Abstract
Mutations affecting proteolipid protein 1 (PLP1), the major protein in central nervous system myelin, cause the X-linked leukodystrophy Pelizaeus–Merzbacher disease (PMD). We describe the neuropathologic findings in a series of eight male PMD subjects with confirmed PLP1 mutations, including duplications, complete gene deletion, missense and exon-skipping. While PLP1 mutations have effects on oligodendrocytes that result in mutation-specific degrees of dysmyelination, our findings indicate that there are also unexpected effects in the central nervous system resulting in neuronal loss. Although length-dependent axonal degeneration has been described in PLP1 null mutations, there have been no reports on neuronal degeneration in PMD patients. We now demonstrate widespread neuronal loss in PMD. The patterns of neuronal loss appear to be dependent on the mutation type, suggesting selective vulnerability of neuronal populations that depends on the nature of the PLP1 disturbance. Nigral neurons, which were not affected in patients with either null or severe misfolding mutations, and thalamic neurons appear particularly vulnerable in PLP1 duplication and deletion patients, while hippocampal neuronal loss was prominent in a patient with complete PLP1 gene deletion. All subjects showed cerebellar neuronal loss. The patterns of neuronal involvement may explain some clinical findings, such as ataxia, being more prominent in PMD than in other leukodystrophies. While the precise pathogenetic mechanisms are not known, these observations suggest that defective glial functions contribute to neuronal pathology.
Introduction
Pelizaeus–Merzbacher disease (PMD) is an X-linked myelin disorder that presents in early childhood with nystagmus, and hypotonia. Over time, ataxia, spastic quadriparesis, and cognitive impairment evolve. Mutations affecting the proteolipid protein 1 (PLP1) gene cause PMD [16, 25, 37]. PLP1 mutations produce a wide spectrum of clinical disorders, from mild X-linked spastic paraparesis type 2 [33], which lacks other neurologic impairments, to severe PMD, with congenital nystagmus, seizures, severe quadriparesis, respiratory and feeding difficulty, and severe cognitive impairment [15].
The PLP1 gene generates two major transcripts that encode PLP1 and DM20, a smaller protein generated by alternative splicing lacking amino acid residues 117–151 [26]. PLP1 is the major structural protein of central nervous system (CNS) myelin, accounting for approximately 50% of myelin protein, but is also present in peripheral myelin comprising less than 1% of the total myelin protein mass [2]. DM20 is expressed at equivalent levels as PLP1 in the peripheral nervous system, but is less abundant than PLP1 in CNS myelin [28]. The functions of PLP1 and DM20 are not fully understood, but likely include a stabilizing role of the myelin sheath after compaction [3].
PMD is characterized pathologically by diffuse hypomyelination [24, 34]. Different mutational mechanisms have different effects on oligodendrocyte viability and myelination. PLP1 duplications most likely cause overexpression of PLP1, which associates with cholesterol and lipids in the late endosomes and lysosomes, and cause oligodendrocyte dysfunction and cell death [20, 21, 31]. PLP1 point mutations associated with severe PMD cause misfolding of PLP1 and DM20, which accumulate in the endoplasmic reticulum (ER), activating ER stress responses that lead to oligodendrocyte apoptosis and severe hypomyelination [10, 11]. PLP1 null mutations, including complete deletion of the gene, paradoxically result in a mild syndrome during childhood, but with a late and relatively rapid progression characterized primarily by length-dependent axonal degeneration in the long motor and sensory tracts [9, 12] but with relative preservation of myelin and without oligodendrocyte loss. The amount of myelin, quantifiable by magnetic resonance imaging as well as by pathologic analysis, is inversely correlated with clinical disease severity [41].
Although the underlying mechanisms are not understood, PLP1 is also important for maintaining axonal integrity in long tracts of the CNS [9] and for peripheral nerve function [8, 35]. Axonal damage in PMD has been associated with PLP1 null mutations in both Plp1 knockout mice as well as in humans who develop length-dependent axonal degeneration [1, 9, 12, 32]. Neuronal involvement in PMD, however, has not been systematically characterized despite historical descriptions of extrapyramidal signs by Pelizaeus [27]. These findings therefore raise the question as to whether neuronal involvement may account for components of the clinical manifestations and for the phenotypic heterogeneity encountered in PMD.
Here, we characterize the extent and neuroanatomic distribution of neuronal involvement in eight patients with PMD caused by different mutations of PLP1 such as: (1) overexpression (due to PLP1 duplication), (2) loss of expression (deletion of the PLP1 gene) or (3) PLP1 misfolding, (missense or small point mutations).
Materials and methods
Case reports
PLP1 duplication cases (cases 1–4)
Cases 1 and 2 were brothers from a family that has been described clinically, and died at 47 and 54 years, respectively [42]. They had classical PMD syndromes, with understandable but dysarthric speech, spastic quadriparesis, and never ambulated independently. Voluntary movements were slow, however, evaluation of rigidity was compromised by severe spasticity.
Case 3 has been previously reported and was a first cousin of cases 1 and 2 [43]. His clinical course was similar to that of his cousins. He expired at age 50.
Case 4 was unrelated to the previous cases, but also had a similar clinical syndrome until his death at age 37 from pulmonary embolism.
PLP1 null mutation cases (including case 5)
Case 5 had a complete deletion of the PLP1 gene and flanking genes on the X chromosome as previously described [17, 30]. He was wheelchair confined from late adolescence and developed progressive spasticity and dysarthria. He had severe spastic quadriplegia during the last 5 years of life and lost speech 2 years before he expired at age 47 years from aspiration pneumonia. Limited postmortem specimens were available from three members of a previously described family that has a frame shift mutation resulting in complete absence of PLP1 expression [8, 9].
PLP1 point mutation cases (cases 6–8)
Cases 6 and 7 had severe PMD and were 23- and 25-year-old brothers who have previously been described [6], and who subsequently were discovered to have a PLP1 point mutation that disrupts splicing, causing in-frame exon 6 skipping, with loss of 22 residues [39].
Case 8 also had severe PMD, and had a missense mutation resulting in substitution of threonine 43 by an isoleucine [29]. He had neonatal respiratory distress, nystagmus, and was never able to speak or to sit. He expired from respiratory failure at age 19.
