Abstract
A novel coronavirus, severe acute respiratory syndrome coronavirus (SARS-CoV), has recently been identified as the causative agent of severe acute respiratory syndrome (SARS). SARS-CoV appears similar to other coronaviruses in both virion structure and genome organization. It is known for other coronaviruses that the spike (S) glycoprotein is required for both viral attachment to permissive cells and for fusion of the viral envelope with the host cell membrane. Here we describe the construction and expression of a soluble codon-optimized SARS-CoV S glycoprotein comprising the first 1,190 amino acids of the native S glycoprotein (S1190). The codon-optimized and native S glycoproteins exhibit similar molecular weight as determined by Western blot analysis, indicating that synthetic S glycoprotein is modified correctly in a mammalian expression system. S1190 binds to the surface of Vero E6 cells, a cell permissive to infection, as demonstrated by fluorescence-activated cell sorter analysis, suggesting that S1190 maintains the biologic activity present in native S glycoprotein. This interaction is blocked with serum obtained from recovering SARS patients, indicating that the binding is specific. In an effort to map the ligand-binding domain of the SARS-CoV S glycoprotein, carboxy- and amino-terminal truncations of the S1190 glycoprotein were constructed. Amino acids 270 to 510 were the minimal receptor-binding region of the SARS-CoV S glycoprotein as determined by flow cytometry. We speculate that amino acids 1 to 510 of the SARS-CoV S glycoprotein represent a unique domain containing the receptor-binding site (amino acids 270 to 510), analogous to the S1 subunit of other coronavirus S glycoproteins.
Severe acute respiratory syndrome (SARS) is a recently described disease that has affected approximately 8,500 people worldwide with a mortality rate of approximately 10% (according to the World Health Organization). The causative agent of SARS is a newly identified coronavirus, SARS-CoV, first isolated by propagation on Vero E6 cells (5, 12, 17). The SARS-CoV genome has been sequenced, and the probable coding regions for viral proteins have been deduced. Like other coronaviruses, SARS-CoV is a positive-strand RNA virus that encodes four main structural proteins, M, N, E, and S (20). Genetic analysis of the coding regions has demonstrated that SARS-CoV is distinct from the three known antigenic groups of coronaviruses (5, 12); however, recent data studying the replicase gene suggest that SARS-CoV may be most related to group 2 coronaviruses (21).
The S glycoprotein, a 1,255-amino-acid type I membrane glycoprotein (20), is the prominent protein present in the viral membrane and presents as the typical spike structure found on all coronaviruses. SARS-CoV S glycoprotein domain structure has been deduced from sequence analysis (20). The S glycoprotein consists of a leader (amino acids 1 to 14), an ectodomain represented by amino acids 15 to 1190, a membrane-spanning domain (amino acids 1191 to 1227), and a short intracellular tail (amino acids 1227 to 1255) (20). The full-length SARS-CoV S glycoprotein has 23 potential N-linked glycosylation sites predicted by sequence analysis (20). For group 2 and group 3 coronaviruses, the S glycoprotein is posttranslationally cleaved into two noncovalently associated subunits, S1 and S2 (6, 15, 22, 23). The motif that leads to cleavage of the subunits in these coronaviruses (15) is not present in SARS-CoV, suggesting that cleavage of the SARS-CoV S glycoprotein does not occur (20).
Although the process by which SARS-CoV penetrates the cellular membrane has not been determined, the mechanism is most likely similar to that described for other coronaviruses. The S glycoprotein interacts with the cellular surface, and for coronaviruses HCoV-229E and mouse hepatitis virus (MHV) amino acids 1 to 547 (2) and 1 to 330 (13), respectively, are required for binding to the cellular receptor. This interaction is predicted to lead to conformational changes in the carboxy-terminal half of the S glycoprotein. This change culminates in fusion of the virus and host cell membranes, allowing for entry of the virus (25-27). Sequence analysis of the SARS-CoV S glycoprotein using the LearnCoil VMF software has predicted the presence of two coiled-coil motifs present at amino acids 900 to 974 and 1148 to 1190. These coiled-coil structures are present in the fusion domain of many varied viruses, including MHV (4, 11, 14) and human immunodeficiency virus type 1 (9), of which entry events have been predicted to occur as described above.
Here we describe the construction and expression of a codon-optimized gene encoding the soluble ectodomain (amino acids 1 to 1190) of the SARS-CoV S glycoprotein. Codon-optimized S glycoprotein (S1190) was secreted into the growth medium and purified by affinity chromatography. Expression levels of secreted S1190 glycoprotein were determined to be approximately 5 mg/liter after purification. The S1190 synthetic S glycoprotein was shown to have an apparent molecular mass of 170 kDa, a size similar to that observed for native S protein expressed in SARS-CoV-infected Vero E6 cells. Purified S1190 protein was readily detected by human SARS convalescent-phase serum (provided by Larry Anderson, Centers for Disease Control and Prevention [CDC]) as determined by Western blot analysis. Synthetic S glycoprotein could also bind to the surface of Vero E6 cells, demonstrating that soluble, codon-optimized S glycoprotein retains the biologic activity present in the native molecule. Carboxy-terminal truncations of S1190 were produced, and it was demonstrated that the amino acids 1 to 510 (S510) are required for binding to Vero E6 cell surfaces. Amino-terminal truncations of the S510 glycoprotein demonstrated that amino acids 270 to 510 contain the minimal receptor-binding domain of the SARS-CoV S glycoprotein.
