Abstract
Electron transfer in cytochrome P450 enzymes is a fundamental process for activity. It is difficult to measure electron transfer in these enzymes because under the conditions typically used they exist in a variety of states. Using nanotechnology based techniques, gold conducting nanopillars were constructed in an indexed array. To each of these nanopillars the P450 enzyme CYP2C9 was attached and conductivity measurements made using conducting probe atomic force microscopy under constant force conditions. The conductivity measurements were made on CYP2C9 alone and with bound substrates, a bound substrate-effector pair, and a bound inhibitor. Fitting of the data with the Poole-Frankel model indicates a correlation between the barrier height for electron transfer and the ease of CYP2C9-mediated metabolism of the bound substrates though the spin-state of iron is not well correlated. The approach described here should have broad application to the measurement of electron transfer in P450 enzymes and other metalloenzymes.
Keywords: Cytochrome P450, Electron Transfer, Conducting Atomic Force Microscopy, Self-Assembled Monolayer
Electron transfer (ET) in cytochrome P450 (P450) enzymes has been extensively studied because of its importance in metabolic processes,1,2 respiratory chains,3–5 and other possible P450 based applications.6–11 Current dogma suggests that substrate binding may serve as a switch and facilitates ET in P450s so that the catalytic cycle can be initiated.1,2,12,13 A corollary to this theory is that the ease of substrate metabolism is proportional to the ease of ET.14–17 Several approaches have been used to measure ET in P450 enzymes including electrochemical-based and other methods18,19 but no definitive approach has been devised for monitoring ET in P450s and therefore limited conclusions have been made regarding a correlation between ET and metabolism.
Substrate binding has often been observed to alter the spin-state of iron in P450s due to changes in its coordination and hence local geometry about iron. Spin-state can be easily determined by absorption spectroscopy and efforts have been made to correlate spin-state and ease of metabolism.12,20,21 For example, many P450 substrates cause the spin-state to shift from low to high spin upon substrate binding (Type I binders). In contrast, many nitrogen containing inhibitors have little effect on spin-state (Type II binders22). This supports the proposal that spin-state and ET may be correlated.1,17 However, there are many substrates that do not cause spin-state changes and, therefore, changes in ET are not necessarily directly correlated to spin-state changes.
Other factors, such as conformational changes,23,24 that may occur upon substrate binding or when the co-enzyme cytochrome P450 reductase binds may also alter the ET process.25–27 Furthermore, in solution P450s form aggregates that may affect ET. Unfortunately, efforts to minimize or eliminate aggregates (e.g., the addition of detergents, salts, and lipids) can alter ET14,28 and therefore aggregates are particularly problematic for solution-based ET studies.
A system that is designed to measure ET on a single enzyme while simultaneously minimizing the effects of aggregation may avoid many of the problems that have heretofore been encountered. In one reported approach, the substrate was bonded to an electrode and a P450 was added and allowed to bind to the substrate. This method may have reduced protein-protein interactions, however, it required the substrate to be modified so that it could be bonded to an electrode via a tether.29 An alternative is to bond the P450 to a planar gold electrode and allow the substrate to bind to the P450, thereby avoiding modification of the substrate.30 Neither of these approaches permits the measurement of ET effects on a single enzyme; rather, they measure ET for a large ensemble.
Nanoscale methods have been used to measure ET on single molecules.31 Scanning probe microscopy,32 mechanical break junctions,33 nanogap electrode techniques,34 and conducting probe atomic force microscopy (CAFM) have all been applied to small molecules and proteins.35 However, in the case of proteins, the state of the protein under study is largely unknown and it may be present as a monomer, oligomer, or some mixture of both. Furthermore, these approaches are not designed to monitor a single protein molecule, to permit variation of the substrate, or to alter other environmental variables. Thus, our goal is to create a platform that will allow ET to be studied on a single P450 in an arrangement that permits examination of the effect of various substrates, co-enzymes, or other changes on the same enzyme molecule with simultaneous monitoring of ET.
Our approach to isolating a P450 and measuring ET is to fabricate a regular array of gold nanopillars and then selectively attach the P450 enzyme to the nanopillar. Nanopillars with lateral dimensions on the order of the diameter of the enzyme likely result in each nanopillar bearing one enzyme. ET can then be measured by completing the circuit with a conducting probe atomic force microscopy tip. To this end we fabricated an array of nanopillars, in the range of 20–40 nm (lateral size), using electron-beam lithography on a doped silicon substrate. Similar arrays have been fabricated by other methods.36,37 The array (21 × 21 elements, periodicity 250 nm) was indexed so that a specific nanopillar could be located, probed, and revisited as needed. Selective attachment of the enzyme to the nanopillar was achieved by attachment of a thiol-based self-assembled monolayer (SAM) that contained an ω-thio carboxylic acid to which the P450 was bonded via the N-terminus of the P450.38 Since thiol groups bond to gold and not silicon when applied in solution,39 the SAM forms only on the gold nanopillars and not on the surrounding silicon surface. Also, we have previously shown that electrical conductivity requires the P450 to be bonded to the SAM covering the gold substrate.30 P450s bound to the SAM behave like an insulator. Finally, a solid platinum CAFM tip was used as the second electrode in the system so that current-voltage (I–V) curves could be measured.
Experimental Section
General
All chemicals were of analytical reagent grade and were used without further purification. Acetone, isopropanol, methyl isobutyl ketone, mercaptoundecanoic acid (MUA), 8-octanethiol (OT), N-((3-dimethylamino)-propyl)-N-ethyl carbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide (NHS), flurbiprofen, dapsone, and phosphate buffered saline (PBS, 40 mM, pH 7.4 containing 154 mM NaCl, prepared from potassium mono and dibasic phosphate) were purchased from Sigma-Aldrich (Milw, WI). Ethanol (100%) was purchased from Pharmco-Aaper (Brookfield, CT). The 300 MIF developers, 495K poly(methyl-methacrylate) and 950K poly(methyl-methacrylate) were obtained from Micro Chem (Newton, MA). Boron-doped [100] silicon wafers were obtained from University Wafer (South Boston, MA). AZ 5214 photoresist was obtained from (AZ Electronic Materials, Capitol Scientific, Austin, TX). CYP2C9 was prepared by expression in an Escherichia coli system, isolated, and purified as described previously.38 A Barnstead Nanopure water purification system (Thermo Scientific, Waltham, MA) supplied the deionized water.
Fabrication of nanopillar arrays
Silicon wafers were cleaned by immersion in 1:10 hydrofluoric acid (49%): deionized water solution for 3 min and rinsed in a cascading bath for 10 min. The wafers were then blown dry with nitrogen gas and stored in an airtight container in a clean room until needed.
