Abstract
Marek's disease, a lymphoproliferative disease of chickens, is caused by an alphaherpesvirus, Marek's disease virus (MDV). This virus encodes a virokine, vIL-8, with general homology to cellular CXC chemokines such as interleukin-8 (IL-8) and Gro-α. To study the function of vIL-8 gene, we deleted both copies of vIL-8 residing in the terminal repeat long and internal repeat long region of the viral genome and generated a mutant virus with vIL-8 deleted, rMd5/ΔvIL-8. Growth kinetics study showed that vIL-8 gene is dispensable for virus replication in cell culture. In vivo, the vIL-8 gene is involved in early cytolytic infections in lymphoid organs, as evidenced by limited viral antigen expression of rMd5/ΔvIL-8. However, the rMd5/ΔvIL-8 virus is unimpaired in virus replication in the feather follicle epithelium. vIL-8 does not appear to be important for establishment of latency, since rMd5/ΔvIL-8 and the wild-type virus have similar viremia titers at 14 days postinfection, a period when the virus titer comes primarily from reactivated latent genomes. Nevertheless, because of the impaired cytolytic infections, the overall transformation efficiency of the virus with vIL-8 deleted is much lower, as reflected by the reduced number of transformed cells at 5 weeks postinoculation and the presence of fewer gross tumors. Importantly, the revertant virus that restored the expression of vIL-8 gene also restored the wild-type phenotype, indicating the deficient phenotypes are results of vIL-8 deletion. One of the interesting differences between the MDV vIL-8 gene and its cellular counterpart is the presence of a DKR (Asp-Lys-Arg) motif instead of ELR (Glu-Leu-Arg) preceding the invariable CXC motif. To study the significance of this variation, we generated recombinant MDV, rMd5/vIL-8-ELR, carrying the ELR motif. Both in vitro and in vivo studies revealed that the DKR motif is as competent as ELR in pathogenesis of MDV.
Marek's disease (MD) is a contagious, lymphoproliferative disease of domestic chickens in which mononuclear infiltration, demyelination of peripheral nerves, and T-cell lymphomas are common features (4). The etiological agent of MD is a lymphotropic, oncogenic herpesvirus, MD virus (MDV). The MDV genome is about 180 kb in length and is classified as an alphaherpesvirus on the basis of DNA sequence homology and genome structure (5, 21). Recently, the complete nucleotide sequences have been determined for all serotypes of MDV (1, 16, 19, 32). The data showed that MDV and other alphaherpesviruses are colinear in the unique long and short regions but differ substantially in the adjacent repeats (19, 30, 32). MDV is grouped into three serotypes: serotype 1 consists of all pathogenic virus strains, serotype 2 comprises the naturally occurring, nononcogenic strains in chickens, and serotype 3 includes the nonpathogenic herpesvirus of turkeys (3, 6, 17, 18). MD incidence has largely been controlled by vaccination with all three serotypes of MDV, often in bi- and multivalent combinations since the 1970s (34, 35). However, there is a continuation of an apparent evolutionary trend of MDV towards greater virulence, which has resulted in recent increased losses from MD in vaccinated flocks (7). A thorough understanding of the genes involved in replication, immune modulation, and oncogenesis holds the key to the development of improved live vaccines, based on targeted mutations of the MDV genome. We have focused on genes specific to serotype 1 of MDV and have developed a cosmid-based recombinant virus approach to study their functions in vivo (29). In this study, we report our findings on vIL-8, a virokine encoded by serotype 1 of MDV.
vIL-8 is located in the repeat region of the MDV genome and, like other virokines of herpesviruses, may be involved in viral replication and/or host immune modulation (24). MDV vIL-8 shares significant homology to cellular CXC chemokines such as interleukin-8 (IL-8) and GRO-α and is the only one found in alphaherpesvirus. Cytomegalovirus, a betaherpevirus, encodes two CXC virokines (i.e., UL146 and −147) (28). Most other virokines belong to the CC family of chemokines. Mutagenesis studies of ELR+ chemokine (e.g., interleukin-8 [IL-8]) and ELR− chemokine (e.g., MIG) revealed that the presence of the ELR motif correlated well with the chemokines' ability to attract neutrophils during inflammation (2) and to induce angiogenesis in tumorigenesis (31).
Previously, we reported the identification of MDV vIL-8 and the initial characterizations of this virokine (20). It was found that vIL-8 has a DKR motif in place of ELR, and in a chemotaxis assay, the major cell types targeted by vIL-8 are mononuclear cells rather than heterophils (chicken equivalent of neutrophils). A vIL-8 deletion mutant in the genetic background of RB1B strain of MDV was constructed by inserting a soluble-modified green fluorescent protein expression cassette at the site of deletion. This ΔvIL-8 virus was found to replicate well in cell culture, but much less so in vivo, and had a weak oncogenic phenotype (26). In this early study, revertant virus was not developed and inadvertent mutations responsible for some of the observed phenotypes cannot be completely ruled out. Nevertheless, the results have provided an important framework for our understanding of the general properties of MDV vIL-8.
In this report, we extend the study of vIL-8 by using the newly established MDV cosmid DNA library (29) to construct recombinant MDVs. This approach permits the efficient construction of revertant virus, which was unattainable by the previous approach (26). We also characterize the in vivo infection course of the mutant virus in more detail. Our results are consistent with the notion that vIL-8 plays an important role in the establishment of early infections, presumably functioning to recruit target cells for MDV infection. To test whether the DKR of vIL-8 is critical for pathogenesis, we also developed a mutant in which DKR was replaced by ELR. The properties of these mutants will be discussed.
MATERIALS AND METHODS
Antibodies.