MATERIALS AND METHODS
Construction of a synthetic gene encoding soluble codon-optimized SARS-CoV spike (S) protein and S protein fragments.
The amino acid sequence of the SARS-CoV (Urbani strain) S protein was obtained from the NCBI database (AAP13441). The soluble portion of the protein was determined to be the first 1,190 amino acids (of 1,255) and, as such, only the DNA encoding this sequence was synthesized. The DNA sequence was codon optimized for mammalian cell expression (1, 16), replacing the natural codons with the following optimum codons: alanine (GCC), arginine (CGC), asparagine (AAC), aspartic acid (GAC), cysteine (TGC), glutamic acid (GAG), glutamine (CAG), glycine (GGC), histidine (CAC), isoleucine (ATC), leucine (CTG), lysine (AAG), methionine (ATG), phenylalanine (TTC), proline (CCC), serine (TCC), threonine (ACC), tryptophan (TGG), tyrosine (TAC), and valine (GTG). Runs of Cs and Gs were avoided, to simplify both synthesis of oligonucleotides as well as PCR conditions. When these stretches of Gs and Cs occurred, suboptimal codons were used. The 5′ end of the gene was modified to include a restriction site for HindIII and an irrelevant upstream overhang to facilitate cloning. The 3′ end of the synthetic gene was similarly modified to include an XbaI site and overhang sequences.
A total of 104 oligonucleotides were obtained (Integrated DNA Technologies; polyacrylamide gel electrophoresis purified) that represented the entire coding region of both the sense and antisense strands of the S protein gene, as well as engineered restriction sites. The most-5′ oligonucleotide of each strand was a 35-mer and all others were 70-mers, resulting in a 35-bp overlap between strands. In essence, the oligonucleotides from the sense strand fully overlapped the oligonucleotides of the antisense strand, leaving no gaps. Construction of the codon-optimized gene was performed as follows. Thirteen groups of oligonucleotides were selected that contained eight oligonucleotides (four sense and four antisense) in each group. PCR was performed on each set in a reaction mixture containing 20 μM deoxynucleoside triphosphates, 30 pmol of end oligonucleotides, 10 pmol of internal oligonucleotides, 1× cloned Pfu reaction buffer (Stratagene), and 1 U of Turbo Pfu (Stratagene). Thirty cycles of thermocycling (95°C for 15 s, 62°C for 30 s, and 68°C for 2 min) were performed, and the PCR products were resolved on 1% agarose gels. Specific products were gel purified (Qiagen) and divided into four separate groups containing either three or four of the first-step PCR products. PCR was again performed on each group, using oligonucleotides corresponding to the most-5′ end of each strand. These four PCR products were resolved on 0.8% agarose gels and gel purified as before. The four PCR products were mixed and amplified using oligonucleotides corresponding to the 5′ end of each strand of the entire synthetic gene. This final amplification yielded the 3,605-bp sequence consisting of the synthetic gene flanked by restriction sites.
The final PCR product encoding the SARS-CoV S glycoprotein gene was digested with HindIII and XbaI and cloned into pcDNA3.1 Myc/His (Invitrogen) in frame with the c-myc and His6 epitope tags. The cloned gene was sequenced to confirm that no errors had been accumulated during the PCR process. Of the four clones sequenced, none had sequence errors and no further genetic manipulations were required.
Once the sequence of the full-length soluble SARS-CoV S glycoprotein gene was confirmed, DNA encoding carboxy-terminally truncated soluble S glycoproteins was synthesized by PCR amplifying the desired fragment from the vector containing the full-length, codon-optimized gene encoding the S glycoprotein. Since the codon-optimized S1190 gene was used as a template for PCR, all truncated constructs were also codon optimized. Truncations were then cloned into pcDNA3.1 Myc/His as described above, and the DNA sequence was confirmed.
N-terminal truncations were also synthesized. PCR was used to amplify the leader sequence of the S1190 gene, containing a 3′ overhang corresponding to downstream sequences. The downstream sequences were then amplified and combined with the leader-overhang PCR product. PCR was again performed to synthesize copies of a gene that consisted of the S1190 leader fused immediately 5′ of the downstream coding region. These constructs essentially created deletions between the leader peptide and the desired downstream sequence.
Cells and cell culture.
HEK-293T/17 and Vero E6 cells, obtained from the American Type Culture Collection, were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 100 IU of penicillin-streptomycin (complete DMEM) at 37°C with 5% CO2. To harvest cells, phosphate-buffered saline (PBS) containing 5 mM EDTA was added to the tissue culture dish and incubated for 5 min at room temperature.
Expression and purification of codon-optimized S glycoproteins.