Pathological analysis
The brains were examined grossly and paraffin-embedded tissues were sectioned and stained with routine hematoxylin– eosin luxol-fast blue (LFB), Nissl stain, and Bielschowsky silver stain. Immunohistochemistry was performed using the immunoperoxidase technique to detect GFAP expression, α-synuclein and phospho-tau positivity. Formal CNS histo-pathologic analyses and scoring of all cases were performed at Wayne State University (AAFS and CRP). A scoring system ranging from 0 to 3 was devised to semi-quantify the extent of neuronal loss and astrogliosis. For neuronal scoring, a score of 0 was given when the neuronal population of a given region was intact, 1 indicated mild neuronal loss, 2 indicated moderate neuronal loss, and a score of 3 was assigned when there was severe or total loss of the neuronal population. Astrogliosis was scored on a scale of 0 to 3 with the use of GFAP immunoperoxidase. A score of 0 was assigned to areas with no astrogliosis, 1 or 2 to areas with mild or moderate astrogliosis, respectively, and 3 to areas with intense astrogliosis.
Results
Characterization of white matter
PLP1 duplication cases (cases 1–4)
The cases showed similar distributions of dysmyelination typical of that reported in other cases with PMD. The white matter was markedly atrophic throughout the CNS. Since the primary defect in PMD is disturbance of myelin formation, we refer to the myelin abnormalities as dysmyelinating, but cannot exclude the possibility that loss of preexisting myelin (albeit abnormal myelin) may also have occurred. The subcortical white matter was diffusely and severely dysmyelinated and contained axonal spheroids. The corpus callosum was thin, markedly dysmyelinated, with astrogliosis. The internal, external and extreme capsules and the anterior commissure were dysmyelinated with astrogliosis and occasional axonal spheroids. The penicillary fibers of the striatum were completely devoid of myelin. In the brainstem, profound dysmyelination was evident in the medial longitudinal fasciculus, medial lemniscus, transverse pontine fibers and in the corticospinal tracts of the pons. In contrast, the arcuate fibers of the medulla were relatively spared. In the spinal cord, the corticospinal tracts, Lissauer’s tract and the dorsal columns and dorsal spinocerebellar tracts were markedly dysmyelinated. However, the spinothalamic and anterior spinocerebellar tracts were relatively spared.
PLP1 null mutation cases (including case 5)
Case 5 as well as the three cases with a PLP1 null syndrome all showed similar patterns of white matter abnormalities. Demarcation of gray and white matter was readily apparent, in contrast to the misfolding and duplication cases. The thinning of white matter was less pronounced than that observed in the duplication and point mutation cases. The staining of white matter with LFB was not as severely reduced as in cases 1–4 or cases 6–8, and the U-fibers were spared. We have previously described the length-dependent axonopathy in the spinal motor and sensory tracts [9].
PLP1 point mutation cases (cases 6–8)
Cases 6 and 7 exhibited marked atrophy of cerebral white matter. The subcortical white matter was diffusely dysmyelinated, showed increased astrogliosis and axonal loss with occasional axonal spheroids. The internal, external and extreme capsules and the anterior commissure showed loss of myelin and occasional axonal spheroids. In contrast to cases 1–4, myelin loss was focal and patchy in the penicillary fibers of the striatum. No phospho-tau positive glia were identified in any of the cases. In the brainstem, the medial longitudinal fasciculus, medial lemniscus and transverse pontine fibers were dysmyelinated, whereas the arcuate fibers in the medulla were spared. In the spinal cord, the spinothalamic and anterior spinocerebellar tracts were relatively spared, whereas the corticospinal tracts, Lissauer’s tract, dorsal columns and dorsal spinocerebellar tracts were markedly dysmyelinated. Case 8 showed sparse staining for myelin. The white matter displayed numerous spheroids. Astrocytosis was present in the cerebral white matter. The internal capsule was pale on LFB staining and the white matter tracts in the pons appeared vacuolated. The pyramidal tracts were small. The cerebellar white matter showed dysmyelination and astrogliosis.
Neuronal loss and gliosis (Table 1)
Table 1.
Quantification of regional CNS neuronal loss and gliosis in Pelizaeus–Merzbacher disease
