Abstract
We investigated the in vitro RNA dimerization properties of the untranslated leader RNA derived from human immunodeficiency virus type 1 variants circulating in an individual with a low viral load and slow disease progression. The leader sequences of these viruses contain highly unusual polymorphisms within the dimerization initiation site (DIS): an insert that abolishes dimerization and a compensatory substitution. The dimerization of leader RNA from late stages of infection is further improved by additional mutations outside the DIS motif that facilitate a secondary structure switch from a dimerization-incompetent to a dimerization-competent RNA conformation.
Disease induction following human immunodeficiency virus type 1 (HIV-1) infection, or the ability to control the infection, is thought to depend on at least three factors: genetic susceptibility of the host, the ability of the host to elicit an effective immune response, and the virulence or fitness of the incoming virus. The magnitude of viral replication, as measured by plasma viral load, is predictive of the rate of disease progression, and asymptomatic infection usually correlates with low to undetectable viral load and CD4+ lymphocyte homeostasis (30, 31). Studies of individuals with nonprogressive HIV-1 infection have provided valuable information on host factors that are important for viral replication. The presence of polymorphisms within the coreceptors CCR5 and CCR2 in asymptomatic patients underscores the importance of these cellular proteins in viral replication (32). Similarly, polymorphisms within viral genes that appear in nonprogressors underscore a role for those proteins in aspects of viral replication or pathogenicity. Deletions and difficult-to-revert polymorphisms associated with delayed disease progression have been identified in the HIV-1 Nef, Gag, Rev, Vpu, and Vpr proteins (1, 4-6, 9, 11, 22, 24, 40, 41, 46).
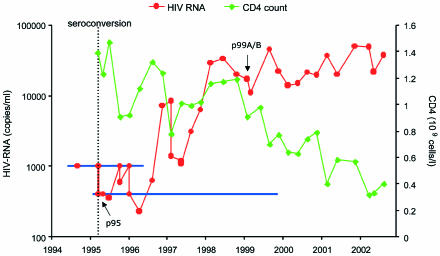
From the Amsterdam HIV-AIDS cohort of homosexual men, we identified individual H0671, who became infected with a subtype B virus in 1995 but who had a low viral load in the first two years of infection despite not receiving antiretroviral therapy (Fig. 1). Individual H0671 entered the Amsterdam Cohort Studies on 5 June 1986 and tested seropositive for HIV-1 specific antibodies on 22 March 1995. The individual is a 48-year-old male who is heterozygous for the Δ32 mutation in the CCR5 coreceptor. For two years after seroconversion, viral RNA in the blood remained below 1,000 copies/ml and then gradually increased towards approximately 50,000 copies/ml. This pattern is rather unusual, as less than 1% of the infected individuals in our cohort had a viral load below 1,000 copies/ml in the first year after seroconversion. The CD4 count of H0671 has remained fairly stable since the time of infection, but a decline was apparent as of 1999, and this decline coincides with the increase in viral load. This atypical disease course may suggest that patient H0671 was infected with a poorly replicating virus that gained replicative potential over time.
FIG. 1.
Slow disease progression in patient H0671. Viral load as measured by viral RNA in the blood is indicated by a red line. Blue lines indicate the detection level of two different assay systems (NucliSens, which was abandoned in 1996, and QT NASBA). The number of CD4-positive cells is indicated in green. A dotted line indicates the time of seroconversion. Arrows indicate time points at which the samples were analyzed in this study.
