Abstract
Many human adenovirus (Ad) serotypes use the coxsackie B virus-Ad receptor (CAR). Recently, CD46 was suggested to be a receptor of species B Ad serotype 11 (Ad11), Ad14, Ad16, Ad21, Ad35, and Ad50. Using Sindbis virus-mediated cDNA library expression, we identify here the membrane cofactor protein CD46 as a surface receptor of species B Ad3. All four major CD46 transcripts and one minor CD46 transcript expressed in nucleated human cells were isolated. Rodent BHK cells stably expressing the BC1 form of CD46 bound radiolabeled Ad3 with a dissociation constant of 0.3 nM, identical to that of CD46-positive HeLa cells expressing twice as many Ad3 binding sites. Pull-down experiments with recombinant Ad3 fibers and a soluble form of the CD46 extracellular domain linked to the Fc portion of human immunoglobulin G (CD46ex-Fc) indicated direct interactions of the Ad3 fiber knob with CD46ex-Fc but not CARex-Fc (Fc-linked extracellular domain of CAR). Ad3 colocalized with cell surface CD46 in both rodent and human cells at the light and electron microscopy levels. Anti-CD46 antibodies and CD46ex-Fc inhibited Ad3 binding to CD46-expressing BHK cells more than 10-fold and to human cells 2-fold. In CD46-expressing BHK cells, wild-type Ad3 and a chimeric Ad consisting of the Ad5 capsid and the Ad3 fiber elicited dose-dependent cytopathic effects and transgene expression, albeit less efficiently than in human cells. Together, our results show that all of the major splice forms of CD46 are predominant and functional binding sites of Ad3 on CD46-expressing rodent and human cells but may not be the sole receptor of species B Ads on human cells. These results have implications for understanding viral pathogenesis and therapeutic gene delivery.
To date, 51 human adenovirus (Ad) serotypes have been identified and classified into six species, A to F (47). The major Ad vectors currently used in clinical applications are derived from species C Ad serotype 2 (Ad2) and Ad5. Their biology is very well characterized (for reviews, see references 27, 33, and 38). Species C entry into epithelial cells occurs after virus binding to the coxsackie B virus-Ad receptor (CAR) (2), followed by engagement of heterodimeric αv-integrins as secondary receptors (49), which facilitate viral endocytosis and signaling in target cells. Although species C Ads very efficiently infect a number of cells and tissues, the lack of CAR or integrin expression may limit their general usefulness for gene therapy.
Unlike most serotypes of species A, C, D, E, and F Ads, the species B Ads were suggested to bind to a different cell surface receptor (6, 32, 43). The species B Ads cause a significant number of infections worldwide. The B1 subspecies, including Ad3, Ad7, Ad16, Ad21, and Ad50, predominantly infect the upper respiratory tract, and the B2 subspecies, including Ad11, Ad14, Ad34, and Ad35, infect the kidneys and the urinary tract (48). Some species B Ads are capable of infecting cells refractory to the well-characterized species C Ads and have a low seroprevalence (34, 46). These characteristics make the species B Ads interesting vectors for gene therapy approaches (14). We demonstrate here that the membrane cofactor CD46 is a receptor for species B Ad3. Our data indicate that CD46 directly binds to the Ad3 fiber knob with subnanomolar affinity and mediates dose-dependent viral transgene expression and cytopathic effects (CPE) in rodent and human cells.
MATERIALS AND METHODS
Virus.
The molecular identity of the Ad3 stock (prototype strain GB, kindly provided by the late T. Adrian, Medizinische Hochschule Hannover, Hannover, Germany) was verified by DNA restriction analysis (1) and DNA sequencing. This Ad3 stock has a single point mutation in the fiber gene resulting in a Tyr-to-Ser change at position 218 of the fiber head (Y218S), relative to the published sequence (39). Ad3 was grown, isolated, radiolabeled, or fluorochrome labeled as described for Ad2 (12, 30). CsCl-purified 3H-labeled Ad3 had a specific radioactivity of ∼2.14 × 105 dpm/μg. Purified virus was judged to be homogeneous by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and negative-staining electron microscopy (EM) analyses. Alexa-488- and Texas red (TR)-labeled Ad3 had the same infectivity as the unlabeled parental virus.
Construction of Ad5-based AdCMV-eGFP, derived from an E1/E3 deletion mutant, has been described elsewhere (28). The chimeric Ad3 fiber- Ad5-based AdCMV-eGFP vector (Ad5/F3) was constructed in a manner similar to that described for Ad5/F35 (37). Briefly, Ad5/F3 contained 45 amino acids of the Ad5 tail fused to 86 amino acids of the Ad3 shaft and 187 amino acids of the Ad3 knob by overlapping PCR (fiber swapping). The chimeric fragment was introduced into transfer plasmid pBL-EcofrgAd5, containing 6,729 bp of the right end of Ad5, including the fiber gene and inverted terminal repeat sequence, by using the unique NdeI and AflII sites. The AdCMV-eGFP-5F3 chimeric virus was generated by homologous recombination of EcofrgAd5/F3 with a pTG-H5dl324-derived plasmid (4), which contained a cytomegalovirus (CMV)-enhanced green fluorescent protein (eGFP) expression cassette in the E1 region and uniquely cutting restriction site SwaI inserted into the deleted fiber region. The resulting construct, pTG-H5dl324-CMV-eGFP-5F3, was digested with PacI to release the viral genome, which was rescued by transfection of helper 911 cells (9). CsCl purification of Ad5-based AdCMV-eGFP and Ad5/F3 yielded 6 × 109 and 2.4 × 108 PFU, respectively, with physical titers of 7.5 × 1011 and 1.6 × 1010 virions/ml, respectively, as determined by previously described methods (15).
Cells and screening of the cDNA library.
BHK-21 cells and all other cells were grown as described previously (8, 19). For stable transfections, the CD46-encoding cDNA clones 28 and 54 or, alternatively, the cDNA encoding CAR (2, 8) was PCR amplified and cloned into pCDNA3 (Invitrogen). For bulk cultures, plates containing >100 G418-resistant clones were trypsinized and enriched for CD46 expression by fluorescence-activated cell sorting on a MoFlo high-speed sorter (Dako-Cytomation, Fort Collins, Colo.). Clonal lines were established by limiting dilution of sorted bulk cultures of BHK cells expressing CD46 (BHK-CD46 cells)-clone 54 (BHK-CD46-cl54 bulk cells). Generation of the Sindbis virus K562 cDNA library, sorting, reverse transcription-PCR, and sequencing were performed as described earlier (19).
