Abstract
The placenta plays a central role in determining the outcome of pregnancy. It undergoes changes during gestation as the fetus develops and as demands for energy substrate transfer and gas exchange increase. The molecular mechanisms that coordinate these changes have yet to be fully elucidated. The study performed a large scale screen of the transcriptome of the rat placenta throughout mid-late gestation (E14.25–E20) with emphasis on characterizing gestational age associated changes in the expression of genes invoved in angiogenic pathways. Sprague Dawley dams were sacrificed at E14.25, E15.25, E17.25 and E20 (n = 6 per group) and RNA was isolated from one placenta per dam. Changes in placental gene expression were identifed using Illumina Rat Ref-12 Expression BeadChip Microarrays. Differentially expressed genes (>2-fold change, <1% false discovery rate, FDR) were functionally categorised by gene ontology pathway analysis. A subset of differentially expressed genes identified by microarrays were confirmed using Real-Time qPCR. The expression of thirty one genes involved in the angiogenic pathway was shown to change over time, using microarray analysis (22 genes displayed increased and 9 gene decreased expression). Five genes (4 up regulated: Cd36, Mmp14, Rhob and Angpt4 and 1 down regulated: Foxm1) involved in angiogenesis and blood vessel morphogenesis were subjected to further validation. qPCR confirmed late gestational increased expression of Cd36, Mmp14, Rhob and Angpt4 and a decrease in expression of Foxm1 before labour onset (P<0.0001). The observed acute, pre-labour changes in the expression of the 31 genes during gestation warrant further investigation to elucidate their role in pregnancy.
Introduction
A successful outcome to pregnancy depends on establishing an effective materno-fetal exchange interface. Two processes critical for materno-fetal exchange are adequate perfusion of the placenta by maternal blood, and formation of the placental villous tree (the exchange surface of the placenta) and its vascular network. Failure to achieve either of these processes is associated with adverse pregnancy outcome, including: miscarriage; intrauterine growth restriction; preeclampsia and preterm delivery. Furthermore, epidemiological studies identify a strong association between compromised placental structure and function, impaired fetal growth and the development of adult-onset diseases [1]–[3].
Development of the vascular network within placental villi during first trimester is a critical process that involves both vasculogenesis and angiogenesis. In humans, vasculogenesis starts during the third week post-conception. Hemangioblastic cell cords differentiate in situ from placental mesenchymal stem cells in the villous cores. The cords elongate through proliferation and cell recruitment, and connect with the vasculature of the developing fetus. A feto-placental circulation starts around 8 weeks of gestation and perfusion of the placental villi by maternal blood occurs 4–6 weeks later. Elongation of the capillaries results in looping of the vessels. Obtrusion of both capillary loops and new sprouts results in the formation of terminal villi.
The placental vasculature continues to develop and adapt during pregnancy in response to the increasing requirements of the fetus. Reynolds et al., [4] reported that the large increase in transplacental exchange, that supports the exponential increase in fetal growth during the last half of gestation, depends primarily on the dramatic growth of the placental vascular beds and the resultant large increases in uterine and umbilical blood flow [5]. The microvasculature of the placenta develops by the process of branching angiogensis that increases capillary numbers and surface densities. In the human placenta, branching angiogenesis, the formation of new vessels through sprouting, occurs from 5 weeks gestation through to 24 weeks gestation, while non-branching angiogenesis, the formation of capillary loops through elongation, predominates thereafter to term.
Available data supports the hypothesis that fetal growth and energy demand regulates placental growth and vascularization. For instance, the activity and expression of some nutrient transporters, in the mouse placenta, is modulated by fetal nutrient demands for growth [6]. Mutant mouse small placentae are capable of increasing glucose and amino acid transfer to meet the nutrient demands of the growing fetus, particularly in late gestation [6]. In human trophoblasts, System A, which includes the amino acid transport genes such as Slc38A1, Slc38A2 and Slc38A4 has been shown to increase as pregnancy progresses, coinciding with increased fetal nutrient demands [7]. During late gestation in rat, glucose transporter genes Slc2A3 and Slc2A1 are upregulated and downregulated respectively, in placenta [6]. The molecular and cellular mechanisms that regulate vascularisation and angiogenesis within the placental villous tree during pregnancy remain to be fully elucidated. Previous studies have established the utility of rat and mouse models to eluciate the molecular and cellular mechanisms involved in placentation and placental development since it is more difficult to obtain human placenta at various gestational stages [8]–[10]. Genome-wide gene expression in rat placenta has been studied in late pregnancy [11] [12]. At embryonic day 18.5 (E18.5), RNA sequencing showed differential gene expression between the rat labyrinth zone, junctional zone and metrial gland [12]. At E21, Buffat et al. reported changes in genome-wide placental gene expression in rats exposed to an isocaloric low protein diet [11]. Goyal et al. (2010), studied global gene expression in placentae from two different rat species at 17.5 days of gestation and found 272 genes differentially expressed [13]. Large scale molecular analysis of the transcriptome of the normal rat placenta across mid to late gestation, however, has not been performed. In particular, gestational variation in the expresssion of genes known to be involved in angiogenesis within the rat placenta is yet to be documented.
The aim of this study, therefore, was to characterize gestational variation in rat placental gene expression using microarray analysis (22,000 gene probes) and specifically identify changes in the molecular pathways involved in angiogenesis. The hypothesis to be tested was that genes involved in regulating angiogenesis are differentially expressed during late gestation compared to late mid-gestation. Placentae were collected at four gestational ages (E14.25; E15.25; E17.25 and E20) and gene expression was compared.
Materials and Methods
Animals and Diets
All animal experiments were performed at the Department of Anatomy and Developmental Biology, Monash University (Melbourne, Australia) with the approval of The School of Biomedical Sciences Animal Ethics Committee of Monash University. Experiments were carried out in accordance with the National Health and Medical Research Council of Australia “Australian Code of Practice for the Care and Use of Animals for Scientific Purposes” (7th edition, 2004).
Sprague Dawley dams were allowed to adapt to the animal house for one week, consuming standard chow diet and water ad libitum prior to start of the study. The animals were maintained in a light controlled environment (12 h light/dark cycle) throughout this study and fed on a standard chow diet. Animals were time mated in a 3 hour period, with male Sprague Dawley rats. This was designated as Day 0 of pregnancy. The rationale of using a 3 h window mating time is to reduce variability of gestational age among the offspring and to maximize the accuracy in staging of gestation. Pregnancy was confirmed at the time of sacrifice. After mating, dams were housed individually. All animals had access to food and water ad libitum throughout this study.
Tissue Collection
The pregnant dams were anaesthetized (Isoflurane Rhodia Australia P/L, VIC, Australia) and sacrificed at embryonic day (E) 14.25, 15.25, 17.25 and 20 (n = 6 per gestational age). Whole placentae were collected from the pregnant dams, weighed and then snap frozen in liquid nitrogen. Tissues were stored at −80°C until processed and analysed.