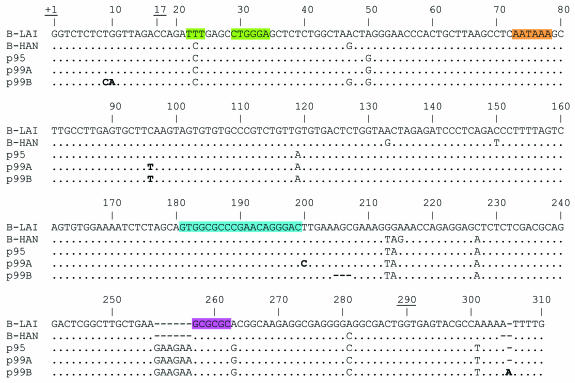
To identify polymorphisms in the viral genome that are potentially responsible for the low viral load at the onset of infection, we performed full genome sequencing of the viruses circulating in patient H0671. Serum samples and peripheral blood mononuclear cells (PBMCs) were collected every 3 months to determine viral load and CD4+ cell counts. Biological clones were generated from the patient PBMCs that were cocultivated with phytohemagglutinin-stimulated healthy donor PBMCs. In total, the complete genome of seven biological clones from early, intermediate, and late stages of infection were sequenced by the novel PALM method (15). We noted highly unusual variations in the otherwise extremely conserved nontranslated leader RNA domain. This untranslated leader contains important RNA motifs that regulate key steps in the virus replication cycle such as that of gene expression (transcription, RNA processing, and translation) and virion-associated processes (genome packaging and reverse transcription) (2). In the patient-derived leader sequence, a 6-nucleotide insert at position 256 and an A263G substitution downstream of the insert are present (Fig. 2; positions are relative to the transcriptional start site in the HIV-1LAI prototype). The GAAGAA insert is a double repeat of the preceding GAA triplet, suggesting that it may have occurred through slippage during reverse transcription. Both the insert and the substitution remain fixed in the virus population throughout the course of infection. The insert is not observed in any of the viral isolates recorded in the Los Alamos HIV sequence database, but the substitution has previously been reported (39).
FIG. 2.
Unusual sequence variation in the HIV-1 untranslated leader RNA. The alignment of the HIV-1 leader RNA sequence as found in the virus population of patient H0671 and two subtype B reference strains, LAI and HAN, are shown. These reference sequences were included to indicate common variation in the leader sequence. Patient-derived sequences are from the time of seroconversion (p95, taken in 1995) and from 1999 (p99A and B), when the viral load had increased. The numbering of the residues corresponds to the LAI isolate, with +1 referring to the transcriptional start site. The termini of transcripts used in this study are underlined (5′, positions +1 and 17; 3′, position 290). Several structure and sequence elements are highlighted: the TAR hairpin bulge and loop (green), the AAUAAA polyadenylation signal (orange), the primer binding site (blue), and the DIS palindrome (pink). Nucleotides in boldface indicate sequence changes uniquely observed in virus isolates from late stages of infection.
These unusual mutations in the patient isolate are located within the dimerization initiation site (DIS) hairpin that is thought to mediate the dimerization of the genomic RNA that is packaged into virions (14, 33-35, 37, 38, 45). DIS-mediated RNA dimerization is well characterized biochemically and occurs through intermolecular base pairing of two loop-exposed palindromes, an interaction referred to as loop-loop kissing (25, 37). However, we recently demonstrated that HIV-1 leader RNA monomers adopt a ground state conformation in which the DIS palindrome is occluded by a long-distance interaction (LDI) with upstream sequences (20, 21), and this renders the RNA dimerization incompetent. Refolding of the RNA into a branched conformation with multiple hairpins (BMH) triggers the exposure of the DIS palindrome, which allows the intermolecular kissing-loop interaction to take place (Fig. 3A). We have suggested that the LDI-to-BMH conformational rearrangement may serve as a molecular switch to coordinate early and late events in the virus replication cycle (20).
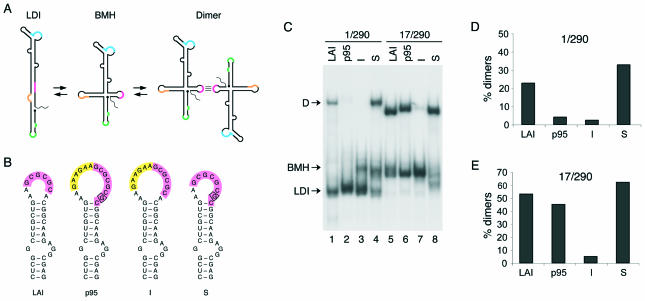
FIG. 3.
RNA dimerization properties of RNA corresponding to the HIV-1 leader as found in patient H0671. (A) Alternative secondary structures of the HIV-1 leader RNA. Several structure and sequence elements are highlighted: the TAR hairpin bulge and loop (green), the AAUAAA polyadenylation signal (orange), the primer binding site (blue), and the DIS palindrome (pink). The most 5′ residue of the transcript is indicated by a dot. The ground state conformation contains an LDI between the poly(A) and DIS domains. The alternate conformation exposes the poly(A) and DIS hairpins and takes the form of a branched structure (BMH). The BMH monomer allows the formation of RNA dimers through the base pairing of the palindromes of two DIS loops. (B) Secondary structure models of the wild-type DIS element of the LAI strain, the p95 patient-derived DIS (I+S), and two mutants containing the individual I or S. The DIS palindrome is shown in pink, the 6-nucloetide insert is shown in orange, and the A263G substitution is boxed. (C) Nondenaturing gel showing the RNA dimerization properties of the 1-290 and 17-290 transcripts. Arrows indicate the dimeric (D) and monomeric (BMH and LDI) RNA. (D) RNA dimer yield for the 1-290 transcripts shown in panel C. (E) RNA dimer yield for the 17-290 transcripts shown in panel C.