| Neuroanatomic systems | Case 1 (47 years) PLP1 dup |
Case 2 (54 years) PLP1 dup |
Case 3 (50 years) PLP1 dup |
Case 4 (37 years) PLP1 dup |
Case 5 (47 years) del PLP1 |
Case 6 (23 years) del exon 6 |
Case 7 (25 years) del exon 6 |
Case 8 (19 years) Thr43>Ile |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NL | G | NL | G | NL | G | NL | G | NL | G | NL | G | NL | G | NL | G | |
| Striatonigral | ||||||||||||||||
| Substania nigra | 3+ | 3+ | 3+ | 2+ | 2+ | 3+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | N/A | N/A |
| Nucleus accumbens | 3+ | 3+ | 2+ | 2+ | 1+ | 2+ | 0 | 0 | 1+ | 1+ | 0 | 0 | N/A | N/A | 0 | 0 |
| Putamen | 3+a | 2+ | 3+a | 2+ | 1+ | 1+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Caudate | 2+b | 2+ | 1+ | 2+ | 2+ | 2+ | 1+ | 0 | N/A | N/A | 0 | 0 | 0 | 0 | 0 | 0 |
| Claustrum | 1+ | 1+ | 0 | 0 | 1+ | 1+ | 0 | 1+ | 1+ | 1+ | 0 | 0 | 1+ | 1+ | N/A | N/A |
| Amygdala | 2+ | 3+d | 2+ | 3+d | 3+ | 3+ | 1+ | 1+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Globus pallidus | 0 | 0 | 0 | 0 | 2+ | 3+ | 1+ | 1+ | 0 | 0 | 0 | 0 | N/A | N/A | 0 | 0 |
| Limbic | ||||||||||||||||
| CA1 | 3+ | 3+ | 1+ | 1+ | 2+ | 2+ | 0 | 0 | 1+ | 1+ | 0 | 0 | 0 | 0 | 0 | 0 |
| CA2 | 3+ | 3+ | 1+ | 1+ | 1+ | 1+ | 0 | 0 | 2+ | 2+ | 0 | 0 | 0 | 0 | 0 | 0 |
| CA3 | 1+ | 1+ | 1+ | 1+ | 0 | 1+ | 0 | 0 | 3+ | 3+ | 0 | 0 | 0 | 0 | 0 | 0 |
| CA4 | 3+ | 3+ | 1+ | 1+ | 1+ | 2+ | 0 | 1+ | 3+ | 3+ | 1+ | 1+ | 1+ | 1+ | 0 | 0 |
| Dentate | 3+ | 3+ | 1+ | 1+ | 0 | 1+ | 0 | 0 | 1+ | 1+ | 0 | 0 | 0 | 0 | 0 | 0 |
| Subiculum | 1+ | 1+ | 0 | 0 | 2+ | 2+ | 0 | 1+ | 1+ | 1+ | 0 | 0 | 0 | 0 | 0 | 0 |
| Prosubiculum | 2+ | 2+ | 1+ | 1+ | 2+ | 2+ | 1+ | 1+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mesocortico-limbic | ||||||||||||||||
| Anterior cingulate | 0 | 1-2+ | 2+b | 2+b | 2+ | 2+c | 0 | 0 | 0 | 1+ | 1+ | 1+ | 0 | 0 | N/A | N/A |
| Entorhinal | 1+e | 1+ | 2+ | 2+ | 1+ | 2+c | 0 | 1+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Insular cortex | 1+ | 2+c | 1+ | 2+c | 1+ | 2+ | 0 | 1+c | N/A | N/A | 1+e | 1+e | 0 | 1+e | 0 | 0 |
| Mammillary bodies | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 0 | 1+ | N/A | N/A | 0 | 0 | N/A | N/A | N/A | N/A |
| Hypothalamus | 1+ | 1+ | 2+ | 1+ | 1+ | 2+ | 0 | 1+ | 3+ | 2+ | 0 | 2+ | N/A | N/A | N/A | N/A |
| Cerebral Cortex | ||||||||||||||||
| Parietal cortex | 1+ | 2+c | 1+ | 2+c | 1+ | 2+ | 0 | 0 | 0 | 1+ | 1+e | 1+e | 0 | 1+e | 0 | 0 |
| Frontal cortex | 1+ | 2+c | 1+ | 2+c | 1+ | 2+c | 0 | 0 | 0 | 1+ | 1+e | 1+e | 0 | 1+e | 0 | 1+ |
| Temporal cortex | 1+ | 2+c | 2+ | 2+c | 1+ | 1+c | 0 | 0 | 0 | 1+ | 1+ e | 1+e | 0 | 1+e | 0 | 1+ |
| Occipital cortex | 0 | 2+c | 1+ | 2+c | 2+ | 3+ | N/A | N/A | 0 | 1+ | 1+e | 1+e | 0 | 1+e | 0 | 0 |
| Thalamic | ||||||||||||||||
| Pulvinar | 3+ | 3+ | 3+ | 3+ | 2+ | 3+ | 0 | 0 | 0 | 0 | 1+ | 1+ | N/A | N/A | N/A | N/A |
| Dorsomedial nucleus | 3+ | 3+ | 3+ | 3+ | 2+ | 2+ | 1+ | 1+ | 3+ | 3+ | 1+ | 1+ | N/A | N/A | 0 | 0 |
| Anterior nucleus | 2+ | 2+ | 3+ | 3+ | 2+ | 2+ | 0 | 0 | N/A | N/A | 1+ | 1+ | N/A | N/A | N/A | N/A |
| Ventrolateral nucleus | 2+ | 3+ | 3+ | 3+ | 2+ | 2+ | 1+ | 0 | 2+ | 2+ | 0 | 1+ | N/A | N/A | 0 | 0 |
| Subthalamic nucleus | 0 | 0 | N/A | N/A | 0 | 0 | N/A | N/A | 1+f | 0 | 0 | 0 | N/A | N/A | 0 | 0 |
| Olivopontocerebellar | ||||||||||||||||
| Cerebellar cortex | 3+ | 3+ | 3+ | 3+ | 2+ | 3+ | 1+ | 1+ | 3+ | 1+ | 3+ | 3+ | 3+ | 3+ | 2+ | 3+ |
| Cerebellar dentate | 3+ | 3+ | 3+ | 3+ | 1+ | 2+ | 0 | 0 | N/A | N/A | 2+ | 2+ | 1+ | 1+ | 1+ | 2+ |
| Inferior olivary nucleus | 2+ | 2+ | 2+ | 2+ | 1+ | 1+ | 1+ | 1+ | 1+ | 1+ | 0 | 0 | 0 | 0 | 1+ | 1+ |
| Pontine nuclei | 1+ | 1+ | 0 | 1+ | 1+ | 2+ | 0 | 0 | 1+ | 1+ | 0 | 0 | 0 | 0 | 0 | 0 |
NL neuronal loss, G gliosis, N/A not available
Primarily inferior,
primarily anterior,
primarily deep layers,
especially cortical nuclei,
focal,
acute neuronal loss
PLP1 duplication cases (cases 1–4)
Severe loss of neurons and astrogliosis in the substantia nigra were observed in cases 1–3 (Table 1; Fig. 1a–c), whereas the ventral tegmental area was relatively preserved. No α-synuclein or tau positive inclusions were identified in any of the cases. No nigral neuronal loss was detected in case 4, the youngest of the duplication cases. The putamen, nucleus accumbens and caudate nucleus all exhibited mild to severe neuronal loss and astrogliosis, while the globus pallidus was spared in cases 1 and 2.
