Abstract
The t(5;17)(q35;q21) APL variant results in expression of a fusion protein linking the N-terminus of nucleophosmin (NPM) to the C-terminus of the retinoic acid receptor alpha (RAR). We have previously shown that NPM-RAR is capable of binding to DNA either as a homodimer or heterodimer with RXR. To determine the biological significance of NPM-RAR/RXR interaction, we developed two mutants of NPM-RAR that showed markedly diminished ability to bind RXR. U937 subclones expressing the NPM-RAR mutants showed significantly less inhibition of vitamin D3/TGFbeta-induced differentiation, compared with NPM-RAR. These results support the hypothesis that RXR interaction is necessary for NPM-RAR-mediated myeloid maturation arrest.
Keywords: acute promyelocytic leukemia, NPM-RAR, RXR, differentiation
INTRODUCTION
Acute promyelocytic leukemia (APL) is characterized by a differentiation blockade at the promyelocytic stage of myeloid maturation [1]. The majority of patients with APL express t(15;17), which juxtaposes in the same reading frame the genes encoding the PML tumor suppressor protein and the retinoic acid receptor alpha (RARA)[2, 3]. RARA is a member of the nuclear hormone receptor family, and binds to consensus DNA sequences (know as the retinoic acid response element, or RARE) in the promoters of target genes. Wild-type RARA preferentially binds to RAREs as a heterodimer with one of the members of the retinoid X receptor family (RXR alpha, beta, gamma). In the absence of ligand, RARA/RXR heterodimers bind to promoters and recruit a corepressor complex containing a histone deacetylase to silence expression of target genes[4, 5]. Ligand binding induces a conformational change in RARA, which disallows interaction with the corepressor and instead reveals the binding domain for a histone acetyl transferase-containing coactivator complex, to initiate transcription of the target genes[1].
The PML-RAR fusion contains the C-terminal 503 amino acid residues of RARA[2, 3], which in RARA encode the DNA binding, RXR interaction, ligand binding, and corepressor and coactivator interaction domains. PML-RAR exhibits high affinity for the corepressor proteins NCoR and SMRT, and in the presence of physiologic levels of its ligand, retinoic acid, fails to release corepressor complexes [6-8]. Potential target genes involved in myeloid differentiation remain repressed, contributing to the leukemic phenotype. Introduction of pharmacologic doses of all-trans retinoic acid (ATRA) induces co-repressor release and co-activator recruitment, as well as degradation of PML-RAR [9], to allow re-expression of the repressed genes. Despite characterization of the APL transcriptosome [10, 11], it remains unclear which target genes are critical to development of the APL phenotype. Indeed, it remains controversial whether the transcriptional targets of PML-RAR are limited to RARA-responsive genes, or whether PML-RAR binds to and regulates promoters other than those that are recognized by RARA, to manifest transcriptional promiscuity[12]. PML-RAR contains the dimerization domains of PML[2, 3], as well as the RXR-binding domain of RARA, and so could potentially form homodimers, or heterodimers with either unrearranged PML or RXR or possibly both. In vitro, PML-RAR homodimers preferentially recognize different RARE DNA sequences than PML-RAR/RXR heterodimers[12-15]. Based on results obtained with artificial constructs that fuse RARA to heterologous dimerization domains, it has been suggested that homodimerization of RARA is sufficient for creation of the APL phenotype[16-20]. Yet, there is also extensive data to support the model that interaction with RXR is essential for the APL phenotype[21-24]. Indeed, ChIP-seq experiments performed by Martens et al. [15] indicate that PML-RAR and RXR colocalize on target promoters.
In an effort to identify the key molecular events that underlie APL, we have been studying the variant translocations found in rare cases of APL. These represent leukemias with similar phenotype, but different genotype, and as such represent “experiments of nature” with which to test mechanistic hypotheses[25]. To date seven variant translocations have been characterized on a molecular basis: t(11;17)q(23;q21) [26] which fuses the PLZF transcriptional repressor to the same C-terminal sequences of RARA as are expressed in PML-RAR; t(5;17)(q35;q21) that joins nucleophosmin (NPM) to RARA[27]; t(11;17)(q13;q21) that fuses NUMA to RARA[28]; STAT5b-RARA[29] described in a patient with der 17; FIP1L1-RAR expressed in t(4;17)((q12;q21) [30]; fusion of RARA with the regulatory subunit of the cyclic adenosine monophosphate dependent protein kinase PRKAR1A on 17q24 [31]; and t(X;17)(p11;q12) which fuses BCOR to RARA[32]. We have focused our studies on t(5;17) NPM-RAR, which is the second most common of the variants, and which responds to the differentiating effect of ATRA[33], similar to t(15;17) APL.