The DIS hairpin is extremely conserved in HIV-1 isolates, and its natural variation is restricted to GCGCGC or GUGCAC palindromes in the loop of the hairpin, and isolates have been reported with a hybrid GUGCGC palindrome (2, 44). This finding suggests that the rare mutations in the patient isolate may have compromised the functionality of the DIS element and, consequently, viral replication. The insert is located immediately upstream of the DIS palindrome and increases the loop size of the hairpin (Fig. 3B). Consequently, the palindrome is shifted away from its central loop-exposed position, which is likely to negatively influence RNA dimerization (13, 23, 38). The substitution is located directly downstream of the palindrome, also in the single-stranded loop, and extends the palindrome from GCGCGC to GCGCGCGC.
To investigate dimerization properties of this unusual DIS element, we synthesized a transcript at positions 1 through 290 (1-290 transcript) corresponding to the untranslated leader RNA as present in patient H0671 at the early stage of infection (p95) with both the insert (I) and the substitution (S). Two mutant transcripts that individually contain the I and S mutations within the DIS motif were made (Fig. 3B). As a wild-type control, we used the RNA of the subtype B LAI isolate. The procedures of RNA synthesis, dimerization, and nondenaturing gel electrophoresis have been described elsewhere (19-21). A representative experiment is shown in Fig. 3C, which demonstrates that the p95 (I+S) and mutant I transcripts produce a low level of RNA dimers. The dimerization of the p95 sample (I+S) is slightly more efficient than that of the I mutant, and this result was reproducibly observed in experiments addressing the temperature dependency of RNA dimerization (results not shown). The dimerization of the S mutant is more efficient than that of the wild-type control (Fig. 3C and D). These results demonstrate that the 6-nucleotide insert is detrimental to DIS-dependent RNA dimerization and that the substitution, which extends the DIS palindrome, enhances RNA dimerization of both LAI and patient-derived RNA.
In a parallel experiment, we analyzed the same set of RNAs in the context of a 5′ truncated transcript at positions 17 to 290 (17-290 transcript). This was done to dissect the complex HIV-1 RNA dimerization mechanism, which depends on the intramolecular LDI-to-BMH secondary structure rearrangement of the leader RNA prior to dimerization (20, 21). The truncation of the 5′ TAR element causes a preference for the BMH conformation in which the DIS palindrome is constitutively exposed (19). In this context, there was indeed a strong increase in the RNA dimer yield compared to that for the full-length transcripts (Fig. 3B, C, and D). Poor dimerization of the I mutant was still apparent, as was the efficient dimerization of the S mutant (Fig. 3C and E). However, the dimerization of p95 (I+S) was significantly improved, albeit slightly less efficiently than the wild-type control. These results confirm the detrimental effect of the insert on RNA dimerization and the compensatory nature of the substitution. Importantly, these results indicate that the patient-derived leader RNA has a dimerization defect. This defect is not absolute, and significant amounts of dimers can be induced when the transcript is forced in the dimerization-competent BMH conformation. The isolated I mutant has a more severe dimerization defect that cannot be rescued by this strategy.