Monoclonal antibody (MAb) H19 (14) reacts specifically to the MDV phosphoprotein 38 (pp38). The rabbit antibodies were generated against a vIL-8 specific peptide, KKLERQHRTRK (J. Kamil, S.-F. Lin, and H.-J. Kung, unpublished observations).
Cells and viruses.
Primary duck embryonic fibroblasts (DEF) were used for virus propagation and DNA transfections. Recombinant viruses were generated from cosmids derived from a very virulent MDV strain, Md5 (33).
Plasmids.
A 3.1-kb fragment (MDV nucleotides 1451 to 4543) containing the entire sequence of the vIL-8 gene was obtained by digesting the SN5 cosmid with BamHI and was cloned into the same site of pUC19, generating the transfer vector pUC19/SN5BamHI. Subsequently, pUC19/SN5BamHI was digested with ClaI (MDV nucleotide 2808) and NcoI (MDV nucleotide 3605), blunt ended, and religated to generate the vIL-8 deletion transfer vector, pUC19/SN5BamHI/ΔvIL-8.
Replacement of the DKR (Asp-Lys-Arg) motif of vIL-8 with the ELR (Glu-Leu-Arg) motif in the plasmid pUC19/SN5BamHI was carried out using the QuikChangeXL site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. The primers used were FP (5′GAGTCTCGCTGTCGAGCTGAGGTGCAAGTGCG3′) and RP (5′CGCACTTGCACCTCAGCTCGACAGCGAGACTC3′), in which the introduced mutations are underlined. The presence of the corresponding mutations in the plasmid pUC19/SN5BamHI/ELR was confirmed by sequencing.
Cosmids.
MDV cosmid clones SN5, P89, SN16, A6, and B40 from the very virulent strain, Md5 (33), encompassing the entire MDV genome, were used to generate recombinant Md5 viruses (29) (Fig. 1). Cosmid clones A6 and SN5, containing a copy of the complete coding sequence of the MDV unique gene vIL-8 in the opposite orientation, were used to introduce vIL-8-specific mutations.
FIG. 1.
Construction of recombinant virus with deletion of the vIL-8 gene. (A) The MDV genome consists of terminal repeat long (TRL) and short (TRS), internal repeat long (IRL) and short (IRS), and unique long (UL) and unique short (US) DNA segments. (B) Schematic representation of overlapping clones generated to reconstitute an infectious virus from a very virulent strain of MDV (Md5). The restriction enzymes used to generate each cosmid clone and their positions are indicated. (C) Cosmids SN5/ΔvIL8 and A6/ΔvIL8 have the vIL8 coding sequences deleted by ClaI and NcoI digestions. The locations of the restriction enzymes used to introduce the deletions are indicated.
The RecA-assisted restriction endonuclease (RARE) cleavage method (15) was used to delete the vIL-8 gene from the SN5 and A6 cosmid DNAs. Briefly, the SN5 and A6 cosmids were incubated with RecA protein, ADP/ATP γs, and two oligonucleotides, vIL-8blkF (5′-GCCCGCATCTCGCAGCCCCCGGATCCGATCCCGCAGACCC-3′) and vIL-8blkR (5′-TCCCCTGCTAGCCCTGCCCTAGGTAATGCATTTTAAATCT-3′), overlapping the two BamHI sites flanking the vIL-8 sequence (MDV nucleotides 1451 to 4543) to protect these sites from methylation. The protected cosmid DNAs were methylated with BamHI methylase, denatured, and digested with BamHI to generate SN5/ΔBamHI and A6/ΔBamHI. These cosmid DNAs were treated with calf intestinal alkaline phosphatase and were purified by phenol-chloroform extraction followed by ethanol precipitation. To introduce the vIL-8 deletion into the SN5 and A6 cosmids, the pUC19/SN5BamHI/ΔvIL-8 transfer vector was digested with BamHI and the SN5BamHI/ΔvIL-8 fragment was ligated to SN5/ΔBamHI and A6/ΔBamHI, generating SN5/ΔvIL-8 and A6/ΔvIL-8, respectively.
Similarly, to introduce the ELR motif into the vIL-8 coding sequence, the pUC19/SN5BamHI/ELR transfer vector was digested with BamHI and the SN5BamHI/ELR fragment was ligated to SN5/ΔBamHI and A6/ΔBamHI, generating SN5/vIL-8-ELR and A6/vIL-8-ELR, respectively.
Transfections.
Parental P89, SN16, and B40 and mutant SN5/ΔvIL-8, A6/ΔvIL-8, SN5/vIL-8-ELR, and A6/vIL-8-ELR cosmid DNAs were digested with NotI and purified by phenol-chloroform extraction and ethanol precipitation. To generate a mutant virus with vIL-8 gene deletion, rMd5/ΔvIL-8, 500 μg of each digested cosmid DNA (P89, SN16, B40, SN5/ΔvIL-8, and A6/ΔvIL-8) along with 2 μg of sheared salmon sperm DNA were used to transfect 5 × 105 DEF in 35-mm-diameter dishes by the calcium phosphate method (25). Four days after transfection, cells were trypsinized, seeded onto a 100-mm dish, and monitored daily for cytopathic effect (CPE). Viral stocks were subsequently made in DEF for further analysis. An MDV mutant virus carrying the ELR motif, rMd5/vIL-8-ELR, was generated in a similar method with P89, SN16, B40, SN5/vIL-8-ELR, and A6/vIL-8-ELR cosmid DNAs. The parental virus rMd5 was generated by cotransfecting the five digested parental cosmid DNAs (SN5, P89, SN16, A6, and B40).
Revertant virus.