All constructs were transfected into HEK-293T/17 cells using Lipofectamine 2000 (Invitrogen) as described by the manufacturer. Briefly, cells were grown to 80% confluence in 150-mm tissue culture dishes in 15 ml of DMEM-10% fetal calf serum (FCS). Thirty micrograms of DNA mixed with 75 μl of Lipofectamine 2000 was added to the cells, and plates were incubated overnight at 37°C. Medium was removed and stored, and fresh complete DMEM was added to the cells. Cells were incubated for an additional 24 h, at which time 3 mM sodium butyrate (Sigma) was added to the medium. An additional 24-h incubation was performed, and supernatants were removed from the plate. This supernatant was combined with the transfection supernatant and filtered using a 0.45-mm-pore-size filter apparatus. Filtered supernatants were mixed with Ni-nitrilotriacetic acid-agarose (Invitrogen) at a ratio of 0.5 ml of agarose for 40 ml of culture supernatant. Supernatant-agarose mixtures were incubated for 2 h on a rocking platform at room temperature. Agarose was removed from the supernatant by column filtration. Beads were washed with PBS, and protein was eluted using 250 mM imidazole. Eluted protein was dialyzed against PBS for 2 h at room temperature and concentrated to 2 ml with an Amicon Centriprep YM-10. Sodium dodecyl sulfate-PAGE (SDS-PAGE) and Coomassie blue staining were used to determine purity of isolated proteins.
SDS-PAGE and Western blotting.
Various concentrations of purified S glycoproteins were mixed with 2× reducing Laemmli sample buffer and boiled for 5 min. Samples were resolved using 12% Novex gels (Invitrogen) for 1.5 h at 200 V. Gels were transferred to Immobilon P (Millipore) as described by the manufacturer, and Western blot analysis was performed. Proteins were detected using the anti-c-myc (9E10) antibody (0.1 μg/ml; Sigma), followed by an anti-mouse immunoglobulin G (IgG)-horseradish peroxidase conjugate (1:5,000; Jackson ImmunoResearch). For detection with human convalescent-phase serum (provided by Larry Anderson, CDC), a dilution of 1:2,000 was used followed by detection with anti-human IgG-horseradish peroxidase (Jackson ImmunoResearch). For detection with mouse serum raised against synthetic S glycoproteins, the method was as described for the anti-c-myc antibody. Membranes were incubated with enhanced chemiluminescence reagent for 1 min and exposed to X-Omat-AR film for various periods of time.
S glycoprotein-binding assay.
Vero E6 or HEK-293T/17 cells were harvested with PBS-5 mM EDTA and aliquoted to microcentrifuge tubes (1 × 106 to 5 × 106 each). Pellets were resuspended in PBS containing 10% fetal bovine serum and various concentrations of the truncated soluble S glycoproteins (0.01 nM to 1 μM). Cells and S glycoprotein were incubated for 1 h at room temperature and washed once in PBS-2% FCS. Pellets were resuspended in 100 μl of PBS-2% FCS containing 10 μg of anti-c-myc (9E10) antibody/ml, incubated for 1 h at 4οC, and washed once in PBS-2% FCS. Pellets were resuspended in 100 μl of PBS-2% FCS containing 5 μl of anti-mouse IgG-phycoerythrin (PE; Jackson ImmunoResearch). Mixtures were incubated at 4°C for 40 min and washed twice, and fluorescence-activated cell sorter (FACS) analysis was performed using a FACScan instrument with CellQuest software (Becton Dickinson).
In order to specifically block S glycoprotein binding to Vero E6 cells, human convalescent-phase serum was incubated with cells and S glycoprotein. Serum concentration never exceeded 10%, and as human serum was diluted, FCS was used to normalize all reaction mixtures to a final concentration of 10% serum. Normal human serum was used as a negative control.
RESULTS
Construction and expression of soluble codon-optimized SARS-CoV S glycoprotein.
The genes that encode viral proteins quite often have poor codon usage, leading to difficulties in producing sufficient quantities of purified recombinant protein (8). To overcome the possible issue of poor codon usage of the S glycoprotein gene, we constructed a synthetic codon-optimized S glycoprotein gene. Analysis of optimal codon usage in mammalian cells has been described elsewhere (1, 16). A codon-optimized gene encoding the first 1,190 amino acids of the SARS-CoV S glycoprotein (S1190) was synthesized and cloned into the mammalian expression vector pcDNA 3.1 Myc/His. The first 1,190 amino acids represent the predicted leader sequence and extracellular domain of the S glycoprotein, excluding transmembrane and intracellular domains. As such, when expressed, the gene product is a secreted, soluble version of the S glycoprotein. The vector used contains two epitope tags, the c-myc and His6 tags. The c-myc tag was exploited for immunoprecipitations and Western blot analysis of proteins, while the His6 tag allowed for native purification of expressed protein.
pcDNA 3.1 Myc/His S1190 was transfected into HEK-293T/17 cells, supernatants were recovered, and S1190 glycoprotein was purified by metal-affinity chromatography. Proteins were eluted from the resin with imidazole, dialyzed, and concentrated. S1190 concentration was determined by both spectrophotometry and bicinchoninic acid, both of which yielded equivalent results (data not shown). It was determined that secreted S1190 was expressed at a level of approximately 5 mg/liter after purification.
To assess purity of the S1190 glycoprotein preparations, proteins were resolved by SDS-PAGE and visualized by Coomassie staining (Fig. 1A). The major band of a relative molecular mass of 170 kDa was observed, and purity of this protein was estimated to be greater than 90%. To ensure that the purified protein was S1190, proteins were resolved using SDS-PAGE and protein identity was determined using Western blot analysis utilizing the anti-c-myc antibody, 9E10. As shown in Fig. 1B, a major band of approximately 170 kDa was observed. The distribution of this band in the gel matrix as well as the larger-than-expected apparent molecular weight suggested that this protein is glycosylated, as expected. The lower-molecular-weight species detected in Fig. 1B are clearly carboxy-terminal fragments of the S1190 protein, as demonstrated by detection with the carboxy-terminal myc tag. It is unclear whether these products represent natural cleavage products or are a consequence of overexpression and purification of the S1190 glycoprotein. In any case, these species represent a very small fraction of the total purified protein.