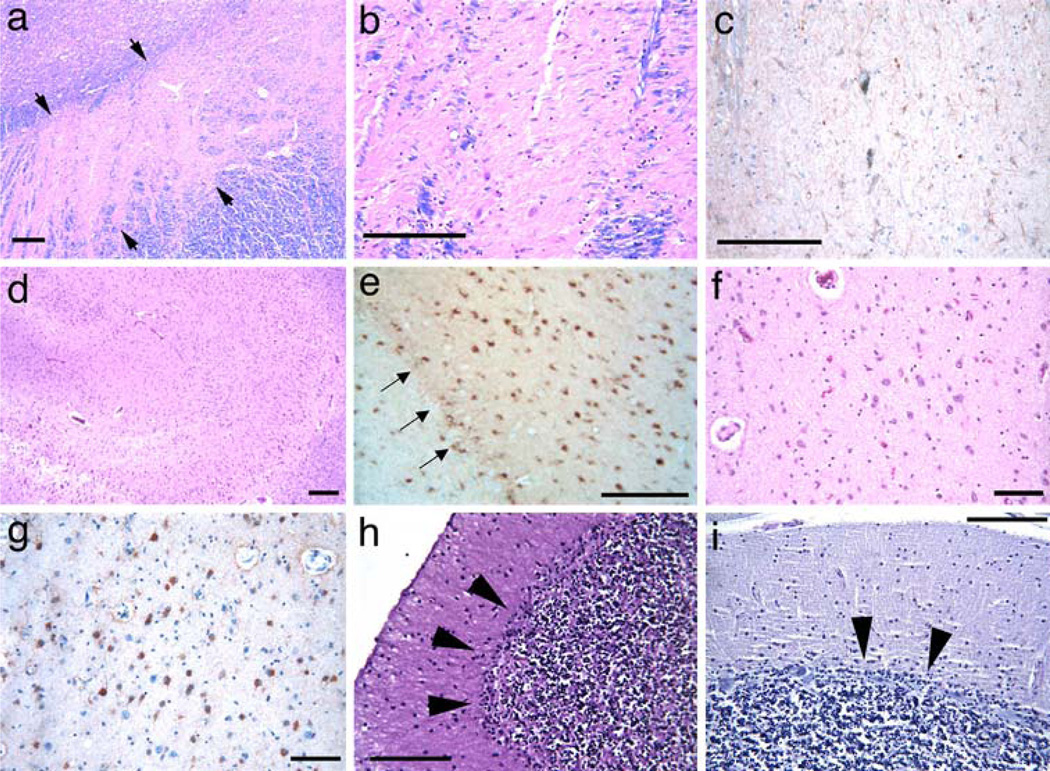
Fig. 1.
Pathologic changes in PLP1 duplication patients. Severe neuronal loss in the pars compacta of the substantia nigra (a HE–LFB stain) and higher magnification in b, accompanied by dense gliosis, as shown by GFAP positivity (c) in case 1. Arrowheads bracket the substantia nigra in a. Extensive hippocampal pyramidal cell loss and loss of dentate fascia neurons in case 1 (d HE–LFB stain). This was associated with dense gliosis also involving the dentate fascia (arrows, e GFAP). Severe neuronal loss (f HE–LFB stain) and gliosis, as indicated by GFAP staining (g) in the central nucleus of the amygdala in case 2. Loss of Purkinje cells in cases 1 (h HE–LFB stain) and 4 (i Nissl stain). Note the Bergmann gliosis in these cases (black arrowheads). Scale bars are 100 µm
In the limbic system, the hippocampi were grossly atrophic with varying neuronal loss (Table 1). CA1, CA2 and CA4, as well as the dentate gyrus, showed neuronal loss and astrogliosis that varied among this group of cases (Fig. 1d, e). CA1 was not preferentially affected. The perforant tract was preserved, whereas the alveus and fimbria showed myelin loss. The youngest case, case 4, showed mild neuronal loss and astrogliosis in the hippocampus (Table 1).
Microscopically, moderate to severe neuronal loss and astrogliosis were present in the deep cortical areas. The anterior cingulate exhibited moderate neuronal loss, whereas the posterior cingulate was relatively spared. The entorhinal and insular cortices showed changes similar to those of the anterior cingulate cortex. Cerebral cortex showed mild neuronal loss in cases 1–3 with moderate gliosis accentuated in deep cortical layers (Table 1). Case 4 showed no cortical abnormalities. The anterior thalamus and amygdala exhibited moderate to severe neuronal loss and astrogliosis (Fig. 1f, g, respectively; Table 1), while the mammillary bodies and hypothalamus showed mild neuronal loss. The dorsomedial and pulvinar complexes as well as the ventroposterolateral nucleus of the thalamus showed extensive neuronal loss (Table 1). The dorsomedial nucleus of the thalamus showed severe neuronal loss and astrogliosis in case 2. Marked neuronal loss of cerebellar cortex and the dentate nucleus were present, with diffuse Purkinje cell and internal granular cell loss and Bergmann gliosis (Table 1; Fig. 1h). Neuronal loss was less severe in case 4 (Table 1), the youngest of the four duplication cases, but there was prominent loss of Purkinje cells (Fig. 1i). Moderate neuronal loss was evident in the inferior olivary nuclei. The pons showed mild loss and astrogliosis in the pontine nuclei and in the central tegmental area. The spinal cord from case 1 showed numerous spheroids and severe neuronal loss in the dorsal column nuclei. The posterior horns and Clarke’s column showed neuronal loss and extensive astrogliosis, whereas the anterior horn structures were relatively preserved. Lumbar dorsal root ganglia showed moderate ganglion cell loss and irregular axonal swellings. Both anterior and dorsal roots showed focal axonal degeneration and myelin breakdown, being more severe in dorsal roots.
PLP1 null mutation cases (including case 5)
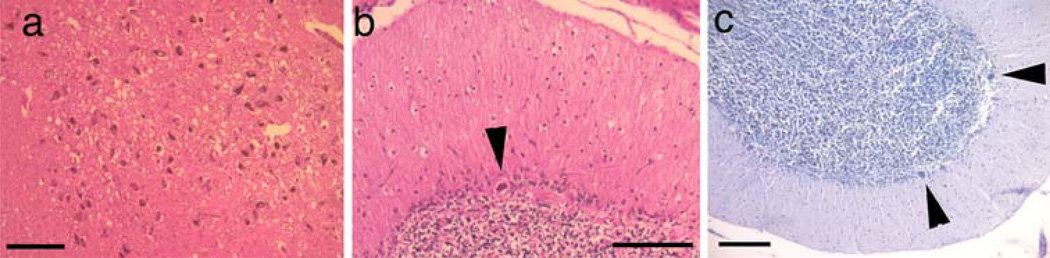
No nigral neuronal loss or astrogliosis was observed (Fig. 2a). In the hippocampus, there was severe neuronal loss, most evident in CA3 and CA4, and less so in CA2 and CA1. Severe neuronal loss was noted in the thalamus, particularly the dorsomedial nucleus and the hypothalamus (Table 1). Severe loss of Purkinje cells was observed in both the PLP1 deletion cases (Fig. 2b) and a case with premature termination mutation (Fig. 2c).
Fig. 2.