RNA Isolation
Total RNA was extracted from 30 mg of pulverized frozen placental tissue (n = 6 placentae per gestational age group, one placenta per dam), using the AllPrep DNA/RNA Mini Kit (Qiagen) as per manufacturers’ instructions. Genomic DNA was removed by On-column Dnase1 treatment. Following extraction, total RNA was quantified via NanoDrop ND-1000 spectrophotometer (Thermo Scientific, DE, USA). RNA quality was verified using an Agilent 2100 Bioanalyzer (VIC, Australia) prior to the analysis. RNA samples that fulfilled the following criteria were selected for microarray analysis: (i) RIN >8.5; (ii) 260/280 ratio >2; (iii) 260∶230>1. RIN (RNA Integrity Number) values were greater than 8.7. For qPCR, the RNA was reverse transcribed using the QuantiTect Reverse transcription kit (Qiagen) using 1 µg of RNA per sample.
Microarray Analysis on Illumina Rat Ref Arrays
For the microarray analysis, 500 ng of total RNA was converted to double stranded cDNA and this was used to generate biotinylated cRNA probes using the Illumina TotalPrep RNA Amplification Kit. Biotin-labelled cRNA were then hybridized to Illumina RatRef-12 Expression BeadChip (San Diego, CA, USA). Slides were scanned on a BeadStation 500 System using Beadscan software Version 3.5.31. No RNA samples were pooled in this analysis, each of the placental samples was analyzed independently. Samples were hybridized into wells at random. Two chips of 12 wells were used for the experiment. Illumina Whole-Genome Gene Expression BeadChips consist of oligonucleotides immobilized to beads held in microwells on the surface of an array substrate. Labelled sample cRNA were detected by hybridization to 50 mer probes on BeadChip. Post washing and staining, BeadChips were scanned on a BeadArray reader. Array experiment readout deposited on ArrayExpress. ArrayExpress Accession number is E-MTAB-1987.
Quantitative Real-Time PCR Validation Experiments
qPCR was used to validate the expression of three placental genes (Ptgs2, Pla2g2a and Nos2) known to display gestational age changes in expression. Each experiment was performed in duplicate and normalized against the expression of β-Actin (Actb) using the ddCt method [14]. RNA was reverse transcribed into complentary DNA strands, using the Quantitect Reverse transcription kit (Qiagen, VIC, Australia) following manufacturers instructions. Primers used for qPCR throughout the study are detailed in Table S1 and were all designed using PRIMER-BLAST/ncbi against Rattus Norvegicus mRNA. qPCR analyses was performed using ABI 6000 using standard SYBR green from Life Technologies (VIC, Australia). Normalized Ct ratios were subjected to ANOVA. Spearman’s Rank Correlation, p<0.05 was used for correlation plots. Prior to statistical analysis of gene expression during gestation, homogeneity of variance and normality were assessed using Bartlett’s and Shapiro-Wilk tests, respectively. Normally distributed and homogenous data were assessed by ANOVA otherwise data were analysed using non-parametic Kruskal-Wallis tests.
BioInformatic Anayses and Statistics
Bioinformatic analysis was performed by using the Illumina Beadstudio and Significance Analysis of Microarry (SAM, Stanford University) software. Data were normalized by performing a probe-intensity transformation and normalization via the Lumi package, Bioconductor. After normalization, differentially expressed placental genes (i.e. >2 fold expression, false discovery rate of <1%) were identified using SAM and were further analysed using Web-based Gene Set Analysis Toolkit (WebGestalt, http://bioinfo.vanderbilt.edu/webgestalt/). Genes were assigned to their respective functional classes based on the Gene Ontology (GO) database. Differences in group means were assessed by post-hoc comparisons (Bonferroni tests).
Normalized expression data were subjected to Principal Component Analyses (PCA, XLSTAT). 2D observation plots were generated for individual gestational age samples and correlation circles for the 31 angiogenic genes analyzed.
Results
Mircoarray Analysis
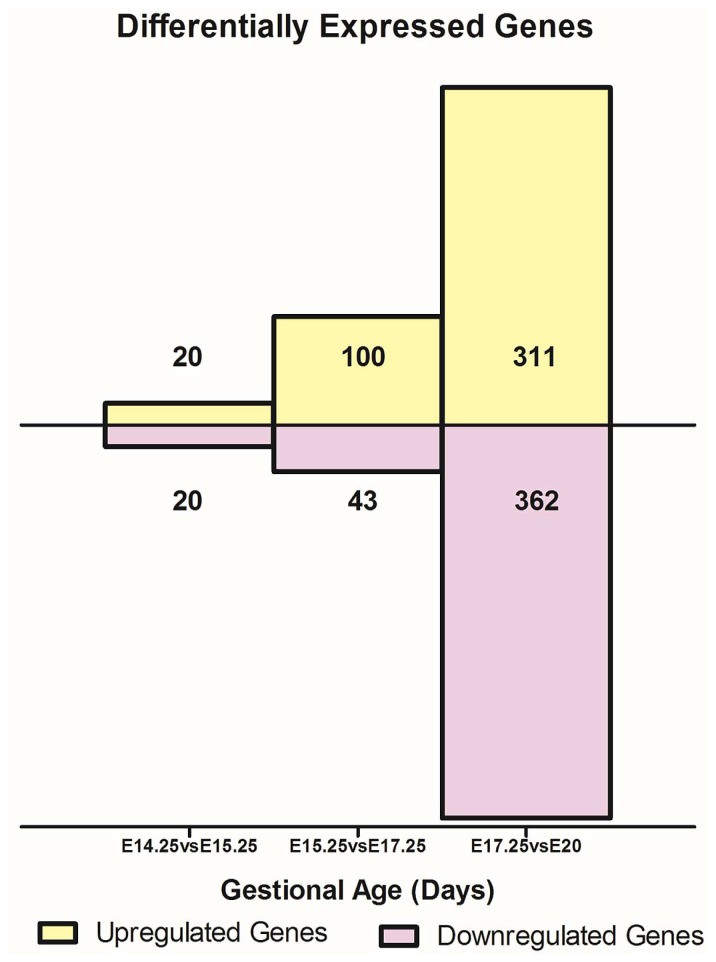
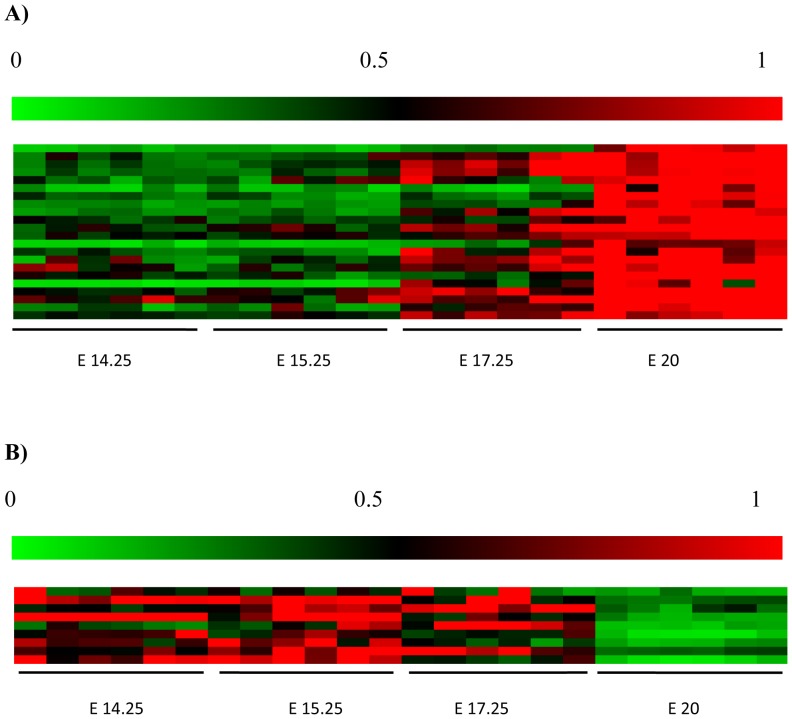
To characterize gestational age dependent changes in gene expression, microarray analysis was performed on placental cDNA obtained from pregnant rats at gestational days E14.25, E15.25, E17.25 and E20. A cutoff of >2 fold change in expression and false discovery rate of less than 1% was used to ascribe the differential expression of genes. Between gestational age E14.25 and E15.25, 40 gene transcripts were differentially expressed, between E15.25 vs E17.25, 143 gene transcripts were differentially expressed and between E17.25 vs E20, 678 were differentially expressed using SAM Analyses (Figure 1). Specific differentially expressed angiogenic genes were then selected from the SAM readout and a heat map (Figure 2) displays expression changes over time.
