Abstract
Herpes simplex virus type 1 (HSV-1) DNA replication is associated with nuclear domains called ND10, which contain host recombination proteins such as RPA, RAD51, and NBS1 and participate in the cell's response to DNA damage. The stages of HSV-1 infection have been described previously. Infected cells at stage IIIa are observed after the initial disruption of ND10 and display nuclear foci, or prereplicative sites, containing the viral single-stranded-DNA-binding protein (UL29), the origin-binding protein (UL9), and the heterotrimeric helicase-primase. At stage IIIb, the viral polymerase, its processivity factor, and the ND10, protein PML, are also recruited to these sites. In this work, RPA, RAD51, and NBS1 were observed predominantly in stage IIIb but not stage IIIa prereplicative sites, suggesting that the efficient recruitment of these recombination proteins is dependent on the presence of the viral polymerase and other replication proteins within these sites. On the other hand, Ku86 was not found in any of the precursors to replication compartments, suggesting that it is excluded from the early stages of HSV-1 replication. Western blot analysis showed that RPA and NBS1 were (hyper)phosphorylated during infection, indicating that infection induces the host response to DNA damage. Finally, RPA, RAD51, and NBS1 were found to be associated with UL29 foci observed in transfected cells expressing UL29 and the helicase-primase heterotrimer and containing intact ND10. The ability to recruit recombination and repair proteins to various subassemblies of viral replication proteins thus appears to depend on several factors, including the presence of the viral polymerase and/or UL9 within prereplicative sites and the integrity of ND10.
Herpes simplex virus type 1 (HSV-1) is a linear double-stranded DNA virus that replicates in the nucleus of the infected cell. Viral DNA synthesis takes place within globular domains called replication compartments (55), which contain the seven essential viral DNA replication proteins: the origin-binding protein (UL9), the single-stranded-DNA-binding protein (UL29 or ICP8), the helicase-primase heterotrimer (UL5/UL8/UL52), the viral polymerase (UL30), and its processivity factor (UL42) (reviewed in reference 75). Due to the lack of a reconstituted in vitro replication system, it has not been possible to determine whether cellular proteins are also involved in HSV-1 DNA replication.
Several lines of evidence indicate that the process of HSV-1 DNA replication is linked to recombination. For example, recombination is a frequent event within the HSV-1 genome as well as between infecting genomes and is stimulated on HSV-1 infection (14, 15, 20, 45, 59, 71, 72). Furthermore, newly replicated DNA is larger than unit length and adopts a highly complex, possibly branched structure (6, 38, 60, 61, 64), the formation of which is presumed to require recombination (reviewed in reference 37). Newly replicated DNA has also undergone genomic inversion (4, 28, 60, 81). Thus, the properties of replicating DNA indicate a likely role for recombination-mediated replication (37, 75). Although recent reports indicate that viral proteins may function to promote recombination (49, 50, 57), it is likely that recombination during HSV-1 infection also may involve cellular recombination pathways (79).
HSV-1 DNA replication is closely associated with a nuclear matrix-bound multiprotein domains called ND10 (also known as promyelocytic leukemia [PML] nuclear bodies), which are defined and organized by the PML protein (1, 23, 42). Upon entry into the nucleus, parental viral genomes are found adjacent to ND10 (42), and the viral immediate-early protein, ICP0, induces the degradation of PML and the dispersal of ND10 and associated proteins (17, 41).
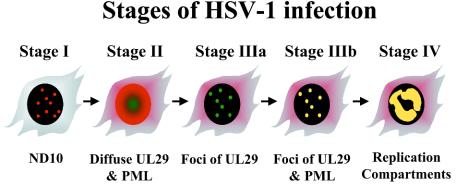
With antibodies directed against UL29 and PML, indirect immunofluorescence (IF) microscopy of HEp-2 cells infected with wild-type or mutant virus revealed the existence of several types of viral protein subassemblies leading to the formation of replication compartments (9, 12, 31) (Fig. 1). Infected cells at stage I possess intact ND10 and display no staining for UL29. Cells at stage II appear at 1 to 2 h postinfection and are characterized by the disruption of ND10 and diffuse nuclear staining for UL29. Stage IIIa foci are defined as a limited number of prereplicative sites which contain five viral replication proteins (UL29, UL5, UL8, UL52, and UL9) (8). Stage IIIa foci, seen in cells infected with virus lacking the viral DNA polymerase, do not contain PML. Viral replication proteins UL29, UL5, UL8, UL52, and UL9 are essential for the formation of stage IIIa prereplicative sites, since mutants deficient in the genes encoding these proteins do not progress beyond stage II (31, 34).
FIG. 1.
Five stages of early HSV-1 infection as defined by immunocytochemical staining of viral UL29 (red) and cellular PML (green). Yellow indicates colocalization of UL29 and at least one isoform of PML. See the text for a description of the diagram.
During wild-type infection, if viral DNA replication is blocked by the addition of the viral polymerase inhibitor phosphonoacetic acid (PAA), stage IIIb prereplicative sites containing all seven essential viral proteins form. These foci are characterized by the redistribution of certain isoforms of PML back to these sites (8). The recruitment of PML to viral prereplicative sites depends on the presence of HSV-1 polymerase within these foci (9). The viral polymerase, in turn, requires an active primase for its recruitment to prereplicative sites (12), suggesting that the formation of replication compartments proceeds by an ordered series of events. The laying down of RNA primers may signal a conformational change at the replication fork which is required for the recruitment of HSV-1 DNA polymerase and transition to the elongation phase of viral DNA replication (12). If polymerase is present, PML can be recruited to these sites (9). Thus, recruitment of PML is dependent on the composition of the prereplicative site and hence the conformation of the replication fork. If viral replication is allowed to precede, all seven essential viral replication proteins, as well as certain isoforms of PML, are found in replication compartments (stage IV).
Transfection studies have also permitted the identification of viral protein subassemblies. Replication compartments can be reconstituted in cells transfected with plasmids encoding an HSV-1 origin of replication and expressing the seven essential viral replication proteins (35, 69, 82). Additionally, transfected cells expressing a subset of the essential viral replication proteins (UL29, UL5, UL8, and UL52) exhibit a limited number of nuclear foci found adjacent to ND10 (33, 70) which are likely to be biologically relevant, since replication compartments are also ND10 associated.
ND10 have been shown to contain host recombination/repair proteins (reviewed in reference 47) and participate in the cell's response to DNA damage (10, 78). ND10 proteins are redistributed in response to DNA damage (10), and it has been proposed that ND10 may store and release proteins in response to certain external insults such as viral infection and DNA damage (47). For instance, the ND10-associated proteins PML, Mre11, γH2AX, RAD50, replication protein A (RPA), and TopBP1 are redistributed to DNA repair foci induced by ionizing radiation (3, 10, 78). The observation that some isoforms of PML are recruited into stage IIIb prereplicative sites (9) may also be an example of redistribution of cellular proteins in response to DNA damage.
In this paper we investigate the hypothesis that replicating viral DNA may induce a cellular response to DNA damage and that host recombination and repair proteins are recruited to HSV-1 DNA in a manner reminiscent of the recruitment of these proteins to sites of cellular DNA damage. We propose that HSV-1 genomes target and disrupt ND10 in order to gain access to certain ND10-associated proteins, including those involved in recombination and repair. In this study, we show that components of the homologous recombination (HR) pathway, RPA, RAD51 and NBS1 were recruited to some types of prereplicative sites but not others. Thus, as with PML (9, 12), recruitment of these HR proteins depends on the composition of the prereplicative sites. Additionally, Ku86, which is a member of the nonhomologous end-joining (NHEJ) pathway, was not recruited to any of the subassemblies of viral proteins. We have also shown that viral infection stimulates a damage response which correlates with the recruitment of components of the HR recombination pathway to viral prereplicative sites.
MATERIALS AND METHODS
Cell lines.
African green monkey kidney fibroblasts (Vero cells) were purchased from the American Type Culture Collection (Manassas, Va.) and maintained as monolayers in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, Calif.) supplemented with 5% fetal bovine serum (Gemini Bio-Products, Woodland, Calif.), penicillin, streptomycin, and amphotericin B (Invitrogen). Cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Reagents and antibodies.