NPM is a nucleolar phosphoprotein that plays a role in regulation of a myriad of cellular functions. It binds nascent ribosomal particles, modulates centrosomal function, and serves as a chaperonin for Rb, p53, c-myc, and a plethora of other proteins (reviewed in [34]). Aside from APL, it is mutated in other hematologic malignancies: as the translocation partner with MLF1 in t(3;5) myelodysplastic syndrome and AML[35]; as the translocation partner of ALK in t(2;5) anaplastic lymphoma[36]; and in non-APL AML as a series of C-terminal mutations which alter its nuclear localization signal[37]. Interestingly, clinical data suggests that expression of the C-terminal mutation may be associated with increased responsiveness to ATRA plus chemotherapy in non-APL AML[38]: whether the mutated NPM is mechanistically involved in the alteration of chemotherapy-sensitivity of these patients has yet to be fully investigated.
In this manuscript we investigate the functional significance of RXR interaction with NPM-RAR towards NPM-RAR-induced myeloid maturation arrest. We have previously shown that NPM-RAR can bind to RXR, and interact with DNA either as a homodimer or a heterodimer with RXR, though NPM-RAR /RXR heterodimers have greater affinity for RAREs[27, 39]. We now show that heterodimer formation with RXR is necessary for NPM-RAR mediated myeloid maturation arrest.
MATERIALS AND METHODS
Plasmids
mRXRb cDNA was a gift of Peirre Chambon. The cloning of NPM-RAR from a cDNA library created from t(5;17) leukemic blasts was described previously[27]. Del20-RARA and D256A-RARA were gifts of R. Koenig. To generate Del20-NPM-RAR and D314A-NPM-RAR, the Sst1-BamH1 fragments encoding the mutated sequences of Del20-RARA and D256A-RARA were subcloned into the corresponding Sst1-BamH1 sites of NPM-RAR. The B19pLTN1 expression vector has previously been described[39]. pIH902, encoding the maltose binding protein, was obtained from NEB (Ipswitch, MA).
Cell lines
COS7 cells were obtained from the ATCC, and grown in DMEM with 10% Fetal Bovine Serum (Invitrogen, Carlsbad, CA), 2mM glutamine, 100 U/ml penicillin, and 100 microg/ml streptomycin. COS cells were transiently transfected with RARA-B19pLTN1 or NPM-RARB19pLTN1 expression plasmids by CaPO4 precipitation, as described[39]. U937 cells were obtained from the ATCC, and grown in RPMI 1640 supplemented with 10% Fetal Bovine Serum (Invitrogen, Carlsbad, CA), 2mM glutamine, 100 U/ml penicillin, and 100 microg/ml streptomycin. Cells were transfected using a BioRad Electroporator (BioRad, Hercules CA) in sterile cuvettes containing 5 ×106 cells with 20 micrograms DNA at 250mV, 960micoF in 200 microliters Optimem (Invitrogen). After electroporation, cells were allowed to rest in nonselective medium for two days before plating at limiting dilution in medium containing 1 mg/ml G418(Invitrogen).
Differentiation
U937 cells were cultured in Teflon bottles (VWR, Bridgeport NJ) in medium containing 250 ng/mlVitamin D3 (Enzo Life Sciences, Farmingdale, NY) and 1 ng/ml TGFbeta (Genzyme, Cambridge, MA) for five days. Cells were incubated with FITC-conjugated monoclonal antibody targeting CD14 or FcR1 (Beckman Coulter, Fullerton CA). Flow cytometry was performed by the UPCI Flow Cytometry Facility using a Coulter Epics XL Flow Cytometer (Beckman Coulter, Fullerton CA). Morphological analysis was performed by staining cytospin preparations of the cells with Hema 3 Stain (Fisher Scientific, PA). Photomicrographs were taken through an Olympus microscope with a 60x oil-immersion objective lens (using the software SPOT 5.1).
NBT staining was performed by mixing cell suspensions with an equal volume of a solution containing nitroblue tetrazolium (2 mg/mL), bovine albumin (17 mg/mL), and TPA (2 μg/mL) at 37°C for 30 min. Following incubation, the medium was discarded and formazan deposits were dissolved by adding 100 μl of dimethyl sulfoxide (DMSO). Optical density was measured at 570 nm.
Co-precipitation
cDNA for NPM-RAR or the mutants was subcloned into the vector PIH902 in the appropriate reading frame to create a fusion protein with Maltose Binding Protein (MBP). MBP-fusion proteins were purified per the manufacturer’s protocol (NEB). 35S-mRXRbeta was generated by in vitro translation (TNT Coupled Reticulocyte Lysate System, Promega, Madison, WI) in the presence of 35S-methionine and 35S-cysteine (GE, Piscataway, NJ). For association studies, 30 micrograms of each MBP-fusion protein and 20 microliters of the RXR in vitro translation products were incubated for 4 h at 4°C in 20mM Tris pH 8, 110mM NaCl, 1mM EDTA, and 0.5% NP-40, and then allowed to bind to amylose resin. After extensive washing with the incubation buffer, the resin was boiled with SDS loading buffer and the eluted products separated by SDS-PAGE. The gels were dried and the radiolabeled proteins imaged by autoradiography.
