Abstract
The envelope glycoproteins of the human immunodeficiency virus and the related simian immunodeficiency virus (SIV) mediate viral entry into host cells by fusing viral and target cell membranes. We have reported expression, purification, and characterization of gp140 (also called gp160e), the soluble, trimeric ectodomain of the SIV envelope glycoprotein, gp160 (B. Chen et al., J. Biol. Chem. 275:34946-34953, 2000). We have now expressed and purified chimeric proteins of SIV gp140 and its variants with the catalytic subunit (C) of Escherichia coli aspartate transcarbamoylase (ATCase). The fusion proteins (SIV gp140-ATC) bind viral receptor CD4 and a number of monoclonal antibodies specific for SIV gp140. The chimeric molecule also has ATCase activity, which requires trimerization of the ATCase C chains. Thus, the fusion protein is trimeric. When ATCase regulatory subunit dimers (R2) are added, the fusion protein assembles into dimers of trimers as expected from the structure of C6R6 ATCase. Negative-stain electron microscopy reveals spikey features of both SIV gp140 and SIV gp140-ATC. The production of the fusion proteins may enhance the possibilities for structure determination of the envelope glycoprotein either by electron cryomicroscopy or X-ray crystallography.
The human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) envelope glycoproteins mediate viral attachment and entry. They are also the principal targets of neutralizing antibodies. The single-chain precursor, gp160, forms a trimer. It is cleaved by a furin-like protease in a late compartment of the export pathway into fragments known as gp120 and gp41 (1, 35). The fragments remain noncovalently associated in a trimeric assembly, even after cleavage, but the interfragment contact is relatively weak, and gp120 dissociates slowly (sheds) from mature virions (22, 23). We refer to the mature, cleaved Env protein (both from HIV and from SIV) as gp120/gp41. We further use the designation gp140 for the part of the gp160 polypeptide chain that lies outside the viral membrane (the ectodomain); after cleavage at the furin sites, gp140 would contain gp120 and the ectodomain of gp41 (Fig. 1).
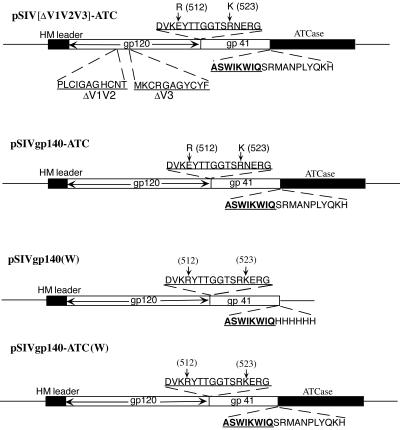
FIG. 1.
Constructs for expression in insect cells of chimeric proteins of SIV gp140 and its variants with the catalytic subunit (C) of E. coli ATCase. Schematic representations are shown for the expression constructs pSIVgp140-ATC, pSIVgp140 ATC(W), pSIV[ΔV1V2V3]-ATC, and pSIVgp140(W). The posttranslational cleavage sites (residues 512 and 523) between gp120 and gp41 were mutated as shown in pSIVgp140-ATC and pSIV[ΔV1V2V3]-ATC, while pSIVgp140(W) and pSIVgp140 ATC(W) contain wild-type cleavage sites. All constructs use the honeybee melittin (HM) secretion signal. The sequences from which the V1, V2, and V3 segments have been deleted and replaced with a GAG linker (in italics) are shown for pSIV[ΔV1V2V3]-ATC. The sequences near cleavage sites are shown in bold. The sequences of the junction between SIVgp140 and the ATCase C chain are also shown, with gp140 sequences in bold and ATCase sequences in regular font.
Interaction of HIV gp120/gp41 with the viral receptor, CD4, induces a conformational change, detected by altered antigenic properties, enhanced proteolytic sensitivity of the gp120 moiety, and enhanced spontaneous dissociation (2, 27, 29). The change increases affinity of gp120/gp41 for the chemokine-binding coreceptor (CXCR4 or CCR5), perhaps by exposing or stabilizing the site of coreceptor binding (34, 40). Interaction with the coreceptor leads to membrane fusion, probably because the coreceptor induces yet another conformational rearrangement in which gp120 dissociates, the fusion protein is exposed, and gp41 refolds into a helical hairpin (5, 36).