Pathologic findings in PLP1 null patients. Normal neuronal density of neurons in the substantia nigra of case 5 with a PLP1 deletion (a HE–LFB stain). Purkinje cell loss in case 5 (b HE–LFB stain with a PLP1 gene deletion and (c Nissl stain) a patient with a frameshift mutation that prevents PLP1 expression. Black arrowheads show the few remaining Purkinje cells. Scale bars are 100 µm
PLP1 point mutation cases (cases 6–8)
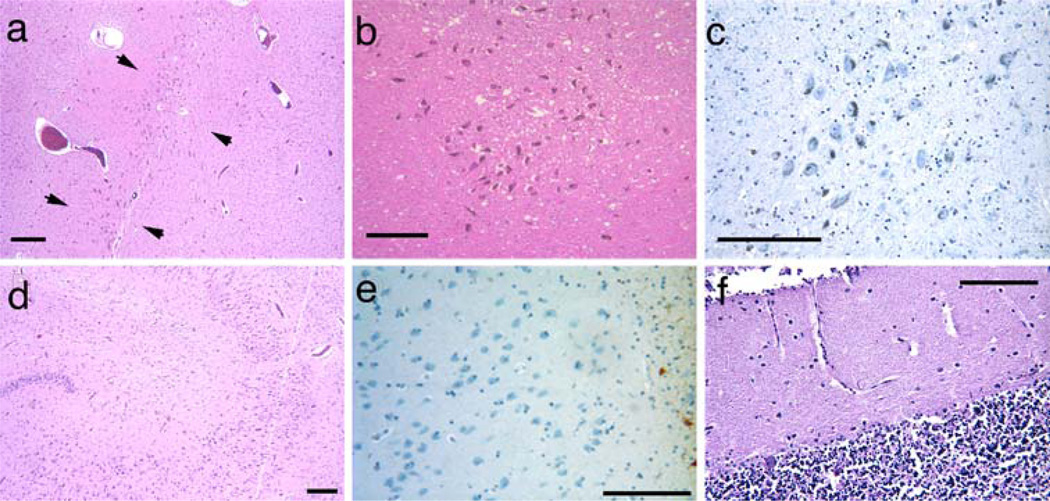
Neither gross hypopigmentation of the substantia nigra nor neuronal loss were detected microscopically in either cases 6 and 7 (Fig. 3a–c). No neuronal loss or gliosis was appreciated in the putamen, caudate nucleus, nucleus accumbens or globus pallidus (Table 1). No hippocampal neuronal loss was observed in the pyramidal cell layer, dentate fascia (Fig. 3d), subiculum or prosubiculum, although there was mild neuronal loss in the end plate (CA4) in both brothers (cases 6 and 7) (Table 1). Case 8 showed no hippocampal neuronal loss. The anterior cingulate and insular cortices showed mild neuronal loss and astrogliosis in the deep layers in case 6 but not in case 7. Focal mild neuronal loss and mild gliosis were evident in other cortical areas in case 6. In case 6, the anterior nucleus of thalamus showed mild neuronal loss and gliosis, whereas the mammillary bodies were spared. Neuronal loss and astrogliosis were evident in case 6 in the anterior, dorsomedial and pulvinar nuclei of the thalamus (Table 1). In none of the cases did the amygdala show appreciable neuronal loss or astrogliosis (Fig. 3e). The pons appeared small, due to white matter atrophy; however, neither neuronal loss nor astrogliosis were appreciated. The cerebellum showed moderate to severe Purkinje cell loss and astrogliosis in all three cases (Table 1; Fig. 3f). The dentate nucleus of the cerebellum showed significant neuronal loss, whereas the inferior olivary nucleus was relatively spared. Spinal cord from case 6 showed similar, though milder, changes to those of case 1.
Fig. 3.
Pathology of PLP1 point mutation cases. Normal neuronal density in the substantia nigra (a HE–LFB stain), higher magnification in b (HE–LFB stain) and lack of gliosis (c GFAP immunostain) in case 6. Arrowheads bracket the substantia nigra in a. Case 7 showed normal neuronal populations in the hippocampal pyramidal cell layer and dentate fascia (d HE–LFB stain). Case 7 showed no appreciable neuronal loss or gliosis in the amygdala (e GFAP immunostain). The cerebellum showed severe Purkinje cell loss in case 8 with a PLP1 missense mutation (f HE–LFB stain). Scale bars are 100 µm
Discussion
There are few reports of the neuropathological changes in PMD patients with confirmed PLP1 mutations [4, 9, 13, 19, 23, 24, 39, 44]. We report the neuropathological findings in eight subjects with PLP1 mutations, which encompass the major mutational categories across the PMD clinical spectrum: gene duplication with overexpression of PLP1, null mutations and point mutations that result in misfolded PLP1. In addition to the widespread dysmyelination in all patients, we found neuronal degeneration of a systemic nature occurring in the PMD patients, albeit with variable regional patterns and severities, depending upon mutation type and possibly patient age (see Table 1). Alternative explanations for neuronal degeneration such as hypoxia or ischemia seem unlikely, since susceptible neuronal populations were not preferentially affected, whereas less susceptible populations such as striatonigral or thalamic neurons were, and none of the patients suffered prolonged hypoxic or hypotensive insults prior to death.
The cerebellum, particularly Purkinje cells, was affected in all cases. Other components of the olivopontocerebellar system were also affected in the majority of cases, irrespective of mutation type. The thalamus, in particular the dorsomedial nucleus, was affected in all examined cases, except for case 8, who died at age 19 years, and was most severe in the duplication and deletion cases. In general, neuronal loss was more widespread and evident in the duplication and null cases, but absent or mild in the severe misfolding cases. Some of these differences may be accounted for, at least in part, by the ages of the patients. The neuronal loss was less severe in the younger patients, suggesting that it is a late phenomenon, rather than a developmental defect, which may account for some of the late clinical deterioration that is typically observed.
Genetic analysis enhances the understanding of normal biological processes as well as disease mechanisms. Null mutations provide the most important insights, especially when the essential roles of the corresponding protein are not understood. The clear loss of neurons in the case with deletion of the entire PLP1 gene establishes an essential role of PLP1 and oligodendrocytes in maintaining not only axonal integrity, but also that of subsets of neurons, such as those of the hippocampus, cerebellum and thalamus. The similar findings in the three cases with a premature termination of the PLP1 gene demonstrate that the neuronal loss results from PLP1 itself, and is not the result of lack of expression of genes flanking the PLP1 gene. While the clinical signs in PMD are assumed to result from the myelin abnormality, our findings raise the possibility that ataxia and extrapyramidal findings, which are more prominent in PMD than in patients with other leukodystrophies, may stem from loss of neuronal function.