To generate a revertant virus from rMd5/ΔvIL-8 containing the vIL-8 gene, transfer vector pUC19/SN5BamHI was digested with BamHI and cotransfected into DEF cells with the purified rMd5/ΔvIL-8 viral DNA. After the CPE was evident, transfected cells were overlayed with 1.25% of Bacto-Agar and more than 400 viral plaques were picked by trypsinization. Cells from each plaque were divided into two aliquots: one was used to reinfect a fresh 60-mm dish of DEF, and the other was used for PCR analysis. Integration of the vIL-8 gene into the rMd5/ΔvIL-8 genome was detected by PCR with primers ClaIF (5′-GGCGCAGCACTGAATAAGCC-3′) and BamHoriR (5′-GGAGTAATCTGCGTT-3′), which would generate 2,200- and 1,400-bp fragments in the revertant and deletion mutant viruses, respectively.
IFA and IHC.
An indirect immunofluorescence assay (IFA) of cosmid-transfected DEF cells was carried out as previously described (13). For immunohistochemistry (IHC), lymphoid organs (thymus, spleen, bursa of Fabricius) and feather follicles of infected and uninfected chickens were embedded in OCT (optimal cutting temperature) compound (Sakura Finetek U.S.A., Inc., Torrance, Calif.), immediately frozen in liquid nitrogen, and stored at −80°C until use. Four- to 8-μm-thick cryostat sections of tissue blocks were prepared, fixed with cold ethanol for 5 min, and air dried. Immunostaining was carried out with the Vectastain ABC kit (Vector Laboratories, Inc., Burlingame, Calif.) as suggested by the manufacturer. For IFA staining, MAb H19, specific for antigen pp38, was used at a working dilution of 1:300 and rabbit serum against vIL-8 was used at a working dilution of 1:50. For IHC staining, MAb H19 was used at a working dilution of 1:3,200.
Growth kinetics.
The growth kinetics of rMd5, rMd5/ΔvIL-8 (clones 1 and 2), and rMd5/vIL-8-ELR (clones 1 and 2) viruses were determined as described previously (12). Briefly, DEF cells seeded on 60-mm plates were inoculated with 100 PFU of the different viruses. On days 1, 2, 3, 4, and 5 postinoculation, the infected cells were trypsinized, fresh DEF cells seeded on 35-mm plates were inoculated with serial dilutions, and plaques of different dilutions were counted 7 days postinfection.
Southern blot.
DNAs from rMd5-, rMd5/ΔvIL-8-, rMd5/vIL-8-ELR-, and rMd5/ΔvIL-8-RV-infected or uninfected DEF were isolated as previously described (29). Five micrograms of each viral DNA was digested with EcoRI or BamHI or double digested with BamHI and SalI. The DNA fragments were then separated on a 1% agarose Tris-borate/EDTA (TBE) gel and transferred to nylon membranes. Two individual [32P]dCTP-labeled DNA probes, one from the total genomic viral DNA (SN5, P89, SN16, A6, and B40 cosmid DNA fragments) and one from the 3.1-kb BamHI fragment containing the vIL-8 gene, were generated by random priming, and hybridization was carried out using standard protocols.
Western blot.
Supernatants from rMd5-, rMd5/ΔvIL-8-, rMd5/vIL-8-ELR-, and rMd5/ΔvIL-8-RV-infected DEF as well as the supernatant from the noninfected DEF were collected 3 days postinfection. Sixteen microliters of each supernatant was separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (15% polyacrylamide) by standard procedures. Proteins were then transferred to nitrocellulose membranes, blocked with 5% skim milk for 1 h at room temperature, and probed with rabbit anti-vIL-8 polyclonal antibody (1:200 diluted in TBS buffer [20 mM Tris-HCl, pH 7.5, 0.8% NaCl]) at 37°C for 1 h. After 3 washes with TBS buffer, horseradish peroxidase-conjugated goat anti-rabbit antibody (1:3,000) was added to the blots and this mixture was incubated at 37°C for 1 h. Following three washes with TBS, antibody-bound specific antigens were detected by incubation with the ECL Western blotting detection reagent (Amersham Bioscience, Little Chalfont, United Kingdom) and exposed to X-ray film.
Pathogenesis studies.
Specific-pathogen-free MD-susceptible progeny (15 × 7)F1 of the Avian Disease and Oncology Laboratory line 15I5 males and line 71 females were used in all the studies. These progeny were free of maternal antibodies against MDV. Chickens were wing banded at hatching and randomly sorted into different experimental groups (17 chickens per group) and held in modified Horsfall-Bauer isolators. Day-old chickens were inoculated subcutaneously with 2,000 PFU of rMd5, rMd5/ΔvIL-8, rMd5/vIL-8-ELR, or rMd5/ΔvIL-8-RV. All of the chickens that died during the trial or were killed at the end of the experiment (8 weeks postinoculation) were necropsied and evaluated for gross tumor incidence.
Viremia assay.
To examine in vivo virus replication and reactivation, five birds from each group were randomly selected and bled at 6, 8, 14, and 35 days postinoculation. Buffy coats were obtained by centrifugation at 500 × g for 5 min. Lymphocytes were then counted and diluted to 106 cells/ml. For each chicken sample, duplicated 35-mm plates of freshly seeded DEF monolayers were inoculated with both 105 and 106 lymphocytes and viral plaques were counted 7 days postinoculation.
RESULTS
Construction of vIL-8 deletion mutant rMd5/ΔvIL-8.