The structure of a truncated and enzymatically deglycosylated form of HIV gp120, complexed with a D1-D2 fragment of CD4 and a monoclonal Fab, has been determined by X-ray crystallography (18). The Fab derives from monoclonal antibody (MAb) 17b, which binds the CD4-induced conformation of gp120 (40). It contacts many of the residues important for coreceptor interaction. The monomeric gp120 fragment lacks 52 N-terminal residues, 19 C-terminal residues, and the V1, V2, and V3 loops. It is, in effect, the receptor-binding core of the Env subunit. The structure of the gp41 ectodomain in what is probably its postfusion state has also been determined (5, 20, 36, 44).
The receptor- and coreceptor-induced transition and the fusion rearrangement imply a succession of conformational states. Some of the transitions may involve only modest changes, while others are clearly very-large-scale rearrangements. The crystal structure of the gp120 core probably corresponds to the liganded receptor-binding fragment as it is released from gp41, that of the gp41 ectodomain, to the postfusion conformation. How the known structure of gp120 would fit onto the “head” of a trimer has been modeled (19), but its conformation as it would appear on the surface of the virus before CD4 binding is currently unknown. The prefusion structure of the gp41 ectodomain is also unknown. If influenza virus HA2 is a suitable precedent (32), gp41 probably undergoes a major refolding transition, and models based on the postfusion structure as a “stalk” may be seriously flawed. There is some evidence from the thermodynamics of CD4 binding for a relatively flexible conformation of gp120 prior to the CD4 interaction (26). Indeed, it is possible that CD4-independent forms of gp120 spontaneously assume the tighter structure normally induced by CD4 (13, 21), but structural data are lacking on this point.
Neutralization by antibodies depends on binding to surfaces available on one or another of the trimeric states of gp120/gp41, and epitopes exposed on monomeric gp120 but occluded in the trimer induce antibodies with poor neutralizing capacity (28). Structural information about the trimer is therefore crucial for rational engineering of recombinant immunogens. Difficulty experienced in preparing soluble gp140 in trimeric form has, however, hampered biochemical and structural studies of the prefusion Env trimer. In an attempt to capture the conformation of the envelope glycoprotein before CD4 binding, we have recently prepared a soluble trimer of SIV gp140 (also called gp160e) (6). We now describe expression and purification of chimeric proteins of SIV gp140 and its variants with the catalytic subunit (C) of Escherichia coli aspartate transcarbamoylase (ATCase). ATCase C is a very stable trimer (3), and its attachment to the C terminus of gp140 is intended to enhance trimer stability. We demonstrate that the fusion protein has ATCase activity, which requires trimerization of the C chain. When ATCase regulatory subunit dimers (R2) are added, the chimera assembles into dimers of trimers, as expected from the structure of the C6R6 ATCase (14). The production of such a fusion protein may enhance the possibilities for structure determination, either by electron cryomicroscopy or by X-ray crystallography, of SIV and HIV Env in their prefusion states.
MATERIALS AND METHODS
Expression of SIV gp140-ATC fusion proteins in insect cells.
pSIV[ΔV1V2V3]-ATC and pSIVgp140-ATC were constructed by fusing the open reading frame (ORF) of the E. coli ATCase C subunit (kindly provided by E. Kantrowitz, Boston College) in frame to the C termini of pFBSIV[ΔV1V2V3]-His1 and pFBSIV-His1 (6), respectively, by standard PCR techniques. The cleavage sites (residue 512 and 523) between gp120 and gp41 in pFBSIV-His1 and pSIVgp140-ATC were changed back to wild-type sequences by PCR-based site-directed mutagenesis to give pSIVgp140(W) and pSIVgp140-ATC(W). Both restriction digestion and DNA sequencing verified all the expression constructs. The proteins were expressed using the Bac-to-Bac system (Invitrogen). Recombinant baculovirus was generated according to the manufacturer's protocol and amplified in Sf9 insect cells in Hink's TNM-FH medium (JRH Biosciences). Expression of the proteins was confirmed by Western blotting using a mixture of MAbs 15E8, 9G3, and 116 (16) against SIV envelope glycoprotein. The optimal amount of virus and postinfection harvest time were determined by small-scale tests in 100-ml spinner flasks. For large-scale protein production, 10 liters of Trichoplusia ni (Hi-5) cells (2 × 106 cells/ml) in Ex-cell 405 medium (JRH Biosciences) was infected at a multiplicity of infection (MOI) of 2.5. At 72 h postinfection, the supernatant was harvested by centrifugation and concentrated to 2 liters in a tangential flow filtration system (ProFlux M 12; Millipore). The concentrated supernatant was clarified by centrifugation.
Protein purification.