Glycerol gelatin, 1,4-diazobicyclo-[2.2.2]octane (DABCO), camptothecin, and PAA were obtained from Sigma Chemical Co. (St. Louis, Mo.). Mouse monoclonal anti-UL29 antibody 39S (65) was obtained from the American Type Culture Collection. Rabbit polyclonal anti-UL29 antibody 367 (63) was supplied by William Ruyechan (State University of New York at Buffalo). Rabbit polyclonal anti-UL29 antibody 3-83 (26) was provided by David Knipe (Harvard Medical School). Mouse monoclonal antibodies against the 32-kDa subunit (71-9A) (16) and the 70-kDa subunit (2H10; also called 70c) (25) of RPA were supplied by Marc Wold (University of Iowa). Monoclonal antibodies Ab-1 (anti-RAD51) and Ab-4 (anti-Ku70), and anti-DNA-PKcs antibody cocktail (Ab-4) against the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) were purchased from NeoMarkers (Fremont, Calif.). Monoclonal antibodies directed against RAD50 (13B3) and MRE11 (12D7) were purchased from GeneTex (San Antonio, Tex.). Rabbit polyclonal anti-RAD51 antibody Ab-1 was purchased from Oncogene (San Diego, Calif.). Rabbit polyclonal antibodies anti-Ku86 (H-300) and anti-RAD52 (H-300) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Rabbit polyclonal anti-NBS1 antibody (NB 100-143) was purchased from Novus Biologicals (Littleton, Colo.). Mouse monoclonal antitubulin was purchased from Sigma.
Initially, fluorescein isothiocyanate-conjugated goat anti-mouse or goat anti-rabbit immunoglobulin and Texas red-conjugated goat anti-mouse or goat anti-rabbit immunoglobulin antibodies (Cappel, Organon Teknika Corporation, Durham, N.C.) were used as secondary antibodies for IF analysis. In order to achieve greater photostability of the fluorophore, subsequent experiments employed secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Molecular Probes, Eugene, Oreg.). All images presented were obtained with the Alexa Fluor conjugates.
Viruses and infections.
Strain KOS was used as wild-type HSV-1. The polymerase null virus HP66 was described previously (36). Subconfluent monolayers of cells were adsorbed for 1 h with 10 PFU per cell and incubated for 5.5 to 6.0 h postadsorption. Mock infections were carried out in parallel. Where indicated, PAA was added to the medium at a concentration of 400 μg/ml at the time of adsorption and maintained throughout the course of infection.
Plasmids and transfections.
Constructs which constitutively express UL5 (pCM-UL5b), UL8 (pCM-UL8), UL29 (pCM-DBP), and UL52 (pCMV-UL52) from the cytomegalovirus immediate-early promoter were described previously (21, 34). Plasmid poriS contains oriS on a 100-bp MspI fragment inserted into the SmaI site of pUC119 (52).
Subconfluent monolayers of cells were transfected with 0.25 μg of each plasmid (1.25 μg total of plasmid DNA) with Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions.
Indirect immunofluorescence.
Vero cells were grown on glass coverslips for 6 to 18 h prior to infection or transfection. At 5.5 to 6.0 h postadsorption or 16 to 18 h posttransfection, cells were washed briefly in phosphate-buffered saline (PBS) supplemented with 1 mM MgCl2 and 0.1 mM CaCl2 (PBS+) and fixed in 4% paraformaldehyde in PBS+ for 10 min. After blocking in 3% normal goat serum (NGS) in PBS, cells were washed in PBS and, for most antibodies, permeabilized for 10 min in 1% Triton X-100 in PBS. When probing with rabbit anti-RAD51 antibody, cells were permeabilized in acetone for 2 min at −20°C, followed by a wash with PBS. Cells were incubated with primary antibodies diluted in 3% NGS for at least 30 min. Antibodies 39S, 71-9A, and 2H10 and the polyclonal antibodies anti-RAD51, anti-NBS1, and anti-Ku86 were used at a concentration of 1:200, while antibody 367 was used at 1:400. After extensive washing with PBS, cells were incubated for 30 min with secondary antibodies diluted in 3% NGS at a concentration of 1:200. After a final wash with PBS, coverslips were mounted in glycerol gelatin containing 2.5% DABCO to retard photobleaching.
Microscopy.
Cells stained for IF were examined with a Zeiss LSM 410 confocal microscope system equipped with an argon-krypton laser, an Axiovert 135 inverted microscope, and a Zeiss 100X Plan Neofluar objective. Alexa Fluor 488 was excited at 488 nm, and Alexa Fluor 594 was excited at 568 nm. The emission filters used were BP505-530 and LP610, respectively. The two channels were scanned individually with settings established by using control slides. Channels were overlaid by computer for merged images. As controls, samples were stained with one primary antibody and both secondary antibodies. No overlap between the optical channels was observed for any of the samples at the settings used. At least 100 UL29-positive cells from a minimum of two independent experiments were counted per host protein examined. Collected images were arranged with Adobe Photoshop 6.0 and labeled with Adobe Illustrator 10.
Tallies of cell populations were also performed with an Olympus BX60 microscope equipped with a mercury burner, a universal reflected-light fluorescent vertical illuminator, and a UPlanFl 100× objective (Olympus America Inc., Melville, N.Y.). A U-MNB filter set (Olympus), in which the long-pass barrier filter was replaced with the band pass D540/40 M filter (Chroma Technology Corp., Rockingham, Vt.), was employed for viewing Alexa Fluor 488-labeled samples, while a Texas Red filter set (Chroma Technology Corp.) was used for Alexa Fluor 594-labeled samples. Controls were performed as described above. Similar results were obtained regardless of the microscope system used.
Western analysis.
Protein expression was examined by Western blot analysis. Vero cells were grown in 100-mm plates to 70% confluence. Cells were either mock infected or infected with wild-type or mutant HSV-1 at a multiplicity of infection of 10. Where indicated, 1 μM camptothecin or 400 μg of PAA per ml was added to the culture medium at the time of adsorption. At the indicated times, whole-cell extracts were prepared as follows. After washing the cells with ice-cold PBS, 500 μl of lysis buffer (25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, 10 mM EDTA) was added to each plate and incubated on ice for 30 min. Cells were then scraped into the buffer and transferred to 1.5-ml centrifuge tubes. After a brief sonication, cell lysates were clarified by centrifugation at 10,000 × g at 4°C for 10 min. The supernatants were transferred to SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer, boiled for 5 min, and subjected to electrophoresis in SDS-12% polyacrylamide gels. The resolved proteins were transferred onto polyvinylidene difluoride membranes by electroblotting at 60 V for 2 h. The membranes were blocked in 5% (wt/vol) nonfat dry milk in TBST (0.2% [vol/vol] Tween 20 in PBS). The membranes were incubated with primary antibodies for at least 1 h. Primary antibodies in TBST were used at the following dilutions: monoclonal anti-RAD51, anti-DNA-PKcs cocktail, and anti-Ku70 at 1:200; anti-RAD51 and antitubulin at 1:1000; 3-83, 71-9A, 2H10, anti-Mre11, and anti-Rad52 at 1:5,000; and anti-NBS1 and anti-Ku86 at 1:10,000. After several washes in TBST, the membranes were incubated with alkaline phosphatase-conjugated secondary antibodies at a dilution of 1:10,000 for 1 to 2 h and developed with alkaline phosphatase color detection according to the manufacturer's instructions (Promega, Madison, Wis.).
RESULTS
Subcellular distribution of RPA in infected cells.
IF of HSV-1-infected cells was used to determine the cellular distribution of host recombination/repair proteins with respect to the viral single-stranded-DNA-binding protein UL29. The viral structures examined include stage IIIa prereplicative sites (observed in cells infected with the polymerase null mutant HP66), stage IIIb prereplicative sites (seen in cells infected with wild-type virus in the presence of PAA), and replication compartments (seen in uninhibited wild-type infection). As stated above, stage IIIa foci do not contain PML, while stage IIIb foci and replication compartments possess at least one isoform of PML. The redistribution of PML back to viral assemblies is dependent on the recruitment of the viral polymerase to these sites (9) which, in turn, is dependent on an active primase (12).