FIG. 1.
Expression and purification of soluble S1190 glycoprotein. Codon-optimized S1190 glycoprotein was cloned into pcDNA3.1 Myc/His and expressed in HEK-293T/17 cells to a level of 5 mg/liter. Protein was purified using metal-affinity chromatography, dialyzed, and concentrated. Purified protein was analyzed by Coomassie staining (A) or Western blotting using the anti-c-myc antibody for detection (B).
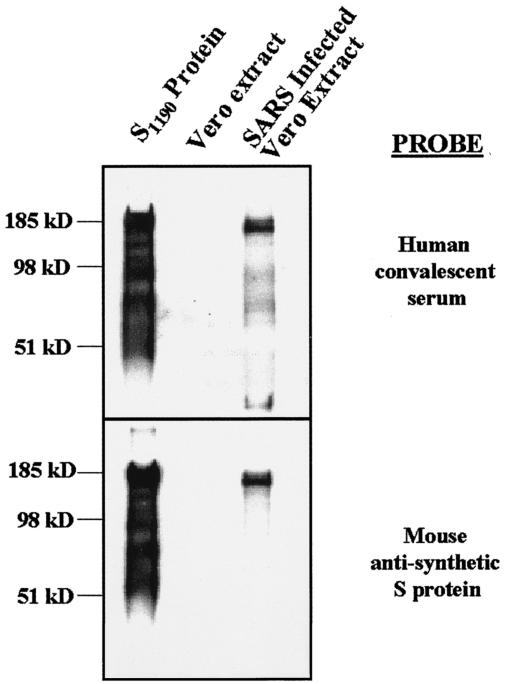
To determine if HEK-293T/17 cells appropriately posttranslationally modify the synthetic S glycoprotein, we attempted to compare the relative molecular weight of the codon-optimized S1190 protein with that observed for native S protein. SARS-CoV-infected Vero cell lysate was obtained from the CDC. Lysate, equivalent to 2 × 104 solubilized infected cells along with 200 ng of codon-optimized S glycoprotein, was resolved using SDS-PAGE. Gels were transferred to solid support, and Western blotting was performed using human SARS patient convalescent-phase serum as a detection reagent. As shown in Fig. 2 (top panel), the main species detected in the SARS-infected Vero E6 cells and S1190 lanes had an apparent molecular mass of approximately 170 kDa. No bands were detected in the uninfected Vero E6 lysate control. Lower-molecular-weight species were again detected in the lane containing S1190 glycoprotein. These bands were not observed in the lane containing native SARS-CoV S glycoprotein. As demonstrated in Fig. 1B, this discrepancy in banding pattern between the two lanes was most likely a function of the amount of protein present in the lane. When smaller quantities of synthetic S1190 glycoprotein were resolved by SDS-PAGE, we only observed the main 170-kDa species. It remains possible, however, that these smaller fragments represent an artifact of overexpression in the HEK-293T/17 cells.
FIG. 2.
The apparent molecular mass of the S1190 protein is similar to that of native S protein. S1190 protein (200 ng), Vero E6 cell extract, and SARS CoV-infected Vero E6 cell extract were resolved by SDS-PAGE. Proteins were transferred to a solid support, and Western blotting was performed using either human SARS convalescent-phase serum (top panel) or mouse anti-synthetic S protein (bottom panel). The major species detected for S1190 and SARS-infected extract in both cases was of an apparent molecular mass of approximately 170 kDa. No signal was observed in the lane containing the Vero E6 cell extract.
To ensure that the proteins observed were in fact the S glycoproteins, we performed Western blot analysis, this time using mouse serum raised against the synthetic S glycoprotein. As shown in Fig. 2 (bottom panel), a major species of approximately 170 kDa was observed in both the S1190 and infected Vero E6 cell lysate lanes. The expected contribution of the transmembrane domain and cytoplasmic tail to the molecular weight of the native S protein is expected to be negligible. These data suggest that codon-optimized S glycoprotein is modified similarly to native S glycoprotein.
Codon-optimized SARS-CoV S glycoprotein binds to Vero E6 cells.
In order for virus to infect target cells, it must first bind to the viral receptor on the cell surface. The protein that mediates this binding is predicted to be the S glycoprotein. Unfortunately, at this time, the cellular receptor for the viral S glycoprotein is not known. However, Vero E6 cells are readily infectible with SARS-CoV in culture and are assumed to express the receptor for the SARS-CoV S glycoprotein.