The expressed proteins with pSIV[ΔV1V2V3]-ATC, pSIVgp140-ATC, and pSIVgp140-ATC(W) were purified by an immunoaffinity chromatography using a MAb 17A11 (10) affinity column (5-ml bed volume), where the MAb 17A11 was cross-linked at 5 mg/ml to GammaBind Plus Sepharose (Pharmacia) with dimethyl pimelimidate (Pierce). The concentrated insect cell supernatants were passed through the column with a flow rate of about 0.5 ml/min. After extensive washing with phosphate-buffered saline, the protein was then eluted with 2.5 M MgCl2, immediately followed by a desalting step using a Sephadex G-25 column. The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The fractions containing SIV gp140 were pooled, concentrated, and further purified by gel filtration chromatography on Superdex 200 or Superose 6 (Pharmacia) with a buffer containing 25 mM Tris-HCl (pH 8.0) and 150 mM NaCl. SIV gp140Δ(V1V2) and gp140Δ(V1V2,V3) proteins were purified following the same procedure. SIV gp140(W) was purified by metal chelate affinity chromatography as previously described (6).
Binding of SIVgp140-ATC(W) to CD4 and antibody Fab fragments.
Preparations of four-domain soluble CD4 (sCD4) and of MAbs and their Fab fragments followed standard procedures (16). SIVgp140-ATC(W) was incubated with sCD4 or Fab in phosphate-buffered saline at room temperature for at least 30 min and then separated from excess unbound ligand by a gel filtration chromatography on Superose 6 or Superdex 200 (Pharmacia). Molecular weights were calculated from the elution volumes of known proteins. Peak fractions were verified to contain both SIVgp140-ATC(W) and CD4 or Fab by SDS-PAGE.
Assembly of SIV(ΔV1V2V3)-ATC and SIVgp140-ATC(W) into dimers of trimers.
The E. coli ATCase regulatory subunit was overexpressed in E. coli cells using expression plasmid pEK244 (a gift from M. Williams and E. Kantrowitz, Boston College) and purified as described elsewhere (12). The protein was dialyzed against 25 mM Tris (pH 8.0) and 0.1 mM zinc acetate overnight. Purified SIVgp140-ATC(W) or SIV(ΔV1V2V3)-ATC protein was incubated with an excess amount of R subunit for several hours at room temperature or overnight at 4°C. The complexes were then separated from excess unbound R dimers by gel filtration chromatography on Superose. Molecular weights were calculated from the elution volumes of known proteins. Peak fractions were verified to contain both SIVgp140-ATC(W) and ATCase R subunit by SDS-PAGE in a 10% gel.
Negative-stain electron microscopy.
For staining with uranyl formate, samples at ∼10 μg/ml were adsorbed to glow-discharged carbon grids for 30 s, followed by washing with 2 drops of deionized water and staining with 2 drops of freshly prepared 0.75% uranyl formate. Images were taken on negative film at a magnification of ×52,000 and a defocus of 1.5 μm using the low-dose procedure with a Philips Tecnai12 electron microscope operated at 120 kV. The images on negative film were then digitized with a Zeiss SCAI scanner (pixel size of 0.4 nm at the specimen level). Individual particles were then selected manually and processed with SPIDER (11).
For staining with silicotungstate, 2 μl of protein sample at 100 μg/ml was adsorbed to a carbon-coated grid. The grid was floated on 1% sodium silicotungstate, pH 7.5, for 2 min and then air dried. Images were taken on film with a Jeol 1200EX microscope operated at 100 kV, under minimum dose, accurate defocus conditions.
RESULTS
Expression and purification of a chimeric protein with SIV gp140 fused to the C subunit of E. coli ATCase.