Immunoreagents and flow cytometry.
Details for the anti-CD46 antibodies used here are listed in Table 1. For cytofluorometric analysis, 2 × 105 cells were incubated with 5 μg of Ad3- Alexa-488, with 1 μg of E4.3 antibody for CD46 detection, or with 1 μg of E1-1 antibody for CAR detection as described previously (8). For eGFP expression analysis, triplicate samples of 2 × 105 cells were infected at multiplicities of infection (MOIs) of 3.3, 10, and 30, washed at 2 h postinfection (p.i.), and analyzed at 2 days p.i.
TABLE 1.
Antibodies used for CD46 staining and blocking
| Antibody | Epitope recognized | Source |
|---|---|---|
| GB-24 | SCR3/4 | T. Seya, Osaka Medical Center, Osaka, Japan |
| M75 | SCR2 | J. Atkinson, Washington University, St. Louis, Mo. |
| E4.3 | SCR1 | Pharmingen Switzerland, Basel, Switzerland |
| MCI20.6 | SCR1 | D. Gerlier, Faculté Médecine Lyon, Lyon, France |
| 13/42 | SCR1 | BMA Biomedicals AG, Augst, Switzerland |
| Tra-2.10 | SCR1 | J. Atkinson |
| MEM-258 | SCR4 | Serotec Ltd., Oxford, United Kingdom |
| Rabbit anti-CD46 (amino acids 35-328) | Not available | Santa Cruz Biotechnology, Santa Cruz, Calif. |
Binding isotherm and Scatchard plots.
Subconfluent cells were detached in phosphate-buffered saline- 20 mM EDTA and washed twice with RPMI medium-0.2% bovine serum albumin (BSA). Triplicate samples of 5 × 105 cells were incubated in 100 μl of RPMI medium-BSA for 30 min at 37°C with or without 2 μg of Ad3 dodecahedron fiber (DF). Note that Ad3 DF and the fiber head, prepared as described previously (7, 10), had the wild-type sequence Y218 (P. Fender, Grenoble, France, personal communication). Cold RPMI medium-BSA with the appropriate amount of virus (1 part 3H-labeled Ad3 plus 9 parts Ad3) was added to a final reaction volume of 200 μl and allowed to react for 90 min. Ad3 concentrations were based on an approximate molecular mass of 150 MDa per particle. The cells were washed twice with RPMI medium-BSA and once with Hanks' balanced salt solution. Cell-associated radioactivity was measured as described earlier (28). Regression analyses of binding data were performed with Prism software (GraphPad Software Inc., San Diego, Calif.).
Blocking of Ad3 binding.
Cells were detached with phosphate-buffered saline- 20 mM EDTA, and 2 × 105 cells were incubated with monoclonal and polyclonal antibodies specific for CD46, anti-human CAR antibody E1-1 (8), soluble CD46ex-Fc (comprising 295 amino acids of the mature extracellular domain fused to 232 amino acids of the human immunoglobulin G1 (IgG1) Fc domain, including the hinge, CH2, and CH3 regions), and CARex-Fc (8) or a 30-fold excess of unlabeled Ad3 on ice for 15 min. Subsequently, 5 μg of Ad3- Alexa-488 or 3H-labeled Ad3 was added in a total volume of 200 μl on ice; after 1 h, the cells were washed and analyzed for bound virus by cytofluorometry or liquid scintillation counting (28). The data were normalized to the amounts of virus bound in the absence of inhibitors and are presented as the mean of three independent experiments and standard deviations. Statistical evaluation was performed with Student's t test. Experiments to assess CPE were carried out by using microtiter plate assays with triplicate input samples. Serial 10-fold dilutions of various virus stocks were plated in a volume of 50 μl in a 96-well dish, starting with a concentration of 6 × 105 viral particles/ml. All dilutions were prepared in cell culture medium. Cells were diluted to 6.7 × 104/ml, and 150 μl of this suspension was added per well (MOIs ranged from 3 × 101 to 3 × 104). Attached cells were fixed with methanol at 72 h p.i. and stained with crystal violet.
Microscopy.
Confocal laser scanning microscopy (CLSM) was performed by using a DM RXA2 TCS SP2 AOBS microscope (Leica Microsystems, Wetzlar, Germany) equipped with an Ar or Ar-Kr laser, an He-Ne 543/594 laser, an He-Ne 633 laser, a diode laser (405 nm), and a ×63 oil immersion objective (N.A. 1.4 PL APO). The pinhole value was 115 μm, airy 1, yielding optical sections of about 0.12 μm, and the step size was set to 0.122 μm with a voxel of 0.233 by 0.233 by 0.122 μm. Image processing was performed with MetaMorph (Universal Imaging Inc., Visitron Inc., Downingtown, Pa.) and Photoshop (Adobe). Transmission EM was performed by using a modification of a previously described protocol (40). Briefly, Ad3 was cold bound to or internalized in BHK-CD46-cl54 bulk cells as described previously (29, 44), and the specimens were fixed in 2% paraformaldehyde (PFA)- 1.5% glutaraldehyde (GA) in 0.1 M sodium cacodylate (pH 7.4) overnight, washed in fixative-free buffer, and postfixed in 1% osmium tetroxide (Electron Microscopy Sciences)- 1.5% potassium ferricyanide at 4°C for 1 h. The specimens were rinsed in 0.1 M sodium cacodylate, contrasted with 1% tannic acid in 0.05 M sodium cacodylate at room temperature for 45 min, washed in 1% sodium sulfate, rinsed in H2O, stained in 2% uranyl acetate in H2O overnight, and embedded in Epon. For preembedding immunogold EM, cells were fixed in 0.2% GA- 2% PFA in 0.1 M sodium cacodylate (pH 7.4) for 20 min, labeled with anti-CD46 antibodies (5 μg/ml in 10% goat serum) for 1 h and goat anti-mouse IgG (5-nm gold; 10 μg/ml; British BioCell International) for 2 h, postfixed in 1.5% GA- 2% PFA, and further processed as described above.
CD46ex-Fc pull-down assays.