Large alignment patterns were created by photolithography, beginning with a pre-coating dehydration bake (100 °C, 10 min and then cooled to rm temp. The wafers were placed in a Laurell WS-400B-6NPP/LITE spin coater (North Wales, PA), AZ 5214 photoresist was dropped onto the surface, the wafer spun (4000 rpm, 60 s), and solvent evaporated by placing on a hot plate (110 °C, 60 s). A chrome-on-glass mask (Figure S1) (Advance Reproductions Corp., Andover, MA) was placed in a MA-6 Mask Aligner (Karl Suss, Garching, Germany), exposed to UV radiation (25 sec, 320 nm, 4 W/m2), and then immersed in 300 MIF Developer (30–60 seconds) to remove uncured photoresist. The wafers were then rinsed in deionized water (5–15 s) and blown dry with nitrogen gas.
A Temescal BJD-2000 system (Edwards Vacuum, Phoenix, AZ) with an Inficon XTC/2 deposition controller (East Syracuse, NY) was used for metal evaporation. Chamber pressures were ≤ 1.0 × 10−5 Torr, samples were rotated (1–2 rpm) and monitored during deposition for metal thickness using a crystal monitor with gold-coated 6 MHz quartz piezoelectric crystals (Kurt J. Lesker Co., Clairton, PA). Deposition rates of 0.3–0.5 Ǻ/s were maintained during the deposition of a titanium adhesion layer (2 nm) and a gold layer (50 nm). After deposition samples were cooled to room temperature before removing from the chamber. Lift-off of the photoresist was performed by placing the samples in acetone and swirling them (Figure S2).
Prior to electron beam lithography, samples were cleaned in an ultrasonic bath (Bransonic 1210, Danbury, CT) with acetone and then isopropanol, 5 min each, blown dry with nitrogen gas, and baked (150 °C, 30 min). After cooling to room temperature samples were spin coated with 495K poly(methyl-methacrylate) (PMMA) (4% in anisole) (7000 rpm, 30 s), placed on a hot plate (180°C, 2 min) to evaporate solvent, and cooled to room temperature. Samples were then spin coated with 950K PMMA (4% in anisole) (7000 rpm, 30 s), placed on a hot plate (180 °C, 2 min) to evaporate solvent, and cooled to room temperature.
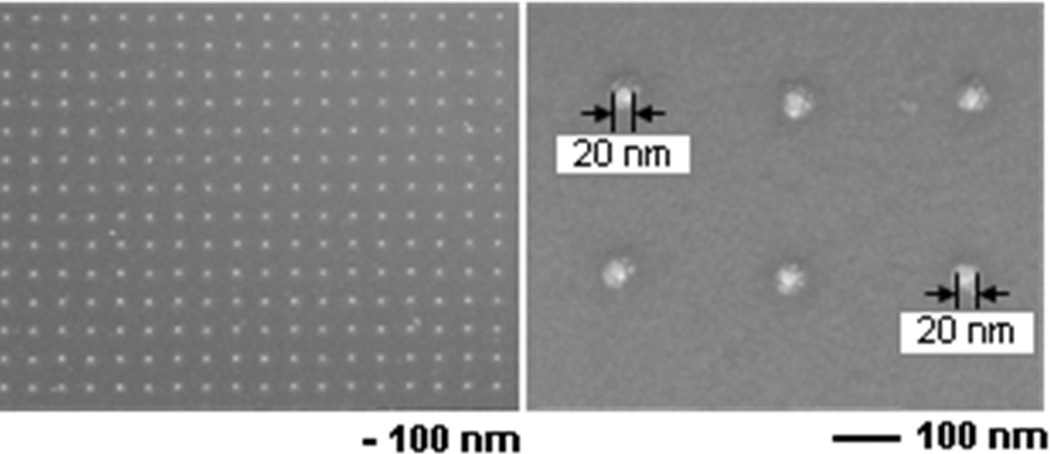
Electron beam lithography was then performed using a JEOL JSM-7600F field emission analytical scanning electron microscope (SEM) (Tokyo, Japan) equipped with Nanometer Pattern Generating System (NPGS) software from JC Nabity Lithography Systems (Bozeman, MT). The pressure inside the chamber was ≤ 9.6 × 10−5 Torr, the accelerating voltage of the electron beam was 30.0 kV, the working distance was 8.0 mm and the probe current 37–40 pA. After focusing, the pattern was written under software control. Upon completion of electron beam lithography, samples were rinsed with a solution of 1:3 methyl isobutyl ketone: isopropyl alcohol (70 s) and then in 100% isopropyl alcohol (20 s). Samples prepared for experimental use were coated with a 2 nm titanium/10 nm gold layer using electron beam evaporation as described above. SEM images of the nanopillar arrays are shown in Figure 1 (see also Figure S3).
Figure 1.
SEM image of nanopillar array (left) and zoom to show size of the individual nanopillars.
Self-assembled monolayer (SAM) preparation and CYP2C9 attachment
Wafers were sonicated in deionized water, ethanol, and acetone, each for 5 min, washed with deionized water, ethanol, immersed in an ethanolic solution of 8-octanethiol (OT) (75 mM) and 11-mercaptoundecanoic acid (MUA) (25 mM) (18 h), and rinsed with ethanol and then PBS, each, three times. Wafers were immersed in PBS containing EDC (2 mM) and NHS (5 mM) for 2 h, and then immersed in a PBS solution containing 50 nM CYP2C9, 40 μM flurbiprofen, and 40 μM dapsone for 24 h. After the CYP2C9 was attached, the wafers were rinsed with PBS. All processes were performed in an argon atmosphere at room temp.
SEM and AFM imaging
SEM imaging was performed using a JEOL JSM-7600F field emission analytical scanning electron microscope with a pressure inside the chamber was ≤ 9.6 × 10−5 Torr. AFM imaging was performed using an Asylum MFP 3D-BIO AFM (Santa Barbara, CA) or a Veeco Multimode scanning probe microscope (Santa Barbara, CA) in tapping mode using Asyelec-01 silicon tips (Asylum Research, Santa Barbara, CA).
Conducting probe atomic force microscopy (CAFM) measurements
Prior to CAFM measurements, the tip cantilever spring constants were measured in accordance with the manufacturer’s instructions on freshly stripped mica for any RMN 25Pt300B tips (Rocky Mountain Nanotechnology, Salt Lake City, UT) used. Spring constant calculations were used to apply equal force each time CAFM measurements were conducted. Alignment marks on each sample were used to take measurements on the same nanopillars for each CAFM measurement. CAFM scans were taken by starting the bias at 0 V, moving linearly to maximum bias (4.5 to 5 V), moving linearly to minimum bias (−4.5 to −5 V), and finally back to 0 V. Curves shown for each sample are the average of at least 5 scans taken from the maximum to the minimum voltage cycle. Between experiments, samples were rinsed in deionized water for at least 1 h to wash small molecules (flurbiprofen, dapsone, or aniline) from the enzyme.