Our investigators have previously expressed and purified SIVmac32H gp140, the ectodomain of the envelope glycoprotein, using both insect cells and CHO-Lec3.2.8.1 cells (6). In the form we expressed, the cleavage sites between gp120 and gp41 were mutated so that they were no longer substrates for furin-like proteases. We have been unable to obtain crystals, however, even using variants with variable domains deleted. We have therefore turned to chimeric proteins by fusing SIVgp140 and its variants with the C subunit of E. coli ATCase. We chose this strategy both to stabilize further the gp140 trimer and to add additional, nonvariable and nonglycosylated protein as a crystallization tag. The E. coli ATCase C subunit was fused directly to the C termini of SIVgp140 and of the species with V1, V2, and V3 deleted to produce chimeric proteins we designate as SIVgp140-ATC and SIV[ΔV1V2V3]-ATC, respectively (Fig. 1). In both cases, two extra amino acid residues (SR) were introduced by a KpnI restriction site between gp140 and ATCase C. We also have expressed proteins SIVgp140(W), SIVgp140-ATC(W), and SIV[ΔV1V2V3]-ATC(W), in which the cleavage sites between gp120 and gp41 have been changed back to the wild-type sequence (Fig. 1 and data not shown). The chimeric proteins were secreted into culture medium from Hi-5 insect cells at levels of 0.5 to 1 mg/liter, similar to those obtained with gp140, indicating that addition of the ATCase C subunit does not affect expression of envelope glycoprotein. Purified SIVgp140(W) containing the wild-type cleavage sites between gp120 and gp41 remained largely uncleaved (data not shown; see Fig. 3), in contrast to what was found for gp140 from CHO cells, where about 50% of the protein was processed into gp120 and gp41 (A. Dessen and D. C. Wiley, unpublished results). Other groups have shown that insect cell-expressed HIV gp160 remains largely uncleaved and that coexpression of furin can enhance cleavage efficiency (24, 41). Sf9 cells do have an endogenous furin-like protease (8), but it is apparently relatively inactive at the processing sites in HIV and SIV Env. When we treated our SIV gp140 with furin in vitro, we obtained the correct cleavage, as well as an additional one, probably in the V3 loop (well-known to be exceptionally protease sensitive).
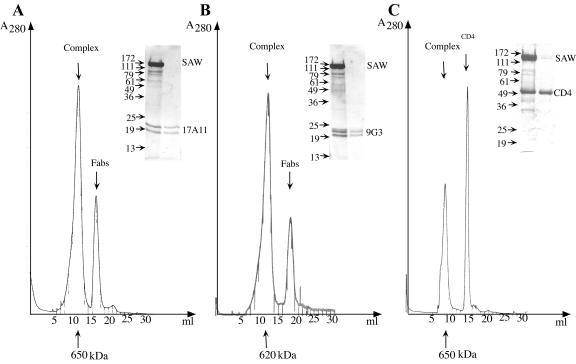
FIG. 3.
Purification of SIVgp140-ATC(W) complexed with CD4 or Fabs by gel filtration chromatography. (A and B) Purified SIV gp140-ATC(W) (SAW) was incubated with Fab fragments generated by papain digestion from either 17A11 or 9G3 neutralizing MAbs; these MAbs recognize epitopes on gp120 and gp41, respectively. The complexes were separated from excess unbound Fabs by gel filtration chromatography on a Superose 6 column. (C) Purified SIV gp140-ATC(W) was incubated with four-domain sCD4 from CHO cells (kindly provided by M. Kim). The complexes were separated from excess unbound CD4 by a gel filtration chromatography on a Superdex 200 column. The apparent molecular mass was calculated based on a standard curve plotted using the elution times of known proteins. The peak fractions were pooled, concentrated, and analyzed by SDS-PAGE in a 10% gel (inset).
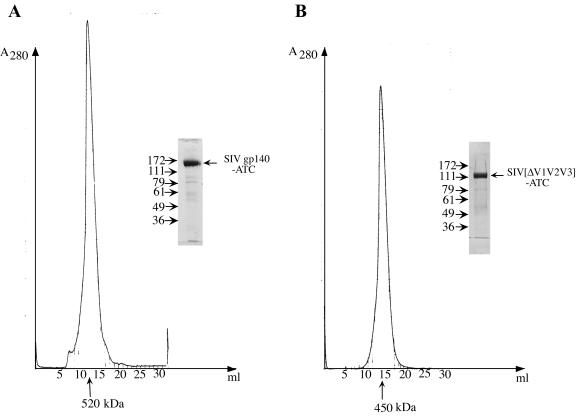
All the fusion proteins were purified from insect cell supernatants by immunoaffinity chromatography using the MAb 17A11 (6). The proteins were eluted with 2.5 M MgCl2, immediately followed by a desalting step; elution with low pH buffer would cause aggregation. The fractions containing the protein were concentrated and further purified by gel filtration chromatography. In Fig. 2, purified SIVgp140-ATC and SIV[ΔV1V2V3]-ATC eluted from the sizing column (Superose 6) with relative molecular masses of 520 and 450 kDa, respectively. Monodispersity of the purified proteins, indicated by the relatively sharp gel filtration peaks, was confirmed by dynamic light scattering, which yielded polydispersities of less than 20% (data not shown). Analysis by SDS-PAGE showed that SIVgp140-ATC migrated as a single protein band with an apparent molecular mass of 160 kDa; SIV[ΔV1V2V3]-ATC migrated at approximately 120 kDa (Fig. 2). The fusion proteins retained ATCase activity as determined by a pH-stat assay (39). For example, SIV[ΔV1V2V3]-ATC has a specific activity of 1.5 × 104 U/μmol, compared to that of the ATCase C subunit trimer of 3.7 × 104 U/μmol (carried out by Mark Williams in the Kantrowitz lab). The slightly lower activity of SIV[ΔV1V2V3]-ATC may have been due to a constraint imposed on the C subunit by covalent linkage to SIV gp140. A trimer of ATCase C is active; a monomer is not (14). Thus, the molecular weight measurement and the activity of the C subunit both indicate that the chimeric protein is properly folded and trimeric.