Southern blots
Southern blotting was performed using standard techniques[27], digesting genomic DNA with EcoR1 and probing with a 32P-radiolabled Neo probe corresponding to the Xho1/Sal1 fragment of pMCI Neo.
Immunoblots
Western blotting was performed as previously described[27]. Immunoblots were probed with a polyclonal rabbit anti-RARA antibody (Santa Cruz Biotechnology, Santa Cruz, CA) recognizing the C-terminus of RARA conserved in NPM-RAR and the mutant constructs.
RESULTS
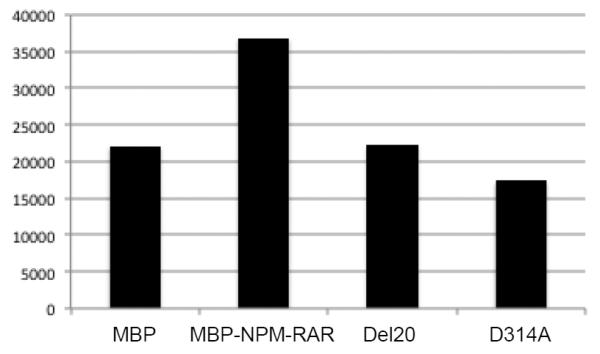
We have previously demonstrated that NPM-RAR binds to RXR in both co-precipitation and DNA binding–electrophoretic mobility shift assays (EMSA)[39]. In order to determine the biologic significance of NPM-RAR/RXR interaction, we engineered two mutants of NPM-RAR based upon prior work of Rosen et al. [40], who had shown that mutants harboring either a point mutation of RARA at residue 256 (changing aspartate to alanine), or a deletion of 20 amino acids in the beginning of the E domain of RARA (amino acids 242-261), decreased RARA affinity for RXR. We generated corresponding mutations in the RARA domains of NPM-RAR - deletion of amino acid residues 300-319 (del20), or point mutation of amino acid 314 changing aspartate to alanine (D314A) (Figure 1). To confirm that the NPM-RAR mutations similarly impeded NPM-RAR interaction with RXR, we incubated radiolabeled RXR with MBP-NPM-RAR or the MBP-mutant, and captured the complexes with amylose-resin. After extensive washing, the complexes were separated by SDS-PAGE (Figure 2A). We found that MBP-NPM-RAR binds to RXR, consistent with our previously published findings. Consistent with Rosen’s observations with the analogous RARA mutants, we observed decreased affinity of the del20 and D314A-NPM-RAR mutants for RXR, equivalent to background levels seen with incubation of radiolabeled RXR with control MBP vector alone (Figures 2A and 2B).
Figure 1.
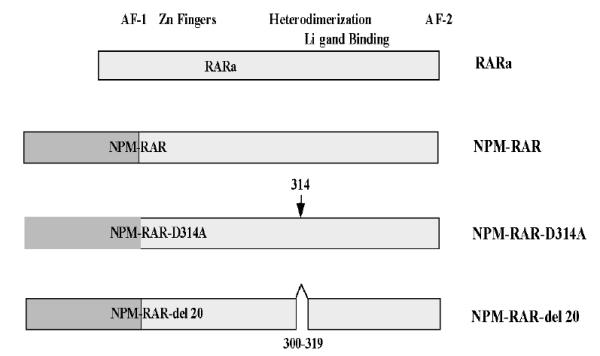
Cartoon illustrating the sites of the NPM-RAR-D314A and -Del20 mutations.
Figure 2. D314A and Del20 mutations decrease NPM-RAR binding with RXR.
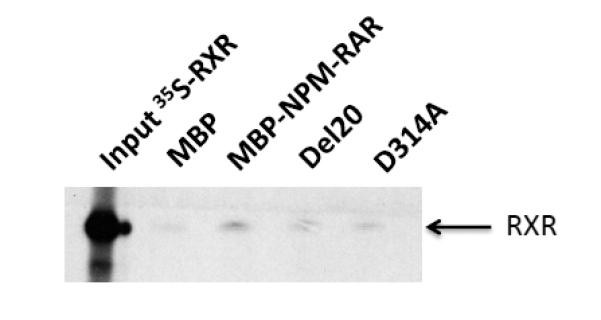
A. Reticulocyte lysates containing 35S-RXR were incubated with MBP, MBP-NPM-RAR, NPM-NPM-RAR-D314A or MBP-NPM-RAR-del20. Complexes were captured on amylose resin, and separated on 8% SDS-PAGE before autoradiography.