A FACS-based assay was developed to measure the ability of codon-optimized soluble S glycoprotein to bind to the Vero E6 cell surface. Briefly, Vero E6 cells were incubated with various concentrations of soluble S1190 glycoprotein to allow for binding. In order to detect S1190 binding to the cell surface, we took advantage of the fact that the soluble S1190 protein is fused to the c-myc epitope tag. S glycoprotein-bound cells were incubated with the anti-c-myc antibody 9E10, and bound anti-c-myc antibody was detected using an anti-mouse-PE-conjugated antibody. Cells were subsequently analyzed by flow cytometry, and the results are shown in Fig. 3A. Soluble synthetic S1190 glycoprotein readily bound to the surface of Vero E6 cells in a dose-dependent manner. Uniform binding was observed for the entire population of Vero E6 cells and not a minor subset (data not shown). To demonstrate specificity of the interaction of S1190 with a possible viral receptor expressed on the surface of Vero E6 cells, we performed the S1190-binding assay using HEK-293T/17 cells. This cell type is not expected to express the SARS CoV receptor, as demonstrated by the inability of this cell to be infected with SARS-CoV in vitro (data not shown). S1190 binding to HEK-293T/17 cell surfaces was not observed at any of the concentrations tested (Fig. 3A). These data demonstrate that soluble synthetic S1190 glycoprotein possesses biological properties expected to be present in the native S glycoprotein.
FIG. 3.
S1190 specifically binds to the surface of Vero E6 cells. (A) Vero E6 cells (diamonds) or HEK-293T/17 cells (squares) were incubated with 1 to 100 nM S1190 protein to determine the ability to bind to cell surfaces. S1190 binding was detected by anti-c-myc antibody followed by anti-mouse IgG-PE. Samples were analyzed by flow cytometry, and mean fluorescence intensity was plotted. (B) Vero E6 cells were incubated with 30 nM S1190 glycoprotein in the presence of increasing concentrations of normal (circles) or SARS convalescent-phase (squares) human serum. Serum concentration was maintained at 10% by the addition of fetal bovine serum. Binding was measured by flow cytometry, and the results were plotted as a percentage of the signal observed in a sample containing no test serum.
To ensure that the binding observed was in fact specific, we attempted to block binding using antibodies specific to the native SARS-CoV S glycoprotein. We obtained a pool of serum from individuals previously infected with SARS-CoV from the CDC. The antibodies present in this serum would be anticipated to disrupt the binding of S glycoprotein to the cellular receptor of the virus. Vero E6 cells were incubated with 30 nM S1190 glycoprotein in the presence of various concentrations of convalescent-phase or normal human serum. S glycoprotein binding was detected using FACS analysis as described above (Fig. 3B). Convalescent-phase serum specifically blocked binding of synthetic S1190 glycoprotein to the surface of Vero E6 cells. In contrast, serum from uninfected individuals had no effect on S1190 binding. Unfortunately, the control serum and convalescent-phase serum were not matched, i.e., serum from the same individual pre- and postexposure. To confirm the result above in a more controlled manner, rabbit serum was also raised against the S1190 glycoprotein. This serum could block the interaction of S1190 glycoprotein with Vero E6 cell surfaces, whereas preimmune rabbit serum could not (data not shown). These data demonstrate that S1190 binding to the surface of Vero E6 cells is indeed specific.
Localization of the SARS-CoV S glycoprotein ligand-binding domain to amino acids 1 to 510.
It is known for other coronaviruses that the amino-terminal half of the S glycoprotein spike contains the sequences responsible for ligand binding. To further characterize the interaction between SARS-CoV S glycoprotein and the Vero E6 cell surface, we created C-terminal truncations of the soluble S1190 glycoprotein. DNA encoding these truncations was synthesized via PCR using S1190 DNA as template. All truncated genes retained the c-myc and His6 tags to simplify detection and purification. Specifically, DNA encoding S350, S490, S590, S690, and S790 was cloned into the mammalian expression vector pcDNA3.1 Myc/His. The constructs, when expressed, contained amino acids 1 through 350, 490, 590, 690, and 790, respectively (Fig. 4). The constructs were transfected into HEK-293T/17 cells as described above, and secreted proteins were purified by metal-affinity chromatography (Fig. 5A). As previously found, all proteins were expressed at levels of >5 mg/liter and appeared to be glycosylated. Purified glycoproteins were incubated with either Vero E6 cells or HEK-293T/17 cells with various concentrations of the S glycoprotein fragments, and FACS analysis was performed. Figure 5B shows the results of each protein at a concentration of 100 nM. S proteins containing at least the first 590 amino acids specifically bound to the surface of Vero E6 cells but not to HEK-293T/17 cells. Binding of S350 and S490 to the cell surface was essentially equivalent for both HEK-293T/17 and Vero E6 cells. This indicates that these regions of the S glycoprotein do not specifically bind to the cell surface. Even at the highest concentrations tested (1 μM), no specific binding was observed for proteins S350 and S490 (data not shown). These data suggest that the first 590 amino acids of the SARS-CoV S protein are required for interaction with the surface of Vero E6 cells.
FIG. 4.
Soluble S glycoproteins. Shown is a schematic of the native SARS CoV S glycoprotein as well as the various S glycoproteins synthesized. Landmarks include the leader peptide (black; amino acids 1 to 14), the predicted ectodomain (white; amino acids 15 to 1190), the transmembrane domain (dark gray; amino acids 1191 to 1227), and the cytoplasmic tail (light gray; amino acids 1228 to 1255). All soluble constructs were C-terminally truncated, and the relative sizes are shown.
FIG. 5.
Expression and binding of C-terminally truncated S proteins. (A) DNA encoding C-terminally truncated S proteins (S350, S490, S590, S690, and S790) was synthesized via PCR using the vector encoding the S1190 glycoprotein as template. PCR products were cloned into pcDNA3.1 Myc/His and expressed in 293T cells. Western blotting was performed using the anti-c-myc antibody for detection. (B) Vero E6 (black bars) or HEK-293T/17 (gray bars) cells were incubated with a 100 nM concentration of each soluble S glycoprotein fragment and detected with antibody directed against the c-myc tag. FACS analysis was performed, and mean fluorescence intensity was plotted.