Results and Discussion
Figure 1 shows scanning electron microscopy (SEM) images of a portion of the array prior to addition of the SAM and P450. Each nanopillar is approximately 20 nm in diameter. The SAM was attached to the nanopillars by treatment with a solution of OT and MUA (3:1) in absolute ethanol. CYP2C9 was bonded to the carboxyl groups of SAM. We have proposed that this is likely via the N-terminus of the enzyme38 though this is not unequivocal nor is the possibility that it may be bonded in several orientations as there are several lysine residues on the surface of CYP2C9. Further, there have been reports that the orientation of heme relative to the electrode surface can impact ET.40 While this question will have to be addressed, the consistency of the data obtained here indicates that orientation is not altering ET, the site of bonding is the same for each CYP2C9, and/or there is sufficient mobility so that the required orientation can be adopted.
An important question is whether the CYP2C9 has retained its in vitro/in vivo activity. P450s have been immobilized in a variety of matrices and often are no longer reduced by CPR and NADPH, although they can be reduced by electrochemical means. However, we have shown that CYP2C9 retains its normal activity with respect to substrate binding and metabolite formation if the attachment process (bonding to MUA on gold) is conducted in the presence of flurbiprofen and dapsone.38 Activity is not retained if only one of these substrates, or neither, is present during bonding. Here, as both substrates were present during bonding, CYP2C9 should retain its endogenous activity.
Another possible issue is related to the fact that our measurements are made in air and not in solution. Thus, the enzyme might be considered to be completely dry. However, this is unlikely because unless extraordinary measures are taken, proteins usually retain a hydration shell. This is evident in the crystal structures of P450s (e.g., CYP2C9) which indicate that these proteins contain water molecules even in single crystal forms which are nominally dry.41 Similar studies have been conducted with proteins present as a monolayer on a gold surface and the conductance data correlates with the biological data.19,42,43 We also note that deuterating azurin protein monolayers also causes changes in the electrical conductivity that are correlated with electron transfer processes measurements made in solution that are presumably biologically relevant, once proton conduction, which only occurs in solution, is taken into account.44
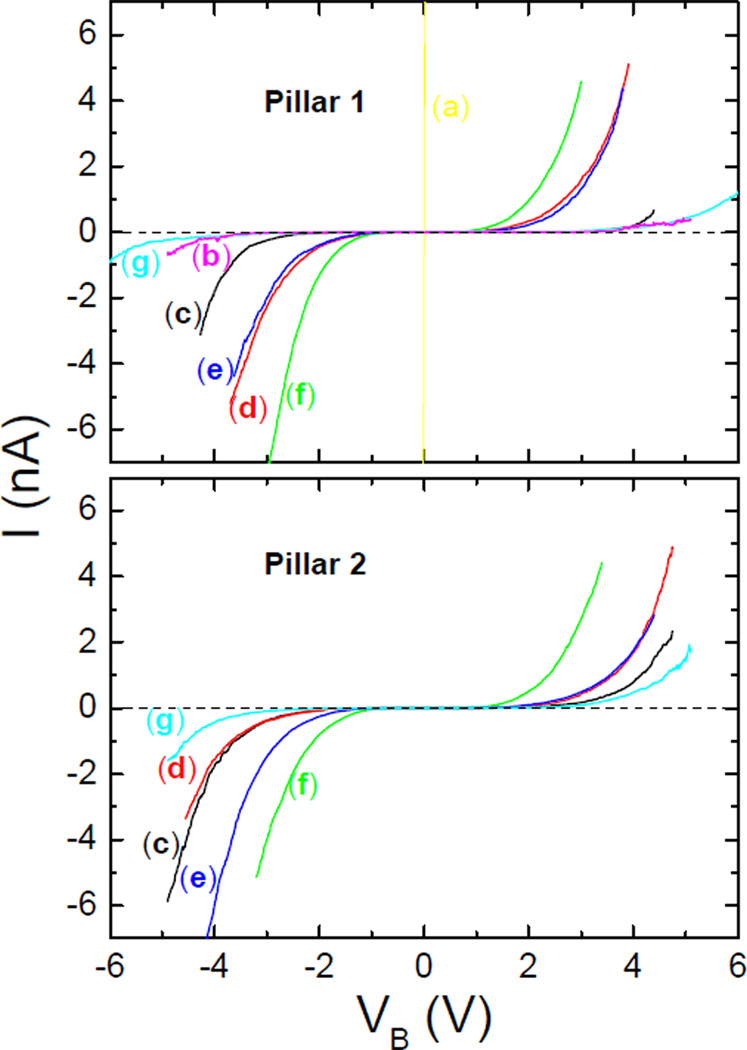
ET was assessed by measuring I–V curves. I–V curves provide conductance information which occurs through pathways known to dominate the ET process.19 To obtain these data, at least five curves were averaged, all of them going from a maximum positive to a minimum negative voltage. As shown in Figure 2, curve (a), the I–V curves for the bare gold nanopillars were ohmic and highly conductive. After forming the OT/MUA SAM on the nanopillars, the behavior of the I–V curve was consistent with the SAM acting as an insulator (Figure 2, curve (b)). However, if driven to sufficiently positive/negative potentials, the nanopillars reverted to exhibiting ohmic behavior (data not shown), implying the SAM removal from the nanopillar. This process appeared to be local to the nanopillar being probed as interrogation of other nanopillars in the array resulted in I–V curves consistent with the presence of the SAM.
Figure 2.
I–V curves measured for two nanopillars of (a) pure gold, (b) SAM, (c) CYP2C9 alone, (d) CYP2C9 with flurbiprofen, (e) CYP2C9 with dapsone, (f) CYP2C9 with both flurbiprofen and dapsone, and (g) CYP2C9 and the inhibitor aniline. The gold and SAM data are not shown for nanopillar 2 for clarity.
Figure 2 also displays a data set obtained on a different nanopillar. Some qualitative differences are observed at positive potentials though the trends remain the same. Larger differences are observed at negative potentials. This was taken into account when the data were fit (see below). The cause for the variability is not known but could be due to differences between nanopillar topographic characteristics which would lead to slight differences in how the AFM tip interacts with the enzyme, or different conformations of proteins on different nanopillars. It should be emphasized that all of the I–V measurements for nanopillar 1 were obtained on the same CYP2C9 and likewise for those made with nanopillar 2. Also important is that the same tip force was used for measurements. While sufficient force must be applied to engage the protein, excessively high forces will compress the enzyme and alter ET.43
Currently, it is not possible to unequivocally demonstrate that there is no more than one P450 on a nanopillar or that the CAFM probe tip is only interacting with one enzyme. The nanopillar lateral dimensions are, on average, 30 nm (range 20–40 nm) and the CYP2C9 is approximately 6 nm in diameter.41 Based on our previous studies we expect there to be 50% or less38 on the nanopillar’s top so there could be up to ~13 CYP2C9 present on each nanopillar though fewer are expected based on the CYP2C9 concentration used. However, the similarities seen when making separate measurements on the same enzyme/nanopillar and from different nanopillars suggest that the measurements are on a single protein. Future work will aim to use indexed nanopillar arrays with nanopillars < 15 nm lateral dimensions to minimize surface area for attachment of the P450.