Because the life cycle of HSV-1 is intimately associated with ND10, we are particularly interested in recombination proteins that are associated with this nuclear domain. For this reason we chose the following host proteins for this investigation: RPA, a heterotrimeric single-stranded-DNA-binding protein that is involved in replication, recombination, and repair (22); RAD51, a member of the RAD52 epistasis group of gene products involved in HR (5); and NBS1, a component of the mammalian MRE11 DNA repair complex (11) that plays an essential role in HR in vertebrate cells (68). Although not reported to be specifically associated with ND10, Ku86, a component of the NHEJ pathway (reviewed in reference 30), was also selected for this study.
It has been shown previously that both the 32- and 70-kDa subunits of RPA (RPA32 and RPA70, respectively) colocalize with UL29 in replication compartments (76). In cells in which HSV-1 replication is inhibited with PAA, RPA70 has also been found in the numerous viral sites specific to the S phase of the cell cycle but not within the less-numerous, S-phase-independent sites (69), which we refer to as stage IIIb foci.
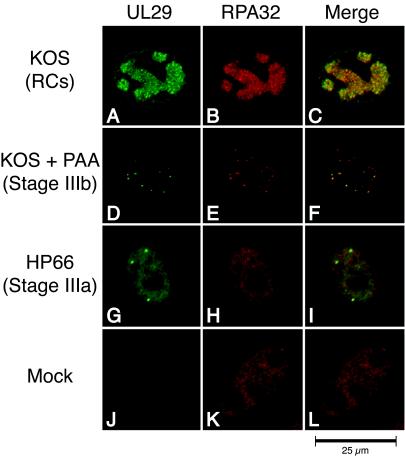
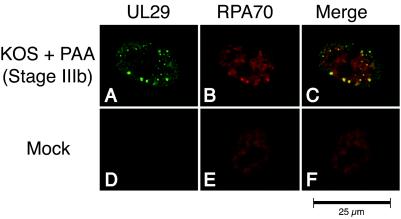
In order to extend these findings, we wanted to determine whether RPA32 colocalizes with UL29 in stage IIIa sites as well as in stage IIIb foci and replication compartments. As expected, no UL29 was observed in mock-infected cells above the background staining of the control (Fig. 2J). In mock-infected cells, RPA32 was diffusely distributed within the nucleus which typically contained fewer than five RPA foci (Fig. 2K). Double staining of Vero cells infected with wild-type HSV-1 indicated that, as previously reported (76), UL29 and RPA32 colocalized within replication compartments (Fig. 2A to C). A significant colocalization of RPA32 and UL29 was observed in cells infected in the presence of PAA (Fig. 2D to F). This is in contrast to results reported by Uprichard and Knipe (69), who observed no RPA staining in the S-phase-independent sites that we refer to as stage IIIb foci. One possibility for this discrepancy is that different anti-RPA antibodies were employed. We therefore repeated the experiment with a monoclonal antibody (2H10) directed against RPA70 (Fig. 3). Again, we observed a significant colocalization of RPA70 and UL29 in stage IIIb foci (Fig. 3A to C) as well as in replication compartments (data not shown). In our hands, RPA, as detected with antibodies against both the 32- and 70-kDa subunits, is recruited to stage IIIb foci.
FIG. 2.
RPA 32-kDa subunit colocalizes with UL29 in replication compartments (RCs) and stage IIIb foci but not stage IIIa foci. Vero cells were infected with KOS in either the absence (A to C) or presence (D to F) of PAA or infected with the polymerase null mutant virus HP66 (G to I). As described in Materials and Methods, cells were double labeled with rabbit anti-UL29 (367) and mouse anti-RPA32 (71-9A) to detect the localization of the HSV-1 major DNA-binding protein UL29 (green) and cellular RPA32 (red). The viral structures examined included replication compartments, stage IIIb foci, and stage IIIa foci. Mock infection in the presence of PAA gave staining patterns similar to that of mock infection in the absence of PAA (J to L and results not shown).
FIG. 3.
RPA 70-kDa subunit colocalizes with UL29 in stage IIIb prereplicative sites of HSV-1-infected cells. Vero cells were mock infected (D to F) or infected with KOS in the presence of PAA (A to C). As described in Materials and Methods, cells were double labeled with rabbit anti-UL29 (367) and mouse anti-RPA70 (2H10) to detect the localization of the HSV-1 major DNA-binding protein UL29 (green) and cellular RPA70 (red). Viral structures shown are stage IIIb foci.
We next asked whether UL29 and RPA colocalize in stage IIIa foci of cells infected with the polymerase null virus HP66. Interestingly, no significant colocalization of RPA and UL29 was observed for stage IIIa foci (Fig. 2G and H). In the minor population of cells in which colocalization of RPA and UL29 was noted, coincident staining was typically faint. This is in contrast to the stage IIIb population, which exhibited a colocalization pattern of UL29 and RPA that was strong and complete.
We have previously shown that PML recruitment to viral replication foci in infected cells is dependent on the presence of viral polymerase within these foci and, thus, presumably on the composition of viral proteins at the replication fork (9). In this study we show that the recruitment of RPA to stage III sites is similarly dependent on the presence of the HSV-1 polymerase and the status of the replication machinery.
Subcellular distribution of RAD51 in infected cells.
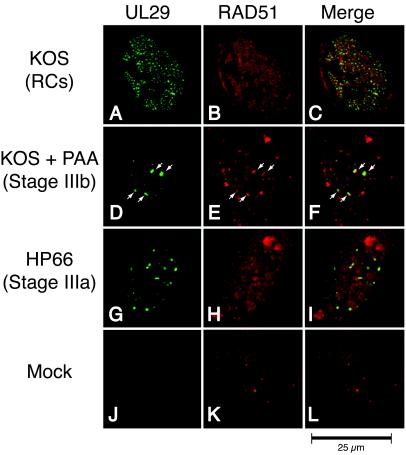
In parallel IF studies, we determined the localization of RAD51, a recombinase involved in HR, with respect to UL29 in mock- and HSV-1-infected cells. As expected, no UL29 staining was observed in uninfected controls with 39S, a monoclonal antibody directed against UL29 (Fig. 4J). Mock-infected cells typically contained fewer than five RAD51 foci per cell, and most of the staining was nuclear (Fig. 4K). Double labeling of KOS-infected cells indicated that 61% of the cells with replication compartments displayed colocalization of RAD51 and UL29 (Fig. 4D to F). Furthermore, while many cells at stage IIIb of infection displayed a subset of RAD51 foci that colocalized with UL29, few cells at stage IIIa displayed this colocalization pattern (Fig. 4G to I). We also noted that the colocalization pattern of RAD51 in stage IIIb foci was neither as strong nor as complete as that of RPA. Nevertheless, the observation that RAD51 can be found predominantly in stage IIIb but not stage IIIa prereplicative sites indicates that, like that of RPA, its recruitment to viral foci may depend on the composition or the conformation of proteins at the replication fork.
FIG. 4.
RAD51 colocalizes predominantly with UL29 in replication compartments (RCs) and stage IIIb foci but not stage IIIa foci. Vero cells were infected with KOS in either the absence (A to C) or presence (D to F) of PAA or infected with the polymerase null mutant virus HP66 (G to I). As described in Materials and Methods, cells were double labeled with mouse anti-UL29 (39S) and rabbit anti-RAD51 to detect the localization of the HSV-1 major DNA-binding protein UL29 (green) and cellular RAD51 (red). Viral structures examined included replication compartments, stage IIIb foci, and stage IIIa foci. Mock infection in the presence of PAA gave staining patterns similar to that of mock infection in the absence of PAA (J to L and results not shown).
Subcellular distribution of NBS1 in infected cells.