Ad3 DF and the dodedahedron penton (DP) of Ad3 were centrifuged at 16,100 × g for 10 min, and the fiber head and CD46ex-Fc were centrifuged at 100,000 × g for 30 min. CD46ex-Fc was incubated with the respective viral proteins in a final volume of 50 μl of binding buffer (BB; 50 mM HEPES-KOH [pH 7.4], 0.5% Triton X-100, 150 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride, 1 μg each of chymostatin, leupeptin, aprotinin, and pepstatin per ml, 0.1% BSA) for 1 h. The ratio of binding partners was 1.1 μg of CD46ex-Fc to 1 μg of DF or DP and 3.3 μg of CD46ex-Fc to 1 μg of fiber head. A total of 450 μl of BSA-free buffer and 20 μl of a 1:1 slurry of protein G-Sepharose 4 Fast Flow (Amersham) were added, and the mixture was incubated under constant agitation for 2 h. The Sepharose beads were sedimented at 3,000 × g for 2 min, washed twice with BB without BSA, and washed twice with BB without BSA and Triton X-100. The pellet was resuspended in 20 μl of doubly concentrated reducing sample buffer and boiled at 95°C for 5 min. A total of 15 μl of the supernatant was separated by SDS- 12% PAGE and silver stained at room temperature as described previously (16). All steps were carried out at 4°C unless indicated otherwise.
RESULTS
Isolation of CD46 cDNAs encoding the Ad3 receptor.
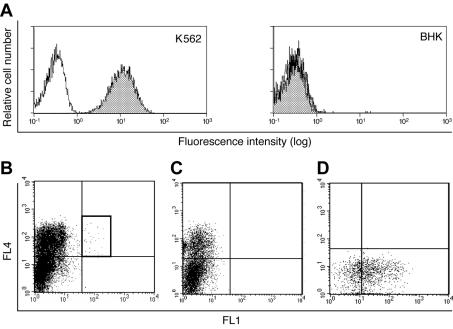
To identify a cell surface receptor(s) for species B1 Ads, we used the Sindbis virus-based cDNA expression system (19) to screen for the binding of Ad3 particles labeled with the fluorophore Alexa-488 to BHK cells. The cDNA library was made from human K562 chronic myelogenous leukemia cells, which efficiently bound Ad3- Alexa-488 (Fig. 1A, left panel). In contrast, rodent BHK cells did not bind significant amounts of Ad3, in agreement with the earlier notion that the receptor for species B Ads is not conserved between rodents and primates (13, 14, 36) (Fig. 1A, right panel). About 0.0024% of Sindbis virus-infected cells expressing cDNAs also bound fluorescent Ad3- Alexa-488 (Fig. 1B), whereas cells infected with Sindbis virus lacking cDNA inserts did not (Fig. 1C). Rescreening resulted in more than 30 positive clones binding Ad3; one of these is shown in Fig. 1D. Of 30 positive clones, 28 contained cDNA fragments of about 3 kb encoding CD46; the other 2 clones represented cloning artifacts. It is known that multiple splice variants of the Ser/Thr-rich domain-encoding exons and cytoplasmic tail exons give rise to four major splice variants, C1, C2, BC1, and BC2, and to additional minor variants (21). Among the 10 inserts that were completely sequenced, the major splice variants C2 and BC1 were found five times and two times, respectively, and the variants BC2 and C1 were found once each. The rare form B1 was found once, whereas the rare forms ABC1 and ABC2, typically expressed in cancer cells, were not recovered.
FIG. 1.
CD46-expressing cells bind Ad3. (A) Flow cytometry of human leukemia K562 cells and rodent BHK cells that were not incubated with (white histograms) or that were incubated with (shaded histograms) Ad3- Alexa-488. (B and C) Single-cell sorting of BHK cells infected with Sindbis virus expressing a K562 cDNA library (B) and control Sindbis virus (C) after staining with propidium iodide (PI), anti-Sindbis virus antibodies (FL4), and Ad3- Alexa-488 (FL1). (D) PI-negative, Sindbis virus- and Ad3-positive cells (gate) were single cell sorted into 24-well plates containing BHK feeder cells and rescreened for Ad3- Alexa-488 binding. One of 30 positive clones is shown.
Ad3 binds with a high affinity to BHK-CD46 cells through direct fiber head-CD46 interactions.
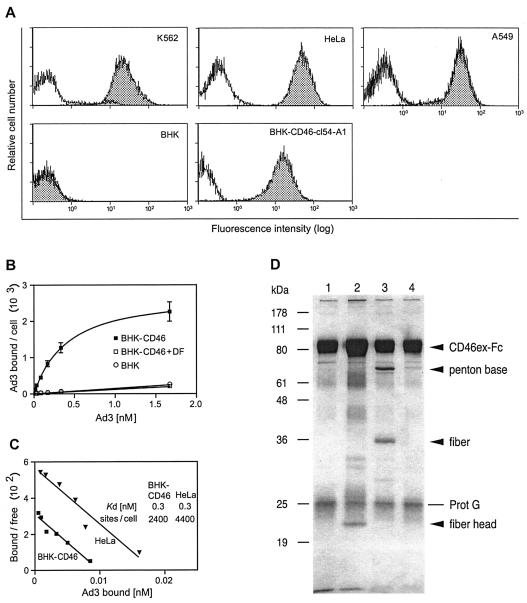
BHK cells stably expressing the BC1 form (cDNA clone 54) were generated. CD46 expression levels of bulk cultures (BHK-CD46-cl54 bulk cells; data not shown) or clonal BHK-CD46-cl54-A1 cells expressed CD46 at intermediate levels, compared to human cell lines, such as K562, HeLa, and A549 (Fig. 2A). The binding of 3H-thymidine-labeled Ad3 to BHK-CD46-cl54 bulk cells was dose dependent and saturable, compared to background binding in the presence of 2 nM Ad3 DF (10) and to parental BHK cells (Fig. 2B). One DF consists of 12 penton base pentamers, each of which contains one trimeric fiber molecule with a distal fiber head at the outside. Scatchard plots of Ad3 binding showed parallel slopes with extrapolated Kd values of 0.3 nM for both BHK-CD46 transfectants and HeLa cells (Fig. 2C). The number of Ad3 binding sites on BHK-CD46-cl54 bulk cells was 2.4 × 103, and that on human cervical carcinoma HeLa cells was 4.4 × 103, consistent with earlier experiments with human nasopharyngeal carcinoma KB cells and human lung carcinoma A549 cells (6). Competition with the recombinant Ad3 fiber knob (7) eliminated more than 75% of Ad3 binding to HeLa cells (mean of two experiments; data not shown), in agreement with earlier experiments (43). Further, CD46ex-Fc was able to pull down both the recombinant Ad3 fiber head and DF but not DP (Fig. 2D). DF was not recognized by CARex-Fc, indicating specific binding of DF to CD46ex-Fc (data not shown). In addition, CD46ex-Fc also pulled down Ad3 but not Ad2 or the fiber head of Ad2, further confirming the specificity of the interaction of CD46 with Ad3 (data not shown). These results indicate that CD46 expression on nonpermissive rodent cells mediates high-affinity and saturable Ad3 binding through a direct interaction of the extracellular CD46 domain with the Ad3 fiber head.
