Abstract
While recent studies have demonstrated that retroviral vectors can be used to stably express short hairpin RNA (shRNA) to inhibit gene expression, these studies have utilized replication-defective retroviruses. We describe the creation of a replication-competent, Gateway-compatible retroviral vector capable of expressing shRNA that inhibits the expression of specific genes.
Retroviral vectors can be used to express short hairpin RNA (shRNA) under the control of an RNA polymerase III (Pol III) promoter, such as U6 or H1, for the purpose of inhibiting gene expression in a sequence-specific manner (3, 9, 15). However, all studies to date have utilized replication-defective retroviruses that require packaging cell lines or helper virus for virus production. A series of replication-competent retroviral vectors have been developed based on the Schmitt Ruppin A (SR-A) strain of Rous sarcoma virus (reviewed in reference 6). These vectors, referred to as RCASBP(A) or RCANBP(A), have several advantages over other retroviral vectors: (i) in avian cells, these vectors are replication competent; (ii) they routinely achieve high titers in avian cells (107 infectious units/ml); and (iii) in mammalian cells, infectious viruses are not produced, allowing for multiple rounds of infection into the same cell while preventing uncontrolled cell-to-cell spread (5, 7). Furthermore, successful infection of mammalian cells by the RCASBP/RCANBP(A) virus requires the use of vectors with non-avian envelope proteins, such as the amphotropic envelope from murine leukemia virus (1), or ectopic expression of the avian retroviral receptor TVA on the cell surface (2, 19). For this study, we used cells expressing TVA to allow cell-specific targeted infection of mammalian cells (5). We demonstrate that RCANBP(A)-mediated delivery of shRNA targeted against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) can specifically reduce GAPDH expression in mammalian cells.
Development of a Gateway-compatible RCANBP(A) viral vector.
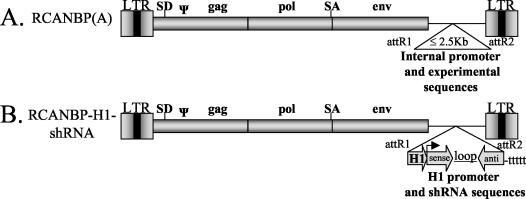
We developed a replication-competent retroviral vector expressing shRNA under the control of the human H1 promoter (Fig. 1). This vector was engineered to be Gateway compatible to facilitate efficient cloning of DNA sequences into the viral vector (construction details available upon request). The Gateway-compatible RCANBP(A) vector (Fig. 1A) was used to generate RCANBP-H1-GAPDH, which contains an RNA Pol III-shRNA cassette consisting of the human H1 promoter upstream of a GAPDH shRNA hairpin-loop sequence (Ambion, Austin, Tex.) (Fig. 1B). This viral vector produced replication-competent virus in the immortalized chicken fibroblast cell line DF-1 (16), as determined by enzyme-linked immunosorbent assay (17), and had a high titer relative to RCASBP(A)AP, which was used as a standard (data not shown).
FIG. 1.
Schematic representation of a Gateway-compatible, replication-competent, retroviral vector capable of delivering shRNA. (A) Gateway-compatible RCANBP(A). (B) The RCANBP-H1-shRNA viral vector has been engineered to express shRNA by introducing an RNA Pol III-shRNA cassette, containing the H1 promoter upstream of the shRNA stem-loop sequence. SA, splice acceptor; SD, splice donor; LTR, long terminal repeat; ψ, packaging signal.
Generation of TVA-positive cell lines.
A stable clonal line, A375-TVA, from the human malignant melanoma cell line A375 (8) was created by transfection of a pCDNA3.1/hygro construct (Invitrogen, Carlsbad, Calif.) containing the TVA gene and selection in medium containing 300-μg/ml hygromycin B. The percentage of TVA-positive cells in this clonal line was assessed by infection with RCASBP(A)-GFP virus (12). FACS analysis indicated that more than 90% of the cells were infected by RCASBP(A)-GFP (data not shown). These data indicate that the A375-TVA cell line is competent for RCASBP/RCANBP(A) infection.
RNAi of GAPDH expression in RCANBP-H1-GAPDH-infected cells.