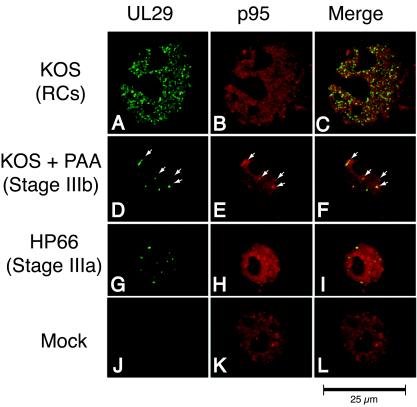
Similar experiments were performed with antibodies directed against UL29 and NBS1, a component of the MRE11 DNA repair complex. Mock-infected cells exhibited nuclear staining of NBS1, with five or fewer foci seen per cell (Fig. 5K). When wild-type-infected cells were double stained with anti-NBS1 and 39S, NBS1 was always seen in replication compartments. Approximately 42% of the cells displaying stage IIIb foci exhibited an association of UL29 and NBS1. In many cases, a subset of NBS1 nuclear foci was seen associated with a subset of UL29 nuclear foci. Only ≈19% of cells at stage IIIa displayed an association of UL29 and NBS1 (Fig. 5G to I). Thus, like PML, RPA, and RAD51, NBS1 is recruited to viral foci predominantly when viral polymerase is present.
FIG. 5.
NBS1 colocalizes predominantly with UL29 in replication compartments (RCs) and stage IIIb foci but not stage IIIa foci. Vero cells were infected with KOS in either the absence (A to C) or presence (D to F) of PAA or infected with the polymerase null mutant virus HP66 (G to I). As described in Materials and Methods, cells were double labeled with mouse anti-UL29 (39S) and rabbit anti-NBS1 to detect the localization of the HSV-1 major DNA-binding protein UL29 (green) and cellular NBS1 (red). Viral structures examined included replication compartments, stage IIIb foci, and stage IIIa foci. Mock infection in the presence of PAA gave staining patterns similar to that of mock infection in the absence of PAA (J to L and results not shown).
As stated above, some stage IIIb foci colocalized with RAD51 (Fig. 4D to F) and some colocalized with NBS1 (Fig. 5D to F). The MRE11 complex, of which NBS1 is a member, and RAD51 play important but distinct roles in HR. Each is able to form foci at sites of DNA damage (39, 56). These two proteins, however, are cytologically distinct and are not found together within any given nuclear focus (44). It is therefore possible that while RAD51 and NBS1 are able to be recruited to stage IIIb foci, their different roles in the response to DNA double-strand breaks may prevent their recruitment to the same viral focus. This may explain the observation that neither RAD51 nor NBS1 is found in stage IIIb foci. In order to test this proposal directly, further experiments will be necessary.
Subcellular distribution of Ku86 in infected cells.
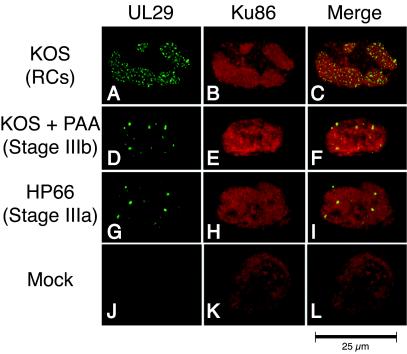
Next we analyzed the localization pattern of Ku86, a DNA end-binding protein that is an essential factor involved in NHEJ. In mock-infected cells, Ku86 exhibited homogenous staining throughout the nucleus (Fig. 6K). KOS-infected cells displayed a redistribution of Ku86 into replication compartments; however, Ku86 was never found in stage IIIb or stage IIIa prereplicative sites (Fig. 6A to I). This is in contrast to the staining patterns of RPA, RAD51, and NBS1 which were observed in stage IIIb foci. These observations indicate that Ku86 is either excluded from or not actively recruited to early viral structures and suggest that NHEJ may not play a role in the early stages of HSV-1 DNA replication.
FIG. 6.
Ku86 colocalizes with UL29 only in replication compartments (RCs). Vero cells were infected with KOS in either the absence (A to C) or presence (D to F) of PAA or infected with the polymerase null mutant virus HP66 (G to I). As described in Materials and Methods, cells were double labeled with mouse anti-UL29 (39S) and rabbit anti-Ku86 to detect the localization of the HSV-1 major DNA-binding protein UL29 (green) and cellular Ku86 (red). Viral structures examined included replication compartments, stage IIIb foci, and stage IIIa foci. Mock infection in the presence of PAA gave staining patterns similar to that of mock infection in the absence of PAA (J to L and results not shown).
Western blot analysis of infected cells.
We performed Western blot analysis of infected cell lysates to determine whether the relocalization of the host proteins observed in IF during infection correlated with any changes in protein levels or posttranslational modifications. In the presence or absence of PAA, cells were either mock infected or infected with either KOS or HP66. At 1 and 7 h postinfection, cells were harvested for immunoblot analysis as described in Materials and Methods.
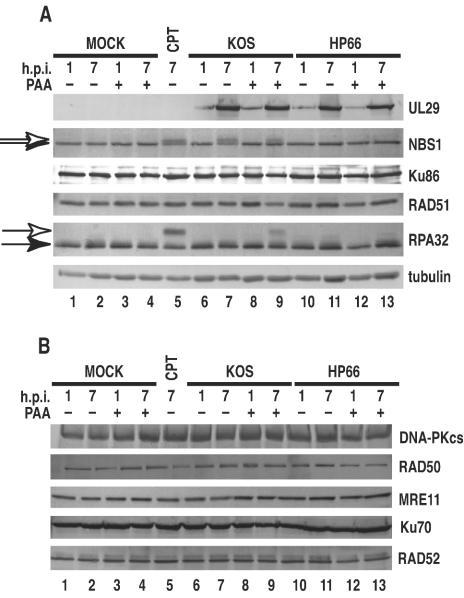
Immunoblotting showed that, for each of the infected cell lysates, UL29 was detected at 1 h postinfection, with increased levels of UL29 seen at 7 h postinfection (Fig. 7A). As expected, UL29 was not detected in any of the mock-infected cell lysates. Compared to mock-infected controls, the levels of RPA32, RPA70, RAD51, NBS1, and Ku86 were unaffected in infected cell lysates harvested at 1 and 7 h postinfection. At 7 h postinfection, a shift in the electrophoretic mobility was noted for NBS1 in lysates prepared from cells infected with KOS in either the absence or presence of PAA (Fig. 7A, lanes 7 and 9). The altered migration of NBS1 has been shown to be due to phosphorylation, which occurs in response to treatment with DNA-damaging agents (39, 44, 48). We confirmed this result by treating Vero cells with the DNA-damaging agent camptothecin at 1 μM for 6 h prior to preparation of cell lysates (Fig. 7A, lane 5). Further evidence that this slower-migrating species of NBS1 is phosphorylated was provided by the observation that it was eliminated by treatment with lambda protein phosphatase (New England Biolabs, Beverly, Mass.) (results not shown).
FIG. 7.
Western blots analysis. Lysates were prepared at the indicated hours postinfection (h.p.i.). In the presence or absence of PAA, Vero cells were either mock-infected or infected with either KOS or the polymerase null mutant virus HP66. As a control for DNA damage, mock-infected cells were also treated with 1 μM camptothecin (CPT) for 6 h prior to harvest. (A) Membranes were probed with antibodies directed against UL29, NBS1, Ku86, RAD51, and RPA32 as described in Materials and Methods. The open arrows indicate the slower-migrating species of NBS1 and RPA32. Results for tubulin are provided as a loading control. (B) Membranes were probed with antibodies directed against RAD52, RAD50, MRE11, and DNA-PKcs.
Phosphorylation of NBS1 has also been observed following infection with adenovirus mutants that form genome concatemers (66). The shift in the migration of NBS1 noted for KOS infection may therefore reflect a DNA damage response of the cell induced by infection with wild-type virus. A shift in the migration of NBS1 was not observed in HP66-infected cells at the same time point. That the mobility shift was noted for NBS1 when the viral polymerase was inhibited (Fig. 7A, lanes 7 and 9) but not when it was absent (Fig. 7A, lanes 11 and 13) suggests that this particular cellular response to DNA damage is dependent on the presence but not the activity of viral polymerase. This correlates with the IF data indicating that recruitment of NBS1 and other recombination proteins to viral foci is dependent on the presence of the viral polymerase. A time course for lysates derived from KOS-infected cells indicated that phosphorylation of NBS1 occurred at approximately 5 h postinfection (data not shown).