To more finely map the critical ligand-binding domain of the SARS-CoV S glycoprotein, we created more soluble constructs covering the sequence between S490 and S590. Specifically, we synthesized DNA that encoded S500, S510, S520, S540, S550, S560, S570, and S580 (nomenclature as described above). S530 was not cloned, since no positive colonies were obtained on the initial screen. The constructs were expressed in HEK-293T/17 cells, and the proteins were purified (Fig. 6A) as described above. A 100 nM concentration of each truncated protein was incubated with Vero E6 cells to determine cell surface interaction. Binding was detected using the anti-c-myc antibody followed by an anti-mouse-PE antibody. Flow cytometry was performed, and the results are shown in Fig. 6B. All proteins containing at least the first 510 amino acids could specifically bind to the surface of Vero E6 cells. Constructs smaller than S510 gave signals equivalent to that seen with secondary antibody alone. Interaction of S glycoprotein fragments with Vero E6 cells was specific, as demonstrated by blocking with convalescent-phase serum (data not shown). These data demonstrate that the first 510 amino acids of the SARS-CoV S protein are both necessary and sufficient for interaction with receptor expressed by Vero E6 cells. The first 510 ± 10 amino acids represent a domain analogous to the S1 domain of other coronavirus S glycoproteins.
FIG. 6.
Amino acids 1 to 510 represent the ligand-binding domain of the SARS CoV S protein. Constructs encoding C-terminally truncated proteins covering the gap between amino acids 490 and 590 (S510, S520, S540, S550, S560, S570, and S580) were synthesized using PCR and cloned into pcDNA3.1 Myc/His. All constructs were expressed in HEK-293T/17 cells and purified by metal-affinity chromatography. (A) Proteins were resolved by SDS-PAGE and Western blot analysis performed in conjunction with detection with the anti-c-myc antibody. (B) All purified S glycoproteins were incubated with Vero E6 cells at a concentration of 100 nM. Binding was measured using the anti-c-myc antibody and FACS analysis. Mean fluorescence intensity was plotted for each construct and compared.
S510 and S1190 have similar affinities for Vero E6 cells.
To ensure that the amino-terminal 510-amino-acid domain represents the entire receptor-binding domain, we attempted to approximately measure the binding kinetics of both S1190 and S510 for Vero E6 cells. S1190 and S510 were incubated with Vero E6 cells at concentrations ranging from 0.01 to 1 μM. As a negative control, S350 was included in the experiment. S glycoprotein binding was detected via flow cytometry as described before (Fig. 7). Although FACS analysis cannot be used to measure the true affinity of protein-protein interactions, it can be used to compare relative affinity of two differing proteins. S1190 and S510 exhibited very similar profiles for binding to the Vero E6 cell surface. These data suggest that S510 binds to Vero E6 cells at least as well as S1190 binds. S350 did not bind specifically to the surface of Vero E6 cells at any concentration tested. All other soluble S glycoproteins containing at least the first 510 amino acids were also tested in this way, and all showed similar binding profiles to the cellular surface (data not shown). These data demonstrate that S510 is indeed the ligand-binding domain of the SARS CoV S protein.
FIG. 7.
S510 and S1190 have comparable binding profiles for Vero E6 cell surfaces. Vero E6 cells were incubated with various concentrations of S1190 (squares), S510 (triangles), and S350 (circles) glycoproteins. Binding was detected with the anti-c-myc antibody, and FACS analysis was performed. Concentration versus mean fluorescence intensity was plotted, and the results for each protein were compared.
Amino acids 270 to 510 comprise the minimal ligand-binding domain of the soluble S glycoprotein.
Amino-terminal truncations of the S510 glycoprotein were synthesized to map the minimal receptor-binding region within the S1 domain of the spike glycoprotein. Specifically, sequences corresponding to the leader peptide were fused to sequences downstream in the S510 coding region, resulting in genes encoding S90-510 (amino acids 90 to 510), S150-510 (amino acids 150 to 510), S210-510 (amino acids 210 to 510), S270-510 (amino acids 270 to 510) (Fig. 8A), S330-510 (amino acids 330 to 510), and S390-510 (amino acids 390 to 510). All constructs were transfected into HEK-293T/17 cells, and the protein was purified by metal-affinity chromatography. Interestingly, only expression of S270-510 was observed, and expression levels were similar to the other S glycoprotein fragments (data not shown). Purified S270-510 was incubated with Vero E6 cells at various concentrations, FACS analysis was performed, and the results are shown in Fig. 8B. S270-510 binding to Vero E6 cells was nearly identical to that observed for S590. S350 showed no specific binding to Vero E6 cells. Both S270-510 and S590 did not demonstrate specific binding to the surface of HEK-293T/17 cells. These data demonstrate that amino acids 270 to 510 contain the minimal domain required for interaction with the surface of Vero E6 cells.
FIG. 8.