Why some neurons appear especially vulnerable to injury in the absence of PLP1 is not clear. Null mutations have been described both in Plp1-knockout mice as well as humans [8, 9, 22, 30]. In man, they are associated with relatively mild disease, characterized by spastic paraparesis, mild cognitive impairment, ataxia, mild dystonia and demyelinating peripheral neuropathy [8]. Mice and humans with Plp1 null mutations develop minor defects in myelin that can be appreciated ultrastructurally [3, 22], but over time neurologic dysfunction with diffuse Wallerian degeneration without significant hypomyelination of central axons ensues [12]. This effect appears to depend upon PLP1, since Wallerian degeneration does not occur in shiverer mice that lack myelin basic protein, another major myelin protein of the central nervous system [12].
Point mutations affecting the PLP1 coding region provide a second mechanism leading to the development of PMD and are responsible for about 20% of cases [14, 25]. A toxic gain-of-function due to PLP1 misfolding, accumulation in the endoplasmic reticulum and activation of the unfolded protein response (UPR) has been proposed that causes varying amounts of oligodendroglial apoptosis and corresponding degrees of dysmyelination [11, 39]. Since the mutant PLP1 associated with severe PMD has been experimentally shown to be completely retained in the endoplasmic reticulum, the loss of Purkinje cells in these cases could conceivably result from a lack of functional PLP1 in oligodendrocytes, causing neuronal loss through a mechanism, still not understood, as occurs in patients with PLP1 null mutations. Therefore, in these patients, we believe that the clinical syndrome results from the presence of abnormal PLP1 and DM20, which results in oligodendrocyte apoptosis, resulting in early dysmyelination, and from both the inability to make full length PLP1 and DM20 and transport it to the cell surface, with resultant late-onset axonal and neuronal degeneration. The relative lack of neuronal loss in the cases with misfolded PLP1 suggests that neuronal loss is a relatively late phenomenon, as the three patients were significantly younger at time of death than were the other patients. Although we cannot exclude a neuroprotective property of misfolded PLP1, this would seem unlikely in view of the cerebellar and hippocampal neuronal loss seen in these cases.
The most extensive neuronal loss was observed in patients with PLP1 duplications. In addition to loss in the regions affected in PLP1 null and misfolded PLP1 cases, these cases had severe loss of neurons in the striatonigral, limbic, thalamic and cortical structures. The clinical and pathologic findings were remarkably similar in the three related subjects. However, no neuronal loss was observed in the substantia nigra in case 4, who was significantly younger than the other three duplication cases. While this may indicate that the neuronal pathology is unrelated to PLP1 overexpression, we believe that this is unlikely, since subject 4 had neuronal loss in other brain regions. Therefore, we believe that the neuronal loss is a late, rather than developmental, phenomenon and is more prominent in certain neuronal subpopulations.
The mechanism(s) underlying neuronal loss in PMD is not known, however, one may speculate that it shares similarities with the axonal pathology described in multiple sclerosis (MS) [40]. Axonal pathology in MS is assumed to be due to perturbations in the oligodendrocyte– axonal interactions, in addition to an immunemediated process directed against neurons [40, 45]. We have demonstrated that Plp1 deficiency perturbs both retrograde and anterograde axonal transport [7]. Interestingly, retrograde transport appeared to be more profoundly affected, and raises the possibility that loss of trophic support from peripheral targets could lead to axonal and, ultimately, neuronal degeneration. The patterns and degree of neuronal loss did not appear to correlate with the patterns of axonal injury, but will require more systematic study to resolve.
It is intriguing that the most severe neuronal loss was observed in the PLP1 duplication cases. It is likely that PLP1 duplication leads to overexpression of PLP1, as has been shown in transgenic mice [20, 31]. Overexpression of PLP1 perturbs the assembly of membrane rafts and leads to accumulation of PLP1 with cholesterol and lipids in the late endosomal/lysosomal compartments [21, 36]. Boucher et al. [5] reported that overexpression of Plp1, but not of DM20, reduces in vitro neuronal viability, either when Plp1-overexpressing cells are co-cultured with neurons, or when conditioned media from such cells is applied to neuronal cultures. The mechanism of this effect may be mediated by acidification of the extracellular environment, an effect observed only with Plp1, but not with Dm20 or the jimpy Plp1 that has a frame shift due to a mutation affecting Plp1 splicing [5, 38].
One possible mechanism underlying the varied systemic neuronal degeneration reported here is activation of innate immune responses, which could potentially lead to downstream neuronal apoptosis. Studies in Plp1 transgenic mice have demonstrated a significant role for secondary immune activation contributing to myelin and/or axonal damage [18]. Our preliminary studies indicate that microglial and inflammatory mediator upregulation also occurs in patients with PLP1 duplications. This possibility is now being actively pursued in the cases reported here.
Here, we have demonstrated that the pathology resulting from various PLP1 mutations is not confined to dysmyelination and axonal degeneration, but neurons as well. Since PLP1 is not expressed in neurons, the neuronal degeneration and loss most likely result from a disturbance in glial–neuronal interactions. These observations raise the possibility that neuronal loss may contribute to neurologic dysfunction in patients with other myelin disorders. Enhanced understanding of oligodendrocyte–axonal interactions could lead to the development of novel treatment strategies, such as neuroprotective agents, for patients afflicted with dysmyelinating disorders.
Acknowledgments
The authors are especially grateful to the families who generously donated specimens for research. J.G. thanks the PMD Foundation, the National Institutes of Health (NS043783), the Children’s Research Center of Michigan, and the National Multiple Sclerosis Society (RG3204) for support. G.M.H. acknowledges the Nemours Foundation and National Institutes of Health grant P20 RR-020173-01 from the National Center for Research Resources. The authors also are grateful to the Alzheimer disease Research Center of the University of Washington for assistance.
Contributor Information
Anders A. F. Sima, Department of Pathology, School of Medicine, Wayne State University, Detroit, MI, USA Department of Neurosurgery, School of Medicine, Wayne State University, Detroit, MI, 48201, USA; Department of Neurosurgery, School of Medicine, Wayne State University, Detroit, MI, USA.
Christopher R. Pierson, Department of Pathology, School of Medicine, Wayne State University, Detroit, MI, USA
Randall L. Woltjer, Department of Pathology, Oregon Health Sciences University, Portland, OR, USA
Grace M. Hobson, Nemours Biomedical Research, A. I. duPont Hospital for Children, Wilmington, DE, USA
Jeffrey A. Golden, Department of Pathology, Children’s Hospital of Philadelphia, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
William J. Kupsky, Department of Pathology, School of Medicine, Wayne State University, Detroit, MI, USA Department of Neurosurgery, School of Medicine, Wayne State University, Detroit, MI 48201, USA; Department of Neurosurgery, School of Medicine, Wayne State University, Detroit, MI, USA.