It is well established that RPA32 is hyperphosphorylated in response to DNA damage (13, 32). Consistent with previous findings, treatment of Vero cells with 1 μM camptothecin for 6 h prior to harvest (Fig. 7A, lane 5) resulted in a shift in the electrophoretic mobility of RPA32. This shift was also observed in cells infected with KOS in the presence of PAA (Fig. 7A, lane 9). Although we have not yet been able to directly demonstrate that the shift is due to hyperphosphorylation by using lambda protein phosphatase treatment, it is the simplest explanation. Furthermore, hyperphosphorylated RPA32 has been shown to be resistant to extraction, remaining within the nucleus (43). We confirmed this result by using extraction conditions compatible with phosphatase activity; we were unable to extract the slowly migrating species of RPA from either camptothecin-treated cells or cells infected with KOS in the presence of PAA (results not shown). Taken together with previous results, the results of this experiment are consistent with the interpretation that the slower-migrating form of RPA32 is due to hyperphosphorylation as a result of DNA damage. This mobility shift was not seen in KOS-infected cells at 1 or 7 h postinfection in the absence of PAA (Fig. 7A, lanes 6 and 7, respectively). Furthermore, a time course experiment indicated that Vero cells infected with KOS in the absence of PAA and harvested hourly up to 7 h postinfection did not exhibit hyperphosphorylation at any of these time points (results not shown). PAA treatment of mock-infected cells did not elicit this response (Fig. 7A, lanes 3 and 4), indicating that PAA is not itself inducing the cellular response to DNA damage. Interestingly, the hyperphosphorylation of RPA32 was not observed in lysates of cells infected with the polymerase null virus (Fig. 7A, lanes 7, 11, and 13). Thus, NBS1 phosphorylation occurred in KOS-infected cells regardless of whether the viral polymerase was inhibited, whereas hyperphosphorylation of RPA32 was noted only in cells infected with KOS under conditions that inhibited the polymerase.
In addition to RPA, RAD51, NBS1, and Ku86, we performed Western blot analysis of other host recombination proteins: Rad52 (an essential protein involved in HR), MRE11 and RAD50 (which, together with NBS1, form the MRE11 DNA repair complex), as well as the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) and Ku70 (which, together with Ku86, participate in NHEJ) (reviewed in reference 30). Figure 7B shows that the levels of these proteins in lysates from infected cells remained unchanged compared to those in mock-infected controls. It has previously been reported that DNA-PKcs is degraded during HSV-1 infection of HeLa S3 cells (29, 53). We show here that, in Vero cells, DNA-PKcs is not degraded during the course of HSV-1 infection, indicating that HSV-1-induced degradation of DNA-PKcs may be cell type specific. Western blot analysis indicates that HSV-1 infection does not affect the steady-state levels of these host recombination-repair proteins but does induce the phosphorylation of NBS1 and RPA32 as a host response to DNA damage.
IF characterization of host recombination proteins in transfected cells expressing the four-protein subassembly of viral replication proteins.
Cells expressing the viral helicase-primase heterotrimer and single-stranded-DNA-binding protein are able to form a four-protein subassembly which localizes adjacent to ND10 (35). Although these foci resemble stage III foci, they are not identical. During these transfection experiments, ND10 are not disrupted because ICP0 is not present. Furthermore, transfection foci do not require the presence of UL9 for their formation, whereas stage IIIa focus formation occurs only if UL29, UL5, UL8, UL52, and UL9 are all present. To examine the relationship between the four-protein transfection foci and the stage III foci observed during infection, we examined whether RPA, RAD51, NBS1, and Ku86 could be observed with the four-protein subassembly.
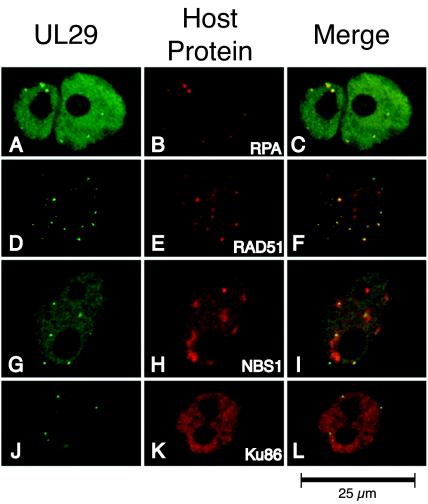
Vero cells were transfected with plasmids encoding oriS, UL29, UL5, UL8, and UL52 and double stained with antibodies directed against UL29 and one of the indicated host recombination proteins as described in Materials and Methods (Fig. 8). Untransfected controls displayed staining patterns similar to that reported for mock-infected controls described above (data not shown). Tallies of UL29-positive cells indicated that RPA, RAD51, and NBS1 each displayed an association with the four-protein subassembly in transfected cells. RPA displayed a complete colocalization with UL29 foci (Fig. 8A to C). Figure 8 also shows that a subset of RAD51 foci associated or colocalized with UL29. NBS1 exhibited an unusual staining pattern (Fig. 8D to F). Most of the UL29 foci were found associated with NBS1; however, rather than a punctate staining pattern, as seen for the other host recombination proteins, NBS1 appeared to surround the four-protein transfection foci (Fig. 8G and H). The significance of this staining pattern is not clear. Finally, Ku86 was not specifically associated with the four-protein subassembly in transfected cells (Fig. 8J to L) and instead displayed a diffuse nuclear staining pattern identical to that seen in mock-transfected controls (data not shown).
FIG. 8.
IF characterization of cellular recombination proteins with respect to the four-protein viral subassembly of transfected cells. Vero cells were transfected with expression plasmids bearing UL29, UL5, UL8, UL5,2 and oriS as described in Materials and Methods. At 16 to 24 h posttransfection, cells were fixed, permeabilized, and double labeled with antibodies to detect the localization of the HSV-1 major DNA-binding protein UL29 (green) and the indicated cellular recombination protein (red). The primary antibodies used were 367 (A), 39S (D, G, and J), 71-9A (B), rabbit anti-RAD51 (E), anti-NBS1 (H), and anti-Ku86 (K). Merged images are shown in panels C, F, I, and L. The foci observed in all four rows represent the four-protein subassembly UL29/UL5/UL8/UL52, which occurs at a limited number of sites within the nucleus of cells that are typically not in the S phase of the cell cycle. In our experience, the polyclonal UL29 antibody 367 stains diffusely within the nucleus in addition to a limited number of UL29 foci (A), whereas the monoclonal antibody 39S primarily detects UL29 within discrete foci (D, G, and J). Some variability in UL29 staining was always noted.
In summary, our findings show that while the recruitment of RPA, RAD51, and NBS1 was compromised at stage IIIa foci in infected cells (Fig. 2, 4, and 5), each protein was found associated with UL29 foci in transfected cells expressing UL29 and the helicase-primase proteins (Fig. 8). The observation that RPA, RAD51, and NBS1 were found at the transfection foci and not stage IIIa prereplicative sites is intriguing and indicates that the conditions for the recruitment of these host factors to prereplicative sites during infection may differ from the conditions found in transfected cells.
DISCUSSION
In this study, immunofluorescence microscopy was used to characterize the distribution of cellular recombination/repair proteins in replication compartments and their precursors (summarized in Table 1). Our finding that RPA, RAD51, NBS1, and Ku86 are recruited to replication compartments confirms and extends previous observations that RPA, PCNA, DNA ligase, polymerase α, p53, retinoblastoma protein, and RNA polymerase II are found in replication compartments (27, 58, 76). Since viral gene expression, DNA replication, and packaging are all believed to occur within replication compartments, it is difficult to know whether the presence of these factors in replication compartments is specific for DNA replication per se.