FIG. 2.
Ad3 binds with a high affinity to CD46-expressing cells through the fiber head. (A) Cytofluorometric analysis of CD46 expression levels in human and hamster cell lines. White histograms show staining with isotype control antibodies, and shaded histograms show CD46-specific staining. The cells used were human K562, HeLa, and A549 cells, parental BHK cells, and stably transfected BHK cells expressing the BC1 isoform of CD46. (B) Binding isotherms of Ad3 incubated with BHK-CD46-cl54 bulk cells or parental BHK cells. Nonspecific binding was determined in the presence of Ad3 DF. Error bar depicts standard error of the mean. (C) Scatchard analysis of Ad3 binding to BHK-CD46-cl54 bulk cells and HeLa cells. The number of binding sites per cell and the Kd values were calculated from the bound and the free virion concentrations determined by subtracting the bound virus from the input virus. (D) Direct interaction of the Ad3 fiber head with CD46ex-Fc. CD46ex-Fc was incubated without any addition (lane 1), with the purified fiber head (lane 2), with DF (lane 3), and with DP (lane 4). CD46ex-Fc was pulled down by protein G (Prot G)-Sepharose, and the SDS eluates were analyzed by SDS-PAGE and silver staining.
Colocalization of Ad3 with CD46 during viral entry.
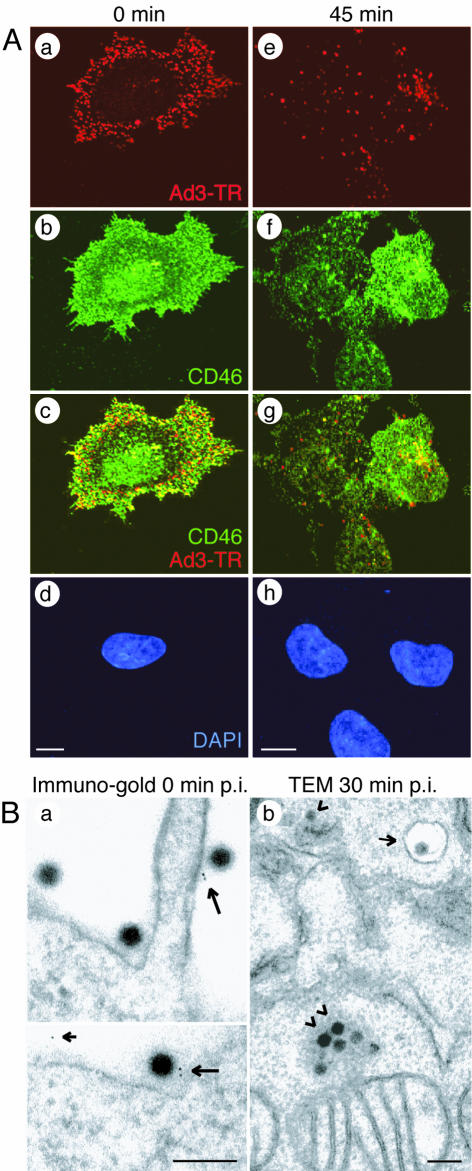
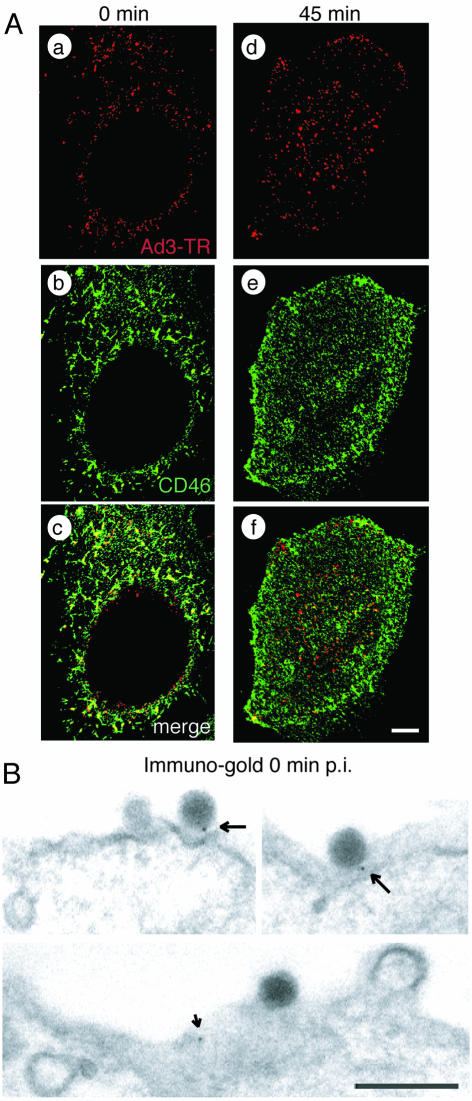
We next analyzed Ad3 interactions with BHK-CD46 cells and human epithelial cells (HeLa cells) positive for CD46 and permissive for Ad3 by light microscopy and EM. TR-labeled Ad3 bound homogeneously to the cell surface at 4°C, and a large fraction colocalized with CD46, as determined by indirect immunofluorescence and single-section CLSM (Fig. 3A and 4A). Overlapping staining of CD46 and staining of TR-labeled Ad3 was particularly prominent in basal and peripheral regions of BHK-CD46 and HeLa cells, consistent with basolateral surface expression of CD46 in certain polarized monkey and human cell lines (23) and both apical and basolateral expression in differentiated human airway epithelial and tracheal tissues (41). Ad3-CD46 colocalization was confirmed by high-resolution indirect immunogold EM depicting gold particles attached to goat anti-mouse IgG near Ad3 and occasionally elsewhere on the cell surface (Fig. 3B and 4B). No gold particles were observed in the absence of anti-CD46 antibodies (data not shown). Upon warming, Ad3 was found in the cytosol and endosomal vesicles of BHK-CD46 cells, partly colocalized with CD46 (Fig. 3A, e to g, and Fig. 3B, b). Similar results were obtained with HeLa cells (Fig. 4A, d to f).
FIG. 3.
