., E. Samoilovich, G. Dunn, A. Lackenby, E. Feldman, A. Heath, E. Svirchevskaya, G. Cooper, M. Yermalovich, and P. D. Minor. 2002. Isolation of an intertypic poliovirus capsid recombinant from a child with vaccine-associated paralytic poliomyelitis. J. Virol. 76:10921-10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minor, P. D., M. Ferguson, D. M. Evans, J. W. Almond, and J. P. Icenogle. 1986. Antigenic structure of polioviruses of serotypes 1, 2 and 3. J. Gen. Virol. 67:1283-1291. [DOI] [PubMed] [Google Scholar]
- 29.Piirainen, L., M. Stenvik, M. Roivainen, J. Eskola, E. C. Beuvery, and T. Hovi. 1999. Randomised, controlled trial with the trypsin-modified inactivated poliovirus vaccine: assessment of intestinal immunity with live challenge virus. Vaccine 17:1084-1090. [DOI] [PubMed] [Google Scholar]
- 30.Pipkin, P. A., D. J. Wood, V. R. Racaniello, and P. D. Minor. 1993. Characterisation of L cells expressing the human poliovirus receptor for the specific detection of polioviruses in vitro. J. Virol. Methods 41:333-340. [DOI] [PubMed] [Google Scholar]
- 31.Pöyry, T., M. Stenvik, and T. Hovi. 1988. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl. Environ. Microbiol. 54:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranta, J., T. Hovi, and E. Arjas. 2001. Poliovirus surveillance by examining sewage water specimens: studies on detection probability using simulation models. Risk Anal. 21:1087-1096. [DOI] [PubMed] [Google Scholar]
- 33.Rico-Hesse, R., M. A. Pallansch, B. K. Nottay, and O. M. Kew. 1987. Geographic distribution of wild poliovirus type 1 genotypes. Virology 160:311-322. [DOI] [PubMed] [Google Scholar]
- 34.Rousset, D., M. Rakoto-Andrianarivelo, R. Razafindratsimandresy, B. Randriamanalina, S. Guillot, J. Balanant, P. Mauclere, and F. Delpeyroux. 2003. Recombinant vaccine-derived poliovirus in Madagascar. Emerg. Infect. Dis. 9:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabin, A. B., and L. R. Boulger. 1973. History of Sabin attenuated poliovirus oral live vaccine strains. J. Biol. Stand. 1:115-118. [Google Scholar]
- 36.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 37.Shulman, L. M., Y. Manor, R. Handsher, F. Delpeyroux, M. J. McDonough, T. Halmut, I. Silberstein, J. Alfandari, J. Quay, T. Fisher, J. Robinov, O. M. Kew, R. Crainic, and E. Mendelson. 2000. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J. Clin. Microbiol. 38:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanway, G., A. J. Cann, R. Hauptmann, P. Hughes, L. D. Clarke, R. C. Mountford, P. D. Minor, G. C. Schild, and J. W. Almond. 1983. The nucleotide sequence of poliovirus type 3 leon 12 a1b: comparison with poliovirus type 1. Nucleic Acids Res. 11:5629-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Avoort, H. G. A. M., B. P. Hull, T. Hovi, M. A. Pallansch, O. M. Kew, R. Crainic, D. J. Wood, M. N. Mulders, and A. M. van Loon. 1995. Comparative study of five methods for intratypic differentiation of polioviruses. J. Clin. Microbiol. 33:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization. 2001. Polio laboratory manual. World Health Organization, Geneva, Switzerland.
- 41.Yang, C.-F., T. Naguib, S.-J. Yang, E. Nasr, J. Jorba, N. Ahmed, R. Campagnoli, H. van der Avoort, H. Shimizu, T. Yoneyama, T. Miyamura, M. Pallansch, and O. Kew. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77:8366-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]