TABLE 1.
Colocalization of UL29a
| Cellular protein | % of population (no. of cells)
|
|||
|---|---|---|---|---|
| KOS (replication compartments) | KOS + PAA (stage IIIb sites) | HP66 (stage IIIa sites) | Transfection with UL29, UL5/UL8/UL52, and oriS (ND10-associated foci) | |
| RPA32 | 95.7 (139) | 91.0 (122) | 14.8 (115) | 91 (100) |
| RAD51 | 61.4 (246) | 76.2 (219) | 48.2 (247) | 75.2 (105) |
| NBS1 | 97.1 (208) | 42.3 (215) | 19.2 (193) | 59.6 (104) |
| Ku86 | 87.2 (125) | 0 (114) | 0 (124) | 0 (104) |
Percentage of the indicated infected or transfected cell population in which UL29 colocalized or associated with the listed host recombination protein. The numbers in parentheses are the number of UL29-positive cells counted for that particular cell population.
In this paper, we focused on the earliest subassemblies of viral DNA replication proteins to better understand whether host factors interact with the DNA synthesis machinery during viral DNA replication. Several observations were made. (i) RPA, RAD51, and NBS1 are recruited predominantly to stage IIIb but not stage IIIa foci; (ii) Ku86 is not recruited to either stage IIIa or stage IIIb foci; (iii) Western blot analysis of Vero cells indicated that the steady-state levels of RPA, RAD51, NBS1, and Ku86 as well as other components of host recombination pathways remain unchanged throughout the course of HSV-1 infection; (iv) during infection with wild-type virus, NBS1 is phosphorylated; (v) in cells infected with wild-type virus in the presence of PAA, RPA is hyperphosphorylated; and (vi) in transfected cells expressing UL29 and the three members of the helicase-primase complex, UL29 foci which associate with RPA, RAD51, and NBS1 but not Ku86 are formed. These results suggest that components of the HR pathway may play a role at the earliest stages of HSV-1 DNA replication. Furthermore, the phosphorylation of NBS1 and RPA suggests that HSV-1 infection may activate components of the DNA damage response. These results add to the increasing body of evidence linking DNA replication, recombination, and repair in several systems (reviewed in reference 77).
Ku86 is not recruited to viral prereplicative sites.
Our finding that Ku86, a component of the NHEJ pathway, was found in neither stage IIIa, stage IIIb, nor transfection foci implies that NHEJ may not be involved in the early stages of viral DNA replication. This conclusion is consistent with recent studies on the circularization of viral genomes in infected cells. It has been proposed that one of the first steps in DNA replication is the circularization of the viral genome, perhaps through the action of cellular recombination proteins (19, 54, 79). Jackson and Deluca, however, demonstrated that HSV-1 genome circularization may be inhibited during lytic infection of Vero cells by the action of ICP0 (24). It is possible that circularization is prevented via inhibition of the NHEJ pathway through ICP0-mediated degradation of DNA-PKcs, which is observed in some cell types (29, 53). We demonstrated, however, that DNA-PKcs is not degraded during HSV-1 infection of Vero cells. Our observation that Ku86 is not recruited to the earliest sites of viral replication may indicate that, in some cell types, HSV-1 inhibits NHEJ via spatial segregation, either by excluding or failing to recruit components of this pathway to viral foci. The possible role of HR during viral DNA replication is described below.
Cellular HR proteins, PML, and viral replication foci during infection.
It has been recognized for some time that HSV-1 DNA replication is closely associated with ND10 (1, 23, 42). The viral immediate-early protein ICP0 localizes to and induces the dispersal of ND10 and associated proteins (17, 41). We have shown previously that, after the initial disruption of ND10, certain isoforms of PML can be redistributed to replication compartments and stage IIIb prereplicative sites but not stage IIIa prereplicative sites (9). Stage IIIa foci, which form in the presence of UL29, the helicase-primase, and UL9, are not competent to recruit PML unless two conditions are met: (i) the helicase-primase is capable of primase activity (12) and (ii) the HSV-1 DNA polymerase is present (9). We have proposed that the recruitment of polymerase to stage IIIb foci requires a conformational change at the replication fork which allows polymerase to be loaded (12). This is reminiscent of the recruitment of bacterial and eukaryotic polymerases, which also require the presence of a primer for efficient recruitment of polymerase to the replication fork (2, 18, 46).
Once polymerase has been loaded, we envisage that the replication fork would be competent to carry out DNA synthesis and recruit cellular proteins such as PML, RPA, RAD51, and NBS1. That stage IIIb foci are observed in the presence of PAA indicates that active DNA synthesis is probably not required for the recruitment of these proteins. However, we cannot rule out that a small amount of DNA synthesis may be taking place in the presence of PAA and may be required for the efficient recruitment of these proteins. Furthermore, it is not clear whether the recruitment of PML, RPA, RAD51, and NBS1 to viral replication forks is actually required for viral replication; however, we are intrigued by the possibility that host recombination and repair proteins may play a role in the generation of the larger-than-unit-length concatemers (reviewed in references 37 and 77).
Recruitment of RPA, RAD51, and NBS1 to HSV-1 replication foci correlates with the DNA damage response.
Although no significant changes in the levels of host recombination proteins were observed, we did note that, on infection, NBS1 is phosphorylated and RPA32 is hyperphosphorylated, indicating a host response to DNA damage. We observed NBS1 phosphorylation starting at ≈5 h postinfection during wild-type infection (Fig. 7A and data not shown). Phosphorylation of NBS1 was also noted for wild-type infection under conditions in which the viral polymerase was inhibited, indicating that the host DNA damage response probably does not require either viral DNA synthesis or late viral gene expression, although we cannot rule out a small amount of DNA synthesis in the presence of PAA. NBS1 phosphorylation did not occur during infection with the polymerase null virus. These results suggest that phosphorylation correlates with the recruitment of polymerase as well as RPA, RAD51, and NBS1 to viral foci (stage IIIb). Taken together, these results suggest that the formation of a replication fork containing the HSV-1 polymerase may be seen as a signal for DNA damage.
It is worth noting that adenovirus, another DNA virus which replicates in the nucleus, has evolved a strategy for replicating its genome that does not involve the formation of larger-than-unit-length concatemers (7, 73). In fact, concatemerization is actively prevented in adenovirus-infected cells by a mechanism which involves the reorganization and degradation of members of the Mre11-Rad50-NBS1 repair complex (66). In the absence of early region 4, the Mre11 repair complex is not degraded, NBS1 is phosphorylated, and concatemers can form (7, 73). Adenovirus has thus evolved to prevent concatemerization of viral DNA by inactivating the Mre11 repair complex (66). It is interesting that herpesviruses have evolved a very different replication strategy, resulting in the generation of longer-than-unit-length viral genomes. Our findings that wild-type infection correlates with NBS1 phosphorylation and that NBS1 is recruited to the earliest sites of viral DNA replication suggest that HSV-1 may utilize the Mre11 repair complex to promote viral DNA concatemerization during replication. It will be of interest to determine whether HSV-1 DNA replication occurs in cell lines which carry mutations in this pathway.
Treatment of cells with DNA-damaging agents such as UV, ionizing radiation, or camptothecin induces a DNA damage response resulting in the hyperphosphorylation of RPA32 (13, 32, 62). RPA32 hyperphosphorylation has been suggested to redirect the functions of RPA from DNA replication towards repair synthesis (40, 51). In this paper, we demonstrate that HSV-1 infection is also able to induce the hyperphosphorylation of RPA32, but only in the presence of PAA. Moreover, infection with the polymerase null virus was unable to induce RPA32 hyperphosphorylation, again suggesting that a primed but inhibited replication fork may be sufficient to transmit the DNA damage signal. That RPA hyperphosphorylation does not occur in HSV-1-infected cells in the absence of PAA suggests that the signal for this type of DNA damage is not transmitted under conditions in which DNA replication occurs and replication compartments form. The nature of the signal communicated by an inhibited viral polymerase is not clear but may be related to conformational changes resembling an uncoupled or stalled replication fork (33). As described above, NBS1 phosphorylation was observed in the presence as well as the absence of PAA, indicating that the signals for these markers of DNA damage are slightly different. Despite these differences, the fact that HSV-1infections can trigger two components of the DNA damage response is significant. Unraveling the role that these proteins play during HSV-1 infection will be of considerable interest.