Ad3 colocalizes with cell surface CD46 of BHK-CD46-cl54-bulk cells. (A) CLSM analyses of TR-labeled Ad3 (red) and CD46 stained with non-function-blocking antibody MCI20.6 (green). Single sections were taken at 0 min (a to d) and at 5 min (e to h) p.i., including 4′,6′-diamidino-2-phenylindole (DAPI)-stained sections (d and h). Scale bars, 5 μm. (B) Transmission EM (TEM). (a) Immunogold staining of CD46 at 0 min p.i. Large arrows indicate Ad3 associated with protein A-gold directed to anti-CD46; the small arrow indicates a gold particle not associated with Ad3. (b) Ad3 internalization at 0 min p.i. Arrowheads indicate clusters of cytosolic Ad3; the small arrow indicates endosomal Ad3. Scale bars, 200 nm.
FIG. 4.
Ad3 colocalizes with cell surface CD46 of HeLa cells. (A) CLSM analyses of TR-labeled Ad3 (red) and CD46 stained with non-function-blocking antibody MCI20.6 (green). Single sections are shown at 0 min (a to c) and at 45 min (d to f) p.i. Scale bar, 5 μm. (B) Transmission EM showing immunogold staining of CD46 at 0 min p.i. Arrows indicate Ad3 associated with protein A-gold directed to anti-CD46; the small arrow indicates a gold particle not associated with Ad3. Scale bar, 200 nm.
CD46 expression mediates binding, gene expression, and CPE.
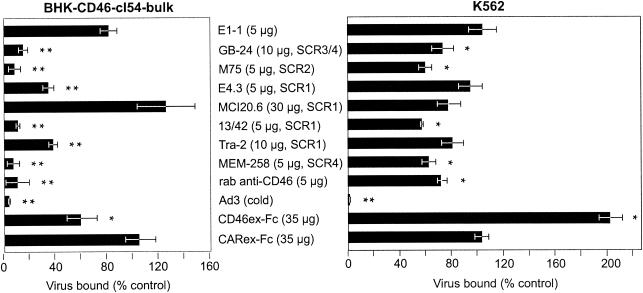
We further tested whether Ad3 binding to BHK-CD46 cells was specific for CD46 by using eight different anti-CD46 antibodies (Table 1) and CD46ex-Fc. Six monoclonal antibodies and one polyclonal antibody to CD46 strongly inhibited either Ad3- Alexa-488 or 3H-labeled Ad3 binding at concentrations that gave saturating antibody binding to BHK-CD46 cells (Fig. 5, left panel, and data not shown). Monoclonal antibodies M75, 13/42, and MEM-258, recognizing epitopes on short consensus region (SCR) 1 (SCR1), SCR2, and SCR4, blocked Ad3 binding by more than 90%. Antibodies GB-24, E4.3, and Tra-2.10, directed to SCR1 and SCR3/4, inhibited binding at intermediate levels of 85, 65, and 62%, respectively. In contrast, antibody MCI20.6, recognizing an SCR1 epitope, and anti-CAR antibody E1-1 had no significant effect on Ad3 binding. The soluble CD46ex-Fc protein inhibited binding in a dose-dependent manner (data not shown), with a maximal inhibition of 43%, whereas the CARex-Fc protein had no effect. The specificity of this assay was further confirmed by including a 30-fold excess of cold Ad3, reducing labeled Ad3 binding by >96%. The anti-CD46 antibodies also inhibited Ad3 binding to K562 cells, albeit to a lesser extent than to BHK-CD46 cells (Fig. 5, right panel). Notably, antibodies M75, 13/42, and MEM-258, which were the most effective blockers of Ad3 binding to BHK-CD46 cells, also conferred maximal blocking in K562 cells amounting to about 40%. The use of a mixture of all seven blocking antibodies did not increase the blocking of Ad3 binding (data not shown). Likewise, CD46ex-Fc did not inhibit Ad3 binding to K562 cells and, surprisingly, somewhat enhanced Ad3 binding (data not shown). This result may be related to the capacity of CD46ex-Fc to form dimers, but additional experiments are required to clarify this observation.
FIG. 5.
CD46-dependent binding of Ad3 revealed by anti-CD46 antibodies and CD46ex-Fc. BHK-CD46-cl54 bulk cells and human K562 cells were incubated with the indicated antibodies, CD46ex-Fc, or CARex-Fc (control), followed by the addition of either Ad3- Alexa-488 or 3H-labeled Ad3. Virus binding was measured by flow cytometry or liquid scintillation counting. The asterisks indicate the level of significance (P values of <0.05 [single asterisks] and <0.005 [double asterisks] for comparisons with the negative controls E1-1 [anti-CAR] and CARex-Fc, respectively). rab, rabbit.
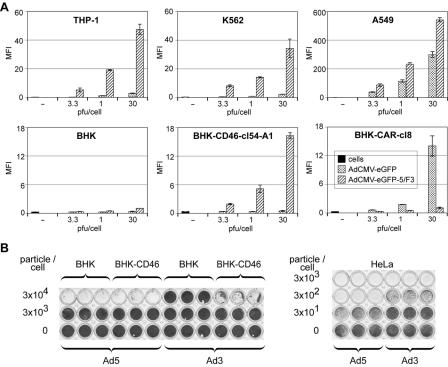
We next measured Ad3-mediated gene delivery to BHK-CD46 transfectants by using Ad5/F3 (Fig. 6A). Ad5/F3-mediated eGFP expression in hematopoietic human K562 and THP-1 cells was 10- to 20-fold higher than eGFP expression from an Ad5 vector, confirming the functionality of Ad5/F3. This finding was in agreement with earlier reports (42). In BHK-CD46-cl54-A1 cells, the eGFP expression of Ad5/F3 increased in a dose-dependent manner to a maximum of 16-fold, compared to that in BHK cells and CAR-expressing BHK cells (BHK-CAR cells). In BHK-CD46-cl28 bulk cells (an additional BC1 cDNA clone) and clone 54 cells, expression increased to an intermediate level of sevenfold (data not shown), correlating with a twofold-lower level of CD46 expression in these cells than in BHK-CD46-cl54-A1 cells. The transgene expression of Ad5/F3 in human A549 cells was about 30-fold higher than that in clonal BHK-CD46 cells. As expected, BHK and BHK-CD46 transfectants remained refractory to Ad5-mediated eGFP expression, whereas eGFP expression from Ad5 in stable BHK-CAR cells was increased about 23-fold, compared to a 300-fold increase in A549 cells.
FIG. 6.