Figure 1. Differentially genes expression during gestation.
A) The total number of differentially expressed genes that were up and down regulated between the 3 gestational groups i.e. E14.25 vs. E15.25, E15.25 vs. E17.25 and E17.25 vs. E20 are displayed as fold change (cut-off >2 and FDR<1%). B) Overall gene expression fold changes from the earliest time point under study E14.25 right up to E20.
Figure 2. Heat Map of relative expression of Angiogenic genes.
Of the 31 differentially expressed genes, 22 were up regulated (A) and 9 were down regulated (B). The heat map diplays data for 6 individual placentae for each gestational age. The genes correspond to the gene list in Table 1, where a change from green, black to red indicates and increase in signal. The colour legend above each map shows relative microarray signal ranges between 0 to 1. Each coloured rectangular box within the heat map represents a separate rat placenta sample. The samples are grouped according to gestational age i.e. E14.25, E15.25, E17.25, E 20.
Quantitative RT-PCR Validation of Gestational-age Dependent Genes
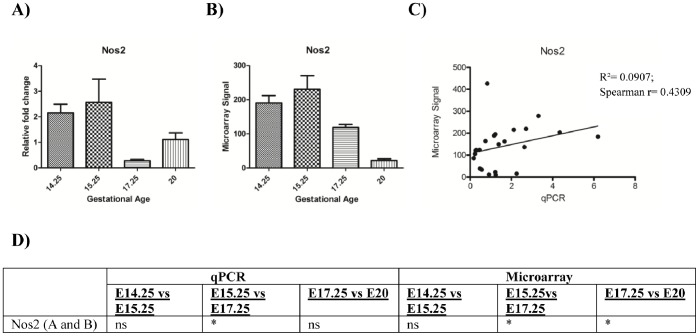
qPCR data were consistent with microarray data and established that both Ptgs2 and Pla2g2a remain relatively constant throughout E14.25, E15.25 and E17.25 and increase dramatically (and also consistent with literature) towards the end of gestation at E20 (Figure 3). Nos2 on the other hand, is downregulated towards E17.25 and E20 (Figure 4 and Figure S2).
Figure 3. Comparison of qPCR and microarray expression data.
qPCR and Microarray data for Ptgs2 are shown in panels A and B respectively; statistical significance (p values) is presented in panel G. C represents linear correlation between qPCR and microarray data for Ptgs2 (p<0.05). qPCR and microarray results for Pla2g2a are shown in panels D and E respectively; statistical significance (p values) are presented in panel G. Panel F indicates the linear correlation (Spearman’s rank correlation) between qPCR and microarray data (p<0.05). Both genes display late gestational increase. Where * = p<0.05, p<0.0005 = **; p<0.0001 = *** and ns = p>0.05 using one-way ANOVA and post-hoc tests (Bonferroni test).
Figure 4. Comparison of qPCR and microarray data for Nos2.
qPCR and microarray results of Nos2 are presented in panels A and B, respectively. Statistical significance (p values) is presented in panel D. Panel C depicts linear correlation (Spearman’s rank correlation) between qPCR and microarray data (p<0.05) Both qPCR and microarray data display a significant decrease in expression with gestational age. (* = p<0.05, p<0.0005 = **; p<0.0001 = *** and ns = p>0.05).
Classification of Genes Involved in Blood Vessel Morphogenesis and Angiogenesis
Genes were classsified into pathways using GO (gene ontology) based upon their Entrez IDs. Within the Blood Vessel Morphogenesis and Angiogenesis pathway a total of 31 differentially expressed genes were identified (GO:0048514 : blood vessel morphogenesis [412 gene products] and GO:0001525 : angiogenesis [333 gene products]). 22 of these genes displayed increased expression and 9 genes decreased expression across the gestational age groups. Between E14.25 and E15.25 none of the 31 genes were differentially expressed, between E15.25 and E17.25, 15 genes were differentially expressed and between E17.25 and E20, 26 genes were differentially expressed (Table 1, Figure 2, Figure S1 and S2). Figure S4 displays the mean centroid analyses of A) upregulated and B) downregulated genes.
Table 1. List of Angiogenic genes identified and pairwise comparison of gene expression between gestational age groups.