FIG. 2.
Gel filtration chromatography of purified SIVgp140-ATC(W) and SIV[ΔV1V2V3]-ATC expressed from insect cells. The chimeric proteins were purified by MAb 17A11 affinity chromatography from supernatants of insect cell cultures infected with recombinant baculoviruses expressing SIV gp140-ATC(W) (A) and SIV[ΔV1V2V3]-ATC (B). The purified proteins were then resolved by gel filtration chromatography using a Superose 6 column. Molecular masses were calculated based on a standard curve plotted using known proteins, which included thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), and bovine serum albumin (67 kDa). Peak fractions were pooled and analyzed by SDS-PAGE (inset).
Complexes of SIV gp140-ATC with CD4 and Fab fragments of neutralizing MAbs.
Binding studies of the fusion proteins with viral receptor, CD4, and with a number of MAbs specific for SIV gp140 provided evidence that the fusion protein is properly folded. In the experiments illustrated in Fig. 3A and B, purified SIV gp140-ATC(W) was incubated with Fab fragments generated by papain digestion from either 17A11 or 9G3 neutralizing MAbs (10, 16). The complexes could be separated from excess unbound Fabs by a gel filtration chromatography on Superose 6. Monoclonal 17A11 recognizes a discontinuous epitope close to the V4 loop of gp120; 9G3 binds a linear gp41 epitope, overlapping that of another MAb, KK41 (16). The KK41 epitope has been mapped to a 20-amino-acid segment encompassing the C-C loop of gp41 (15). Despite the proximity of the ATCase C subunit to gp41 in the fusion protein, the observation that the gp41 epitope of 9G3 is still accessible supports the notion that addition of ATCase C chain does not cause major distortions in the gp140 portion of the fusion protein. Purified SIV gp140-ATC(W) also forms a tight complex with four-domain sCD4 expressed from CHO cells. As shown in Fig. 3C, the complexes were separated from excess unbound CD4 by gel filtration chromatography on Superdex 200. In summary, the trimeric fusion proteins retained the properties of both SIV gp140 and ATCase C.
Assembly of SIV[ΔV1V2V3]-ATC and SIVgp140-ATC(W) into dimers of trimers.
Intact ATCase is a dodecamer in which three regulatory subunit (R chain) dimers clamp two C trimers into a C6R6 assembly (14). We reasoned that if the ATCase C chain were folded, the fusion protein should assemble into dimers of trimers. The structure of C6R6 ATCase shows that, in the fusion protein, the SIV gp140 moiety will lie on the opposite side of the trimer from the interfaces at which the two C trimers associate when ATCase regulatory subunit dimers (R2) are added.
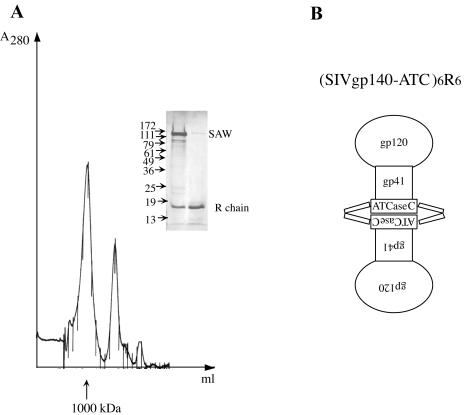
We incubated purified SIVgp140-ATC(W) with ATCase regulatory subunit dimers (R2) prepared from an E. coli strain in which R chain is overexpressed. The complexes were separated from excess unbound R dimers by a gel filtration chromatography on a Superose 6 column. The complex eluted at a position corresponding to a mass of approximately 1,000 kDa, consistent with the expected size of a dimer of trimers. The peak fractions were pooled, concentrated, and analyzed by SDS-PAGE in a 10% gel. The peak contained both SIV gp140-ATC(W) and R chain, as expected for (SIV gp140-ATC)6R6, schematically shown in Fig. 4B. The same result was obtained with SIV[ΔV1V2V3]-ATC when R2 was added (data not shown).
FIG. 4.