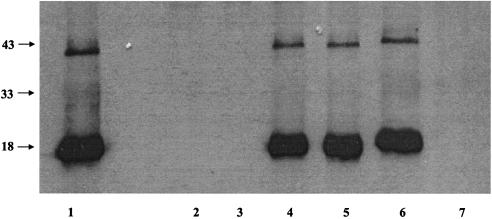
In order to determine the role of MDV vIL-8 in viral replication and pathogenesis, we constructed a virus, rMd5/ΔvIL-8, in which the entire coding sequence of the vIL-8 gene was deleted (Fig. 1C). Cosmids SN5/ΔvIL-8 and A6/ΔvIL-8, lacking the entire coding sequence of vIL-8, were transfected, along with parental SN16, P89, and B40, into DEF and observed for CPE. To confirm the deletion of vIL-8 gene, transfected cells showing CPE were examined by IFA with MAb H19 (anti-pp38) and rabbit anti-vIL-8 polyclonal sera. As expected, rMd5 virus expressed both pp38 and vIL-8 while rMd5/ΔvIL-8 expressed only pp38 (data not shown). Similarly, Western blot analysis of rMd5- and rMd5/ΔvIL-8-infected DEF supernatants indicated that an 18-kDa band, corresponding to vIL-8, was present in supernatants from rMd5-infected cells but absent in supernatants from rMd5/ΔvIL-8-infected cells (Fig. 2). To verify that rMd5/ΔvIL-8 had the expected genome structure, a Southern blot of rMd5 and rMd5/ΔvIL-8 genomic DNA digested with EcoRI was performed. As shown in Fig. 3A, both viruses showed no detectable difference in the pattern of DNA fragments, suggesting that there were no gross rearrangements in the rMd5/ΔvIL-8 genome (Fig. 3A, lanes 5 and 6). In addition, Southern blot analysis of viral DNA digested with BamHI and probed with the 3.1-kb fragment purified from pUC19/SN5BamHI resulted in a 3.1-kb fragment in rMd5 virus and a 2.3-kb fragment in the rMd5/ΔvIL-8 DNA, reflecting the deletion of 798 bp spanning the vIL-8 gene (Fig. 3B, lanes 5 and 6).
FIG. 2.
Western blot analysis of supernatant from rMd5/ΔvIL-8-RV (lane 1), rMd5/ΔvIL-8 (clones 1 and 2 in lanes 2 and 3), rMd5 (lane 4), rMd5/vIL-8-ELR (clones 1 and 2 in lanes 5 and 6), and mock infected DEF (lane 7) using anti-vIL-8 rabbit polyclonal sera. The vIL-8 protein is about 18 kDa.
FIG. 3.
Southern blot analysis of DNA isolated from recombinant MDVs. (A) Viral DNA was digested with EcoRI and probed with all five radiolabeled cosmids. (B) Viral DNA was digested with BamHI (lanes 1 to 6) or double digested with BamHI/SalI (lanes 1′ to 6′) and hybridized with the 3.1-kb MDV BamHI fragment containing the vIL8 gene. Lanes: 1 and 1′, uninfected DEF; 2 and 2′, rMd5; 3, 4, 3′, and 4′, rMd5/vIL-8-ELR (clones 1 and 2); 5, 6, 5′, and 6′, rMd5/ΔvIL-8 (clones 1 and 2). BamHI single digestion produces a 3.1-kb band in both rMd5 and rMd5/vIL-8-ELR viruses and a 2.3-kb fragment in rMd5/ΔvIL-8 virus. BamHI/SalI double digestion results in two bands (1.8 and 1.3 kb) in rMd5 and a single band (3.1 kb) in rMd5/vIL-8-ELR, due to loss of the SalI site.
In vitro and in vivo replication properties of rMd5/ΔvIL-8.
To determine if vIL-8 plays a role in the in vitro replication of MDV in DEF, the growth kinetics of rMd5, rMd5/ΔvIL-8-1 (clone 1), and rMd5/ΔvIL-8-2 (clone 2) were compared. As seen in Fig. 4, viral titers at all time points tested were very similar for all three viruses, indicating that expression of vIL-8 is dispensable for viral replication in cell culture.
FIG. 4.
In vitro growth kinetics of rMd5, rMd5/ΔvIL-8 (clones 1 and 2), and rMd5/vIL-8-ELR (clones 1 and 2) viruses. DEF were infected with approximately 100 PFU of the indicated viruses, and at 1, 2, 3, 4, and 5 days postinfection, the cells were harvested and their titers were determined on fresh DEF. The experiment was performed in duplicate, and the titer is indicated as PFU for each 60-mm dish. Error bars in the figure show the standard deviation of the mean.
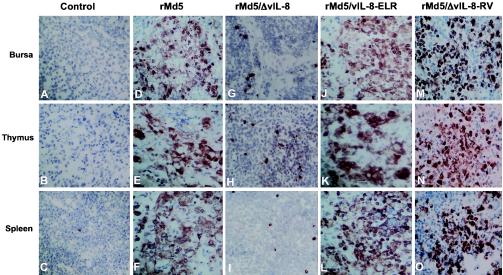
To examine if vIL-8 plays a role in the in vivo viral replication, day-old 15 × 7 chickens were inoculated with rMd5 and rMd5/ΔvIL-8 viruses. Six days postinoculation, lymphoid organs (bursa of Fabricius, thymus, and spleen) from three chickens from each group were collected and examined for virus replication by immunohistochemistry. As shown in Fig. 5, there was a high level of expression of pp38 in the lymphoid organs of rMd5-inoculated chickens (Fig. 5D, E, and F). However, there was a significantly lower level of pp38 expression in rMd5/ΔvIL-8-infected chickens (Fig. 5G, H, and I). These results indicated that the vIL-8 gene is important for early cytolytic infection in lymphoid organs.
FIG. 5.
Immunohistochemistry of lymphoid organs (the tissues in each column from top to bottom represent bursa, thymus, and spleen) of 15 × 7 MDV maternal antibody-negative chickens 6 days after inoculation with control (A, B, and C), rMd5 (D, E, and F), rMd5/ΔvIL-8 (G, H, and I), rMd5/vIL-8-ELR (J, K, and L), or rMd5/ΔvIL-8-RV (M, N, and O). MAb against pp38 (H19) was used for the staining. Antigen expression in lymphoid organs is severely impaired only in rMd5/ΔvIL-8, showing that vIL-8 is involved in early cytolytic infection in lymphocytes.