It is important to recognize the conditions under which the I–V measurements were made. All measurements were conducted in air though the bonded CYP2C9 is likely to have waters associated with it. Consequently, bound substrates, effectors, or inhibitors are not dissociating from the enzyme which would lead to an ensemble of states. At the same time, it is possible that different orientations of bound substrates may be present that would have variable effects on the I–V curve (e.g., aniline binding via the nitrogen or rotated 180 degrees).45 Second, iron is not undergoing a formal reduction though it is participating in electron transduction. Related is the concern that electron flow is simply over the surface of the enzyme. However, we have shown differential I–V behavior between Apo myoglobin and myoglobin indicating the participation of iron in the latter case.46 Finally, because the measurements were made in air, iron is never formally reduced. Consequently, oxygen will not bind, and oxidation of substrates will not occur.
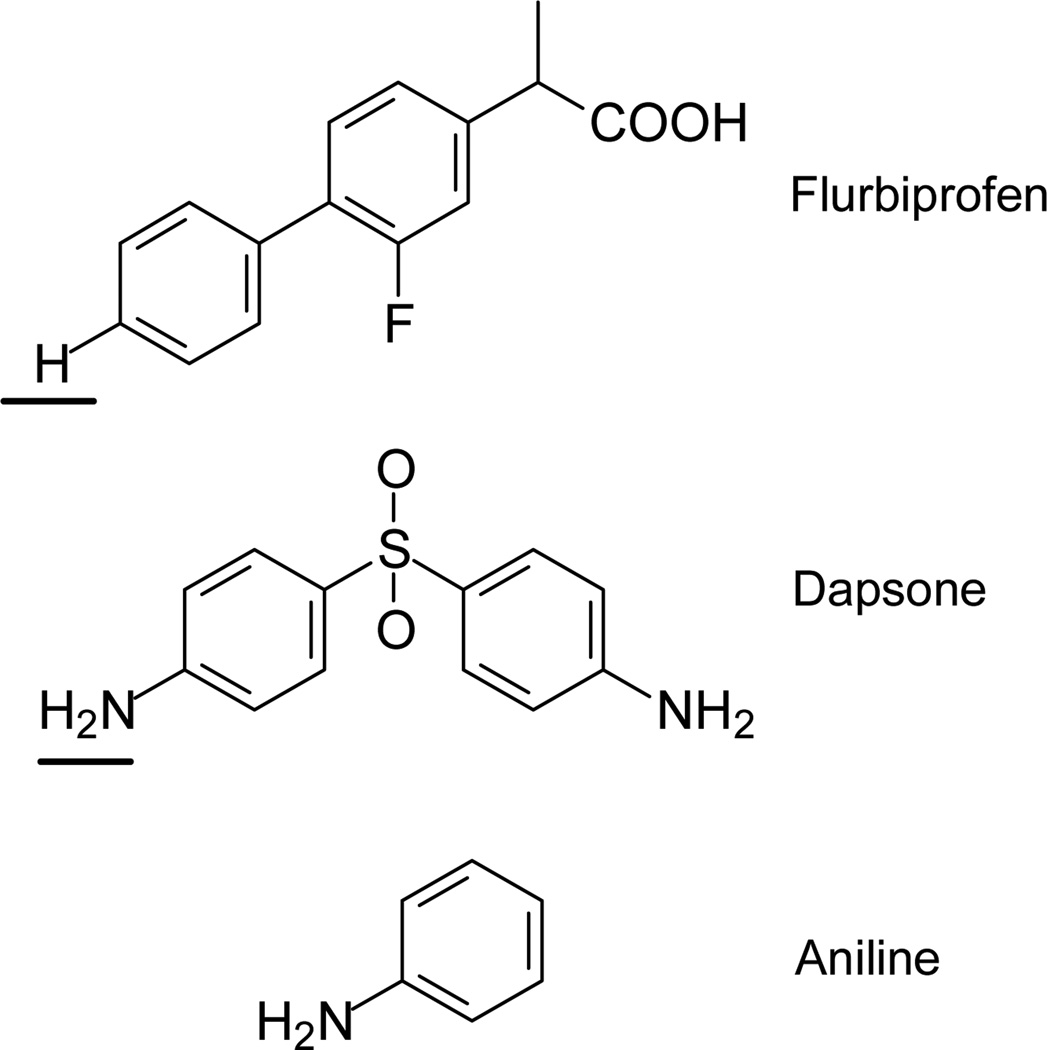
Qualitatively, the I–V curve obtained following bonding of CYP2C9 to the SAM (Figure 2, curve (c)), demonstrates that it is no longer completely insulating as suggested by the decrease in length of the flat portion of the curve over which no current flows. Subsequently, the nanopillars were treated with a solution of flurbiprofen (Chart 1) and I–V measurements repeated which gave Figure 2, curve (d). An additional decrease in the length of the flat portion of the I–V curve, relative to CYP2C9 alone, is observed indicating that ET was easier to achieve in the presence of flurbiprofen. Flurbiprofen is a CYP2C9 substrate that is known to cause a shift in spin state from 3% to 45% high-spin upon binding (Type I binder) depending upon the concentration.20 Hence the increase in ease of ET transfer is expected.
Chart 1. Structures of Substrate and Inhibitors Studied*.
*The site of metabolism is underlined.
When both flurbiprofen and dapsone are present in the active site of CYP2C9, dapsone acts as an effector molecule to position the substrate flurbiprofen closer to the heme iron.21 The simultaneous presence of flurbiprofen and dapsone in the active site is known to increase the rate of metabolism of flurbiprofen. In addition, a further shift in the spin-state ratio of up to 90% high spin can be observed.20 Thus, it is expected that ET will be even easier with both molecules present than when only flurbiprofen is present. The measured I–V curve in Figure 2, curve (f), is consistent with this hypothesis. In addition, the results described thus far are in agreement with the proposed correlation between spin-state and the ease of reduction of the heme iron due to a change in the conformation of the porphyrin ring as a result of substrate binding. They also are aligned with the theory that the rate of P450 mediated metabolism is accelerated as ET becomes easier.
The effect of dapsone (Chart 1) alone on ET was measured for two reasons. First, dapsone is a substrate for CYP2C9, albeit metabolized more slowly than flurbiprofen. Second, dapsone alone does not produce any measurable changes in iron spin state. The I–V curve measured for CYP2C9-dapsone, Figure 2, curve (e), indicates that reduction is slightly more difficult than for flurbiprofen alone, consistent with the metabolism data. However, in this case the result indicates that ET and spin-state are not necessarily correlated and therefore some other effect must be altering ET when dapsone binds. It may be that when dapsone binds to the heme iron, (e.g., via the NH2 group of dapsone), the heme group retains its hexa-coordinate geometry, spin-state is unchanged, and, due to the electron withdrawing nature of the sulfone group of dapsone, iron becomes easier to reduce than the substrate-free CYP2C9.