Transgene expression by chimeric Ad5/F3 and CPE of wild-type Ad3 in BHK-CD46 cells. (A) Ad-mediated eGFP expression in permissive human cells and CD46- or CAR-transfected rodent cells. The indicated cells were incubated with eGFP-expressing Ad5 or Ad5/F3 at different MOIs, and eGFP expression was analyzed at 2 days p.i. by flow cytometry. Results are shown as the mean fluorescence intensity (MFI). (B) CPE of wild-type Ad3 in CD46-transfected BHK cells and human HeLa cells and control infections of BHK-CD46 and HeLa cells with eGFP-expressing Ad5. Cells were infected with 10-fold dilutions of virus (3 × 101 to 3 × 104 particles/cell) for 3 days, fixed with methanol, and stained with crystal violet.
We next measured the CPE of Ad3 in BHK-CD46 cells and found that there is about a 1-log-unit increase in the sensitivity of BHK-CD46 cells over that of native BHK cells (Fig. 6B). Control infections with Ad5 showed no difference in CPE between BHK and BHK-CD46 cells. Human cells were up to 2 log units more sensitive to Ad5 and at least 1 log unit more sensitive to Ad3 than BHK-CAR or BHK-CD46 cells to Ad5 or Ad3, respectively (Fig. 6B), consistent with the eGFP expression data for Ad5/F3 (Fig. 6A). Together, these experiments corroborate the binding data and demonstrate that parental BHK cells are refractory to Ad3 infection, whereas BHK-CD46 transfectants develop Ad3-mediated CPE, implying that CD46 supports Ad3 infection.
DISCUSSION
Our expression cloning approach identified a receptor of species B Ad3, the membrane cofactor CD46. The screening experiments indicated that all four major splice variants, BC1, BC2, C1, and C2, and the rare splice form B1 are capable of binding Ad3, similar to what has been described for the binding of measles virus (24). Ad3 binding to rodent cells expressing the most common isoform, BC1, was of high affinity and saturable; the yield was about 2,400 virus binding sites per rodent cell, compared to 4,400 per HeLa cell, consistent with a previous report on HeLa cells (6). We found that Ad3 binding to CD46-expressing rodent cells was inhibited by a panel of anti-CD46 monoclonal and polyclonal antibodies and by a recombinant extracellular domain of CD46, supporting the notion that CD46 represents a major Ad3 binding site in these cells. In human cells, CD46 is also an important Ad3 receptor, as indicated by colocalization data at the light microscopy and EM levels and by antibody inhibition experiments, although the latter were less efficient than those with rodent cells. Noteably, Ad3 has the same subnanomolar affinity for BHK-CD46 cells as for HeLa cells and enters and gives rise to transgene expression and CPE in both cell types. Further, in vitro pull-down experiments indicated that Ad3 directly binds the extracellular domain of CD46 through contact with the fiber head.
Nonetheless, a recent study had suggested that a particular Ad3 isolate did not bind to Chinese hamster ovary cells expressing the C2 isoform of CD46 (11). We therefore checked the DNA sequence of our Ad3 stock and found that the fiber sequence was identical to the published sequence (39), except for a point mutation (Y218S) in the fiber head. This mutation, however, is unlikely to affect virus binding to CD46, since the recombinant Y218 fiber directly bound to CD46ex-Fc (like our Y218S Ad3) and Y218 DF abolished the binding of our Ad3 to HeLa and BHK-CD46 cells as efficiently as nonlabeled competitor Y218S Ad3. These data strongly argue that all of the splice variants of CD46 can serve as Ad3 binding sites, at least on rodent cells. However, Ad5/F3-mediated gene expression and wild-type Ad3-mediated CPE in rodent cells were lower than those in human epithelial cells (A549 and HeLa cells). Similarly, Ad5-mediated eGFP expression was lower in BHK-CAR cells than in fully permissive A549 cells. It is unknown whether rodent cells lack an unidentified factor for effective Ad-mediated transgene expression, for example, a secondary receptor stimulating endocytosis and nuclear transport (26, 45). It is interesting, however, that the level of Ad3 infection was reduced about threefold in human M21-L12 cells lacking αvβ3- and αvβ5-integrins compared to integrin-positive M21-L4 cells and that infection of M21-L4 cells was inhibited by integrin-specific antibodies and RGD peptides blocking penton base interactions with integrins (25). Whether the association of CD46 with α1-integrins, as reported for HeLa cells (22), facilitates Ad3 infection needs to be investigated. It is possible that human cells express additional Ad3 receptors unrelated to CD46, as suggested by our finding of relatively weak Ad3 binding inhibition with anti-CD46 antibodies in K562 cells. The nature of such hypothetical binding sites is unclear. It is also conceivable that glycosylation may modulate Ad3 binding. Interestingly, CD46 is heavily N and O glycosylated, depending on the tissue type, and the removal of N-linked but not O-linked glycans or sialic acid abolished the binding of measles virus strain Edmonston, which also uses CD46 as a receptor (17).
In terms of CD46 binding and infection, it appears that our Ad3 behaves more like species B Ad11 and Ad35 than Ad7. The binding of both Ad3 and Ad11 to human cells was partially blocked by anti-CD46 antibodies, with 20 to 30% inhibition for Ad11 (35) and about 40% inhibition for Ad3 (this study), or by small interfering CD46 RNAs, which reduced Ad35 binding by about 40% (11). The soluble CD46 receptor also inhibited the transgene expression of Ad5/F35 (11). The binding of Ad7 to human cells was apparently unaffected by anti-CD46 antibodies (35). In contrast, Ad3, Ad7, and Ad11 binding to CD46-expressing rodent cells was inhibited by anti-CD46 antibodies, and infection of CD46 BC1 isoform-expressing Chinese hamster ovary cells was possible with Ad11 but not with Ad7; these results suggested that Ad7 may bind CD46 but not use it for infection. It is unknown, however, whether Ad7 infects human cells with the same efficiency as CD46-expressing rodent cells. Whether Ad3, Ad7, Ad11, Ad35, and other Ad serotypes all use the same binding site on CD46 remains to be investigated.
The species B Ads are considerable human pathogens. Ad3 and Ad7 are responsible for a significant proportion of Ad infections worldwide and cause infections of the upper respiratory tract, including acute febrile and severe respiratory illness, whereas the less abundant Ad11 and Ad35 cause infections of the urinary tract (34). The identification of Ad3 receptor CD46 has medical implications for the development of new antiviral agents. Besides, species B Ads are interesting gene transfer vehicles, as they use a receptor distinct from the common Ad receptor CAR. Since CAR is expressed on many but not all cell types and is down regulated on many cancer cells, conventional therapeutic Ad vectors have limitations. These limitations can be overcome in part by swapping the fiber protein of species C serotypes with that of species B serotypes. Accordingly, fiber-swapped Ad vectors have an extended tropism compared to species C vectors and are able to efficiently infect hematopoietic and dendritic cells (13, 14, 18, 31, 37). In addition, several species B Ads have a low seroprevalence, which makes them attractive for in vivo applications (46).