Transmission of rMd5/ΔvIL-8.
MDV transmission takes place by virus replication in the feather follicle and release of infectious virus in the dander. To examine if vIL-8 is necessary for virus transmission, viral replication in the feather follicle of chickens infected with rMd5 and rMd5/ΔvIL-8 was examined 2 weeks postinoculation. As shown in Fig. 6, both rMd5 (panel A) and rMd5/ΔvIL-8 (panel B) viruses had similar levels of pp38 expression in the feather follicle, suggesting that rMd5/ΔvIL-8 can be transmitted horizontally like parental MDV. In addition, sentinel chickens housed in the same isolator as rMd5/ΔvIL-8-inoculated chickens developed high titers of anti-MDV antibodies (data not shown), indicating that feather follicle epithelium (FFE) replication was not reduced and contact transmission was not abrogated with the deletion of the vIL-8.
FIG. 6.
Immunohistochemical analysis of FFE cells from inoculated chickens. FFE cells were sampled at 2 weeks postinoculation. All of the recombinant viruses rMd5 (A), rMd5/ΔvIL-8 (B), and rMd5/vIL-8-ELR (C) expressed viral antigen in FFEs, indicating that the second lytic infection is not impaired in either the vIL-8 gene deletion or vIL-8 gene mutations. No viral antigen was detected in the control chickens (D).
Latency entry and reactivation of rMd5/ΔvIL-8.
To examine if vIL-8 affects MDV latency and reactivation, peripheral blood lymphocytes were cocultivated with DEF at 6, 8, 14, and 35 days postinoculation. As shown in Fig. 7, both rMd5 and rMd5/ΔvIL-8 viruses reached a peak virus titer 8 days postinoculation, which was followed by a decreased virus titer at 14 days and a subsequent increase at 35 days postinoculation. Interestingly, rMd5/ΔvIL-8 virus showed a statistically significant (Student's t test, P < 0.001) lower viral titer than rMd5 at 35 days postinoculation, while the differences observed at 8 and 14 days were not significant. This may be due to reduced number of transformed lymphocytes in rMd5/ΔvIL-8-infected chickens compared to parental rMd5-infected chickens at later stages of disease.
FIG. 7.
Viral titers at 6, 8, 14, and 35 days postinoculations in peripheral blood lymphocytes of 15 × 7 chickens inoculated with rMd5, rMd5/ΔvIL-8, and rMd5/vIL-8-ELR. Five chickens from each experimental group were tested, and titrations were performed in duplicate. The titer is indicated as PFU/106 peripheral blood lymphocytes. Error bars in the figure show the standard deviation of the mean.
Oncogenicity of rMd5/ΔvIL-8.
In order to determine if the deletion of vIL-8 affects the pathogenic properties of MDV, chickens inoculated with rMd5 or rMd5/ΔvIL-8 were examined for gross tumors and mortality for a period of 8 weeks. As indicated in Table 1, the incidence of mortality was significantly lower in the group inoculated with rMd5/ΔvIL-8 (4.3%) than in the group inoculated with rMd5 (88.2%). In addition, in agreement with the low level of replication of rMd5/ΔvIL-8 in lymphoid tissues, at termination, no atrophy of the bursa of Fabricius and thymus was observed in this group of chickens compared with massive atrophy in the rMd5-inoculated group (data not shown). Similarly, the tumor incidence in the group inoculated with rMd5/ΔvIL-8 was much lower (17.6%) than that observed in the group inoculated with rMd5 (76.7%). These data altogether indicate that the deletion of the vIL-8 gene significantly decreases the virulence of the recombinant virus, rMd5/ΔvIL-8.
TABLE 1.
Comparison of pathogenicities of rMd5, rMd5/ΔvIL-8, rMd5/vIL-8-ELR, and rMd5/ΔvIL-8-RV in 15 × 7 MDV maternal antibody-negative chickensa
| Virusb | No. of chickens that died/no. tested (%) | No. of chickens with tumor/no. tested (%) |
|---|---|---|
| Mock | 0/17 (0) | 0/17 (0) |
| rMd5 | 15/17 (88.2) | 13/17 (76.7) |
| rMd5/ΔvIL-8 | 1/17 (4.3)* | 3/17 (17.6)* |
| rMd5/vIL-8-ELR | 16/17 (94.1) | 15/17 (88.2) |
| rMd5/ΔvIL-8-RV | 17/17 (100) | 13/17 (76.7) |
This experiment was repeated two times separately. *, Student's t test (P ≤ 0.001) analysis indicated value was significantly different from those for the other groups.
All chickens were inoculated with 2,000 PFU of the indicated viruses.
Construction and biological properties of revertant virus rMd5/ΔvIL-8-RV.
To verify that the phenotypic changes observed in the in vivo replication and pathogenesis of rMd5/ΔvIL-8 were only due to the deletion of vIL-8, we generated a revertant virus, rMd5/ΔvIL-8-RV, by cotransfection of rMd5/ΔvIL-8 viral DNA with a transfer vector, pUC19/SN5BamHI, containing the vIL-8 gene. Revertant viruses were selected by plaque purification and screened for the presence of the vIL-8 gene by PCR. In addition, expression of vIL-8 in supernatant of rMd5/ΔvIL-8-RV-infected cells was confirmed by Western blot (Fig. 2, lane 1). As shown in Table 1, the pathogenic properties of the revertant rMd5/ΔvIL-8-RV virus were very similar to those of parental virus with regard to mortality (100%) and tumor incidence (76.7%). These results confirm that vIL-8 plays an important role in MDV pathogenesis.