The fusion protein assembles into dimers of trimers when adding ATCase regulatory subunit dimers (R2). (A) Purified SIV gp140-ATC(W) (SAW) was incubated with ATCase regulatory subunit dimers (R2) prepared from an E. coli strain that overexpresses R subunit (kindly provided by E. Kantrowitz and M. Williams). The complexes were separated from excess unbound R dimers by a gel filtration chromatography on a Superose 6 column. The apparent molecular mass was calculated based on a standard curve plotted using the elution times of known proteins. The peak fractions were pooled, concentrated, and analyzed by SDS-PAGE in a 10% gel (inset). (B) Schematic diagram showing the dimers of trimers of the fusion proteins.
Images of SIV gp140 and SIV gp140-ATC from negative-stain electron microscopy.
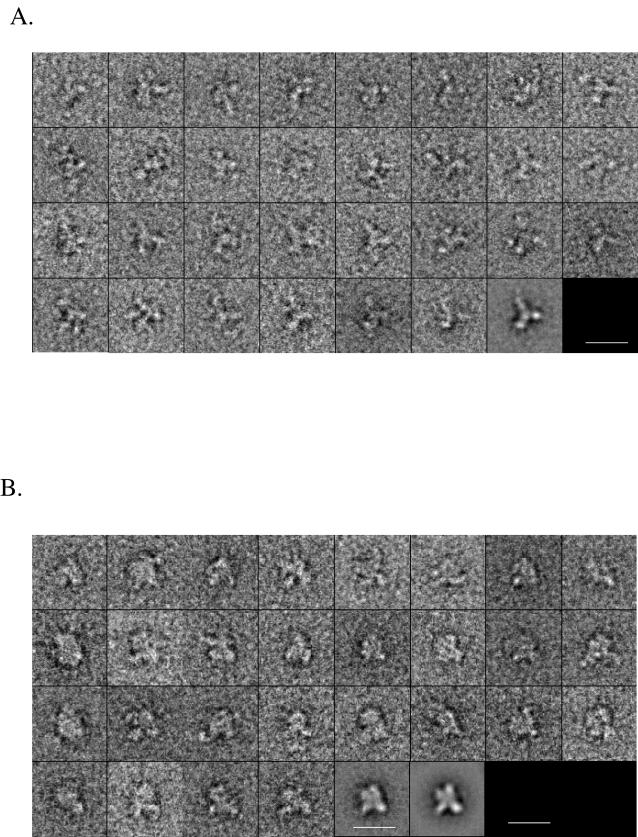
To gain some qualitative structural information, we recorded images of both SIV gp140 and SIV gp140-ATC on glow-discharged, carbon-coated grids using uranyl formate as stain. SIV gp140 was expressed and purified from CHO-Lec3.2.1.8 cells, while SIV gp140-ATC was prepared from insect cells. The images were digitized as described in Materials and Methods. Images of over 3,000 molecules were selected and analyzed using the program suite SPIDER (11). The images were classified into sets, each of which represented the view of many molecules that lay the same way on the grid. Class averages were obtained by rotating and translating the individual images to superimpose on each other. A gallery of images in one class set, along with the average, is shown for SIV gp140 and SIV gp140-ATC in Fig. 5A and B, respectively. Both molecules appeared quite spiky. They can be described as a stalk with three branches projecting from one end. The images presumably represented a trimeric gp41 or gp41/ATCase (the stalk) associated with three gp120 subunits (the projections).
FIG. 5.
Electron microscopic analysis of SIV gp140 and SIV gp140-ATC. Purified SIV gp140 from CHO-Lec3.2.1.8 cells (A) and SIV gp140-ATC from insect cells (B) were negatively stained with uranyl formate. All images are selected raw images except for the last image. Over 2,000 raw images were grouped and averaged to give two-dimensional projections with an enhanced signal-to-noise ratio, as shown in the last two boxes, using SPIDER. Bar, 20 nm.