The effect of a CYP2C9 inhibitor on ET was also examined. Inhibitors are thought to act by binding in the active site thereby blocking access to it. Inhibitors of CYP2C9 do not usually behave as Type I binders,22 rather they behave as Type II binders. Typically, these inhibitors can coordinate to iron through a heteroatom and keep it hexa-coordinate. Consequently, neither the spin-state nor the redox potential of iron is predicted to change. Here aniline (Chart 1), a known CYP2C9 inhibitor, was selected.22 In contrast to the CYP2C9 bound to substrates or CYP2C9 alone, the I–V curve measured for the CYP2C9-aniline complex, shown in Figure 2, curve (g), indicates that ET was significantly more difficult than CYP2C9 alone. This is consistent with a stabilization of iron in the low spin-state.
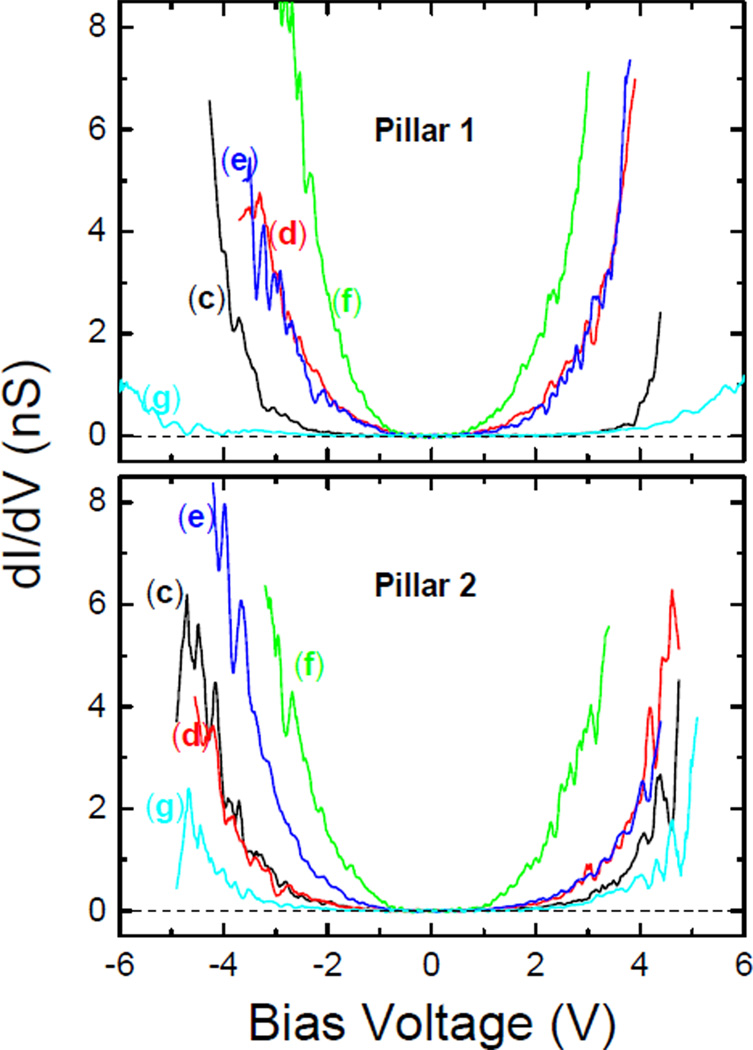
A quantitative analysis of the data was undertaken to analyze the I–V curves. We first note that in all measurements we found that the conductivity near bias voltage VB=0 was zero, and remained zero (to within the noise floor of the data, ±3 pA) until the conductance turned on. In fact, it was not possible to fit the data at small bias using the Simmons model47–49 because the conductance was not ohmic at a small bias. This is evident in Figure 3, which shows the derivative of the I–V curves, dI/dVB, as a function of bias voltage. Clearly, dI/dVB=0 for all the samples and bias voltages smaller than 0.6 V. This means that the tunneling barrier through which electrons travelled from the tip to the gold nanopillar was large in energy. Therefore, performing the typical analyses of CAFM data from proteins obtained at low bias using the Simmons model43,50,51 was not possible in this case. We attempted to use the Fowler-Nordheim (FN) model,52 where electrons tunnel due to field emission, but the fits to the data generated effective electron masses and energy barriers34,35 that were many orders of magnitude off from reasonable values of the electron mass and ~1 eV, respectively. Therefore, we investigated whether it was possible to understand the I–V characteristics using the Poole-Frenkel (PF) emission model, which consists of electrons conducting from one localized state to another through an insulating layer53 and is observed in insulators and metal/insulator interfaces with high densities of traps.37–42 One could surmise that in this case localized states occur in various functional groups of the protein.
Figure 3.
Differential conductance dI/dV as a function of bias voltage. The data are labeled using the same scheme as in Figure 2.
In the PF model the current I is expected to follow the relation
| (1) |
where V is the applied (bias) voltage, q is the charge of an electron, ϕB is the effective voltage barrier that the electron must overcome to move from one localized state to another, d is the distance across which the voltage is applied, εo is the permittivity of free space, and εs is the relative permeability of the material (in this case CYP2C9) at high frequencies, assuming that there is no local polarization induced. Also, k is Boltzmann’s constant, T is the absolute temperature, and C is a constant that depends on the intrinsic mobility of the charge carriers, the effective area of the electrical contact, and the effective distance d across which V is applied. Taking the natural logarithm of both sides of equation (1) yields
| (2) |
so that a plot of ln(I/V) as a function of V1/2 should yield a straight line with an intercept component proportional to ϕB. The slope should be sensitive to changes in the protein’s effective size d and its effective dielectric constant. Because the value of C is unknown, however, it is difficult to obtain an absolute value for ϕB, but by assuming that C remains unchanged upon binding of different substrates, it is possible to obtain a relative change in ϕB.