Construction and replicative properties of rMd5/vIL-8-ELR.
Many CXC chemokines contain an ELR motif that has been associated with chemoattraction of neutrophils. MDV vIL-8, however, carries a DKR motif in place of ELR and attracts mononuclear cells instead of heterophils (chicken neutrophils) (26). In order to determine the significance of the DKR motif to the biological properties of vIL-8, we generated a recombinant MDV, rMd5/vIL-8-ELR, in which the DKR motif was replaced with ELR. Using site-directed mutagenesis and RARE cleavage, we generated two cosmids, SN5/vIL-8-ELR and A6/vIL-8-ELR, in which the nucleotides CAA were changed to GCT, resulting in two amino acid substitutions in the vIL-8 gene from DK (Asp-Lys) to EL (Glu-Leu). Interestingly, these mutations resulted in the deletion of a SalI site in the vIL-8 gene, and this feature was used for the selection of recombinant clones prior to sequencing. Transfection of DEF with cosmids P89, SN16, B40, SN5-ELR, and A6-ELR resulted in a recombinant virus with in vitro growth properties similar to those of parental virus rMd5 (Fig. 4). Immunofluorescence analysis of rMd5/vIL-8-ELR-infected DEF (data not shown) and Western blot analysis of supernatants from these infected cells (Fig. 2, lanes 5 and 6) indicated that vIL-8-ELR was expressed and secreted like parental virus. In addition, Southern blot analysis of EcoRI (Fig. 3A, lanes 3 and 4)- or BamHI-digested rMd5/vIL-8-ELR viral DNA (Fig. 3B, lanes 3 and 4) showed the same DNA pattern as the parental virus, while the BamHI-SalI double-digested DNA (Fig. 3B, lanes 3′ and 4′) showed the loss of the SalI restriction enzyme site, indicating the presence of the introduced mutations.
Pathogenic properties of rMd5/vIL-8-ELR.
To examine if the DKR-to-ELR mutation of MDV vIL-8 had any effect on pathogenicity, rMd5 or rMd5/vIL-8-ELR was inoculated into day-old 15 × 7 chickens, and its effects on viremia, early cytolytic infection, mortality, and tumor induction were examined. As shown in Fig. 7, chickens infected with rMd5/vIL-8-ELR presented similar viremia titers to those in rMd5-infected chickens at all four time points tested. Similarly, viral replication in lymphoid organs (bursa of Fabricius, thymus, and spleen) at 6 days postinoculation, determined by pp38 expression, showed levels similar to those of rMd5 (Fig. 5), indicating that the mutations did not have any effect on the early cytolytic infection of rMd5/vIL-8-ELR. In addition, mortality (94.1%) and tumor incidence (88.2%) induced by rMd5/vIL-8-ELR were very similar to those observed with rMd5 (88.2 and 76.7%, respectively), indicating that the DKR-to-ELR mutation had no effect on the pathogenic properties of the virus (Table 1).
DISCUSSION
In this report, we describe a detailed analysis of pathogenesis of three recombinant MD viruses: (i) vIL-8 knockout virus, rMd5/ΔvIL-8; (ii) its revertant, rMd5/vIL-8-RV; and (iii) rMd5/vIL-8-ELR, which carries ELR motif in the vIL-8 gene. The use of a cosmid-based strategy significantly facilitated the construction of these viruses (29). Our results are consistent with an early report that vIL-8 is not required for in vitro replication but plays an important role in the in vivo propagation and pathogenesis (26). The gross MD tumor incidence caused by the vIL-8 deletion mutant is down to 17.6% of infected birds (from 76.7% of the wild-type value). Importantly, the revertant virus completely restores the pathogenicity, conclusively demonstrating the crucial role vIL-8 plays in this process. The vIL-8-ELR mutant has a pathogenic pattern similar to that of the wild type, indicating ELR does not significantly shift the tropism.
While all the mutant viruses replicate equally well on fibroblasts in vitro, the infection patterns in vivo are quite different. We have conducted a time course analysis of infections in different tissues. MDV induces an early phase of cytolytic infection in lymphoid organs during the first week, which is followed by latency entry and reactivation, resulting in the second phase of cytolyic infections occurring around 2 weeks. We found that rMd5/ΔvIL-8 virus is significantly impaired in the early phase of cytolytic infections in lymphoid organs (Fig. 5). It is well documented that reduction or absence of early cytolytic infection correlates with absence or reduced incidence of lymphomas (8-10, 27). This may account for the low virulence and lymphoma incidence of the vIL-8 deletion mutant. The role of vIL-8 in second lytic infection of MDV was studied at 14 days postinfection, which correlates with the beginning of the MDV latency. We showed that rMd5/ΔvIL-8 virus replicated well in peripheral blood lymphocytes and FFE. The FFE is the only site for productive infection resulting in cell-free infectious viral particles, which are transmitted to contacted birds. It thus seems that vIL-8 plays a minor role, if any, in the second lytic infection phase as well as virus shedding.
Although the development of latency in MDV is not fully understood and we also are not sure how latently infected lymphocytes result in plaque formation in vitro, we assume that latent infection is prerequisite for this process and that a reduction in the proportion of latent infection of lymphocytes in peripheral blood would reduce plaque counts (viremia titer). It is also known that MDV can be reactivated by DEF cells cocultivated with lymphocytes isolated from infected chickens (11) and form virus plaques; therefore, viremia titers in chickens can reflect both the degree of virus reactivation from latency and the number of latently infected cells. It has been shown that a successful cytolytic infection of B and T cells is a prelude to latent infection and transformation of T cells. In our experiments, both parental and vIL-8 deletion viruses reached a peak in viremia at 8 days postinfection, with viral titers very similar for both viruses. It is believed that viral titers measured at this early stage of infection are a combination of an active cytolytic infection and possibly some reactivation from latency, while viral titers measured at 14 days and beyond are likely to be a measure of reactivation from latency. Although both viruses showed similar viremia patterns at 14 and 35 days postinoculation, significantly lower viral titers were observed for the vIL8 deletion mutant virus at 35 days postinoculation. These results suggest that deletion of the vIL-8 gene has no effect in latency and that the difference in reactivation observed at 35 days postinoculation, as indicated by a lower viral titer, is a consequence of the reduced number of transformed cells, as it is also confirmed by the lower tumor incidence.