B. Densitometric quantitation of the signal intensity of the bands in Figure 2A (arbitrary units).
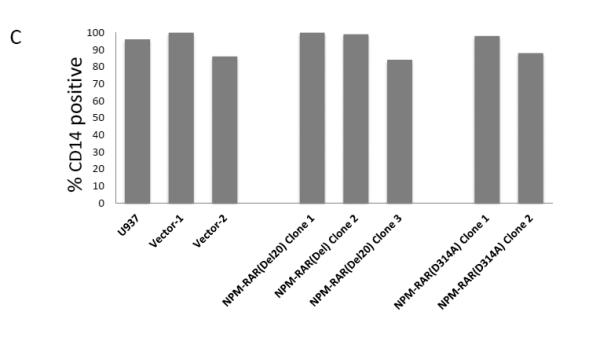
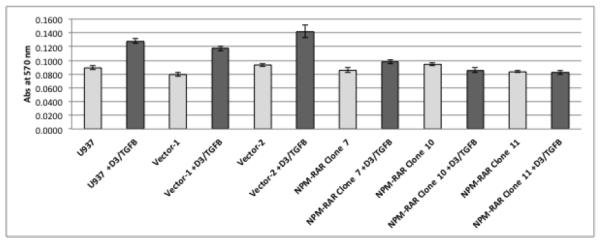
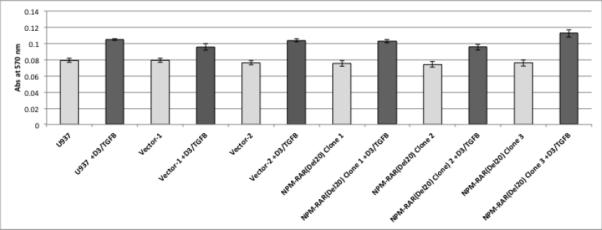
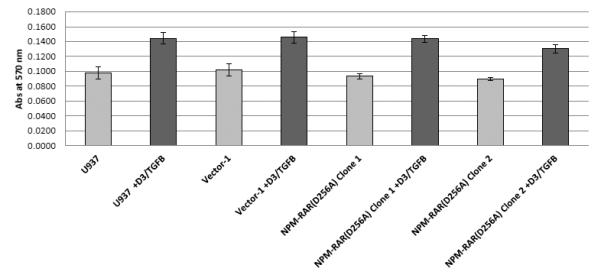
To assess the contribution of NPM-RAR/RXR interaction towards NPM-RAR mediated differentiation blockade, we expressed NPM-RAR, or the D314A or del20 mutants of NPM-RAR, in U937 cells. U937 cells represent a human monoblastic leukemic cell line capable of differentiation into mature monocytes and macrophages by Vitamin D3 and TGFbeta. Independent clones were identified by Southern blot analysis of integration sites (not shown), and expression of appropriately sized proteins confirmed by immunoblotting (Figures 3A and 4A and B). Differentiation of the U937 clones was assessed by flow cytometric analysis for acquisition of the CD14 differentiation marker after 5 days of culture in medium containing 250 ng/ml Vitamin D3 and 1 ng/ml TGFbeta. As previously reported[41], we found that expression of NPM-RAR retarded differentiation into mature monocytes after five days of stimulation with Vitamin D and TGFß (Figure 3B) (p=0.0017). The degree of suppression compared to control varied amongst clones from 50% to 90%. Retardation of differentiation was also confirmed by morphologic analysis (Figure 5A) and NBT staining (Figure 6A), as well as flow cytometric acquisition of the differentiation markers CD11b and FcR1 (not shown). In contrast, clones expressing the D314A or del20 mutants of NPM-RAR, deficient in the ability to bind RXR, did not manifest a differentiation blockade, and showed a differentiation response similar to vector transfected control clones or U937 parental cells (Figures 4C, 5B and C, and 6B and C).
Figure 3. NPM-RAR blocks U937 differentiation.
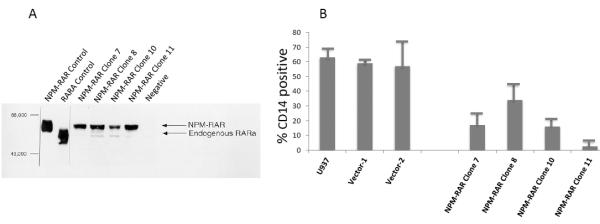
A. Immunoblot of lysates of four independent stably-transfected U937 NPM-RAR clones and vector transfected clone. Control lanes represent lysate of COS cells transiently transfected with NPM-RAR or RARA. Immunbolot was probed with an anti-RARA antibody directed against the C-terminus of RARA.
B. Parental U937, and independent clones stably expressing NPM-RAR, or empty vector, were grown in medium containing 250 ng/ml Vitamin D3 and 1 ng/ml TGFbeta for 5 days. Expression of the CD14 differentiation marker was assessed by flow cytometry. Two-tailed t-test comparing the control versus NPM-RAR clones resulted in a p value of 0.0017.
Figure 4. NPM-RAR-D314A and del20 mutants do not disrupt U937 differentiation.