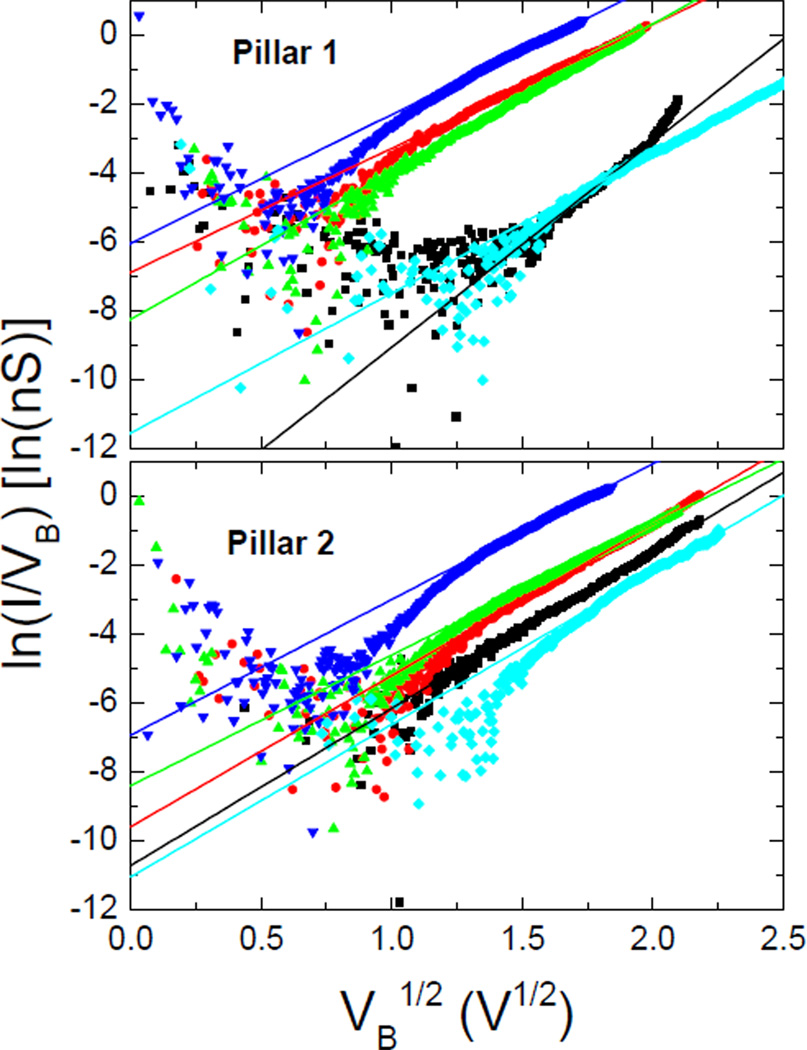
Figure 4 shows such a plot for the positive bias data for both nanopillars. The data were fit to equation 2 for voltages greater than 1 V. Data from smaller voltages did not fit the model very well and it is entirely possible that another mechanism is at play in that region. From inspecting the plot, it is clear that the slope is very similar in all samples, except for the CYP2C9 alone data of nanoillar 1. This means that d and εs are similar for all runs, as expected. The large differences in intercept should be due to the changes in ϕB. The quantitative results are shown in Table I, where the ΔϕB is the shift of barrier height from the CYP2C9 alone runs. The fractional change δd/do, where do is the size of the CYP2C9 enzyme on its own was also calculated. Because the slope of the PF graph (equation 2) should go as m~d−1/2, δd/do~-2 δm/mo, where δm/mo is the relative change in slope. This assumes that the relative permittivity of the protein remains unchanged.
Figure 4.
Poole-Frenkel plot for the data obtained for positive bias voltages for nanopillars 1 and 2 using the same color code as in Figure 2. The symbols are the data and the lines are fits to a straight line.
Table 1.
Results from the Fit to the I–V data with the Poole-Frenkel Model.a
| 2C9 | Flur | Dap | Aniline | Flur/Dap | |
|---|---|---|---|---|---|
| Pillar 1 | |||||
| Slope | 5.96 | 3.59 | 4.28 | 4.05 | 3.73 |
| Intercept | −15.01 | −6.87 | −8.24 | −11.54 | −6.03 |
| ΔϕBb | 0 | −0.21 | −0.17 | −0.09 | −0.23 |
| Δd/do | 0 | 0.79 | 0.56 | 0.64 | 0.74 |
| Pillar 2 | |||||
| Slope | 4.55 | 4.39 | 3.79 | 4.43 | 3.93 |
| Intercept | −10.71 | −9.59 | −8.39 | −11.05 | −6.93 |
| ΔϕBb | 0 | −0.03 | −0.06 | 0.01 | −0.10 |
| Δd/do | 0 | 0.07 | 0.33 | 0.05 | 0.27 |
The units are ln(nS)/V1/2 for the slope, ln(nS) for the y-intercept, eV for ΔϕB, and dimensionless for Δd/do. 2C9, CYP2C9; Flur, Flurbiprofen; Dap, Dapsone.
A value of T=300 K was used to calculate ΔϕB.
Although the magnitude of the effects is different in the two naopillars, the trend is the same. Addition of flurbiprofen and dapsone tend to lower the barrier height by similar amounts. Addition of aniline, on the other hand, tends to not lower the barrier nearly as much, and in the case of nanopillar 2, it appears that the barrier remains essentially unchanged. Addition of flurbiprofen and dapsone tends to lower the barrier height by an amount toor greater than either flurbiprofen or dapsone alone by. Regarding the width of the barrier, the trend is that addition of any substrate tends to increase the barrier width.
It is important to note that use of the PF model in this situation is somewhat arbitrary and a model based on thermionic emission from a Schottky barrier may work equally well, especially at lower bias voltages.54 In order to determine the correct ET mechanism, it would be necessary to measure the conductance as a function of temperature, which is not possible at the present time. Nevertheless, the trends described above are consistent with what is already known about the general properties of CYP2C9 and how it interacts with the selected substrates. At the same time, the ease of P450 reduction and spin-state are not well correlated. In particular, both dapsone and aniline coordinate to iron and do not alter spin state, yet ET is much easier for dapsone and more difficult for aniline, an inhibitor, so ET, alone, is a better predictor for whether a substrate will be metabolized by a P450.
The apparent lack of correlation to spin-state may be due to several factors. First, the spin-state data we are referring to is for CYP2C9 in solution. Our measurements are being conducted on an immobilized CYP2C9 and we do not have direct information regarding the spin-state, though indirect data suggests the spin-state observed in solution is preserved on the nanopillars.38 It has also been found that some P450s may contain more than one binding site and these different binding locations affect spin-state to different degrees.55 Whether this is the case for, for example, dapsone has not known. Further, the degree of spin-state conversion is also dependent upon external conditions such as the presence of detergents56,57 or the concentration of the substrate under study.17 Finally, effects due to changes in protein conformation, to the extent they are related to ET pathway(s) from the surface to the heme center, may alter the ease of ET and metabolism and are not necessarily related to spin-state.58,59
One final consideration concerning ET and spin-state arises from differences between dapsone, a substrate, and aniline, an inhibitor. Neither have an effect on spin state but dapsone has a lower barrier to ET. This may be due to differences in the electron density at the coordinating nitrogen, which is greater for aniline than dapsone (e.g., pKa dapsone = 1.3, aniline 4.6) and therefore suggests that the substrate may play a significant role in modulating ET.
In summary, the platform is quite robust as wafers bearing nanopillar arrays could be removed from the AFM, washed free of one substrate, immersed in a solution containing the same or different substrate(s), I–V measurements made, and the process repeated. As mentioned above, some care had to be exercised so as to not apply too large of a bias voltage. When this occurs the SAM and enzyme were removed, resulting in a large ohmic conductance, although the effects were only local and adjacent nanopillars could still be used.