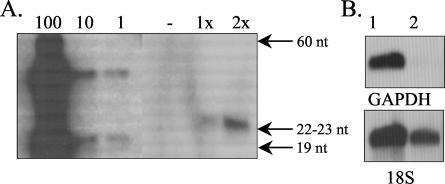
A375-TVA cells were infected over 2 days with the RCANBP-H1-GAPDH virus in the presence of 8-μg/ml Polybrene (Sigma, St. Louis, Mo.), and RNA was extracted after one or two rounds of infection. Polyacrylamide gel electrophoresis-Northern analysis showed that both the one- and two-round-infected A375-TVA cells produced antisense GAPDH RNA, and this RNA was not seen in uninfected cells (Fig. 2A). Comparatively, our retroviral system appears to be more efficient at expressing shRNA than the other retroviral systems, as the RCANBP-H1-GAPDH virus produced 100 to 300 fmol of shRNA per μg of cellular RNA analyzed (Fig. 2A), whereas other retroviral vectors express at most 3 fmol of shRNA per μg of cellular RNA (18). Northern analysis of GAPDH message in A375-TVA cells infected with RCANBP-H1-GAPDH virus showed that the GAPDH message was drastically reduced after only one round of infection (Fig. 2B), and this effect was maintained over four rounds of infection. This reduction in GAPDH message levels is not apparent on day 5 (data not shown). Analysis of cell proliferation over five rounds of RCANBP-H1-GAPDH infection indicated that there were fewer RCANBP-H1-GAPDH-infected cells relative to uninfected cells. However, cell number began to recover after 5 days, and by 13 days, it had recovered to the level seen in the uninfected cells (data not shown). GAPDH is an important ATP-generating enzyme in glycolysis, and a decrease in activity leads to ATP depletion and cell death (4). The initial lag in cell number may be due to cell death associated with the loss of GAPDH expression. Recovery of both cell number and GAPDH message after 5 days in RCANBP-H1-GAPDH-infected A375-TVA cells was associated with loss of TVA-positive cells in culture (data not shown). Infection of A375-TVA cells with a virus containing an unrelated shRNA had no effect on cell growth (data not shown).
FIG. 2.
Ability of RCANBP-H1-GAPDH to express GAPDH shRNA and to reduce GAPDH expression in A375-TVA cells. (A) Polyacrylamide gel electrophoresis-Northern analysis of GAPDH shRNA expression using the sense strand as an RNA probe. A375-TVA cells were uninfected (− [control]) or were infected with RCANBP-H1-GAPDH for one (1x) or two (2x) rounds. Samples were normalized for total RNA content, and 70 μg of total cellular RNA was analyzed per sample. On the sense strand, GAPDH shRNA (19 nucleotides [nt]) was used at 100, 10, and 1 pmol as a size marker and for quantitation of shRNA production. The upper band likely corresponds to hairpin-loop formation (47 nt) of the shRNA prior to loop digestion. (B) Northern blot analysis of GAPDH message in A375-TVA cells that were uninfected (lane 1) or infected with RCANBP-H1-GAPDH (lane 2) for one round. Blots were stripped and reprobed with 18S RNA to ensure equal loading of RNA.
In this report, we describe the development of a replication-competent retroviral vector, RCANBP-H1-GAPDH, containing shRNA sequences specific for human GAPDH under the control of the human H1 promoter. This vector was replication competent in DF-1 cells and expressed GAPDH shRNA after infection of the human melanoma cell line, A375 (Fig. 2A), which expressed the viral receptor TVA. Infection of A375-TVA cells resulted in reduction of GAPDH message after only one round of infection. These data indicate that the RCANBP(A)/TVA system can be used for RNA interference (RNAi)-based silencing of specific genes in avian and mammalian cells in vitro. This retroviral vector takes advantage of the tissue- and cell-specific targeted infection by the RCANBP(A)/TVA system and avoids the disadvantages of replication-defective retroviruses. RNAi studies are rapid, cost-effective, and can be easily adapted for studies in numerous organisms. This delivery system should greatly enhance the utility of RNAi-based studies in both avian and mammalian systems in vivo and in cell culture. RCASBP/RCANBP-based retroviral vectors have been used to study viral replication, avian development, and cancer in mice (5, 7, 10, 11, 13, 14). The majority of these studies have analyzed gain-of-function phenotypes by delivering and overexpressing the gene(s) of interest. This study extends the utility of the RCANBP(A)/TVA system to include loss-of-function analyses of specific genes in vitro as well as in vivo.