| Gene Identifiers | E14.25 vs E15.25 | E15.25 vs E17.25 | E17.25 vs E20 | E14.25 vs E20 | |||||
| Gene name | Entrez ID | Fold change | p value | Fold change | p value | Fold change | p value | Fold change | p value |
| Up- regulated 22 genes | |||||||||
| Bmp4 | 25296 | – | ns | – | ns | 3.8 | *** | 6.77 | *** |
| Vegfa | 83785 | – | ns | <2 | * | – | ns | 3.04 | *** |
| Mmp14 | 81707 | – | ns | <2 | *** | – | ns | 3.44 | *** |
| Acvrl1 | 25237 | – | ns | 2.8 | *** | – | ns | 3.42 | *** |
| Edn1 | 24323 | – | ns | – | ns | <2 | *** | 3.61 | *** |
| IL18 | 29197 | – | ns | – | ns | 6.4 | *** | 6.74 | *** |
| Gpx1 | 24404 | – | ns | – | ns | 3.7 | *** | 3.69 | *** |
| Id1 | 25261 | – | ns | – | ns | <2 | *** | 4.27 | *** |
| Cd36 | 29184 | – | ns | 2.8 | *** | <2 | *** | 4.87 | *** |
| Hs6st1 | 316325 | – | ns | – | ns | <2 | *** | <2 | *** |
| Emcn | 295490 | – | ns | <2 | ** | <2 | *** | 2.94 | *** |
| Rhob | 64373 | – | ns | – | ns | <2 | *** | 2.31 | *** |
| Prl7d1 | 84377 | – | ns | – | ns | <2 | *** | 9.29 | *** |
| Angptl4 | 362850 | – | ns | 3.1 | *** | 3.7 | *** | 6.93 | *** |
| Sox18 | 311723 | – | ns | <2 | ** | – | ns | <2 | *** |
| Klf5 | 84410 | – | ns | <2 | * | <2 | ** | <2 | *** |
| Pf4 | 360918 | – | ns | – | ns | 3.0 | *** | 2.38 | *** |
| Cav1 | 25404 | – | ns | 6.7 | *** | <2 | *** | 15.46 | *** |
| Sgpl1 | 286896 | – | ns | – | ns | <2 | *** | 2.64 | *** |
| Notch4 | 406162 | – | ns | – | ns | <2 | *** | <2 | *** |
| Tek | 89804 | – | ns | <2 | * | 2.2 | *** | 4.72 | *** |
| Tgm2 | 56083 | – | ns | <2 | ** | <2 | *** | 2.34 | *** |
| Down-regulated 9 genes | |||||||||
| Itgb2 | 309684 | – | ns | – | ns | >0.5 | ** | >0.5 | *** |
| ItgaV | 296456 | – | ns | – | ns | >0.5 | * | 0.41 | *** |
| Itga4 | 311144 | – | ns | >0.5 | * | 0.4 | *** | ns | *** |
| Foxm1 | 58921 | – | ns | >0.5 | ** | 0.4 | *** | 0.20 | *** |
| Anpep | 81641 | – | ns | – | ns | 0.2 | *** | 0.2 | *** |
| Gbx2 | 114500 | – | ns | – | ns | 0.2 | *** | 0.13 | *** |
| Atg5 | 365601 | – | ns | >0.5 | ** | – | ns | >0.5 | *** |
| Cited2 | 114490 | – | ns | – | ns | 0.5 | *** | 0.5 | *** |
| Nos2 | 24599 | – | ns | >0.5 | * | 0.2 | *** | 0.16 | *** |
E14.25 vs E15.25 - no significant differences in gene expression. E15.25 vs E17.25–15 of 31 genes were differentially expressed. E17.25 vs E20–26 of 31 genes were differentially expressed. E14.25 vs E20 shows gene expression changes for each gene from E14.25 vs E20. Significance was ascribed where gene expression was changed by <2 (* = p<0.05, p<0.0005 = **; p<0.0001 = *** and ns = p>0.05). The order of the genes listed in the table corresponds to the Heat Map on Figure 2.
Quantitative RT-PCR of Angiogenic Genes
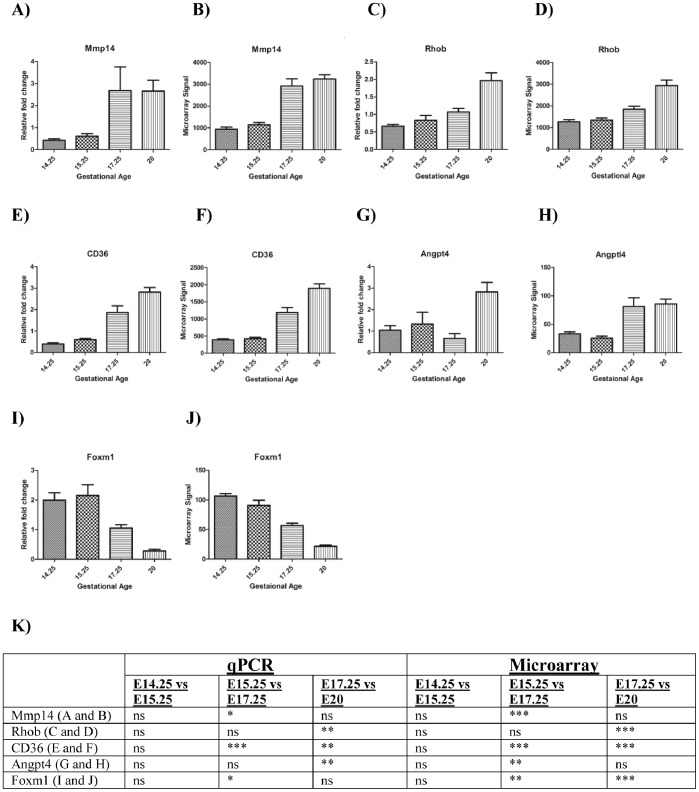
The expression of five angiogenic genes (Mmp14, CD36, Angpt4, Rhob and Foxm1) was confirmed by qPCR (Figure 5). For all genes examined, a significant correlation between microarray and qPCR data was established (Spearman’s Rank Correlation, p<0.05).
Figure 5. qPCR validation of 5 Angiogenic genes.
For Mmp14, qPCR are microarray data are presented in panels A and B, respectively; for Rhob in panels C and D; for CD36 in panels E and F; for Angpt4 in panels G and H; for Foxm1 in panels I and J. Statistical significance (p values) is presented in panel K. Where p<0.05 = *; p<0.0005 = **; p<0.0001 = *** as assessed by one way ANOVA and post-hoc tests (Bonferroni test). The x-axes in all graphs represent the 4 gestational age groups. Y-axes for qPCR graphs denote relative fold change normalised to β-actin. The y-axes for the microarray bar charts denote microarray hybridisation signals.
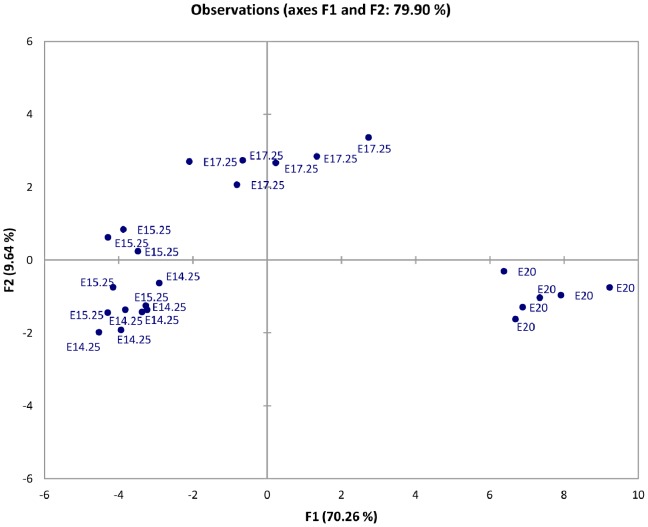
Principal Component Analysis
PCA of placental angiogenic gene expression separated data into three distinct groups. E14.25 and E15.25 samples clustered together, while E17.25 and E20 samples partitioned independently (with F1 accounting for up to 70% of sample variation, Figure 6). These data are consistent with the hypothesis that the expression profile of Blood Vessel Morphogenesis and Angiogenesis genes in the rat placenta changes significantly after 70% of gestation is reached and continues to change thoughout the remainder of gestation. Gestational age based PCA analyses are presented in Figure S3. Furthermore, PCA indicates that the biological variation within the gestational age category for these genes is small (n = 6). This conclusion was additionally supported by ANOVA where the variance in microarray data was partitioned between, gene (p<0.00001), gestational age (p<0.00001) and biological replicates (p>0.9).
Figure 6. Principal Component Analysis (PCA).