Ad3 adds to a growing list of pathogens binding to CD46. For example, CD46 is known to be a receptor for measles virus strain Edmonston, human herpesvirus 6, Neisseria gonorrhoeae, N. meningitidis, Helicobacter pylori, and Streptococcus pyogenes (reviewed in reference 20). CD46 belongs to a family of complement activation regulators (21). The biological role of CD46 is to prevent complement activation on autologous tissue by binding C3b and C4b and by acting simultaneously as a cofactor for proteolytic factor 1. CD46 consists of four amino-terminal SCRs of 60 amino acids each, one to three Ser/Thr-rich domains, a short region of unknown function, one transmembrane domain, and a cytoplasmic tail. The antibody blocking data suggest that Ad3 may bind to all four SCR domains, a suggestion which would be in agreement with the finding that three different receptors of the complement activation regulator family (CD21, CD55, and CD46) can recognize viral particles through two or more SCR domains (for a review, see reference 3). The first two SCR domains of CD46 are trimers (3), indicating that CD46 would be well suited for tight interactions with a trimeric ligand, such as the Ad3 fiber (5). Together with evidence from recent reports suggesting that CD46 is a receptor for species B Ad11 and Ad35 (11, 35), our data corroborate the notion that CD46 is a common receptor for species B Ads. Given that CD46 is expressed on all human cells except for erythrocytes, we expect that viruses using CD46 as a receptor might have evolved broad infectivity tropism, allowing them to utilize cells and infection pathways that are not accessible to other Ad serotypes.
Acknowledgments
We thank W. Schaffner, Institute of Molecular Biology, University of Zürich, Zürich, Switzerland, for continuous encouragement; C. Torres-de los Reyes and Monika Straub for technical assistance; P. Fender and J. Chroboczek for generous gifts of Ad3 DFs and Ad3 fiber heads; F. Ochsenbein for help with graphic designs; E. Niederer (Biomedical Center ETH Zürich) for help with fluorescence-activated cell sorting analyses; and S. Rusconi (University of Fribourg, Fribourg, Switzerland) for plasmid pTG-H5dl324.
We thank W. Schaffner for financial support. This work was supported by the Kanton Zürich (S.H. and U.F.G.) and the Swiss National Science Foundation (U.F.G.).
REFERENCES
- 1.Adrian, T., G. Wadell, J. C. Hierholzer, and R. Wigand. 1986. DNA restriction analysis of adenovirus prototypes 1 to 41. Arch. Virol. 91:277-290. [DOI] [PubMed] [Google Scholar]
- 2.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 3.Casasnovas, J. M., M. Larvie, and T. Stehle. 1999. Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J. 18:2911-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chroboczek, J., R. W. H. Ruigrok, and S. Cusack. 1995. Adenovirus fiber, vol. I. Springer-Verlag KG, Berlin, Germany. [DOI] [PubMed]
- 6.Defer, C., M. T. Belin, M. L. Caillet-Boudin, and P. Boulanger. 1990. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J. Virol. 64:3661-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durmort, C., C. Stehlin, G. Schoehn, A. Mitraki, E. Drouet, S. Cusack, and W. P. Burmeister. 2001. Structure of the fiber head of Ad3, a non-CAR-binding serotype of adenovirus. Virology 285:302-312. [DOI] [PubMed] [Google Scholar]
- 8.Ebbinghaus, C., A. Al-Jaibaji, E. Operschall, A. Schoeffel, I. Peter, U. F. Greber, and S. Hemmi. 2001. Functional and selective targeting of adenovirus to high-affinity Fcγ receptor I-positive cells using a bispecific hybrid adaptor. J. Virol. 75:480-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fallaux, F. J., O. Kranenburg, S. J. Cramer, A. Houweling, H. Van Ormondt, R. C. Hoeben, and A. J. Van Der Eb. 1996. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 7:215-222. [DOI] [PubMed] [Google Scholar]
- 10.Fender, P., R. W. Ruigrok, E. Gout, S. Buffet, and J. Chroboczek. 1997. Adenovirus dodecahedron, a new vector for human gene transfer. Nat. Biotechnol. 15:52-56. [DOI] [PubMed] [Google Scholar]
- 11.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 12.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 13.Havenga, M. J., A. A. Lemckert, J. M. Grimbergen, R. Vogels, L. G. Huisman, D. Valerio, A. Bout, and P. H. Quax. 2001. Improved adenovirus vectors for infection of cardiovascular tissues. J. Virol. 75:3335-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havenga, M. J., A. A. Lemckert, O. J. Ophorst, M. van Meijer, W. T. Germeraad, J. Grimbergen, M. A. van Den Doel, R. Vogels, J. van Deutekom, A. A. Janson, J. D. de Bruijn, F. Uytdehaag, P. H. Quax, T. Logtenberg, M. Mehtali, and A. Bout. 2002. Exploiting the natural diversity in adenovirus tropism for therapy and prevention of disease. J. Virol. 76:4612-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemmi, S., R. Geertsen, A. Mezzacasa, I. Peter, and R. Dummer. 1998. The presence of HCAR (human coxsackievirus and adenovirus receptor) is associated with efficient adenovirus-mediated transgene expression in human melanoma cell cultures. Hum. Gene Ther. 9:2363-2373. [DOI] [PubMed] [Google Scholar]
- 16.Heukeshoven, J., and R. Dernick. 1988. Improved silver staining procedure for fast staining in PhastSystem development unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis 9:28-32. [DOI] [PubMed] [Google Scholar]
- 17.Iwata, K., T. Seya, Y. Yanagi, J. M. Pesando, P. M. Johnson, M. Okabe, S. Ueda, H. Ariga, and S. Nagasawa. 1995. Diversity of sites for measles virus binding and for inactivation of complement C3b and C4b on membrane cofactor protein CD46. J. Biol. Chem. 270:15148-15152. [DOI] [PubMed] [Google Scholar]
- 18.Knaan-Shanzer, S., I. Van Der Velde, M. J. Havenga, A. A. Lemckert, A. A. De Vries, and D. Valerio. 2001. Highly efficient targeted transduction of undifferentiated human hematopoietic cells by adenoviral vectors displaying fiber knobs of subgroup B. Hum. Gene Ther. 12:1989-2005. [DOI] [PubMed] [Google Scholar]