The I–V data are well correlated with the known metabolic and kinetic behavior of the substrates, inhibitors and substrate-effector pairs. Significant differences in the ET properties were observed between the free enzyme and substrate-bound complex (or free enzyme and inhibitor-bound complex). The electrical conductivity data indicate that the effective energy barrier height and width are altered by the substrates in a way that correlates with the known metabolic activity of the enzyme, suggesting that ET in the metabolic redox process is related to the electrical conductivity of the enzyme. To show an unequivocal relationship between ET and the metabolic redox process, careful measurements with other proteins with different electron transfer rates would have to be performed, but this is beyond the scope of this paper. Our data also seem to indicate that spin state and ET are not necessarily correlated, although other factors such as the binding pose, conformation of the surrounding protein, or the rate of spin-inter-conversion,21 subsequent to iron reduction, may mitigate the observed rate. The results obtained here with the substrate-effector pair are especially interesting as they indicate this method may be useful for identifying substrate interactions which can be difficult to detect otherwise. Also of interest is the effect on the ease of reduction by the inhibitor aniline. It is generally thought that inhibitors simply bind and block access to the active site. However, our data suggest that the inhibitors may also make it more difficult to reduce the P450 enzyme.
While more than one P450 molecule may be present on a nanopillar, the total number must be small due (~13 or fewer) due to the available area on the nanopillar surface. Therefore the technique described here is likely capable of single molecule detection. By varying the lateral size of the nanopillars, it should be possible to study the effects of protein-protein interactions on metabolic activity and electron transfer processes in the future. Efforts are currently underway to create nanopillars with lateral dimensions that are below 15 nm. Finally, the nanopillar platform can serve as the basis for experiments that can directly measure redox potentials of P450 without many of the interferences that plaque such measurements now. By its very nature, individual enzymes rather than ensembles can be probed. By using solution methods such as electrochemical scanning tunneling microscopy, redox potentials can be measured.60,61 The provision for simultaneous imaging will allow direct determination of the number of protein molecules present. Conformational effects can also be examined. For example, the effect of tip force on electron transfer will provides an entry point for such studies.43 Overall, the platform will allow the P450 mechanism to be probed in ways that were heretofore not possible.
Supplementary Material
ACKNOWLEDGMENT
We gratefully acknowledge the NSF (grant EPS-1003907) and NIH (GM-086891) for their support. Use of the WVU Shared Research Facilities is acknowledged. Part of this work was performed by DL with the support of the Rosi and Max Varon Visiting Professorship at the Weizmann Institute of Science in Israel sponsored by Prof. D. Cahen. We thank A. Vilan and L. Sepunaru for useful discussions.
ABBREVIATIONS
- ET
electron transfer
- P450
cytochrome P450
- CAFM
conducting probe atomic force microscopy
- SAM
self-assembled monolayer
- SEM
scanning electron microscopy
- OT
octane thiol
- MUA
mercapto undecanoic acid
- CYP2C9
cytochrome P450 2C9
- CPR
cytochrome P450 reductase
- NADPH
nicotinamide adenine dinucleotide phosphate, reduced
- AFM
Atomic Force Microscopy
- PF
Poole-Frankel model
Footnotes
ASSOCIATED CONTENT
Additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. / All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.Hlavica P. Curr. Drug Metab. 2007;8:594–611. doi: 10.2174/138920007781368881. [DOI] [PubMed] [Google Scholar]
- 2.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. J. Biol. Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 3.Volkov AN, van Nuland NAJ. PLoS Comput. Biol. 2012;8:e1002807. doi: 10.1371/journal.pcbi.1002807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray HB, Winkler JR. Ann. Rev. Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 5.Gray HB, Winkler JR. Proc. Natl Acad. Sci, USA. 2005;102:3534–3539. doi: 10.1073/pnas.0408029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair E, Greaves J, Farmer PJ. J. Am. Chem. Soc. 2004;126:8632–8633. doi: 10.1021/ja0488333. [DOI] [PubMed] [Google Scholar]
- 7.Gilardi G, Fantuzzi A. Trends Biotechnol. 2001;11:468–476. doi: 10.1016/s0167-7799(01)01769-3. [DOI] [PubMed] [Google Scholar]
- 8.Kanaeva IP, Kanaeva IP, Gnedenko OV, Pozdnev VF, Shumyantseva VV, Samenkova NF, Kuznetsova GP, Tereza AM, Schmid RD, Archakov AI. J. Mol. Recognit. 2001;14:196. doi: 10.1002/jmr.532. [DOI] [PubMed] [Google Scholar]
- 9.Shumyantseva VV, Ivanov YD, Bistolas N, Scheller FW, Archakov AI, Wollenberger U. Anal. Chem. 2004;76:6046–6052. doi: 10.1021/ac049927y. [DOI] [PubMed] [Google Scholar]
- 10.Bernhardt R. J. Biotechnol. 2006;124:128–145. doi: 10.1016/j.jbiotec.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 11.Fantuzzi A, Capria E, Mak LH, Dodhia VR, Sadeghi SJ, Collins S, Somers G, Huq E, Gilardi G. Anal. Chem. 2010;82:10222–10227. doi: 10.1021/ac102480k. [DOI] [PubMed] [Google Scholar]
- 12.Eyer CS, Backes WL. Arch. Biochem. Biophys. 2006;293:231–240. doi: 10.1016/0003-9861(92)90390-i. [DOI] [PubMed] [Google Scholar]
- 13.Todorovic S, Jung C, Hildebrandt P, Murgida DH. J. Biol. Inorg. Chem. 2006;11:119–127. doi: 10.1007/s00775-005-0054-9. [DOI] [PubMed] [Google Scholar]