HR proteins are associated with the four-protein transfection foci.
RPA, RAD51, and NBS1 were found associated with transfection foci in cells expressing UL29 and the helicase-primase in the absence of infection. This is somewhat surprising because, in the context of infection, stage IIIa foci, which contain these viral replication proteins plus UL9, are compromised in their recruitment of RPA, RAD51, and NBS1. Stage IIIa prereplicative sites form in the absence of polymerase and an active primase. These findings suggest that viral replication precursors that form during transfection and infection differ in fundamental ways. As mentioned above, one difference between stage IIIa foci and the transfection foci is that UL9 is required for the formation of the former but not the latter. It is possible that the presence of UL9 in stage IIIa foci may mask a site or sites needed for the recruitment of host factors. Another major difference is that the transfection foci form in the presence of intact ND10, while stage IIIa foci form only after the ICP0-induced disruption of ND10.
It is possible that ND10 disruption and the degradation of some ND10 proteins free up a site on the nuclear matrix or a matrix-associated factor necessary for the formation of stage IIIa foci. These foci are incompetent for recruitment of PML and other cellular factors until a more complete replication fork is assembled with primers and HSV-1 polymerase. According to this scenario, there may be two ways of recruiting RPA and other cellular factors to viral foci: in the context of transfection, RPA, RAD51, and NBS1 may be recruited to transfection foci which form adjacent to ND10 because of protein-protein interactions, perhaps between UL29 and RPA. In the context of infection, on the other hand, RPA, RAD51, and NBS1 may be recruited to a primed replication fork which contains the HSV-1 DNA polymerase.
We propose that the four-protein subassembly seen in transfected cells represents a biologically significant scaffold. Thus, in addition to the biochemical and enzymatic activities of UL29 and the helicase-primase, the four proteins may play an important structural role. This hypothesis is supported by studies identifying the HSV-1 proteins that provide helper function to adeno-associated virus. Heilbronn and colleagues identified UL29 and the helicase-primase as the minimal HSV-1 proteins capable of supporting adeno-associated virus replication in cell culture (74). Furthermore, the adeno-associated virus Rep protein colocalizes with UL29 and the helicase-primase in adeno-associated virus replication centers (67). Mutant genes defective in helicase or primase activity retain the ability to form transfection foci (12; N. Biswas and S. K. Weller, unpublished data) and still support adeno-associated virus replication (67). We propose that the ability of UL29 and the helicase-primase to form a scaffold which is able to recruit cellular proteins such as RPA is responsible for the helper activity of these four HSV-1 proteins.
Recombination and ND10.
It is noteworthy that RPA, RAD51, and NBS1 have each been shown to be ND10 associated (3, 10, 80), while Ku86 has not. This is of interest in light of our observation that RPA, RAD51, and NBS1 but not Ku86 resemble PML in their ability to localize to stage IIIb but not stage IIIa foci, whereas Ku86 does not colocalize to either. During infection, ND10 are disrupted, and subsequently PML and other ND10-associated proteins appear to be recruited back to UL29 foci. The significance of the re-recruitment of PML is not clear, but we have previously proposed that PML may play a role in the recruitment of other cellular proteins involved in DNA synthesis (9). It has recently been shown that PML and ND10-associated recombination/repair proteins are involved in the host response to DNA damage (3, 10, 78). We have shown that HSV-1 infection induces a response to DNA damage, and it is possible that the re-recruitment of PML to prereplicative sites may be related to this response. We have also proposed that, like bacteriophages lambda and T4, HSV-1 genomes may replicate via a recombination-mediated mechanism (77). As with phage lambda, both viral and cellular recombination proteins may be involved in this process. We have recently shown that HSV-1 encodes a recombinase comprised of UL29 and the alkaline nuclease (57).
In this report, we present evidence consistent with the involvement of host recombination and repair proteins in viral DNA replication. Interestingly, HSV infection appears to be able to inactivate the NHEJ pathway while utilizing the Mre11 repair complex as well as a virally encoded recombinase. Thus, HSV-1 may employ overlapping and partially redundant recombination-repair mechanisms in the generation of concatemeric replication intermediates during replication of its genome.
Acknowledgments
We thank the members of our laboratory for helpful comments on the manuscript. We thank William Ruyechan for the polyclonal anti-UL29 antibody 367, David Knipe for the polyclonal anti-UL29 antibody 3-83, and Marc Wold for the monoclonal anti-RPA antibodies 71-9A and 2H10.
This research was supported by Public Health Service grant AI21747. D.E.W. was supported by NIH training grant F32AI054042.
REFERENCES
- 1.Ascoli, C. A., and G. G. Maul. 1991. Identification of a novel nuclear domain. J. Cell Biol. 112:785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ason, B., J. G. Bertram, M. M. Hingorani, J. M. Beechem, M. O'Donnell, M. F. Goodman, and L. B. Bloom. 2000. A model for Escherichia coli DNA polymerase III holoenzyme assembly at primer/template ends. DNA triggers a change in binding specificity of the gamma complex clamp loader. J. Biol. Chem. 275:3006-3015. [DOI] [PubMed] [Google Scholar]
- 3.Barr, S. M., C. G. Leung, E. E. Chang, and K. A. Cimprich. 2003. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr. Biol. 13:1047-1051. [DOI] [PubMed] [Google Scholar]
- 4.Bataille, D., and A. Epstein. 1994. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology 203:384-388. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, P., and S. C. West. 1998. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 23:247-251. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Porat, T., A. S. Kaplan, B. Stehn, and A. S. Rubenstein. 1976a. Concatemeric forms of intracellular herpesvirus DNA. Virology 69:547-560. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 8.Burkham, J., D. M. Coen, C. B. Hwang, and S. K. Weller. 2001. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J. Virol. 75:2353-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkham, J., D. M. Coen, and S. K. Weller. 1998. The ND10 protein PML is recruited to HSV-1 prereplicative sites and replication compartments in the presence of the viral DNA polymerase. J. Virol. 72:10100-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carbone, R., M. Pearson, S. Minucci, and P. G. Pelicci. 2002. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 21:1633-1640. [DOI] [PubMed] [Google Scholar]
- 11.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates, 3rd, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 12.Carrington-Lawrence, S. D., and S. K. Weller. 2003. Recruitment of polymerase to herpes simplex virus type 1 replication foci in cells expressing mutant primase (UL52) proteins. J. Virol. 77:4237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carty, M. P., M. Zernik-Kobak, S. McGrath, and K. Dixon. 1994. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J. 13:2114-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutch, R. E., V. Bianchi, and I. R. Lehman. 1995. Herpes simplex virus type 1 DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J. Virol. 69:3084-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutch, R. E., R. C. Bruckner, E. S. Mocarski, and I. R. Lehman. 1992. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J. Virol. 66:277-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erdile, L. F., M. S. Wold, and T. J. Kelly. 1990. The primary structure of the 32-kDa subunit of human replication protein A. J. Biol. Chem. 265:3177-3182. [PubMed] [Google Scholar]
- 17.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang, L., M. J. Davey, and M. O'Donnell. 1999. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 4:541-553. [DOI] [PubMed] [Google Scholar]
- 19.Garber, D. A., S. M. Beverley, and D. M. Coen. 1993. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology 197:459-462. [DOI] [PubMed] [Google Scholar]
- 20.Hayward, G. S., R. J. Jacob, S. C. Wadsworth, and B. Roizman. 1975. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc. Natl. Acad. Sci. USA 72:4243-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heilbronn, R., and H. zur Hausen. 1989. A subset of herpes simplex virus replication genes induces DNA amplification within the host cell genome. J. Virol. 63:3683-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iftode, C., Y. Daniely, and J. A. Borowiec. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34:141-180. [DOI] [PubMed] [Google Scholar]