Principal components F1 vs. F2 for individual gestational age groups (n = 6, E14.25, E15.25, E17.25 and E20) for all 31 angiogenic genes are plotted to identify variation in gene expression. Day E14.25 and E15.25 samples cluster together, indicating a similar genes expression profile between these 2 groups. Samples within the E17.25 and E20 groups, however, display independent clustering, indicative of significantly altered gene expression profiles. Biological replicates within each gestational age group are tightly clustered.
Discussion
The aim of this study was to identify mid-late gestation changes in the expression of genes in the whole rat placenta using microarray analysis. In particular, this study focused on changes in genes involved in angiogenic pathways. During mid-late gestation, fetal demand for energy substrates and gas exchange increases dramatically in the absence of a concomitant increase in placental mass. An increase in branching angiogenesis contributes to placental accommodation during this later gestational period. The data obtained establish that the expression of approximately 7% of genes profiled (22,000) within the placenta change significantly between E14.25 and E20 days of gestation. Of this 7%, 42% of genes increased expression and 58% decreased expression. A subset of 31 genes known to be involved in the Blood Vessel Morphogenesis pathways were identified, of which ∼70% displayed increased expression with gestational age. RNA sequencing of different rat placental regions at E18.5 showed that the blood vessel development pathway was prominent in genes that were differentially expressed in the labyrinth zone versus the junctional zone and the metrial gland [12]. In the metrial gland, the vascular development pathway was enriched [12]. These results indicate that the vasculature in the rat placenta is developing late into gestation.
The microarray expression data for eight genes were orthogonally validated by qPCR. These genes included three genes previously known to display gestational age and labour-associated changes and five genes identified in this study that displayed altered expression during gestation. Previously, we and others, have identified late gestation and pre-labour changes in the expression of enzymes involved in the biosynthesis of eicosanoids (i.e. prostaglandin synthase 2, Ptgs2 ([15]–[18] and phospholipase A2 (Pla2a) [19]) and in nitric oxide (i.e. inducible nitric oxide synthase, Nos2 [20]). Consistent with these observations, the gene expression of prostaglandin synthase 2 and secretory Pla2a increased (from E17.25 to E20) and Nos2 decreased (from E15.25 to E20) during gestation (as determined by both microarray analysis and real-time PCR). This finding is consistent with current literature. Furthermore, the data for Ptgs2 are consistent with the literature where expression is known to increase prior to the onset of labour [10], [12], [13]). Phospholipases hydrolyze phospholipids into fatty acids. Phospholipase A2 is an important mediator of arachidonic acid formation, which are the substrates of eicosanoids such as prostaglandins. It has been reported that there is a relative abundance of phospholipase protein in late pregnancy [19].
Of the 31 Blood Vessel Morphogenesis pathway genes that displayed differental expression during gestation in this study, the expression of Cd36, Mmp14, Angpt4, Rhob and Foxm1 was confirmed by qPCR on same samples. In all cases, concordance between microarray and qPCR data was confirmed. These five genes were chosen on the basis of their function and the fold changes shown in Table 1.
CD36 is a, cell membrane scavenger receptor involved in angiogenesis, inflammation and lipid metabolism [21]. In microvascular endothelial cells it functions as a receptor for Thrombospondin-1 (TSP-1) [22]. CD36 has been implicated in inflammatory processes via the activation of phospholipases and the formation of prostaglandins [23] and the cellular uptake of fatty acids via the activation of peroxisome proliferator–activated receptor γ [24]. In this study, Cd36 expression increased by 2.8 fold during E15.25 to E17.25 days of gestation and remains significantly elevated till labour onset. Interestingly, increased expression of Cd36 mRNA and protein is observed in cord blood from late pregnancy compared with early pregnancy samples.
Mmp14 encodes for MMP14 (EC 3.4.24.80, also known as MT1-MMP) which is a key extracellular matrix-remodeling enzyme that promotes collagen remodeling. MMP14 is a dominant cell-surface protease required for endothelial cell tube morphogenesis, invasion and for the creation of vascular guidance tube [23]. However, there is only limited information about the specific role of MMP14 role in placental angiogenesis. In mouse placentae, MMP14 is strongly expressed in the labyrinth region [25]. This region of the placenta is critical for the formation of syncytiotrophoblast and the subsequent formation of fetal vessels. In human placentae, immunoreactive MMP14 is expressed in term and preterm syncytiotrophoblast cells, where it may contribute to the cleavage and release of endoglin [26] and in the endothelium of feto-placental vessels. Endoglin has been suggested to be a major factor in the development of preeclampsia [27]. Placental expression of MMP14 is greater in pregnancies complicated by gestational diabetes mellitus [28] and, in vitro it is induced by insulin and IGF-II in vitro [28] and HIF-1α [24]. In this study, Mmp14 mRNA expression increased from E15.25 to E17.25 (the peak of trophoblast invasion takes place at this stage) [29]. Another gene that is involved in prostaglandin regulation is Sgpl1 (SGPL1, EC 4.1.2.27) which catalizes the irreversible degradation of the sphingosine-1-phosphate (S1P) [30]. S1P is a bioactive lipid mediator that promotes cell proliferation, survival, migration, adherence, inflammation and angiogenesis [31]. SGPL1, thus, regulates the available pool of S1P and inhibits its signalling activities. SGPL1 may regulate S1P-induced Ptgs2 expression in the rat myometrium [32]. In this study, Sgpl1 expression increased 2.6 fold during gestation from E14.25 to E20.
Angiopoeitin like Protein 4 (EC 2.6.1.2) expression was low at E14.25 and E15.25 and increases at E17.25 remaining constant till E20. Over the gestational period assessed in this study, Angpt4 gene expression increased ∼7-fold. Angpt4, like the other angiopoietins 1 and 2 has been reported to play a role in angiogenesis [33]. Yamakawa et al., reported that ANGPT4 increased endothelial cell migration and tube formation in vitro and reduced vascular leakage [34]. Angiogenic effects of ANGPT4 have also been observed in glioblastoma [35]. To date little is known about the biological functions of ANGPT4 in the placenta. However, ANGPT1 has been shown to potentiate VEGF activity and work together to increase the luminal diameter of blood vessels in sheep placenta [36]. In this study, the receptor for ANGPT1, ANGPT2 [36] and ANGPT4, TEK (Tek) showed an increase in placental expression during from E15.25 to E17.25 and further from E17.25 to E20 (fold change 2.2) of gestation. TEK has been implicated in the regulation of angiogenesis and cell proliferation, migration and survival [36]. Tek expression was significantly, positively correlated (p<0.05) with Angpt4 expression during gestation, (upregulated prior to labour) indicating the possibility that they are working together to promote late gestational angiogenesis. TIE 1 is the other receptor for ANGPT4 and in our Microarray study, interestingly, Tie1 expression does not change from E14.25 right up to E20 (Data not shown).