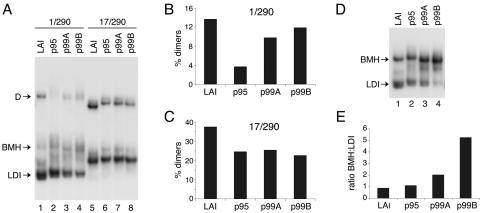
A comparison of the viral sequences from early (p95) and late (p99A and p99B) stages of infection revealed a number of additional sequence changes within the untranslated leader RNA but outside the DIS motif (Fig. 2). Both late sequences have a C96U substitution. In addition, p99A has a U200C substitution, and P99B contains a 3-nucleotide deletion at positions 205 to 207 and three substitutions within the TAR element (nucleotides 1 through 57). The latter changes do not significantly alter the secondary structure of TAR and are similar to sequence variation in other HIV-1 isolates. This is also the case for the variability of the A-rich sequence spanning positions 301 to 305. Interestingly, the C96U substitution common to the p99A and p99B sequences destabilizes the LDI structure by introducing a weak G-U base pair instead of the natural G-C base pair. This may improve the RNA dimerization capacity of the untranslated leader by favoring the dimerization-competent BMH conformation, which exposes the DIS hairpin. The additional sequence changes in p99A and p99B may also affect the LDI-to-BMH equilibrium and subsequent dimerization. We synthesized RNA corresponding to wild-type LAI, p95, p99A, and p99B sequences, and RNA dimerization was monitored in the context of the 1-290 and 17-290 transcripts. A significant difference between the early and late samples is observed when the 1-290 transcript is used. Both p99 samples produce two- to threefold more dimers than the p95 sample (Fig. 4A and B). Interestingly, this difference in dimerization efficiency is lost when the 17-290 transcript is used (Fig. 4C). This finding supports the idea that the late samples have improved dimerization through a shift in the LDI-BMH equilibrium because the differential dimerization is absent in transcripts that are already in the BMH conformation. To directly test whether the LDI-BMH equilibrium is altered, we analyzed the 1-290 transcripts under conditions at which the differentially migrating LDI and BMH conformations coexist during electrophoresis (20). This result is achieved by incubating the RNA in Tris buffer (10 mM Tris-HCl, pH 7.5) prior to electrophoresis. Indeed, the p99A and p99B samples demonstrated a significant shift towards the BMH conformation compared to wild-type LAI and the p95A samples (Fig. 4D and E).
FIG. 4.
RNA dimerization properties of patient-derived HIV-1 leader RNA from early and late stages of infection. (A) Nondenaturing gel showing the dimerization properties of early (p95) and late (p99A and p99B) patient-derived HIV-1 leader RNA in the context of 1-290 and 17-290 transcripts. Arrows indicate the dimeric (D) and monomeric (BMH and LDI) RNA. (B) RNA dimer yield for the 1-290 transcripts shown in panel A. (C) RNA dimer yield for the 17-290 transcripts shown in panel A. (D) Analysis of the LDI-BMH equilibrium of the LAI and patient-derived leader RNA. Nondenaturing gel showing the LDI and BMH conformations of the LAI, p95, p99A, and p99B transcripts (positions 1 to 290). (E) Ratio of BMH to LDI conformers as determined by quantification of the gel shown in panel D.
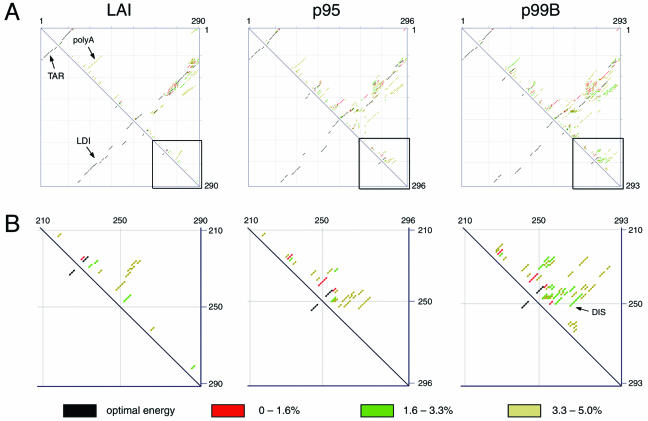
We used the Mfold program to analyze the LDI and BMH structures of the different transcripts in further detail (29, 47, 48). Standard settings were used, corresponding to 1 M NaCl at 37°C, with a 5% suboptimality range. Figure 5A shows the Mfold-generated dot plot representations for wild-type LAI, p95, and p99B leader sequences. In this dot plot, each individual dot represents a base pair of which the residues are defined by the x and y axes of the graph. Within each dot plot, the upper-right triangle shows all predicted base pairs from the 5% most stable structures, and the likelihood of base pair formation is indicated by color coding (Fig. 5A). The lower-left triangle shows the single lowest energy structure, which includes the TAR hairpin and the extended LDI helix for wild-type LAI and p95 sequences. The DIS hairpin is not predicted for either sequence, which is in accord with the preference for the LDI conformation (Fig. 5A and B). The early p95 (I+S) sequence does produce a number of small helices within the DIS region, but none of these correspond to the correctly folded DIS hairpin (Fig. 5B). However, the late p99B sequence shows a marked increase in possible foldings within the DIS domain at the expense of the LDI helix, even though the differences between the early (p95) and late (p99) sequences are not within the DIS motif itself (2). Importantly, correct folding of the DIS hairpin is predicted for the late sequences (Fig. 5B), and this coincides with a higher probability of folding the poly(A) hairpin, which is another hallmark of the BMH conformation. Thus, the structure prediction confirms that the additional changes in the leader sequence of the late samples cause improved folding of the BMH conformation, thereby exposing the DIS hairpin for dimerization.