The ELR mutant rMd5/vIL-8-ELR has both in vitro and in vivo properties similar to pathogenic parental virus, rMd5, an initially surprising finding. In mammals, ELR+ CXC chemokines engage CXCR2, a G-protein-coupled receptor expressed in endothelial cells. We speculated that the presence of ELR in vIL-8 would induce angiogenic activity, thus facilitating more aggressive tumor growth. The fact it did not suggests the interaction of chicken chemokine and its respective receptor is significantly different, the tissue distribution of chicken CXCR2 is different from that of mammals, or infection of MDV induces a significant level of cellular angiogenic factors such as cellular IL-8 or VEGF, such that the contribution by vIL-8 is inconsequential. At present, we have little knowledge about the chicken CXCRs and their expression patterns. We also do not know whether vIL-8 is able to trigger signals necessary for angiogenesis. A possible scenario is that vIL-8 is able to bind certain receptors and attract target cells but unlike its cellular counterpart lacks the ability to trigger intracellular signals required for the proliferation of endothelial cells. Further investigations are required to sort out these questions. It is known, however, that MDV infection results in the release of cellular IL-8 homologs 9E3/CEF and K60 (36), both of which contain the ELR motif, and at least for 9E3/CEF, its angiogenic effect has been demonstrated (22, 23). At the same time, ELR-containing chemokines are known to be chemoattractants for neutrophils, which might reduce virus load and impede tumorigenesis. Our results would argue that the replacement of DKR by ELR does not change significantly its tropism toward target cells. We note that vIL-8 has a significantly long carboxy-terminal domain, which is also considered important in chemotactic functions. The presence of this domain may diminish its ability to attract or activate neutrophils, even in the presence of ELR.
In summary, our finding is most consistent with a model that the MDV-encoded vIL-8 gene is involved in early phase of cytolytic infections—presumably the recruitment of B or T lymphocytes. Deletion of this gene has less impact on either virus reactivation from latency or virus shedding. Impaired early cytolytic infection due to the deletion of vIL-8 leads to weak activation of T cells, resulting in reduced numbers of target cells for transformation and significantly decreased pathogenicity and tumor incidence.
Acknowledgments
This work was supported by a U.S. Egg and Poultry Association Project grant to S.M.R. (project 356). This work was also supported by grants from NIH (CA46613) and USDA (02390 and 02220) to H.-J.K.
We thank I. Gimeno for helpful discussions and R. Witter, B. Lupiani, and R. Silva for critical reading of the manuscript.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baggiolini, M., B. Dewald, and B. Moser. 1997. Human chemokines: an update. Annu. Rev. Immunol. 15:675-705. [DOI] [PubMed] [Google Scholar]
- 3.Becker, Y., Y. Asher, E. Tabor, I. Davidson, M. Malkinson, and Y. Weisman. 1992. Polymerase chain reaction for differentiation between pathogenic and non-pathogenic serotype 1 Marek's disease viruses (MDV) and vaccine viruses of MDV-serotypes 2 and 3. J. Virol. Methods 40:307-322. [DOI] [PubMed] [Google Scholar]
- 4.Biggs, P. M. 1975. Marek's disease—the disease and its prevention by vaccination. Br. J. Cancer 31(Suppl. 2):152-155. [PMC free article] [PubMed] [Google Scholar]
- 5.Buckmaster, A. E., S. D. Scott, M. J. Sanderson, M. E. Boursnell, N. L. Ross, and M. M. Binns. 1988. Gene sequence and mapping data from Marek's disease virus and herpesvirus of turkeys: implications for herpesvirus classification. J. Gen. Virol. 69:2033-2042. [DOI] [PubMed] [Google Scholar]
- 6.Bülow, V., and P. M. Biggs. 1975. Differentiation between strains of Marek's disease. Avian Pathol. 6:395-403. [DOI] [PubMed] [Google Scholar]
- 7.Calnek, B., and R. L. Witter. 1997. Neoplastic diseases: Marek's disease, p. 369-413. In B. Calnek (ed.), Disease of poultry, 10th ed. Iowa State University Press, Ames, Iowa.