- 23.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss, 3rd, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, S. A., and N. A. DeLuca. 2003. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. USA 100:7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenny, M. K., U. Schlegel, H. Furneaux, and J. Hurwitz. 1990. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem. 265:7693-7700. [PubMed] [Google Scholar]
- 26.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaBoissiere, S., and P. O'Hare. 2000. Analysis of HCF, the cellular cofactor of VP16, in herpes simplex virus-infected cells. J. Virol. 74:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 29.Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70:7471-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieber, M. R. 1999. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells 4:77-85. [DOI] [PubMed] [Google Scholar]
- 31.Liptak, L. M., S. L. Uprichard, and D. M. Knipe. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative structures. J. Virol. 70:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, V. F., and D. T. Weaver. 1993. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol. Cell. Biol. 13:7222-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukonis, C. J., J. Burkham, and S. K. Weller. 1997. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J. Virol. 71:4771-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukonis, C. J., and S. K. Weller. 1996. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J. Virol. 70:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virology 71:2390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcy, A. I., P. D. Olivo, M. D. Challberg, and D. M. Coen. 1990. Enzymatic activities of overexpressed herpes simplex virus DNA polymerase purified from recombinant baculovirus-infected insect cells. Nucleic Acids Res. 18:1207-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marintcheva, B., and S. K. Weller. 2001. A tale of two HSV-1 helicases: roles of phage and animal virus helicases in DNA replication and recombination. Prog. Nucleic Acid Res. Mol. Biol. 70:77-118. [DOI] [PubMed] [Google Scholar]
- 38.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type-1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell Biol. 17:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mass, G., T. Nethanel, and G. Kaufmann. 1998. The middle subunit of replication protein A contacts growing RNA-DNA primers in replicating simian virus 40 chromosomes. Mol. Cell. Biol. 18:6399-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 42.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 43.McHugh, M. M., X. Yin, S. R. Kuo, J. S. Liu, T. Melendy, and T. A. Beerman. 2001. The cellular response to DNA damage induced by the enediynes C-1027 and neocarzinostatin includes hyperphosphorylation and increased nuclear retention of replication protein a (RPA) and trans inhibition of DNA replication. Biochemistry 40:4792-4799. [DOI] [PubMed] [Google Scholar]
- 44.Mirzoeva, O. K., and J. H. Petrini. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morse, L. S., L. Pereira, B. Roizman, and P. A. Schaffer. 1978. The use of intertypic recombinants for analysis of gene organization in herpes simplex virus. IARC Sci. Publ. 24:41-61. [PubMed] [Google Scholar]
- 46.Mossi, R., R. C. Keller, E. Ferrari, and U. Hubscher. 2000. DNA polymerase switching: II. Replication factor C abrogates primer synthesis by DNA polymerase alpha at a critical length. J. Mol. Biol. 295:803-814. [DOI] [PubMed] [Google Scholar]
- 47.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 48.Nelms, B. E., R. S. Maser, J. F. MacKay, M. G. Lagally, and J. H. Petrini. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280:590-592. [DOI] [PubMed] [Google Scholar]
- 49.Nimonkar, A. V., and P. E. Boehmer. 2003. The herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) promotes strand invasion. J. Biol. Chem. 278:9678-9682. [DOI] [PubMed] [Google Scholar]
- 50.Nimonkar, A. V., and P. E. Boehmer. 2002. In vitro strand-exchange promoted by the herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) and DNA helicase-primase. J. Biol. Chem. 277:15182-15189. [DOI] [PubMed] [Google Scholar]
- 51.Oakley, G. G., S. M. Patrick, J. Yao, M. P. Carty, J. J. Turchi, and K. Dixon. 2003. RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry 42:3255-3264. [DOI] [PubMed] [Google Scholar]
- 52.Olivo, P. D., N. J. Nelson, and M. D. Challberg. 1988. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc. Natl. Acad. Sci. USA 85:5414-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poffenberger, K. L., and B. Roizman. 1985. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J. Virol. 53:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 56.Raderschall, E., E. I. Golub, and T. Haaf. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA 96:1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reuven, N. B., A. E. Staire, R. S. Myers, and S. K. Weller. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rice, S. A., M. C. Long, V. Lam, and C. A. Spencer. 1994. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J. Virol. 68:988-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarisky, R. T., and P. C. Weber. 1994. Requirement for double-strand breaks but not for specific DNA sequences in herpes simplex virus type 1 genome isomerization events. J. Virol. 68:34-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Severini, A., A. R. Morgan, D. R. Tovell, and L. J. Tyrrel. 1994. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology 200:428-435. [DOI] [PubMed] [Google Scholar]
- 61.Severini, A., D. G. Scraba, and D. L. J. Tyrrel. 1995. Branched structures in the replicative intermediates of herpes simplex virus type 1 DNA. J. Virol. 70:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shao, R. G., C. X. Cao, H. Zhang, K. W. Kohn, M. S. Wold, and Y. Pommier. 1999. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 18:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shelton, L. S., A. G. Albright, W. T. Ruyechan, and F. J. Jenkins. 1994. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J. Virol. 68:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shlomai, J., A. Friedmann, and Y. Becker. 1976. Replicative intermediates of herpes simplex virus DNA. Virology 69:647-659. [DOI] [PubMed] [Google Scholar]
- 65.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 67.Stracker, T. H., G. D. Cassell, P. Ward, Y.-M. Loo, B. van Breukelen, S. D. Carrington-Lawrence, R. K. Hamatake, P. C. van der Vliet, S. K. Weller, T. Melendy, and M. D. Weitzman. 2004. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 78:441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tauchi, H., J. Kobayashi, K. Morishima, D. C. van Gent, T. Shiraishi, N. S. Verkaik, D. vanHeems, E. Ito, A. Nakamura, E. Sonoda, M. Takata, S. Takeda, S. Matsuura, and K. Komatsu. 2002. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 420:93-98. [DOI] [PubMed] [Google Scholar]
- 69.Uprichard, S. L., and D. M. Knipe. 1997. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229:113-125. [DOI] [PubMed] [Google Scholar]
- 70.Uprichard, S. L., and D. M. Knipe. 1996. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J. Virol. 70:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weber, P. C., M. D. Challberg, N. J. Nelson, M. Levine, and J. C. Glorioso. 1988. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell 54:369-381. [DOI] [PubMed] [Google Scholar]
- 72.Weber, P. C., M. Levine, and J. C. Glorioso. 1990. Recombinogenic properties of herpes simplex virus type 1 DNA sequences resident in simian virus 40 minichromosomes. J. Virol. 64:300-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weindler, F. W., and R. Heilbronn. 1991. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J. Virol. 65:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weller, S. K. 1995. Herpes simplex virus DNA replication and genome maturation, p. 189-213. In B. S. G. M. Cooper and R. Temin (ed.), Implications of the DNA provirus: Howard Temin's scientific legacy. ASM Press, Washington, D.C.
- 76.Wilcock, D., and D. P. Lane. 1991. Localization of p53, retinoblastoma and host replication porteins at sites of viral replication in herpes-infected cells. Nature 349:429-431. [DOI] [PubMed] [Google Scholar]
- 77.Wilkinson, D. E., and S. K. Weller. 2003. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life 55:451-458. [DOI] [PubMed] [Google Scholar]
- 78.Xu, Z. X., A. Timanova-Atanasova, R. X. Zhao, and K. S. Chang. 2003. PML colocalizes with and stabilizes the DNA damage response protein TopBP1. Mol. Cell. Biol. 23:4247-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yao, X. D., and P. Elias. 2001. Recombination during early herpes simplex virus type 1 infection is mediated by cellular proteins. J. Biol. Chem. 276:2905-2913. [DOI] [PubMed] [Google Scholar]
- 80.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175-4179. [PubMed] [Google Scholar]
- 81.Zhang, X., S. Efstathiou, and A. Simmons. 1994. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology 202:530-539. [DOI] [PubMed] [Google Scholar]
- 82.Zhong, L., and G. S. Hayward. 1997. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J. Virol. 71:3146-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]