Galen M. Schauer, Department of Genetics, Kaiser Permanente Medical Center, Oakland, CA, USA Department of Pathology, Kaiser Permanente Medical Center, Oakland, CA, USA.
Thomas D. Bird, Department of Neurology, University of Washington School of Medicine, Seattle, WA, USA
Robert P. Skoff, Department of Anatomy and Cell Biology, School of Medicine, Wayne State University, Detroit, MI, USA
James Y. Garbern, Email: jgarbern@med.wayne.edu, Department of Pathology, School of Medicine, Wayne State University, Detroit, MI 48201, USA; Center for Molecular Medicine and Genetics, Wayne State University, Detroit, MI, USA.
References
- 1.Anderson TJ, Schneider A, Barrie JA, et al. Late-onset neurodegeneration in mice with increased dosage of the proteolipid protein gene. J Comp Neurol. 1998;394:506–519. doi: 10.1002/(sici)1096-9861(19980518)394:4<506::aid-cne8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 2.Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- 3.Boison D, Stoffel W. Disruption of the compacted myelin sheath of axons of the central nervous system in proteolipid protein-deficient mice. Proc Natl Acad Sci USA. 1994;91:11709–11713. doi: 10.1073/pnas.91.24.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond C, Si X, Crisp M, et al. Family with Pelizaeus–Merzbacher disease/X-linked spastic paraplegia and a nonsense mutation in exon 6 of the proteolipid protein gene. Am J Med Genet. 1997;71:357–360. [PubMed] [Google Scholar]
- 5.Boucher SE, Cypher MA, Carlock LR, Skoff RP. Prote-olipid protein gene modulates viability and phenotype of neurons. J Neurosci. 2002;22:1772–1783. doi: 10.1523/JNEUROSCI.22-05-01772.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carango P, Funanage VL, Quiros RE, Debruyn CS, Marks HG. Overexpression of DM20 messenger RNA in two brothers with Pelizaeus–Merzbacher disease. Ann Neurol. 1995;38:610–617. doi: 10.1002/ana.410380409. [DOI] [PubMed] [Google Scholar]
- 7.Edgar JM, McLaughlin M, Yool D, et al. Oligodendroglial modulation of fast axonal transport in a mouse model of hereditary spastic paraplegia. J Cell Biol. 2004;166:121–131. doi: 10.1083/jcb.200312012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbern JY, Cambi F, Tang XM, et al. Proteolipid protein is necessary in peripheral as well as central myelin. Neuron. 1997;19:205–218. doi: 10.1016/s0896-6273(00)80360-8. [DOI] [PubMed] [Google Scholar]
- 9.Garbern JY, Yool DA, Moore GJ, et al. Patients lacking the major CNS myelin protein, proteolipid protein 1, develop length-dependent axonal degeneration in the absence of demyelination and inflammation. Brain. 2002;125:551–561. doi: 10.1093/brain/awf043. [DOI] [PubMed] [Google Scholar]
- 10.Gow A, Friedrich VL, Jr, Lazzarini RA. Many naturally occurring mutations of myelin proteolipid protein impair its intracellular transport. J Neurosci Res. 1994;37:574–583. doi: 10.1002/jnr.490370504. [DOI] [PubMed] [Google Scholar]
- 11.Gow A, Lazzarini RA. A cellular mechanism governing the severity of Pelizaeus–Merzbacher disease. Nat Genet. 1996;13:422–428. doi: 10.1038/ng0896-422. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths I, Klugmann M, Anderson T, et al. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science. 1998;280:1610–1613. doi: 10.1126/science.280.5369.1610. [DOI] [PubMed] [Google Scholar]
- 13.Harding B, Ellis D, Malcolm S. A case of Pelizaeus–Merzbacher disease showing increased dosage of the proteolipid protein gene. Neuropathol Appl Neurobiol. 1995;21:111–115. doi: 10.1111/j.1365-2990.1995.tb01036.x. [DOI] [PubMed] [Google Scholar]
- 14.Hodes ME, Pratt VM, Dlouhy SR. Genetics of Pelizaeus–Merzbacher disease. Dev Neurosci. 1993;15:383–394. doi: 10.1159/000111361. [DOI] [PubMed] [Google Scholar]
- 15.Hudson LD, Puckett C, Berndt J, Chan J, Gencic S. Mutation of the proteolipid protein gene PLP in a human X chromosome-linked myelin disorder. Proc Natl Acad Sci USA. 1989;86:8128–8131. doi: 10.1073/pnas.86.20.8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue K, Osaka H, Sugiyama N, et al. A duplicated PLP gene causing Pelizaeus–Merzbacher disease detected by comparative multiplex PCR. Am J Hum Genet. 1996;59:32–39. [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue K, Osaka H, Thurston VC, et al. Genomic rearrangements resulting in plp1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet. 2002;71:838–853. doi: 10.1086/342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ip CW, Kroner A, Bendszus M, et al. Immune cells contribute to myelin degeneration and axonopathic changes in mice overexpressing proteolipid protein in oligodendrocytes. J Neurosci. 2006;26:8206–8216. doi: 10.1523/JNEUROSCI.1921-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwaki A, Muramoto T, Iwaki I, et al. A missense mutation in the proteolipid protein gene responsible for Pelizaeus-Merzbacher disease in a Japanese family. Hum Mol Genet. 1993;2:19–22. doi: 10.1093/hmg/2.1.19. [DOI] [PubMed] [Google Scholar]
- 20.Kagawa T, Ikenaka K, Inoue Y, et al. Glial cell degeneration and hypomyelination caused by overexpression of myelin proteolipid protein gene. Neuron. 1994;13:427–442. doi: 10.1016/0896-6273(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 21.Karim SA, Barrie JA, McCulloch MC, et al. PLP overexpression perturbs myelin protein composition and myelination in a mouse model of Pelizaeus–Merzbacher disease. Glia. 2007;55:341–351. doi: 10.1002/glia.20465. [DOI] [PubMed] [Google Scholar]