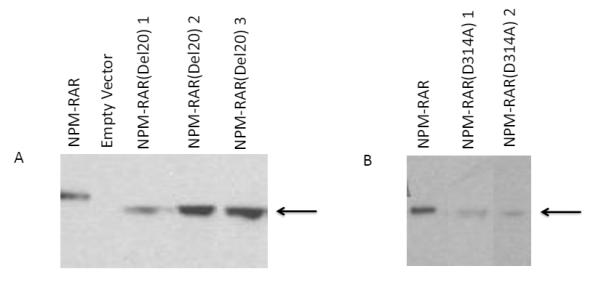
A. Immunoblot of lysates of NPM-RAR-del20 expressing U937 clones, probed with an anti-RARA antibody directed against the C-terminus of RARA.
B. Immunoblot of lysates of NPM-RAR-D314A expressing U937 clones, probed with an anti-RARA antibody directed against the C-terminus of RARA.
C. Parental U937 and clones stably expressing NPM-RAR-D314A, NPM-RAR-del20, or empty vector, were grown in medium containing 250 ng/ml Vitamin D3 and 1 ng/ml TGFbeta for 5 days. Expression of the CD14 differentiation marker was assessed by flow cytometry.
Figure 5.
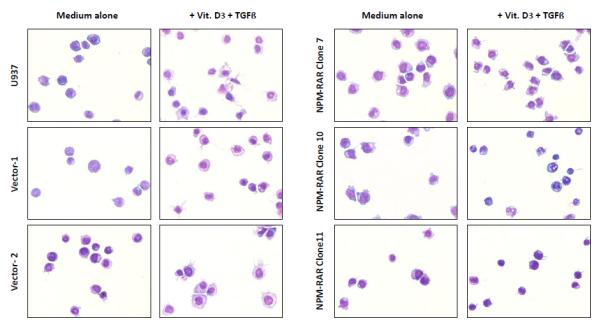
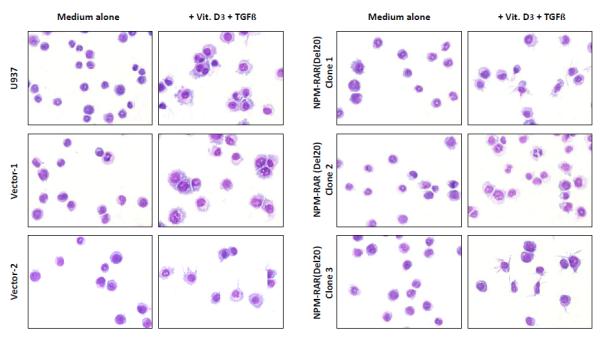
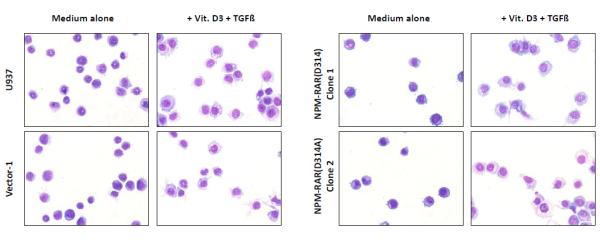
Morphologic analysis of differentiation of U937, Vector transfected, NPM-RAR, NPM-RAR(Del20), and NPM-RAR(D314A) clones. Cells were grown in medium with or without 250 ng/ml Vitamin D3 and 1 ng/ml TGFbeta for 5 days. Cells were applied to glass slides by cytopsin, and stained with Fisher Hema Stain kit. A. NPM-RAR B. NPM-RAR(Del20) C. NPM-RAR(D314A)
Figure 6.
NBT Staining. Cells were grown in medium with or without 250 ng/ml Vitamin D3 and 1 ng/ml TGFbeta for 5 days, and acquisition of oxidases assessed by NBT assay. Each data point represents the mean of one experiment done in triplicate +/− SD; each experiment was performed twice, with similar results. A. NPM-RAR B. NPM-RAR(Del20) C. NPM-RAR(D314A)
DISCUSSION
NPM-RAR represents an important model with which to test proposed mechanisms underlying APL leukemic transformation. NPM-RAR represents the second most common variant of APL[25]. We have previously shown that ectopic expression of NPM-RAR in mice leads to leukemic transformation[42]. Like t(15;17) APL, t(5;17) APL exhibits ATRA-sensitivity[33]. In this manuscript we have shown that NPM-RAR mutants that weakly bind RXR fail to impede Vitamin D3/TGFb-induced differentiation of U937 cells. Our data indicates that binding to RXR is necessary for the NPM-RAR-mediated differentiation blockade.
The U937 model that we employed for these studies has been widely used as a model system for study of the effects of leukemic fusion proteins, such PML-RAR[43], in myeloid differentiation. It should be pointed out that U937 cells differentiate along the monocytic lineage, and thus do not represent an optimal model for APL differentiation arrest. Indeed, as hematopoietic cells differentiate into granulocytes their expression of RXR decreases [44], as opposed to monocytic lineages. We do not feel that these differences alter the interpretation of our results.