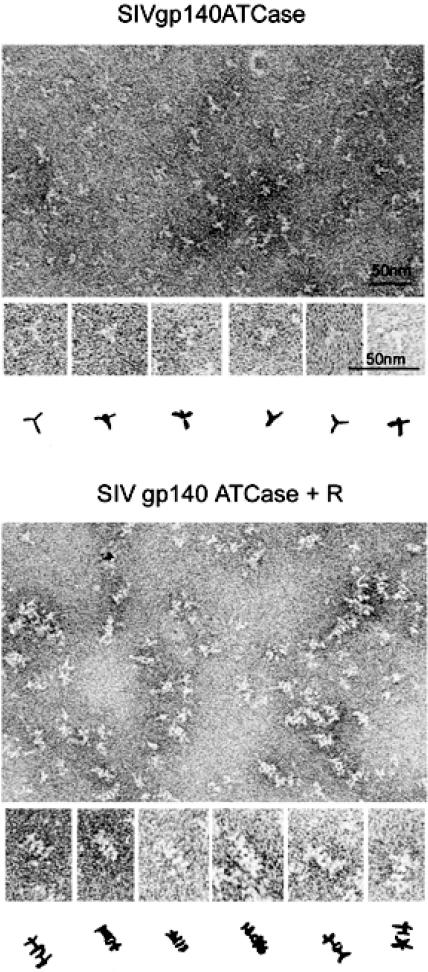
We also obtained, by a slightly different approach using 1% silicotungstate as stain, images of SIV gp140-ATC and SIV gp140-ATC linked by the ATCase R chain (Fig. 6). The gp140-ATCase images are similar to those in Fig. 5A. The images of the R chain linked trimers show many “side” views of four-layer particles, which we interpret as the ATCase heterohexamer in the center (two layers) flanked by gp140 at either end (Fig. 4B).
FIG. 6.
Electron microscopic analysis of SIV gp140-ATC and SIV gp140-ATC linked by ATCase regulatory chains. The proteins were negatively stained with 1% silicotungstate. Beneath each field are six selected images, together with interpretations as hand-drawn line diagrams. The bars in the upper image represent 50 nm. The magnifications in the lower image are the same as those in the corresponding parts of the upper image.
DISCUSSION
Expression of a stably trimeric fusion protein with a familiar and well-characterized threefold “base” has several advantages for analysis of the SIV and HIV envelope glycoproteins. First, we can be confident that the membrane-proximal segments are forming trimer contacts, because the ATCase, which requires oligomerization for enzymatic function, is catalytically active. Second, we can regard the ATCase part of the chimera as a crystallization tag or structure tag. It provides a well-behaved C-terminal component of known characteristics and proven stability. Third, the trimeric ATCase base may permit useful manipulation for future studies by electron microscopy, as suggested by the images in Fig. 6.
Two other approaches to stabilizing gp140 trimers have been described, both involving rod-like extended structures (4, 31, 42, 43). Fusion of a GCN4-based trimerization motif or a T4 fibritin-derived trimerization motif to HIV-1 gp140 yields secreted trimers, but these appear to be more prone to dissociation or aggregation than the SIV gp140-ATC trimers reported here. Indeed, when we fused the ATCase catalytic subunit to the C terminus of HIV-1 gp140, we obtained trimers that tended strongly to aggregate when concentrated, unlike the SIV fusion protein (B. Chen, unpublished results). We ascribe the aggregation to the propensity of the HIV-1 part of the chimeric trimer to come apart. The ATCase part of the fusion protein will still hold the trimer together, but exposed gp140 surfaces will interact at high concentration with complementary surfaces on other trimers and cause association.
The distance from the N terminus of the ATCase C-subunit to the threefold axis is about 28 Å, substantially larger than the corresponding distances for the N termini of the GCN4 coiled-coil (about 8 Å) and the fibritin segment (about 7 Å). It is hard to assess the impact of these differences on the geometry and stability of the chimeric protein, because we do not yet know how the transmembrane segments of gp160 are organized. In the postfusion trimer, 18 amino acid residues intervene between the C terminus of a chain in the well-known six-helix bundle and the transmembrane segment to which it would connect (9). These residues, which include two well-studied epitopes and part of the T-20 fusion inhibitor (25, 33, 37), could have extended or even flexible conformations, allowing the protein to adapt to a variety of trimerization tags. When we deleted nine of these residues from a chimera with ATCase, we obtained protein with a slightly higher tendency to aggregate and somewhat lower ATCase activity. We also attempted to use PCNA (proliferating cell nuclear antigen) as a trimerization tag (17), but the PCNA portion of that chimera apparently could not tolerate some of the relatively harsh conditions we needed to use for purification.