Ras homolog family member B, Rhob has not been extensively studied in the placenta. The data obtained in this study (both microarray and qPCR) establish that Rhob expression gradually increases during gestation (from E14.25 to E20), by a fold change of 2.3. Recently, RHOB (EC 3.6.1.47) was shown to regulate endothelial cell migration, sprouting, and capillary morphogenesis [37], although the mechanism by which RHOB regulates angiogenesis is not well understood. Howe and Addison (2012) concluded that RHOB plays a significant role in VEGF-induced endothelial cell morphogenesis, in part, by negatively modulating the activity of RHOA, [37]. Both RHOA (Ras homolog family member A) and RHOB are essential downstream effectors of VEGF signalling in the angiogenic process. siRNA-inhibition of RhoB results in increased RHOA activation in response to VEGF (vascular edothelial growth factor) stimulation [37]. Vegf was also seen to upregulate the expression of RhoB [26]. In our study, we see that both RhoB and Vegf are up regulated towards late gestation, first Vegf expression increases at E15.25 and then Rhob expression increases downstream at E17.25, indicating a positive correlation. Placental expression of Vegfa was increased 3-fold between E14.25 and E20 in the present study. VEGF is known to be an important regulator of angiogenesis in placenta [38], [39] and its increased expression during late gestation has been previously reported [5], while its receptor Flk (i.e VegfR2) is downregulated from E14.25 vs E20, prior to labour (Microarray data not shown).NOTCH-4 is a modulator of angiogenesis in human placenta and regulates placental cell fate [40]–[42]. Similar to RHOB expression, Notch signalling acts downstream of VEGF. Notch signalling helps to regulate endothelial cell morphogenesis via activation of MMPs [43]. In this study, Notch-4 expression doubled between E15.25 and E20, where its expression is elevated prior to term/labour.
The transcriptional regulator Foxheadbox M1 (Foxm1) was expressed during late midgestation (E14.25–E15.25, as assessed by both microarray and qPCR) but decreased 5-fold by E20. Little, however, is known about FOXM1 function in angiogenesis in the placenta. Interestingly, the onset of labour in humans is associated with decreased placental expression of Foxheadbox O4, (both FoxO4 mRNA and protein) where it may function as a negative regulator of Ptgs2 expression and prostaglandin biosynthesis [44]. In cancer development and progression, FOXM1 has been implicated in regulating the expression of the human endothelial cell caveolae-marker CAV-1 and Foxm1 expression has also been linked to growth of glioma cells in tumour angiogenesis [45], [46]. Placental Cav-1 mRNA expression increased during gestation in this study. Caveolin has been reported to facilitate VEGF/NO-mediated angiogenesis [47]. Our data show, Cav-1 expression increased 6.7 fold between E15.25 and E17.25 and further increased till E20. CAV-1 is known to interact with FGFR1which modulates FGF2 related placental angiogenesis [48]. Unlike in cancer progression we show that an increase in Cav-1 and Vegf expression is negatively correlated with Foxm1 expression. An increase in FOXM1 could have a negative effect on CAV-1 expression or its expression is perhaps acutely switched off prior to labour onset. Figure S5 shows CAV-1 protein localised intensively around placental blood vessels indicating its important role in Angiogenesis.
Angiogenic genes of significance that were upregulated (based on microarray data alone) during gestation included: bone morphogenetic protein 4 (Bmp4); transglutaminase (Tgm EC 2.3.2.13); sphingosine-1 phosphate lyase (Sgpl1, EC 4.1.2.27); endomucin (Emcn); Interleukin-18 (IL-18) and glutathione peroxidase 1 (Gpx1, EC 1.11.1.9).
Microarray analysis also identifed a suite of Blood Vessel Morphogenesis pathway genes that were downregulated during late gestation, prior to labor onset. These included, Itga4, Itgb2, Anpep and Cited2. The integrin subunits alpha 4 (Itga4) and beta 2 (Itgb2) have been previously identified in placentae [26], [28] [49]. ITGA4 forms a heterodimer with integrin beta 1 to function as a receptor for collagen. Increased expression of ITGA4 has been implicated in inhibition of endothelial cell migation during angiogeneis [49]. In this study, Itga4 gene expression decreases during gestation (from E15.25 right up to E20) and Itgb2 expression decreases acutely prior to labour onset. Cited2 gene expression also decreases during late gestation from E17.25 to E20 by two fold. In the mouse placenta, lack of CITED2 is characterized by disorganization of the placental fetal vasculature and a fewer trophoblast giant cells, spongiotrophoblasts and glycogen cells [50]. Hence Cited2 is important for normal placental vascularisation.
Additionaly, during late gestation in rat, glucose transporter genes Slc2A3 and Slc2A1 are upregulated and downregulated respectively, in placenta [6]. This is consistent with our microarray data where Slc2A3 expression increases from E15.25 to E17.25 and further increases at E20 and Slc2A1 is increased from E15.25 to E17.25 and then gets downregulated at E20 (data not shown). Amino acid transport genes such as Slc38A1, Slc38A2 and Slc38A4 have been shown to increase with gestation, coinciding with increased fetal nutrient demands [7], which is also consistent with our microarray data. We can relate the Angiogenic expression changes in this study to the expression of several transporter genes. In conclusion, the data obtained in this study extends our understanding of placental genes that may contribute to placental vascular accommodation during mid-late gestation and late-gestation, in particular, those involved in regulating placental angiogenesis. Several of these gene products work together either directly or indirectly. However, we are not able to ascertain that the pre labour associated changes seen in these 32 angiogenic genes, has any link with the actual labour process, even though some of the changes are acute. Further studies need to be carried out to prove a link between some or all of these genes with the onset or trigger of labour. The study identified gestational age-dependent changes in placental gene expression within the Blood Vessel Morphogenesis pathways that have not previously been characterized in rat placenta. Furthermore, it confirms and extends data for genes previously reported to display gestational and potential labor-associated changes. We also observed that, most expression changes for these genes, occurred between E17.25 and E20, prior to labour onset. One caveat that must be considered in interpreting the results obtained in this study is that mRNA was extracted from whole placental tissue. Therefore, it is not possible to attribute observed changes in gene expression to specific cell types that comprise the rat placenta. Nevertheless, the data obtained identify lead candidate genes for subsequent cell specific analyses (e.g. based cell selection using laser capture microscopy) and provide opportunity to further elucidate functional pathways that may be of significance in mid-late placental function in both normal and pathological pregnancies.
Supporting Information
Gestational Variation in Angiogenic gene expression of 22 genes that display a late gestational increase. The x-axis indicates the gestational ages whereas the y-axis indicates the signal obtained from the microarray hybridisation using the Illumina Bead reader. These 22 genes had SAM p values <0.0001.
(TIF)
Gestational Variation in Angiogenic gene expression of 9 genes that display a late gestational decrease. The x-axis indicates the gestational ages in days whereas the y-axis indicates the signal obtained from the microarray hybridisation using the Illumina Bead reader. SAM p values were <0.0001.
(TIF)
Two Dimensional PCA Observational plots for Gestational Age Comparison Groups: A) E14.25 vs. E15.25. PCA plot F1 vs. F2. B) E15.25 vs. E17.25. PCA plot F1 vs. F2. C) E17.25 vs E20. PCA plot F1 vs. F2. Data were analyzed using Pearson Correlation Matrix (XLSTAT, p<0.05).