The D314A and del20 mutations are outside the “CoR box” domain that binds corepressors and coactivators, and are outside the Zn-finger DNA binding domain of NPM-RAR. Analogous mutations in wild-type RARA described by Rosen et al., which served as the model for these constructs, showed minimal effects on other functional domains of RARA, and did not interfere with DNA interaction nor ligand binding[40].
Our observations indicating the dependency on RXR for NPM-RAR function would be consistent with either of two models: first, that NPM-RAR binds RAREs as a heterodimer with RXR, or second, that NPM-RAR sequesters RXR, and deprives RXR from interacting with other binding partners. RXR forms heterodimers with Vitamin D receptor, thyroid hormone receptor, and peroxisome proliferated-activated receptors, amongst others, which themselves activate diverse transcriptional pathways. Intranuclear localization of RXR is altered in cells expressing APL fusion proteins[45], and Dong has shown that APL fusion protein expression decreases the mobility of RXR[46], consistent with a sequestration model. Taken together with our previously published EMSA results which indicate that RXR increases the affinity of NPM-RAR for DNA[39], our current data would favor a model in which RXR is necessary for appropriate NPM-RAR DNA interaction.
Our work is consistent with observations in models using other RAR fusion proteins which suggest that heterodimerization with RXR is essential for myeloid differentiation blockade. Martens et al. [15] used ChIP-seq to demonstrate that PML-RAR and RXR colocalize to target promoters. Zeisig et al., using an shRNA approach to downmodulate RXR expression, showed that RXR expression is necessary for colony formation by PML-RAR-expressing hematopoietic progenitors[22]. Similarly, Sukhai showed that expression of a mutated RXR abrogated APL-fusion protein leukemia[47]. Zhu demonstrated that PML-RAR mutants incapable of interacting with RXR fail to induce leukemia[23], and Qiu found similar dependence of the PRKAR1A-RAR fusion for RXR[24]. Our observations extend these findings using the APL variant NPM-RAR, and suggests that NPM-RAR/RXR interaction is necessary for the myeloid differentiation blockade that characterizes APL.
Whether the DNA-binding interactions of APL fusion proteins is mediated through RAR-fusion homodimers or RAR-fusion/RXR heterodimers remains controversial, and several groups have shown that forced homodimerization of RARA can lead to leukemic transformation[16-18]. Indeed, the N-terminal sequences within NPM-RAR have been shown to mediate dimerization[48], and we have previously demonstrated that NPM-RAR can form homodimers[39]. Zeisig[22] has proposed a model that might reconcile the homodimer-heterodimer controversy, suggesting that higher-order homotetrameric RARA complexes recruit RXR. Such a model could also be applicable to the case of NPM-RAR.
ACKNOWLEDGEMENTS
The authors would like to thank Drs. Keonig and Chambon for the kind gifts of reagents, and Anuja Chattopadhyay for helpful discussions.
FUNDING: Supported by NIH R01 CA67346 and P30 CA047904
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST None
AUTHORS’ CONTRIBUTIONS: EAR, SLP, and IA performed the experiments and edited the manuscript. RLR designed the experiments and wrote the manuscript.
REFERENCES
- 1.Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–215. [PubMed] [Google Scholar]
- 2.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–84. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 3.Kakizuka A, Miller WH, Jr., Umesono K, Warrell RP, Jr., Frankel SR, Murty VV, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–74. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 4.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 5.Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, et al. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–80. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 6.Guidez F, Ivins S, Zhu J, Soderstrom M, Waxman S, Zelent A. Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARalpha underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood. 1998;91:2634–42. [PubMed] [Google Scholar]
- 7.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–8. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 8.Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr., Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–4. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 9.Licht JD. Acute promyelocytic leukemia--weapons of mass differentiation. N Engl J Med. 2009;360:928–30. doi: 10.1056/NEJMcibr0810371. [DOI] [PubMed] [Google Scholar]