S270-510 binds to Vero E6 cells. (A) Schematic representation of the S510 and S270-510 glycoproteins. The black box (amino acids 1 to 14) represents the S glycoprotein leader sequence. (B) Vero E6 cells or HEK-293T/17 cells were incubated with S350, S590, or S270-510 at concentrations ranging from 0.1 nM to 1 μM. Binding of S glycoproteins was detected using the anti-c-myc antibody followed by anti-mouse IgG conjugated to PE. FACS analysis was performed, and mean fluorescence was plotted as a function of S glycoprotein concentration.
DISCUSSION
Understanding the biochemistry by which SARS-CoV infects target cells is of paramount importance in preventing infection and death associated with SARS. The S glycoprotein, which mediates viral entry, is an obvious protein for study to approach inhibiting viral infection. Here we describe the synthesis and expression of codon-optimized SARS-CoV S glycoprotein. Codon optimization has many benefits over traditional cloning techniques, the most obvious of which is the yield of protein obtained. We have expressed the full-length ectodomain of the S glycoprotein (S1190) at a level of approximately 5 mg/liter. This yield is greater than typically seen for native viral glycoproteins expressed in mammalian cells (8). We have not formally compared the two expression systems, but it is our experience that codon optimizing of viral glycoprotein genes for mammalian cells greatly increases expression levels. At this time, we have the ability to purify >10 mg of S1190 protein at one time, allowing for diverse studies to be undertaken.
Comparisons between S1190 glycoprotein and native SARS-CoV S glycoprotein were performed. The relative molecular weight of the S1190 glycoprotein was essentially identical to that of native S glycoprotein as determined by SDS-PAGE and Western blotting. S1190 protein did, however, demonstrate proteolytic breakdown products not observed in the native protein (Fig. 2). One explanation for this difference is the amount of protein tested in the assay. Significantly more S1190 protein was resolved on the gel than the native S glycoprotein-containing viral lysate. It is possible that these smaller S glycoprotein fragments are present in virally infected cells, but this Western blotting is not sensitive enough to detect them. When quantities of S1190 glycoprotein comparable to that of native glycoprotein in the viral lysate were resolved by SDS-PAGE, we did not see the smaller S glycoprotein fragments (Fig. 1). It is also possible that overexpression of S glycoprotein in mammalian cells leads to degradation of a portion of the expressed S glycoprotein. In any case, the majority of the codon-optimized S1190 has an apparent molecular weight that is equivalent to that of native S glycoprotein.
It has been shown that SARS-CoV can readily infect Vero E6 cells in culture (5, 12, 17). The receptor for the SARS-CoV S glycoprotein has not been identified, but one can assume that it is expressed on the surface of Vero E6 cells. S1190 protein bound to the surface of Vero E6 cells in a dose-dependent manner, and specific antibodies blocked this interaction. These data suggest that soluble S1190 glycoprotein possesses some of the biologic activities present in the native S glycoprotein, specifically receptor binding.
The S glycoprotein of transmissible gastroenteritis virus has been shown to interact not only with the receptor to mediate viral entry but also with sialic acid (18). The latter interaction is not required for fusion but may aid in enteropathogenesis (10). It is a formal possibility that the interaction of soluble SARS-CoV S1190 glycoprotein with Vero E6 cell surfaces is mediated not solely by receptor, but in combination with carbohydrate residues on the Vero E6 cell surface. The interaction of S1190 with ligands other than the cellular receptor could complicate the analysis of S1190 binding to Vero E6 cell surfaces. Identification of the SARS-CoV cellular receptor will allow us to clarify this issue. In any case, the binding of S1190 is specific to the permissive Vero E6 cells.
We have determined that the first 510 amino acids of the SARS CoV S glycoprotein contain the entire ligand-binding domain. Domain structures of the SARS-CoV S protein can now be deduced. For many coronaviruses, such as MHV, the S protein is cleaved into the ligand-binding subunit (S1) and the membrane fusion subunit (S2) (6, 15, 22, 23). The receptor-binding domain of the MHV spike protein has been mapped to amino acids 1 to 330 (13). These amino acids are contained within the S1 region. The ligand-binding domain of a coronavirus that does not express a cleaved S glycoprotein, HCoV-229E, has also been mapped. The first 547 amino acids of the HCoV-229E S protein are required for binding to the receptor hAPN (2). For this viral S glycoprotein, the first 547 amino acids were termed the S1 domain, the designation based on ligand-binding capability and not evidence of physically distinct subunits. Sequence analysis (20) as well as data described herein (Fig. 2) suggest that, analogous to HCoV-229E, SARS-CoV S glycoprotein is not cleaved into S1 and S2 subunits. Interestingly, a domain nearly identical in size to the HCoV-229E S1 domain contains the ligand-binding domain of SARS-CoV S glycoprotein. Since the first 510 amino acids of SARS-CoV S glycoprotein encompass the entire receptor-binding domain, we propose that amino acids 1 to 510 be termed S1 and amino acids 511 to 1190 be called S2.
N-terminal truncation of the S510 glycoprotein demonstrated that amino acids 270 to 510 represent the minimal receptor-binding domain. S270-510 was the only amino-terminal truncation of the S1 domain that could be expressed in HEK-293T/17 cells. S90-510, S150-510, S210-510, S330-510, and S390-510 expression levels were below our detection limits. It is unclear why these truncated constructs were not expressed. The most likely explanation is that sequences were not present in these glycoproteins to ensure proper folding. This misfolding may have prevented secretion into the medium or resulted in degradation of the various proteins. It is possible that a smaller domain than amino acids 270 to 510 confers the ligand binding capacity of the S glycoprotein, but we believe this is unlikely due to our inability to express smaller fragments. We speculate that S270-510 was expressed and secreted, since it represents an intact receptor-binding domain that possesses the appropriate sequences required for proper protein folding.