- 8.Calnek, B. W. 1972. Effects of passive antibody on early pathogenesis of Marek's disease. Infect. Immun. 6:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calnek, B. W., J. C. Carlisle, J. Fabricant, K. K. Murthy, and K. A. Schat. 1979. Comparative pathogenesis studies with oncogenic and nononcogenic Marek's disease viruses and turkey herpesvirus. Am. J. Vet. Res. 40:541-548. [PubMed] [Google Scholar]
- 10.Calnek, B. W., K. A. Schat, M. C. Peckham, and J. Fabricant. 1983. Field trials with a bivalent vaccine (HVT and SB-1) against Marek's disease. Avian Dis. 27:844-849. [PubMed] [Google Scholar]
- 11.Calnek, B. W., W. R. Shek, and K. A. Schat. 1981. Latent infections with Marek's disease virus and turkey herpesvirus. J. Natl. Cancer Inst. 66:585-590. [PubMed] [Google Scholar]
- 12.Cohen, J. I., and K. E. Seidel. 1993. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc. Natl. Acad. Sci. USA 90:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui, Z. Z., L. F. Lee, E. J. Smith, R. L. Witter, and T. S. Chang. 1988. Monoclonal-antibody-mediated enzyme-linked immunosorbent assay for detection of reticuloendotheliosis viruses. Avian Dis. 32:32-40. [PubMed] [Google Scholar]
- 14.Cui, Z. Z., D. Yan, and L. F. Lee. 1990. Marek's disease virus gene clones encoding virus-specific phosphorylated polypeptides and serological characterization of fusion proteins. Virus Genes 3:309-322. [DOI] [PubMed] [Google Scholar]
- 15.Ferrin, L. J., and R. D. Camerini-Otero. 1991. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science 254:1494-1497. [DOI] [PubMed] [Google Scholar]
- 16.Izumiya, Y., H. K. Jang, M. Ono, and T. Mikami. 2001. A complete genomic DNA sequence of Marek's disease virus type 2, strain HPRS24. Curr. Top. Microbiol. Immunol. 255:191-221. [DOI] [PubMed] [Google Scholar]
- 17.Kaaden, O. R., A. Scholz, A. Ben-Zeev, and Y. Becker. 1977. Isolation of Marek's disease virus DNA from infected cells by electrophoresis on polyacrylamide gels. Arch. Virol. 54:75-83. [DOI] [PubMed] [Google Scholar]
- 18.Lee, L. F., X. Liu, and R. L. Witter. 1983. Monoclonal antibodies with specificity for three different serotypes of Marek's disease viruses in chickens. J. Immunol. 130:1003-1006. [PubMed] [Google Scholar]
- 19.Lee, L. F., P. Wu, D. Sui, D. Ren, J. Kamil, H. J. Kung, and R. L. Witter. 2000. The complete unique long sequence and the overall genomic organization of the GA strain of Marek's disease virus. Proc. Natl. Acad. Sci. USA 97:6091-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, J. L., S. F. Lin, L. Xia, P. Brunovskis, D. Li, I. Davidson, L. F. Lee, and H. J. Kung. 1999. MEQ and V-IL8: cellular genes in disguise? Acta Virol. 43:94-101. [PubMed] [Google Scholar]
- 21.Lupiani, B., L. F. Lee, and S. M. Reddy. 2001. Protein-coding content of the sequence of Marek's disease virus serotype 1. Curr. Top. Microbiol. Immunol. 255:159-190. [DOI] [PubMed] [Google Scholar]
- 22.Martins-Green, M. 2001. The chicken chemotactic and angiogenic factor (cCAF), a CXC chemokine. Int. J. Biochem, Cell Biol. 33:427-432. [DOI] [PubMed] [Google Scholar]
- 23.Martins-Green, M., and T. Kelly. 1998. The chicken chemotactic and angiogenic factor (9E3 gene product): its angiogenic properties reside in the C-terminus of the molecule. Cytokine 10:819-830. [DOI] [PubMed] [Google Scholar]
- 24.McGeoch, D. 1989. The genomes of the human herpesviruses: contents, relationships, and evolution. Annu. Rev. Microbiol. 43:235-265. [DOI] [PubMed] [Google Scholar]
- 25.Morgan, R. W., J. L. Cantello, and C. H. McDermott. 1990. Transfection of chicken embryo fibroblasts with Marek's disease virus DNA. Avian Dis. 34:345-351. [PubMed] [Google Scholar]
- 26.Parcells, M. S., S.-F. Lin, R. L. Dienglewicz, V. Majerciak, D. R. Robinson, H.-C. Chen, Z. Wu, G. R. Dubyak, P. Brunovskis, H. D. Hunt, L. F. Lee, and H.-J. Kung. 2001. Marek's disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol. 75:5159-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payne, L. N., and M. Rennie. 1973. Pathogenesis of Marek's disease in chicks with and without maternal antibody. J. Natl. Cancer Inst. 51:1559-1573. [DOI] [PubMed] [Google Scholar]
- 28.Penfold, M. E., D. J. Dairaghi, G. M. Duke, N. Saederup, E. S. Mocarski, G. W. Kemble, and T. J. Schall. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. USA 96:9839-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy, S. M., B. Lupiani, I. M. Gimeno, R. F. Silva, L. F. Lee, and R. L. Witter. 2002. Rescue of a pathogenic Marek's disease virus with overlapping cosmid DNAs: use of a pp38 mutant to validate the technology for the study of gene function. Proc. Natl. Acad. Sci. USA 99:7054-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva, R. F., L. F. Lee, and G. F. Kutish. 2001. The genomic structure of Marek's disease virus. Curr. Top. Microbiol. Immunol. 255:143-158. [DOI] [PubMed] [Google Scholar]
- 31.Strieter, R. M., P. J. Polverini, D. A. Arenberg, and S. L. Kunkel. 1995. The role of CXC chemokines as regulators of angiogenesis. Shock 4:155-160. [DOI] [PubMed] [Google Scholar]
- 32.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2000. The genome of a very virulent Marek's disease virus. J. Virol. 74:7980-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witter, R., J. M. Sharma, and A. M. Fadly. 1980. Pathogenicity of variant Marek's disease virus isolants in vaccinated and unvaccinated chickens. Avian Dis. 24:210-232. [Google Scholar]
- 34.Witter, R. L. 1991. Attenuated revertant serotype 1 Marek's disease viruses: safety and protective efficacy. Avian Dis. 35:877-891. [PubMed] [Google Scholar]
- 35.Witter, R. L. 2001. Protective efficacy of Marek's disease vaccines. Curr. Top. Microbiol. Immunol. 255:57-90. [DOI] [PubMed] [Google Scholar]
- 36.Xing, Z., and K. A. Schat. 2000. Expression of cytokine genes in Marek's disease virus-infected chickens and chicken embryo fibroblast cultures. Immunology 100:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]