- 10.Yuan W, Payton JE, Holt MS, Link DC, Watson MA, DiPersio JF, et al. Commonly dysregulated genes in murine APL cells. Blood. 2007;109:961–70. doi: 10.1182/blood-2006-07-036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoemme C, Peerzada A, Behre G, Wang Y, McClelland M, Nieselt K, et al. Chromatin modifications induced by PML-RARalpha repress critical targets in leukemogenesis as analyzed by ChIP-Chip. Blood. 2008;111:2887–95. doi: 10.1182/blood-2007-03-079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauksdottir H, Privalsky ML. DNA recognition by the aberrant retinoic acid receptors implicated in human acute promyelocytic leukemia. Cell Growth Differ. 2001;12:85–98. [PMC free article] [PubMed] [Google Scholar]
- 13.Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P. PMLRAR homodimers: distinct DNA binding properties and heteromeric interactions with RXR. Embo J. 1993;12:3171–82. doi: 10.1002/j.1460-2075.1993.tb05986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segalla S, Rinaldi L, Kilstrup-Nielsen C, Badaracco G, Minucci S, Pelicci PG, et al. Retinoic acid receptor alpha fusion to PML affects its transcriptional and chromatin-remodeling properties. Mol Cell Biol. 2003;23:8795–808. doi: 10.1128/MCB.23.23.8795-8808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martens JH, Brinkman AB, Simmer F, Francoijs KJ, Nebbioso A, Ferrara F, et al. PML-RARalpha/RXR Alters the Epigenetic Landscape in Acute Promyelocytic Leukemia. Cancer Cell. 2010;17:173–85. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 16.Lin RJ, Evans RM. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol Cell. 2000;5:821–30. doi: 10.1016/s1097-2765(00)80322-6. [DOI] [PubMed] [Google Scholar]
- 17.Minucci S, Maccarana M, Cioce M, De Luca P, Gelmetti V, Segalla S, et al. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol Cell. 2000;5:811–20. doi: 10.1016/s1097-2765(00)80321-4. [DOI] [PubMed] [Google Scholar]
- 18.Kwok C, Zeisig BB, Dong S, So CW. Forced homo-oligomerization of RARalpha leads to transformation of primary hematopoietic cells. Cancer Cell. 2006;9:95–108. doi: 10.1016/j.ccr.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Peres L, Honore N, Nasr R, Zhu J, de The H. Dimerization-induced corepressor binding and relaxed DNA-binding specificity are critical for PML/RARA-induced immortalization. Proc Natl Acad Sci U S A. 2006;103:9238–43. doi: 10.1073/pnas.0603324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Occhionorelli M, Santoro F, Pallavicini I, Gruszka A, Moretti S, Bossi D, et al. The self-association coiled-coil domain of PML is sufficient for the oncogenic conversion of the retinoic acid receptor (RAR) alpha. Leukemia. 2011;25:814–20. doi: 10.1038/leu.2011.18. [DOI] [PubMed] [Google Scholar]
- 21.Kamashev D, Vitoux D, De The H. PML-RARA-RXR oligomers mediate retinoid and rexinoid/cAMP cross-talk in acute promyelocytic leukemia cell differentiation. J Exp Med. 2004;199:1163–74. doi: 10.1084/jem.20032226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeisig BB, Kwok C, Zelent A, Shankaranarayanan P, Gronemeyer H, Dong S, et al. Recruitment of RXR by homotetrameric RARalpha fusion proteins is essential for transformation. Cancer Cell. 2007;12:36–51. doi: 10.1016/j.ccr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, Nasr R, Peres L, Riaucoux-Lormiere F, Honore N, Berthier C, et al. RXR is an essential component of the oncogenic PML/RARA complex in vivo. Cancer Cell. 2007;12:23–35. doi: 10.1016/j.ccr.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Qiu JJ, Lu X, Zeisig BB, Ma Z, Cai X, Chen S, et al. Leukemic transformation by the APL fusion protein PRKAR1A-RAR{alpha} critically depends on recruitment of RXR{alpha} Blood. 2010;115:643–52. doi: 10.1182/blood-2009-07-232652. [DOI] [PubMed] [Google Scholar]
- 25.Redner RL. Variations on a theme: the alternate translocations in APL. Leukemia. 2002;16:1927–32. doi: 10.1038/sj.leu.2402720. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, et al. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. Embo J. 1993;12:1161–7. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redner RL, Rush EA, Faas S, Rudert WA, Corey SJ. The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion. Blood. 1996;87:882–6. [PubMed] [Google Scholar]
- 28.Wells RA, Catzavelos C, Kamel-Reid S. Fusion of retinoic acid receptor alpha to NuMA, the nuclear mitotic apparatus protein, by a variant translocation in acute promyelocytic leukaemia. Nat Genet. 1997;17:109–13. doi: 10.1038/ng0997-109. [DOI] [PubMed] [Google Scholar]