(TIF)
Mean Centroid of Angiogenesis. A) for upregulated genes B) for down regulated genes using Kruskal-Wallist test of Mean Centroid. A)E20 vs E14.25 = ***, E20 vs E15.25 = *** and E20 vs E17.25 = ns and B)E20 vs E14.25 = *, E20 vs E15.25 = *** and E20 vs E17.25 = ns. (* = p<0.05, p<0.0005 = **; p<0.0001 = *** and ns = p>0.05).
(TIF)
Immunohistochemical Localization of CAV-1 and BMP4. Intense staining for CAV-1 around placental blood vessels (A and B), BMP4 (C and D) in endothelial cells lining blood vessels and Rabbit IgG Isotype Negative Control (E). Haemotoxylin staining of nuclei shown in blue.
(TIF)
List of Primer oligonucleotide sequences used for qPCR experiments.
(TIF)
Acknowledgments
The assistance of Mr. Nick Matigan, Eskitis Institute, Griffith University, Australia with the Microarray hybridisation experiments and Mr. Nileshkumar R. Patel for figure formatting is greatly appreciated.
Funding Statement
GER was in receipt of an NHMRC Principal Research Fellowship. MDN received support from the Royal Brisbane and Women’s Hospital Foundation. JAA was a Monash Fellow and Heart Foundation Fellow. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Belkacemi L, Nelson DM, Desai M, Ross MG (2010) Maternal undernutrition influences placental-fetal development. Biology of reproduction 83: 325–331. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey KM (2002) The role of the placenta in fetal programming-a review. Placenta 23 Suppl A: S20–27. [DOI] [PubMed]
- 3. Godfrey KM, Barker DJ (2001) Fetal programming and adult health. Public health nutrition 4: 611–624. [DOI] [PubMed] [Google Scholar]
- 4. Reynolds LP, Redmer DA (2001) Angiogenesis in the placenta. Biology of reproduction 64: 1033–1040. [DOI] [PubMed] [Google Scholar]
- 5. Reynolds LP, Borowicz PP, Vonnahme KA, Johnson ML, Grazul-Bilska AT, et al. (2005) Animal models of placental angiogenesis. Placenta 26: 689–708. [DOI] [PubMed] [Google Scholar]
- 6. Constancia M, Angiolini E, Sandovici I, Smith P, Smith R, et al. (2005) Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proceedings of the National Academy of Sciences of the United States of America 102: 19219–19224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuruvilla AG, D’Souza SW, Glazier JD, Mahendran D, Maresh MJ, et al. (1994) Altered activity of the system A amino acid transporter in microvillous membrane vesicles from placentas of macrosomic babies born to diabetic women. The Journal of clinical investigation 94: 689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aplin J (2000) Maternal influences on placental development. Seminars in Cell & Developmental Biology 11: 115–125. [DOI] [PubMed] [Google Scholar]
- 9. Xie H, Zou L, Zhu J, Yang Y (2011) Effects of netrin-1 and netrin-1 knockdown on human umbilical vein endothelial cells and angiogenesis of rat placenta. Placenta 32: 546–553. [DOI] [PubMed] [Google Scholar]
- 10. Mizutani T, Yoshino M, Satake T, Nakagawa M, Ishimura R, et al. (2004) Identification of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-inducible and -suppressive genes in the rat placenta: Induction of interferon-regulated genes with possible inhibitory roles for angiogenesis in the placenta. Endocrine Journal 51: 569–577. [DOI] [PubMed] [Google Scholar]
- 11. Buffat C, Mondon F, Rigourd V, Boubred F, Bessieres B, et al. (2007) A hierarchical analysis of transcriptome alterations in intrauterine growth restriction (IUGR) reveals common pathophysiological pathways in mammals. The Journal of pathology 213: 337–346. [DOI] [PubMed] [Google Scholar]
- 12. Shankar K, Zhong Y, Kang P, Blackburn ML, Soares MJ, et al. (2012) RNA-seq analysis of the functional compartments within the rat placentation site. Endocrinology 153: 1999–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goyal R, Yellon SM, Longo LD, Mata-Greenwood E (2010) Placental gene expression in a rat ‘model’ of placental insufficiency. Placenta 31: 568–575. [DOI] [PubMed] [Google Scholar]
- 14. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 15. Rice GE, Payne MJ, Wong MH, Thorburn GD (1992) Immunoreactive prostaglandin G/H synthase content increases in ovine cotyledons during late gestation. Placenta 13: 429–437. [DOI] [PubMed] [Google Scholar]
- 16. Gaffney RC, Rice GE, Brennecke SP (1990) Is human labour triggered by an increase in the rate of synthesis of prostaglandin G/H synthase? Reprod Fertil Dev 2: 603–606. [DOI] [PubMed] [Google Scholar]
- 17. Rice GE, Wong MH, Hollingworth SA, Thorburn GD (1990) Prostaglandin G/H synthase activity in ovine cotyledons: a gestational profile. Eicosanoids 3: 231–236. [PubMed] [Google Scholar]
- 18. Bennett PR, Henderson DJ, Moore GE (1992) Changes in expression of the cyclooxygenase gene in human fetal membranes and placenta with labor. American Journal of Obstetrics and Gynecology 167: 212–216. [DOI] [PubMed] [Google Scholar]
- 19. Farina MG, Billi S, Leguizamon G, Weissmann C, Guadagnoli T, et al. (2007) Secretory and cytosolic phospholipase A2 activities and expression are regulated by oxytocin and estradiol during labor. Reproduction 134: 355–364. [DOI] [PubMed] [Google Scholar]
- 20. Suzuki T, Ikeda Y, Yoshikawa H, Tanaka K, Morita H, et al. (2009) Gestational Changes in Production of NO and Expression of NOS mRNA Isoforms in the Rat Placenta. Journal of Veterinary Medical Science 71: 495–498. [DOI] [PubMed] [Google Scholar]
- 21. Febbraio M, Hajjar DP, Silverstein RL (2001) CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. The Journal of clinical investigation 108: 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Silverstein RL, Febbraio M (2009) CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Science signaling 2: re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuda O, Jenkins CM, Skinner JR, Moon SH, Su X, et al. (2011) CD36 Protein Is Involved in Store-operated Calcium Flux, Phospholipase A2 Activation, and Production of Prostaglandin E2. Journal of Biological Chemistry 286: 17785–17795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tontonoz P, Nagy L, Alvarez JGA, Thomazy VA, Evans RM (1998) PPAR gamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 93: 241–252. [DOI] [PubMed] [Google Scholar]
- 25. Szabova L, Son MY, Shi J, Sramko M, Yamada SS, et al. (2010) Membrane-type MMPs are indispensable for placental labyrinth formation and development. Blood 116: 5752–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aplin JD (1993) Expression of Integrin Alpha-6-Beta-4 in Human Trophoblast and Its Loss from Extravillous Cells. Placenta 14: 203–215. [DOI] [PubMed] [Google Scholar]
- 27. Venkatesha S, Toporsian M, Lam C, Hanai J, Mammoto T, et al. (2006) Soluble endoglin contributes to the pathogenesis of preeclampsia. Nature medicine 12: 642–649. [DOI] [PubMed] [Google Scholar]