FIG. 5.
RNA secondary structure prediction of the LAI and patient-derived untranslated leader RNA. (A) Dot plot representation generated for the 1-290 leader sequence by the Mfold program. In the dot plot for the wild-type LAI sequence, the positions of the TAR, poly(A) (polyA), and LDI duplexes are indicated with arrows. Note that the p95 transcript is 6 nucleotides longer than the LAI RNA due to the insert in the DIS domain. The p99B RNA contains this insert plus an additional deletion, making it only 3 nucleotides longer than the LAI RNA. Color coding indicates probability for each base pair in the upper-right triangles, as shown below the plots. The lower-left triangles show the optimal fold. (B) Close-up view of the structure prediction in the DIS domain, which is indicated by a box in panel A. The LAI and p95 sequences show no solutions with the properly folded DIS hairpin. The p99B sequence shows a proper folding of the DIS hairpin, which is indicated by an arrow.
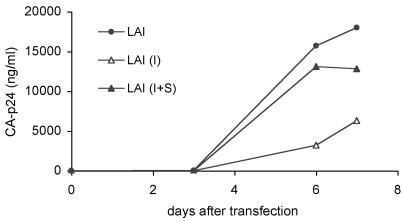
We next investigated the effect of these rare DIS mutations on virus replication by constructing mutant molecular clones of the LAI isolate (LAI I and LAI I+S). Unexpectedly, no gross replication defects were observed for any of these mutants, both in a T-cell line (SupT1) and PBMCs. However, in transfection experiments in SupT1 cells, we did observe improved replication of the I+S mutant compared to that for the I mutant (Fig. 6). No differences in viral growth were apparent upon transfection of PBMCs (data not shown), which is consistent with the recent observation that DIS mutants have rather healthy phenotypes in these cells (18). We performed additional virus competition experiments to substantiate the difference in replicative potential between the I and I+S mutants. In two independent experiments, SupT1 cells were cotransfected with equal amounts of each molecular clone, and the virus population was sequenced 4 weeks (experiment 1) or 6 weeks (experiment 2) after transfection. We consistently observed that viruses containing both the insert and the substitution (I+S) dominated the viral culture by outcompeting viruses containing only the insert. This result supports the notion that the substitution compensates for the defect imposed by the insert. Interestingly, the same A263G substitution was observed upon the prolonged culturing of a mutant virus in which the palindrome was truncated to GC. In this case, the A263G substitution partially repaired the palindrome to GCGC (3). The HIV-1 sequence database also contains two related virus isolates with the A263G substitution in the context of a DIS element with an additional G257A substitution that truncates the palindrome (accession numbers AF538304 and AF538307). Again, the A263G substitution may be a second-site escape mutation that repairs damage to the palindromic sequence. An additional isolate from a long-term nonprogressor in the database has the A263G substitution in an otherwise wild-type DIS element (39). Thus, this virus contains an extended GCGCGCGC palindrome, which was previously selected as a revertant of a DIS mutated HIV-1 variant (3).
FIG. 6.
The effect of leader RNA polymorphisms on virus replication. Replication of the wild-type HIVLAI virus and two mutant viruses that contain I and I+S as found in virus isolates in patient H0671. SupT1 cells were transfected with 1 μg of the virus molecular clones, and replication was monitored by measuring CA-p24 production by enzyme-linked immunosorbent assay (10).
We tried to identify a putative precursor virus that contained only the insert from blood samples taken prior to seroconversion of patient H0671, but we did not detect such a virus. Instead, these samples contained leader sequences identical to those of the p95 isolate. Thus, it is possible that patient H0671 was initially infected with a virus containing both the insert and the substitution and that the compensatory substitution may already have occurred in the donor.