- 29.Arnould C, Philippe C, Bourdon V, Gr goire MJ, Berger R, Jonveaux P. The signal transducer and activator of transcription STAT5b gene is a new partner of retinoic acid receptor alpha in acute promyelocytic-like leukaemia. Hum Mol Genet. 1999;8:1741–9. doi: 10.1093/hmg/8.9.1741. [DOI] [PubMed] [Google Scholar]
- 30.Kondo T, Mori A, Darmanin S, Hashino S, Tanaka J, Asaka M. The seventh pathogenic fusion gene FIP1L1-RARA was isolated from a t(4;17)-positive acute promyelocytic leukemia. Haematologica. 2008;93:1414–6. doi: 10.3324/haematol.12854. [DOI] [PubMed] [Google Scholar]
- 31.Catalano A, Dawson MA, Somana K, Opat S, Schwarer A, Campbell LJ, et al. The PRKAR1A gene is fused to RARA in a new variant acute promyelocytic leukemia. Blood. 2007;110:4073–6. doi: 10.1182/blood-2007-06-095554. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto Y, Tsuzuki S, Tsuzuki M, Handa K, Inaguma Y, Emi N. BCOR as a novel fusion partner of retinoic acid receptor alpha in a t(X;17)(p11;q12) variant of acute promyelocytic leukemia. Blood. 2010;116:4274–83. doi: 10.1182/blood-2010-01-264432. [DOI] [PubMed] [Google Scholar]
- 33.Redner RL, Corey SJ, Rush EA. Differentiation of t(5;17) variant acute promyelocytic leukemic blasts by all-trans retinoic acid. Leukemia. 1997;11:1014–6. doi: 10.1038/sj.leu.2400661. [DOI] [PubMed] [Google Scholar]
- 34.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 35.Yoneda-Kato N, Look AT, Kirstein MN, Valentine MB, Raimondi SC, Cohen KJ, et al. The t(3;5)(q25.1;q34) of myelodysplastic syndrome and acute myeloid leukemia produces a novel fusion gene, NPM-MLF1. Oncogene. 1996;12:265–75. [PubMed] [Google Scholar]
- 36.Morris SW, Kirstein MN, Valentine MB, Dittmer K, Shapiro DN, Look AT, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1995;267:316–7. doi: 10.1126/science.267.5196.316-b. [DOI] [PubMed] [Google Scholar]
- 37.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 38.Schlenk RF, Dohner K, Kneba M, Gotze K, Hartmann F, Del Valle F, et al. Gene mutations and response to treatment with all-trans retinoic acid in elderly patients with acute myeloid leukemia. Results from the AMLSG Trial AML HD98B. Haematologica. 2009;94:54–60. doi: 10.3324/haematol.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redner RL, Chen JD, Rush EA, Li H, Pollock SL. The t(5;17) acute promyelocytic leukemia fusion protein NPM-RAR interacts with co-repressor and co-activator proteins and exhibits both positive and negative transcriptional properties. Blood. 2000;95:2683–90. [PubMed] [Google Scholar]
- 40.Rosen ED, Beninghof EG, Koenig RJ. Dimerization interfaces of thyroid hormone, retinoic acid, vitamin D, and retinoid X receptors. J Biol Chem. 1993;268:11534–41. [PubMed] [Google Scholar]
- 41.Kamel-Reid S, Zhang T, Wells RA. Expression of NPM-RARalpha fusion gene in hematopoietic cells confers sensitivity to troglitazone-induced apoptosis. Oncogene. 2003;22:6424–35. doi: 10.1038/sj.onc.1206696. [DOI] [PubMed] [Google Scholar]
- 42.Rego EM, Ruggero D, Tribioli C, Cattoretti G, Kogan S, Redner RL, et al. Leukemia with distinct phenotypes in transgenic mice expressing PML/RAR alpha, PLZF/RAR alpha or NPM/RAR alpha. Oncogene. 2006;25:1974–9. doi: 10.1038/sj.onc.1209216. [DOI] [PubMed] [Google Scholar]
- 43.Grignani F, Ferrucci PF, Testa U, Talamo G, Fagioli M, Alcalay M, et al. The acute promyelocytic leukemia-specific PML-RAR alpha fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell. 1993;74:423–31. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- 44.Taschner S, Koesters C, Platzer B, Jorgl A, Ellmeier W, Benesch T, et al. Down-regulation of RXRalpha expression is essential for neutrophil development from granulocyte/monocyte progenitors. Blood. 2007;109:971–9. doi: 10.1182/blood-2006-04-020552. [DOI] [PubMed] [Google Scholar]
- 45.Dyck JA, Maul GG, Miller WH, Jr., Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–43. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 46.Dong S, Stenoien DL, Qiu J, Mancini MA, Tweardy DJ. Reduced intranuclear mobility of APL fusion proteins accompanies their mislocalization and results in sequestration and decreased mobility of retinoid X receptor alpha. Mol Cell Biol. 2004;24:4465–75. doi: 10.1128/MCB.24.10.4465-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sukhai MA, Thomas M, Xuan Y, Chan LS, Hamadanizadeh SA, Zhang T, et al. Evidence of functional interaction between NuMA-RARalpha and RXRalpha in an in vivo model of acute promyelocytic leukemia. Oncogene. 2008;27:4666–77. doi: 10.1038/onc.2008.106. [DOI] [PubMed] [Google Scholar]
- 48.Liu QR, Chan PK. Formation of nucleophosmin/B23 oligomers requires both the amino- and the carboxyl-terminal domains of the protein. Eur J Biochem. 1991;200:715–21. doi: 10.1111/j.1432-1033.1991.tb16236.x. [DOI] [PubMed] [Google Scholar]