Expression and purification of large quantities of S1190, S510, and S270-510 glycoproteins will be important for identifying the SARS-CoV cellular receptor and for crystallization studies of the SARS-CoV S glycoprotein. S1190 crystallization would give a better understanding of the mechanism by which the S glycoprotein binds to and fuses with susceptible cells. Also, the S510 and S270-510 glycoproteins present the opportunity to determine the exact structure of the ligand-binding site of the S glycoprotein.
Finally, for other coronaviruses, such as transmissible gastroenteritis virus, MHV, and HCoV-229E, neutralizing epitopes are typically present in the S glycoprotein (2, 3, 7, 19, 24). Neutralizing antibodies directed against the S glycoprotein are reactive to either the S1 receptor-binding domain or hydrophobic residues located in the S2 region. The antibodies specific for S2 are predicted to interfere with fusion of the viral and host cell envelopes. We suggest that these codon-optimized S glycoprotein domains are appropriate targets for monoclonal antibody development or as vaccine candidates.
Acknowledgments
We thank Israel Lowy and Robert Graziano (Medarex, Inc.) and John Sullivan, Robert Finberg, Katherine Luzuriaga, Thomas Greenough, and Mohan Somasundaran (University of Massachusetts Medical School) for thoughtful scientific discussions pertaining to this work. We also thank Hector Hernandez for critical review of the manuscript.
This work was conducted as part of a collaborative development agreement between MBL and Medarex, Inc., with support from the National Institute of Allergy and Infectious Diseases (NO1-AI-65315).
REFERENCES
- 1.Bikker, J. A., S. Trumpp-Kallmeyer, and C. Humblet. 1998. G-protein coupled receptors: models, mutagenesis, and drug design. J. Med. Chem. 41:2911-2927. [DOI] [PubMed] [Google Scholar]
- 2.Bonavia, A., B. D. Zelus, D. E. Wentworth, P. J. Talbot, and K. V. Holmes. 2003. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel, C., and P. J. Talbot. 1990. Protection from lethal coronavirus infection by affinity-purified spike glycoprotein of murine hepatitis virus, strain A59. Virology 174:87-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Groot, R. J., W. Luytjes, M. C. Horzinek, B. A. van der Zeijst, W. J. Spaan, and J. A. Lenstra. 1987. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 196:963-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 6.Frana, M. F., J. N. Behnke, L. S. Sturman, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 56:912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godet, M., J. Grosclaude, B. Delmas, and H. Laude. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 68:8008-8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 9.Jones, P. L., T. Korte, and R. Blumenthal. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273:404-409. [DOI] [PubMed] [Google Scholar]
- 10.Krempl, C., B. Schultze, H. Laude, and G. Herrler. 1997. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 71:3285-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krueger, D. K., S. M. Kelly, D. N. Lewicki, R. Ruffolo, and T. M. Gallagher. 2001. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J. Virol. 75:2792-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 13.Kubo, H., Y. K. Yamada, and F. Taguchi. 1994. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 68:5403-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo, Z. L., and S. R. Weiss. 1998. Mutational analysis of fusion peptide-like regions in the mouse hepatitis virus strain A59 spike protein. Adv. Exp. Med. Biol. 440:17-23. [DOI] [PubMed] [Google Scholar]
- 15.Luytjes, W., L. S. Sturman, P. J. Bredenbeek, J. Charite, B. A. van der Zeijst, M. C. Horzinek, and W. J. Spaan. 1987. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirzabekov, T., N. Bannert, M. Farzan, W. Hofmann, P. Kolchinsky, L. Wu, R. Wyatt, and J. Sodroski. 1999. Enhanced expression, native purification, and characterization of CCR5, a principal HIV-1 coreceptor. J. Biol. Chem. 274:28745-28750. [DOI] [PubMed] [Google Scholar]
- 17.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pensaert, M., P. Callebaut, and J. Vergote. 1986. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet. Q. 8:257-261. [DOI] [PubMed] [Google Scholar]
- 19.Pike, B. V., and D. J. Garwes. 1979. The neutralization of transmissible gastroenteritis virus by normal heterotypic serum. J. Gen. Virol. 42:279-287. [DOI] [PubMed] [Google Scholar]
- 20.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 21.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stern, D. F., and B. M. Sefton. 1982. Coronavirus proteins: biogenesis of avian infectious bronchitis virus virion proteins. J. Virol. 44:794-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sturman, L. S., and K. V. Holmes. 1983. The molecular biology of coronaviruses. Adv. Virus Res. 28:35-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sune, C., G. Jimenez, I. Correa, M. J. Bullido, F. Gebauer, C. Smerdou, and L. Enjuanes. 1990. Mechanisms of transmissible gastroenteritis coronavirus neutralization. Virology 177:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai, J. C., B. D. Zelus, K. V. Holmes, and S. R. Weiss. 2003. The N-terminal domain of the murine coronavirus spike glycoprotein determines the CEACAM1 receptor specificity of the virus strain. J. Virol. 77:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White, J. M. 1992. Membrane fusion. Science 258:917-924. [DOI] [PubMed] [Google Scholar]
- 27.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]