We also performed transfection experiments to compare the replication of virus molecular clones containing the leader RNA from the early and late stages of infection, but these mutants showed a level of replication that is very similar to that of the wild-type LAI isolate. It is surprising that the differences in RNA dimerization between the early and late virus isolates that we measured in vitro do not translate into differences in virus replication. However, discrepancies between in vitro RNA dimerization and cell culture infection studies are common. Since the first demonstration that the DIS hairpin drives dimerization in vitro (37), many studies have confirmed the kissing-loop dimerization mechanism (8, 14, 20, 21, 23, 27, 28, 33, 34, 45). Virus replication studies, on the other hand, show enormous variability in the extent to which mutations in the DIS domain affect replication and the RNA dimer content of progeny virions. Some severe DIS mutants affect virus replication marginally and yield viruses with dimeric genomes (3, 7, 18, 26, 42, 43), whereas other DIS mutants have predominantly monomeric genomes and a severe replication defect (7, 26, 36, 43). In one study, a mutant with a deleted DIS palindrome contained a normal dimeric RNA genome and had a mild replication defect (3). It is possible that the cell culture system does not accurately mimic the restraints imposed on RNA dimerization by the natural course of infection. Indeed, it was recently demonstrated that the phenotype of DIS mutants varies with the cell type used in cell culture infections (18).
We have shown here that the leader RNA from viruses circulating in patient H0671 contain rare mutations in the noncoding DIS domain that inhibit the formation of RNA dimers in vitro. Leader RNA from later stages of infection, when the viral load in the patient had risen, showed improved RNA dimerization due to a shift in the LDI-BMH equilibrium that favored the dimerization-competent BMH conformation. These results are consistent with virus evolution towards replication-improved species from a defective parental virus in patient H0671. The ability to correlate reversion pathways of mutant viruses in cell culture with an adjustment of the LDI-BMH equilibrium has previously been shown (19, 20). Whether the rare mutations in the leader RNA of the patient H0671 virus population directly contributed to the low viral load at the initial stage of infection and the subsequent rise in viral load is not apparent at this point. This will require a reliable assay system to investigate the role of the DIS element in vivo. Nonetheless, our results suggest that the slow disease progression of patient H0671 may be due in part to impaired fitness of the incoming virus with a suboptimal DIS element. It is possible that factors other than the viral DIS defect contributed to the slow disease progression in this patient. We already mentioned that H0671 is heterozygous for the Δ32 deletion in the CCR5 gene that encodes an important coreceptor for HIV-1 entry. There is also evidence for a rigorous cytotoxic T-lymphocyte response in this patient, followed by the evolution of cytotoxic T-lymphocyte escape variants (15). Another peculiarity is the fact that this patient was infected with a zidovudine-resistant virus variant with the 215Y mutation in the RT enzyme. The idea that this mutation may have negatively influenced viral fitness is underscored by the evolution of this amino acid, first to 215D and subsequently to 215N (12, 16, 17). These findings, combined with the DIS defect reported in this study, suggest that the attenuation of the viral quasispecies in patient H0671 may be due to multiple genetic defects.
Acknowledgments
We thank M. Bakker and S. Jurriaans for clinical samples and M. Bakker for patient data analysis. Clinical samples were obtained through the Amsterdam Cohort Study (ACS) on HIV-AIDS, and we thank the participants for their cooperation over many years. We thank Wim van Est for the artwork.
This work was sponsored by The Netherlands Organization for Scientific Research (NWO-CW) and the ZonMW-VIVI program.