- 28. Pfarrer C, Hirsch P, Guillomot M, Leiser R (2003) Interaction of integrin receptors with extracellular matrix is involved in trophoblast giant cell migration in bovine placentomes. Placenta 24: 588–597. [DOI] [PubMed] [Google Scholar]
- 29. Dube E, Gravel A, Martin C, Desparois G, Moussa I, et al. (2012) Modulation of Fatty Acid transport and metabolism by maternal obesity in the human full-term placenta. Biology of reproduction 87: 14. [DOI] [PubMed] [Google Scholar]
- 30.Aguilar A, Saba JD (2011) Truth and consequences of sphingosine-1-phosphate lyase. Advances in enzyme regulation. [DOI] [PMC free article] [PubMed]
- 31. Santulli P, Marcellin L, Noel JC, Borghese B, Fayt I, et al. (2012) Sphingosine pathway deregulation in endometriotic tissues. Fertility and sterility 97: 904–911. [DOI] [PubMed] [Google Scholar]
- 32. Serrano-Sanchez M, Tanfin Z, Leiber D (2008) Signaling pathways involved in sphingosine kinase activation and sphingosine-1-phosphate release in rat myometrium in late pregnancy: role in the induction of cyclooxygenase 2. Endocrinology 149: 4669–4679. [DOI] [PubMed] [Google Scholar]
- 33. Lee HJ, Cho CH, Hwang SJ, Choi HH, Kim KT, et al. (2004) Biological characterization of angiopoietin-3 and angiopoietin-4. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 18: 1200–1208. [DOI] [PubMed] [Google Scholar]
- 34. Yamakawa M, Liu LX, Date T, Belanger AJ, Vincent KA, et al. (2003) Hypoxia-inducible factor-1 mediates activation of cultured vascular endothelial cells by inducing multiple angiogenic factors. Circulation research 93: 664–673. [DOI] [PubMed] [Google Scholar]
- 35. Brunckhorst MK, Wang H, Lu R, Yu Q (2010) Angiopoietin-4 promotes glioblastoma progression by enhancing tumor cell viability and angiogenesis. Cancer research 70: 7283–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Borowicz PP, Arnold DR, Johnson ML, Grazul-Bilska AT, Redmer DA, et al. (2007) Placental growth throughout the last two thirds of pregnancy in sheep: vascular development and angiogenic factor expression. Biology of reproduction 76: 259–267. [DOI] [PubMed] [Google Scholar]
- 37. Howe GA, Addison CL (2012) RhoB controls endothelial cell morphogenesis in part via negative regulation of RhoA. Vascular cell 4: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Torry DS, Torry RJ (1997) Angiogenesis and the expression of vascular endothelial growth factor in endometrium and placenta. American journal of reproductive immunology 37: 21–29. [DOI] [PubMed] [Google Scholar]
- 39. Charnock-Jones DS, Kaufmann P, Mayhew TM (2004) Aspects of human fetoplacental vasculogenesis and angiogenesis. I. Molecular regulation. Placenta 25: 103–113. [DOI] [PubMed] [Google Scholar]
- 40. Cobellis L, Mastrogiacomo A, Federico E, Schettino MT, De FM, et al. (2007) Distribution of Notch protein members in normal and preeclampsia-complicated placentas. Cell and tissue research 330: 527–534. [DOI] [PubMed] [Google Scholar]
- 41. De FM, Cobellis L, Giraldi D, Mastrogiacomo A, Perna A, et al. (2007) Expression and distribution of notch protein members in human placenta throughout pregnancy. Placenta 28: 118–126. [DOI] [PubMed] [Google Scholar]
- 42. Zhao WX, Lin JH (2012) Notch signaling pathway and human placenta. International journal of medical sciences 9: 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Funahashi Y, Shawber CJ, Sharma A, Kanamaru E, Choi YK, et al. (2011) Notch modulates VEGF action in endothelial cells by inducing Matrix Metalloprotease activity. Vascular cell 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lim R, Riley C, Barker G, Rice GE, Lappas M (2012) Human labour is associated with decreased cytoplasmic FoxO4. Placenta 33: 52–59. [DOI] [PubMed] [Google Scholar]
- 45. Huang C, Qiu Z, Wang L, Peng Z, Jia Z, et al. (2012) A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer research 72: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, et al. (2008) FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer research 68: 8733–8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sonveaux P, Martinive P, DeWever J, Batova Z, Daneau G, et al. (2004) Caveolin-1 expression is critical for vascular endothelial growth factor-induced ischemic hindlimb collateralization and nitric oxide-mediated angiogenesis. Circulation research 95: 154–161. [DOI] [PubMed] [Google Scholar]
- 48. Feng L, Liao WX, Luo Q, Zhang HH, Wang W, et al. (2012) Caveolin-1 orchestrates fibroblast growth factor 2 signaling control of angiogenesis in placental artery endothelial cell caveolae. Journal of cellular physiology 227: 2480–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weinlander K, Naschberger E, Lehmann MH, Tripal P, Paster W, et al. (2008) Guanylate binding protein-1 inhibits spreading and migration of endothelial cells through induction of integrin alpha4 expression. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 22: 4168–4178. [DOI] [PubMed] [Google Scholar]
- 50. Withington SL, Scott AN, Saunders DN, Lopes FK, Preis JI, et al. (2006) Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Developmental biology 294: 67–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gestational Variation in Angiogenic gene expression of 22 genes that display a late gestational increase. The x-axis indicates the gestational ages whereas the y-axis indicates the signal obtained from the microarray hybridisation using the Illumina Bead reader. These 22 genes had SAM p values <0.0001.
(TIF)
Gestational Variation in Angiogenic gene expression of 9 genes that display a late gestational decrease. The x-axis indicates the gestational ages in days whereas the y-axis indicates the signal obtained from the microarray hybridisation using the Illumina Bead reader. SAM p values were <0.0001.
(TIF)
Two Dimensional PCA Observational plots for Gestational Age Comparison Groups: A) E14.25 vs. E15.25. PCA plot F1 vs. F2. B) E15.25 vs. E17.25. PCA plot F1 vs. F2. C) E17.25 vs E20. PCA plot F1 vs. F2. Data were analyzed using Pearson Correlation Matrix (XLSTAT, p<0.05).
(TIF)
Mean Centroid of Angiogenesis. A) for upregulated genes B) for down regulated genes using Kruskal-Wallist test of Mean Centroid. A)E20 vs E14.25 = ***, E20 vs E15.25 = *** and E20 vs E17.25 = ns and B)E20 vs E14.25 = *, E20 vs E15.25 = *** and E20 vs E17.25 = ns. (* = p<0.05, p<0.0005 = **; p<0.0001 = *** and ns = p>0.05).
(TIF)
Immunohistochemical Localization of CAV-1 and BMP4. Intense staining for CAV-1 around placental blood vessels (A and B), BMP4 (C and D) in endothelial cells lining blood vessels and Rabbit IgG Isotype Negative Control (E). Haemotoxylin staining of nuclei shown in blue.
(TIF)
List of Primer oligonucleotide sequences used for qPCR experiments.
(TIF)