- 22.Klugmann M, Schwab MH, Pu¨hlhofer A, et al. Assembly of CNS myelin in the absence of proteolipid protein. Neuron. 1997;18:59–70. doi: 10.1016/s0896-6273(01)80046-5. [DOI] [PubMed] [Google Scholar]
- 23.Koeppen AH, Robitaille Y. Pelizaeus–Merzbacher disease. J Neuropathol Exp Neurol. 2002;61:747–759. doi: 10.1093/jnen/61.9.747. [DOI] [PubMed] [Google Scholar]
- 24.Koeppen AH, Ronca NA, Greenfield EA, Hans MB. Defective biosynthesis of proteolipid protein in Pelizaeus–Merzbacher disease. Ann Neurol. 1987;21:159–170. doi: 10.1002/ana.410210208. [DOI] [PubMed] [Google Scholar]
- 25.Mimault C, Giraud G, Courtois V, et al. Proteolipoprotein gene analysis in 82 patients with sporadic Pelizaeus–Merzbacher disease: duplications, the major cause of the disease, originate more frequently in male germ cells, but point mutations do not. The Clinical European Network on Brain Dysmyelinating Disease. Am J Hum Genet. 1999;65:360–369. doi: 10.1086/302483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nave KA, Lai C, Bloom FE, Milner RJ. Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc Natl Acad Sci USA. 1987;84:5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelizaeus F. Über eine eigenthümliche Form Spastischer La¨hmung mit Cerebralerscheinungen auf hereditärer Grundlage (Multiple Sklerose) Arch Psychiatr Nervenkrankh. 1885;16:698–710. [Google Scholar]
- 28.Pham-Dinh D, Birling MC, Roussel G, Dautigny A, Nussbaum JL. Proteolipid DM-20 predominates over PLP in peripheral nervous system. Neuroreport. 1991;2:89–92. doi: 10.1097/00001756-199102000-00006. [DOI] [PubMed] [Google Scholar]
- 29.Pratt VM, Boyadjiev S, Green K, Hodes ME, Dlouhy SR. Pelizaeus–Merzbacher disease caused by a de novo mutation that originated in exon 2 of the maternal great-grandfather of the propositus. Am J Med Genet. 1995;58:70–73. doi: 10.1002/ajmg.1320580114. [DOI] [PubMed] [Google Scholar]
- 30.Raskind WH, Williams CA, Hudson LD, Bird TD. Complete deletion of the proteolipid protein gene (PLP) in a family with X-linked Pelizaeus–Merzbacher disease. Am J Hum Genet. 1991;49:1355–1360. [PMC free article] [PubMed] [Google Scholar]
- 31.Readhead C, Schneider A, Griffiths I, Nave KA. Premature arrest of myelin formation in transgenic mice with increased proteolipid protein gene dosage. Neuron. 1994;12:583–595. doi: 10.1016/0896-6273(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 32.Rosenfeld J, Freidrich VL., Jr Axonal swellings in jimpy mice: does lack of myelin cause neuronal abnormalities? Neu-roscience. 1983;10:959–966. doi: 10.1016/0306-4522(83)90233-6. [DOI] [PubMed] [Google Scholar]
- 33.Saugier-Veber P, Munnich A, Bonneau D, et al. X-linked spastic paraplegia and Pelizaeus–Merzbacher disease are allelic disorders at the proteolipid protein locus. Nat Genet. 1994;6:257–262. doi: 10.1038/ng0394-257. [DOI] [PubMed] [Google Scholar]
- 34.Seitelberger F. Neuropathology and genetics of Pelizaeus–Merzbacher disease. Brain Pathol. 1995;5:267–273. doi: 10.1111/j.1750-3639.1995.tb00603.x. [DOI] [PubMed] [Google Scholar]
- 35.Shy ME, Hobson G, Jain M, et al. Schwann cell expression of PLP1 but not DM20 is necessary to prevent neuropathy. Ann Neurol. 2003;53:354–365. doi: 10.1002/ana.10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simons M, Kramer EM, Macchi P, et al. Overexpression of the myelin proteolipid protein leads to accumulation of cholesterol and proteolipid protein in endosomes/lysosomes: implications for Pelizaeus–Merzbacher disease. J Cell Biol. 2002;157:327–336. doi: 10.1083/jcb.200110138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sistermans EA, de Coo RF, de Wijs IJ, van Oost BA. Duplication of the proteolipid protein gene is the major cause of Pelizaeus–Merzbacher disease. Neurology. 1998;50:1749–1754. doi: 10.1212/wnl.50.6.1749. [DOI] [PubMed] [Google Scholar]
- 38.Skoff RP, Bessert DA, Cerghet M, et al. The myelin proteolipid protein gene modulates apoptosis in neural and non-neural tissues. Cell Death Differ. 2004;11:1247–1257. doi: 10.1038/sj.cdd.4401498. [DOI] [PubMed] [Google Scholar]
- 39.Southwood CM, Garbern J, Jiang W, Gow A. The unfolded protein response modulates disease severity in Pelizaeus–Merzbacher disease. Neuron. 2002;36:585–596. doi: 10.1016/s0896-6273(02)01045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapp BD, Peterson J, Ransohoff RM, et al. Axonal transection in the lesions of multiple sclerosis. N Engl J Med. 1998;338:278–285. doi: 10.1056/NEJM199801293380502. [DOI] [PubMed] [Google Scholar]
- 41.van der Knaap MS, Valk J. Magnetic resonance of myelin, myelination, and myelin disorders. New York: Springer; 2005. p. 1084. [Google Scholar]
- 42.Watanabe I, Patel V, Goebel HH, et al. Early lesion of Pelizaeus–Merzbacher disease: electron microscopic and biochemical study. J Neuropathol Exp Neurol. 1973;32:313–333. doi: 10.1097/00005072-197304000-00010. [DOI] [PubMed] [Google Scholar]
- 43.Wilkus RJ, Farrell DF. Electrophysiologic observations in the classical form of Pelizaeus–Merzbacher disease. Neurology. 1976;26:1042–1045. doi: 10.1212/wnl.26.11.1042. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto T, Nanba E, Zhang H, et al. Jimpymsd mouse mutation and connatal Pelizaeus–Merzbacher disease. Am J Med Genet. 1998;75:439–440. [PubMed] [Google Scholar]
- 45.Yin X, Baek RC, Kirschner DA, et al. Evolution of a neuroprotective function of central nervous system myelin. J Cell Biol. 2006;172:469–478. doi: 10.1083/jcb.200509174. [DOI] [PMC free article] [PubMed] [Google Scholar]