Acknowledgments
We thank Stacie Loftus for advice and Eric Holland for the SPKEQ8TVA construct. We thank Stephen Hughes for advice, for providing the RCANBP(A) vector, and for critical review of the manuscript.
This work was supported by the Van Andel Research Institute. S. L. Holmen is a Pfizer Fellow of the Life Sciences Research Foundation.
REFERENCES
- 1.Barsov, E. V., and S. H. Hughes. 1996. Gene transfer into mammalian cells by a Rous sarcoma virus-based retroviral vector with the host range of the amphotropic murine leukemia virus. J. Virol. 70:3922-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, P., J. A. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. [DOI] [PubMed] [Google Scholar]
- 3.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [DOI] [PubMed] [Google Scholar]
- 4.Choi, D. S., Y. B. Kim, Y. H. Lee, S. H. Cha, and D. E. Sok. 1995. Glyceraldehyde-3-phosphate dehydrogenase as a biochemical marker of cytotoxicity by vinyl sulfones in cultured murine spleen lymphocytes. Cell Biol. Toxicol. 11:23-28. [DOI] [PubMed] [Google Scholar]
- 5.Federspiel, M. J., P. Bates, J. A. Young, H. E. Varmus, and S. H. Hughes. 1994. A system for tissue-specific gene targeting: transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc. Natl. Acad. Sci. USA 91:11241-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Federspiel, M. J., and S. H. Hughes. 1997. Retroviral gene delivery. Methods Cell Biol. 52:179-214. [PubMed] [Google Scholar]
- 7.Fisher, G. H., S. Orsulic, E. Holland, W. P. Hively, Y. Li, B. C. Lewis, B. O. Williams, and H. E. Varmus. 1999. Development of a flexible and specific gene delivery system for production of murine tumor models. Oncogene 18:5253-5260. [DOI] [PubMed] [Google Scholar]
- 8.Giard, D. J., S. A. Aaronson, G. J. Todaro, P. Arnstein, J. H. Kersey, H. Dosik, and W. P. Parks. 1973. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 51:1417-1423. [DOI] [PubMed] [Google Scholar]
- 9.Hemann, M. T., J. S. Fridman, J. T. Zilfou, E. Hernando, P. J. Paddison, C. Cordon-Cardo, G. J. Hannon, and S. W. Lowe. 2003. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 33:396-400. [DOI] [PubMed] [Google Scholar]
- 10.Holland, E. C., and H. E. Varmus. 1998. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proc. Natl. Acad. Sci. USA 95:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmen, S. L., D. W. Salter, W. S. Payne, J. B. Dodgson, S. H. Hughes, and M. J. Federspiel. 1999. Soluble forms of the subgroup A avian leukosis virus [ALV(A)] receptor Tva significantly inhibit ALV(A) infection in vitro and in vivo. J. Virol. 73:10051-10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis, B. C., N. Chinnasamy, R. A. Morgan, and H. E. Varmus. 2001. Development of an avian leukosis-sarcoma virus subgroup A pseudotyped lentiviral vector. J. Virol. 75:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logan, M., and C. Tabin. 1998. Targeted gene misexpression in chick limb buds using avian replication-competent retroviruses. Methods 14:407-420. [DOI] [PubMed] [Google Scholar]
- 14.Orsulic, S., Y. Li, R. A. Soslow, L. A. Vitale-Cross, J. S. Gutkind, and H. E. Varmus. 2002. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell 1:53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, M. Zhang, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 16.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 17.Smith, E. J., A. Fadly, and W. Okazaki. 1979. An enzyme-linked immunosorbent assay for detecting avian leukosis-sarcoma viruses. Avian Dis. 23:698-707. [PubMed] [Google Scholar]
- 18.Stewart, S. A., D. M. Dykxhoorn, D. Palliser, H. Mizuno, E. Y. Yu, D. S. An, D. M. Sabatini, I. S. Chen, W. C. Hahn, P. A. Sharp, R. A. Weinberg, and C. D. Novina. 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9:493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varmus, H. E., T. Padgett, S. Heasley, G. Simon, and J. M. Bishop. 1977. Cellular functions are required for the synthesis and integration of avian sarcoma virus-specific DNA. Cell 11:307-319. [DOI] [PubMed] [Google Scholar]