REFERENCES
- 1.Alexander, L., E. Weiskopf, T. C. Greenough, N. C. Gaddis, M. R. Auerbach, M. H. Malim, S. J. O'Brien, B. D. Walker, J. L. Sullivan, and R. C. Desrosiers. 2000. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J. Virol. 74:4361-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acids Res. Mol. Biol. 54:1-34. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout, B., and J. L. B. van Wamel. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binley, J. M., X. Jin, Y. Huang, L. Zhang, Y. Cao, D. D. Ho, and J. P. Moore. 1998. Persistent antibody responses but declining cytotoxic T-lymphocyte responses to multiple human immunodeficiency virus type 1 antigens in a long-term nonprogressing individual with a defective p17 proviral sequence and no detectable viral RNA expression. J. Virol. 72:3472-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birch, M. R., J. C. Learmont, W. B. Dyer, N. J. Deacon, J. J. Zaunders, N. Saksena, A. L. Cunningham, J. Mills, and J. S. Sullivan. 2001. An examination of signs of disease progression in survivors of the Sydney Blood Bank Cohort (SBBC). J. Clin. Virol. 22:263-270. [DOI] [PubMed] [Google Scholar]
- 6.Carl, S., R. Daniels, A. J. Iafrate, P. Easterbrook, T. C. Greenough, J. Skowronski, and F. Kirchhoff. 2000. Partial “repair” of defective NEF genes in a long-term nonprogressor with human immunodeficiency virus type 1 infection. J. Infect. Dis. 181:132-140. [DOI] [PubMed] [Google Scholar]
- 7.Clever, J. L., and T. G. Parslow. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clever, J. L., M. L. Wong, and T. G. Parslow. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70:5902-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938-1941. [DOI] [PubMed] [Google Scholar]
- 10.Das, A. T., B. Klaver, B. I. F. Klasens, J. L. B. van Wamel, and B. Berkhout. 1997. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 71:2346-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, V. A. Lawson, S. Crowe, A. Maerz, S. Sonza, J. Learmont, J. S. Sullivan, A. Cunningham, D. Dwyer, D. Dowton, and J. Mills. 1995. Genomic structure of an attenuated quasi species of HIV-1 from blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 12.de Ronde, A., M. van Dooren, L. van der Hoek, D. Bouwhuis, E. de Rooij, B. van Gemen, R. de Boer, and J. Goudsmit. 2001. Establishment of new transmissible and drug-sensitive human immunodeficiency virus type 1 wild types due to transmission of nucleoside analogue-resistant virus. J. Virol. 75:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dirac, A. M., H. Huthoff, J. Kjems, and B. Berkhout. 2002. Requirements for RNA heterodimerization of the human immunodeficiency virus type 1 (HIV-1) and HIV-2 genomes. J. Gen. Virol. 83:2533-2542. [DOI] [PubMed] [Google Scholar]
- 14.Ennifar, E., P. Walter, B. Ehresmann, C. Ehresmann, and P. Dumas. 2001. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 8:1064-1068. [DOI] [PubMed] [Google Scholar]
- 15.Geels, M. J., M. Cornelissen, H. Schuitemaker, K. Anderson, D. Kwa, J. Maas, J. T. Dekker, E. Baan, F. Zorgdrager, R. van den Burg, M. van Beelen, V. V. Lukashov, T.-M. Fu, W. Paxton, L. van der Hoek, S. A. Dubey, J. W. Shiver, and J. Goudsmit. 2003. Identification of sequential viral escape mutants associated with altered T-cell responses in a human immunodeficiency virus type 1-infected individual. J. Virol. 77:12430-12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goudsmit, J., A. de Ronde, E. de Rooij, and R. de Boer. 1997. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J. Virol. 71:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goudsmit, J., A. de Ronde, D. D. Ho, and A. S. Perelson. 1996. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J. Virol. 70:5662-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill, M. K., M. Shehu-Xhilaga, S. M. Campbell, P. Poumbourios, S. M. Crowe, and J. Mak. 2003. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J. Virol. 77:8329-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huthoff, H., and B. Berkhout. 2001. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 29:2594-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huthoff, H., and B. Berkhout. 2001. Two alternating structures for the HIV-1 leader RNA. RNA 7:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huthoff, H., and B. Berkhout. 2002. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry 41:10439-10445. [DOI] [PubMed] [Google Scholar]
- 22.Iversen, A. K., E. G. Shpaer, A. G. Rodrigo, M. S. Hirsch, B. D. Walker, H. W. Sheppard, T. C. Merigan, and J. I. Mullins. 1995. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J. Virol. 69:5743-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jossinet, F., J.-C. Paillart, E. Westhof, T. Hermann, E. Skripkin, J. S. Lodmell, C. Ehresmann, B. Ehresmann, and R. Marquet. 1999. Dimerization of HIV-1 genomic RNA of subtypes A and B: RNA loop structure and magnesium binding. RNA 5:1222-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchhoff, F., T. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 25.Laughrea, M., and L. Jette. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464-13474. [DOI] [PubMed] [Google Scholar]