- 19.Koller, D., C. Ruedl, M. Loetscher, J. Vlach, S. Oehen, K. Oertle, M. Schirinzi, E. Deneuve, R. Moser, M. Kopf, J. E. Bailey, W. Renner, and M. F. Bachmann. 2001. A high-throughput alphavirus-based expression cloning system for mammalian cells. Nat. Biotechnol. 19:851-855. [DOI] [PubMed] [Google Scholar]
- 20.Lindahl, G., U. Sjobring, and E. Johnsson. 2000. Human complement regulators: a major target for pathogenic microorganisms. Curr. Opin. Immunol. 12:44-51. [DOI] [PubMed] [Google Scholar]
- 21.Liszewski, M. K., T. W. Post, and J. P. Atkinson. 1991. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu. Rev. Immunol. 9:431-455. [DOI] [PubMed] [Google Scholar]
- 22.Lozahic, S., D. Christiansen, S. Manie, D. Gerlier, M. Billard, C. Boucheix, and E. Rubinstein. 2000. CD46 (membrane cofactor protein) associates with multiple beta1 integrins and tetraspans. Eur. J. Immunol. 30:900-907. [DOI] [PubMed] [Google Scholar]
- 23.Maisner, A., M. K. Liszewski, J. P. Atkinson, R. Schwartz-Albiez, and G. Herrler. 1996. Two different cytoplasmic tails direct isoforms of the membrane cofactor protein (CD46) to the basolateral surface of Madin-Darby canine kidney cells. J. Biol. Chem. 271:18853-18858. [DOI] [PubMed] [Google Scholar]
- 24.Maisner, A., J. Schneider-Schaulies, M. K. Liszewski, J. P. Atkinson, and G. Herrler. 1994. Binding of measles virus to membrane cofactor protein (CD46): importance of disulfide bonds and N-glycans for the receptor function. J. Virol. 68:6299-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathias, P., T. Wickham, M. Moore, and G. Nemerow. 1994. Multiple adenovirus serotypes use αv-integrins for infection. J. Virol. 68:6811-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier, O., K. Boucke, S. Vig, S. Keller, R. P. Stidwill, S. Hemmi, and U. F. Greber. 2002. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin mediated uptake. J. Cell Biol. 158:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier, O., and U. F. Greber. 2003. Adenovirus endocytosis. J. Gene Med. 5:451-462. [DOI] [PubMed] [Google Scholar]
- 28.Nagel, H., S. Maag, A. Tassis, F. O. Nestle, U. F. Greber, and S. Hemmi. 2003. The alphavbeta5 integrin of hematopoietic and nonhematopoietic cells is a transduction receptor of RGD-4C fiber-modified adenoviruses. Gene Ther. 10:1643-1653. [DOI] [PubMed] [Google Scholar]
- 29.Nakano, M. Y., K. Boucke, M. Suomalainen, R. P. Stidwill, and U. F. Greber. 2000. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 74:7085-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano, M. Y., and U. F. Greber. 2000. Quantitative microscopy of fluorescent adenovirus entry. J. Struct. Biol. 129:57-68. [DOI] [PubMed] [Google Scholar]
- 31.Rea, D., M. J. Havenga, M. van Den Assem, R. P. Sutmuller, A. Lemckert, R. C. Hoeben, A. Bout, C. J. Melief, and R. Offringa. 2001. Highly efficient transduction of human monocyte-derived dendritic cells with subgroup B fiber-modified adenovirus vectors enhances transgene-encoded antigen presentation to cytotoxic T cells. J. Immunol. 166:5236-5244. [DOI] [PubMed] [Google Scholar]
- 32.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell, W. C. 2000. Update on adenovirus and its vectors. J. Gen. Virol. 81:2573-2604. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz, H., R. Wigand, and W. Heinrich. 1983. Worldwide epidemiology of human adenovirus infections. Am. J. Epidemiol. 117:455-466. [DOI] [PubMed] [Google Scholar]
- 35.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shayakhmetov, D. M., Z. Y. Li, S. Ni, and A. Lieber. 2002. Targeting of adenovirus vectors to tumor cells does not enable efficient transduction of breast cancer metastases. Cancer Res. 62:1063-1068. [PubMed] [Google Scholar]
- 37.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 74:2567-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 2112-2137. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 39.Signas, C., G. Akusjarvi, and U. Pettersson. 1985. Adenovirus 3 fiber polypeptide gene: implications for the structure of the fiber protein. J. Virol. 53:672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simionescu, N., and M. Simionescu. 1976. Galloylglucoses of low molecular weight as mordant in electron microscopy. I. Procedure, and evidence for mordanting effect. J. Cell Biol. 70:608-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinn, P. L., G. Williams, S. Vongpunsawad, R. Cattaneo, and P. B. McCray, Jr. 2002. Measles virus preferentially transduces the basolateral surface of well-differentiated human airway epithelia. J. Virol. 76:2403-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson, S. C., M. Rollence, J. Marshall-Neff, and A. McClelland. 1997. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J. Virol. 71:4782-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson, S. C., M. Rollence, B. White, L. Weaver, and A. McClelland. 1995. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J. Virol. 69:2850-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suomalainen, M., M. Y. Nakano, K. Boucke, S. Keller, R. P. Stidwill, and U. F. Greber. 1999. Microtubule-dependent minus and plus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 144:657-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trotman, L. C., N. Mosberger, M. Fornerod, R. P. Stidwill, and U. F. Greber. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3:1092-1100. [DOI] [PubMed] [Google Scholar]
- 46.Vogels, R., D. Zuijdgeest, R. van Rijnsoever, E. Hartkoorn, I. Damen, M. P. de Bethune, S. Kostense, G. Penders, N. Helmus, W. Koudstaal, M. Cecchini, A. Wetterwald, M. Sprangers, A. Lemckert, O. Ophorst, B. Koel, M. van Meerendonk, P. Quax, L. Panitti, J. Grimbergen, A. Bout, J. Goudsmit, and M. Havenga. 2003. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 77:8263-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wadell, G. 1994. Adenoviruses, p. 1-7. In R. G. Webster and A. Granoff (ed.), Encyclopedia of virology, vol. 1. Academic Press, Inc., New York, N.Y.
- 48.Wadell, G. 2000. Adenoviruses, p. 307-327. In A. J. Zuckerman, J. E. Banatvala, and J. R. Pattison (ed.), Principles and practice of clinical virology, 4th ed. John Wiley & Sons, Inc., New York, N.Y.
- 49.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]