- 14.Johnson DL, Lewis BC, Elliot DJ, Miners JO, Martin LL. Biochem. Pharmacol. 2005;69:1533–1541. doi: 10.1016/j.bcp.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Fleming BD, Johnson DL, Bond AM, Martin LL. Expert Opin. Drug Metab. Toxicol. 2006;2:581–589. doi: 10.1517/17425255.2.4.581. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Z, Gunner MR. Proteins. 2009;75:719–734. doi: 10.1002/prot.22282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das A, Grinkova YV, Sligar SG. J. Am. Chem. Soc. 2007;129:13778–13779. doi: 10.1021/ja074864x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mak PJ, Denisov IG, Grinkova YV, Sligar SG, Kincaid JR. J. Am. Chem. Soc. 2012;133:1357–1366. doi: 10.1021/ja105869p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ron I, Supunaru L, Itzhakov S, Belenkova T, Friedman N, Pecht I, Sheves M, Cahen D. J. Am. Chem. Soc. 2010;132:4131–4140. doi: 10.1021/ja907328r. [DOI] [PubMed] [Google Scholar]
- 20.Locuson CW, Gannett PM, Tracy TS. Arch. Biochem. Biophys. 2006;449:115–129. doi: 10.1016/j.abb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Anderson JLR, Chapman SK. Dalton Trans. 2005:13–24. doi: 10.1039/b413046d. [DOI] [PubMed] [Google Scholar]
- 22.Locuson CW, Hutzler JM, Tracy TS. Drug Metab. Disp. 2007;35:614–622. doi: 10.1124/dmd.106.012609. [DOI] [PubMed] [Google Scholar]
- 23.Tezcan FA, Winkler JR, Gray HB. J. Am. Chem. Soc. 1998;120:13383–13388. [Google Scholar]
- 24.Manchester JI, Ornstein RL. Biochimie. 1996;78:714–722. doi: 10.1016/s0300-9084(97)82529-1. [DOI] [PubMed] [Google Scholar]
- 25.Hamdane D, Xia C, Im S-C, Zhang H, Kim J-JP, Waskell L. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnan S, Wasalathanthri D, Zhao L, Schenkman JB, Rusling JF. J. Am. Chem. Soc. 2011;133:1459–1465. doi: 10.1021/ja108637s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sevrukova IF, Li H, Zhang H, Peterson JA, Poulos TL. Proc. Natl Acad. Sci, USA. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamazaki H, Ueng Y-F, Shimada T, Guengerich FP. Biochem. 1995;34:8380–8389. doi: 10.1021/bi00026a020. [DOI] [PubMed] [Google Scholar]
- 29.Wei J, Liu H, Dick AR, Yamamoto H, He Y, Waldeck DH. J. Am. Chem. Soc. 2002;124:9591–9599. doi: 10.1021/ja025518c. [DOI] [PubMed] [Google Scholar]
- 30.Yang M, Kabulski JL, Wollenberg L, Chen X, Lederman D, Tracy TS, Gannett PM, Wu N. Drug Metab. Disp. 2009;37:892–899. doi: 10.1124/dmd.108.025452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishida T, Mizutani W, Liang T-Z, Azehara H, Miyake K, Sasaki S, Tokumoto H. Ann. N. Y. Acad. Sci. 2006;1006:164–186. doi: 10.1196/annals.1292.011. [DOI] [PubMed] [Google Scholar]
- 32.Alessandrini A, Corni S, Facci P. Phys. Chem. Chem. Phys. 2006;8:4383–4397. doi: 10.1039/b607021c. [DOI] [PubMed] [Google Scholar]
- 33.Kergueris C, Bourgoin J-P, Palacin S. Phys. Rev. B. 1999;59:12505–12513. [Google Scholar]
- 34.Park J, Paupathy AN, Goldsmith JI, Chang C, Yaish Y, Petta JR, Rinkoski M, Sethna JP, Abruna HD, McEuen PL, Ralph DC. Nature. 2002;417:722. doi: 10.1038/nature00791. [DOI] [PubMed] [Google Scholar]
- 35.Zhao J, Davis JJ. Colloids Surface B. 2005;40:189–194. doi: 10.1016/j.colsurfb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Clement N, Patriarche G, Smaali K, Vaurette F, Nishiguchi K, Troadec D, Fujiwara A, Vuillaume D. Small. 2013;7:2607–2613. doi: 10.1002/smll.201100915. [DOI] [PubMed] [Google Scholar]
- 37.Smaali K, Clement N, Patriarche G, Vuillaume D. ACS Nano. 2012;6:4639–4647. doi: 10.1021/nn301850g. [DOI] [PubMed] [Google Scholar]
- 38.Gannett PM, Kabulski J, Perez FA, Liu Z, Lederman DL, Locuson CW, Ayscue RR, Thomsen NM, Tracy TS. J. Am. Chem. Soc. 2006;128:8374–8375. doi: 10.1021/ja0608693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan C, Zharnikov M, Golzhauser A, Grunze M. Langmuir. 2000;16:6208–6215. [Google Scholar]
- 40.Chen X, Ferrigno R, Yang J, Whitesides GM. Langmuir. 2002;18:7009–7015. [Google Scholar]
- 41.Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. J. Biol. Chem. 2004;279:35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 42.Pudney CR, Khara B, Johannissen LO, Scrutton NS. PLoS Biology. 2011;9:e1001222. doi: 10.1371/journal.pbio.1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J, Davis JJ, Sansom MSP, Hung A. J. Am. Chem. Soc. 2004;126:5601–5609. doi: 10.1021/ja039392a. [DOI] [PubMed] [Google Scholar]
- 44.Amdursky N, Pecht I, Sheves M, Cahen D. Proc. Natl Acad. Sci, USA. 2013;110:507–512. doi: 10.1073/pnas.1210457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson J, Dahal UP, Rock D, Peng C-C, Schenk JO, Joswig-Jones C, Jones JP. Arch. Biochem. Biophys. 2011;511:69–79. doi: 10.1016/j.abb.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li D, Gannett PM, Lederman D. Nanotechnology. 2012;23 doi: 10.1088/0957-4484/23/39/395705. 395705 (10 pp). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmons JG. J. Appl. Phys. 1963;34:1793–1803. [Google Scholar]
- 48.Simmons JG. J. Appl. Phys. 1963;34:2581–2590. [Google Scholar]
- 49.Simmons JG. J. Appl. Phys. 1963;34:238–239. [Google Scholar]
- 50.Davis JJ, Wang N, Morgan A, Zhang T, Zhao J. Faraday Discuss. 2006;131:167–179. doi: 10.1039/b507854g. [DOI] [PubMed] [Google Scholar]
- 51.Vilan A. J. Chem. Phys. C. 2007;111:4431–4444. [Google Scholar]
- 52.Fowler RH, Nordheim L. P Roy Soc Lond A Mat. 1928;119:173–181. [Google Scholar]
- 53.Frenkel J. Phys. Rev. 1938;54:647–648. [Google Scholar]
- 54.Rhoderick EH, Williams RH. Metal-Semiconductor Contacts. 2nd ed. Oxford: Clarendon Press; 1988. [Google Scholar]
- 55.Roberts AG, Atkins WM. Arch. Biochem. Biophys. 2007;463:89–101. doi: 10.1016/j.abb.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hosea NA, Miller GP, Guengerich FP. Biochem. 2000;39:5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- 57.Harlow GR, Halpert JR. Proc. Natl Acad. Sci, USA. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cojocaru V, Balali-Mood K, Sansom MSP, Wade RC. PLoS Comput. Biol. 2011;7:1–14. doi: 10.1371/journal.pcbi.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeuken LJC. Biochim. Biophys. Acta. 2003;1604:67–76. doi: 10.1016/s0005-2728(03)00026-4. [DOI] [PubMed] [Google Scholar]
- 60.Alessandrini A, Salerno M. Appl. Phys. Lett. 2005;86 133902 (3 pages). [Google Scholar]
- 61.Artes JM, Diez-Perez I, Sanz F, Gorostiza P. ACS Nano. 2011;5:2060–2066. doi: 10.1021/nn103236e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.