- 26.Laughrea, M., N. Shen, L. Jette, and M. A. Wainberg. 1999. Variant effects of non-native kissing-loop hairpin palindromes on HIV replication and HIV RNA dimerization: role of stem-loop B in HIV replication and HIV RNA dimerization. Biochemistry 38:226-234. [DOI] [PubMed] [Google Scholar]
- 27.Lodmell, J. S., C. Ehresmann, B. Ehresmann, and R. Marquet. 2000. Convergence of natural and artificial evolution on an RNA loop-loop interaction: the HIV-1 dimerization initiation site. RNA 6:1267-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodmell, J. S., C. Ehresmann, B. Ehresmann, and R. Marquet. 2001. Structure and dimerization of HIV-1 kissing loop aptamers. J. Mol. Biol. 311:475-490. [DOI] [PubMed] [Google Scholar]
- 29.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 30.Mellors, J. W. 1996. Closing in on human immunodeficiency virus-1. Nat. Med. 2:274-275. [DOI] [PubMed] [Google Scholar]
- 31.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 32.Michael, N. L. 1999. Host genetic influences on HIV-1 pathogenesis. Curr. Opin. Immunol. 11:466-474. [DOI] [PubMed] [Google Scholar]
- 33.Mujeeb, A., J. L. Clever, T. M. Billeci, T. L. James, and T. G. Parslow. 1998. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 5:432-436. [DOI] [PubMed] [Google Scholar]
- 34.Muriaux, D., P. Fosse, and J. Paoletti. 1996. A kissing complex together with a stable dimer is involved in the HIV-1 LAI RNA dimerization process in vitro. Biochemistry 35:5075-5082. [DOI] [PubMed] [Google Scholar]
- 35.Muriaux, D., P.-M. Girard, B. Bonnet-Mathoniere, and J. Paoletti. 1995. Dimerization of HIV-1 LAI RNA at low ionic strength. J. Biol. Chem. 270:8209-8216. [DOI] [PubMed] [Google Scholar]
- 36.Paillart, J.-C., L. Berthoux, M. Ottmann, J.-L. Darlix, R. Marquet, B. Ehresmann, and C. Ehresmann. 1996. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 70:8348-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paillart, J.-C., E. Skripkin, B. Ehresmann, C. Ehresmann, and R. Marquet. 1996. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 93:5572-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paillart, J.-C., E. Westhof, C. Ehresmann, B. Ehresmann, and R. Marquet. 1997. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J. Mol. Biol. 270:36-49. [DOI] [PubMed] [Google Scholar]
- 39.Quiñones-Mateu, M. E., A. Mas, T. L. de Lera, V. Soriano, J. Alcamí, M. M. Lederman, and E. Domingo. 1998. LTR and tat variability of HIV-1 isolates from patients with divergent rates of disease progression. Virus Res. 57:11-20. [DOI] [PubMed] [Google Scholar]
- 40.Salvi, R., A. R. Garbuglia, A. Di Caro, S. Pulciani, F. Montella, and A. Benedetto. 1998. Grossly defective nef gene sequences in a human immunodeficiency virus type 1-seropositive long-term nonprogressor. J. Virol. 72:3646-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawai, E. T., M. S. Hamza, M. Ye, K. E. S. Shaw, and P. A. Luciw. 2000. Pathogenic conversion of live attenuated simian immunodeficiency virus vaccines is associated with expression of truncated Nef. J. Virol. 74:2038-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen, N., L. Jetté, C. Liang, M. A. Wainberg, and M. Laughrea. 2000. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J. Virol. 74:5729-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen, N., L. Jetté, M. A. Wainberg, and M. Laughrea. 2001. Role of stem B, loop B, and nucleotides next to the primer binding site and the kissing-loop domain in human immunodeficiency virus type 1 replication and genomic-RNA dimerization. J. Virol. 75:10543-10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.St. Louis, D. C., D. Gotte, E. Sanders-Buell, D. W. Ritchey, M. O. Salminen, J. K. Carr, and F. E. McCutchan. 1998. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection. J. Virol. 72:3991-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theilleux-Delalande, V., F. Girard, T. Huynh-Dinh, and J. Paoletti. 2000. The HIV-1 LAI RNA dimerization: thermodynamic parameters associated with the transition from the kissing complex to the extended dimer. Eur. J. Biochem. 267:2711-2719. [DOI] [PubMed] [Google Scholar]
- 46.Wang, B., Y. C. Ge, P. Palasanthiran, S. H. Xiang, J. Ziegler, D. E. Dwyer, C. Randle, D. Dowton, A. Cunningham, and N. K. Saksena. 1996. Gene defects clustered at the C terminus of the vpr gene of HIV-1 in long-term nonprogressing mother and child pair: in vivo evolution of vpr quasispecies in blood and plasma. Virology 223:224-232. [DOI] [PubMed] [Google Scholar]
- 47.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]
- 48.Zuker, M., and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.