The strikingly four-pronged images of the chimera in negative stain exhibited considerable variability in the relative directions of the projections. Many of the images were not views that one might expect, for example, from a regular, tetrahedrally directed array of spokes. Much of this variability could be due to adsorption forces that flatten the molecule as it adheres to the carbon substrate. We believe, however, that a certain amount of inherent flexibility probably contributes as well. As suggested above, trimeric gp41/ATCase presumably makes up most of one prong. The initial residues of gp120 (those preceding the gp120 core in the gp160 polypeptide chain) may also be part of this prong, as well as some of the residues at the very end of gp120 (those following the gp120 core). The link between the trimeric stalk and the three gp120 spokes would then be the two segments of polypeptide chain, close together in space, that lead into and away from the gp120 core (near residues 64 and 502, respectively). The images suggest that the body of the gp120 core may not be tightly pinned down to the gp41 stalk, unlike influenza virus HA1, for example, which closes in tightly around the apex of the HA2 trimer in the prefusion state of the hemagglutinin (7, 38). It is possible, of course, that some of the gp140 molecules in our sample had been triggered into the CD4-induced, fusion-active conformation and that the heterogeneity of the images came in part from a heterogeneity of states of the molecule. CD4 binding induces dissociation of gp120 (from cleaved gp120/gp41 precursor), so the CD4-triggered state of an uncleaved trimer might be expected to have loosely attached gp120 appendages. Unlike laboratory-adapted strains of HIV-1, however, SIV does not shed gp120 spontaneously (30), and our preparation has not been exposed to conditions we think might mimic CD4 binding. We therefore believe that the spikey structures represent the mature, unliganded conformation.
Current models for association of gp120 in the gp160 trimer are based on the liganded structure of gp120 (19). Evidence for extensive conformation changes upon CD4 and coreceptor binding and the images shown here suggest that the contacts made by gp120 cores within a trimer could be less intimate than hitherto suggested.
Acknowledgments
We thank Margaret Pietras for technical assistance, Evan Kantrowitz and Mark Williams of Boston College for providing plasmids encoding the ATCase C subunit and an E. coli strain overexpressing the ATCase R subunit and for performing ATCase activity assays, and also Mikyung Kim for sCD4.
This research was supported by the Innovation Grant Program for Approaches in HIV Vaccine Research (S.C.H. and D.C.W.), by the Medical Research Council (United Kingdom), by grant AI43649 (to E.L.R.), and by the Howard Hughes Medical Institute (HHMI). This work was also supported by Scholar Award 70529-28-RF from the American Foundation for AIDS Research to B.C. S.C.H. is and D.C.W. was an Investigator of the HHMI.
REFERENCES
- 1.Allan, J. S., J. E. Coligan, F. Barin, M. F. McLane, J. G. Sodroski, C. A. Rosen, W. A. Haseltine, T. H. Lee, and M. Essex. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091-1094. [DOI] [PubMed] [Google Scholar]
- 2.Bachelder, R. E., J. Bilancieri, W. Lin, and N. Letvin. 1995. A human recombinant Fab identifies a human immunodeficiency virus type 1-induced conformational change in cell surface-expressed CD4. J. Virol. 69:5734-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beernink, P. T., J. A. Endrizzi, T. Alber, and H. K. Schachman. 1999. Assessment of the allosteric mechanism of aspartate transcarbamoylase based on the crystalline structure of the unregulated catalytic subunit. Proc. Natl. Acad. Sci. USA 96:5388-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 6.Chen, B., G. Zhou, M. Kim, Y. Chishti, R. E. Hussey, B. Ely, J. J. Skehel, E. L. Reinherz, S. C. Harrison, and D. C. Wiley. 2000. Expression, purification, and characterization of gp160e, the soluble, trimeric ectodomain of the simian immunodeficiency virus envelope glycoprotein, gp160. J. Biol. Chem. 275:34946-34953. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 8.Cieplik, M., H. D. Klenk, and W. Garten. 1998. Identification and characterization of Spodoptera frugiperda furin: a thermostable subtilisin-like endopeptidase. Biol. Chem. 379:1433-1440. [DOI] [PubMed] [Google Scholar]
- 9.Douglas, N. W., G. H. Munro, and R. S. Daniels. 1997. HIV/SIV glycoproteins: structure-function relationships. J. Mol. Biol. 273:122-149. [DOI] [PubMed] [Google Scholar]
- 10.Edinger, A. L., M. Ahuja, T. Sung, K. C. Baxter, B. Haggarty, R. W. Doms, and J. A. Hoxie. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74:7922-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 12.Hack, E. S., T. Vorobyova, J. B. Sakash, J. M. West, C. P. Macol, G. Herve, M. K. Williams, and E. R. Kantrowitz. 2000. Characterization of the aspartate transcarbamoylase from Methanococcus jannaschii. J. Biol. Chem. 275:15820-15827. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, T. L., C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 96:6359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kantrowitz, E. R., and W. N. Lipscomb. 1988. Escherichia coli aspartate transcarbamylase: the relation between structure and function. Science 241:669-674. [DOI] [PubMed] [Google Scholar]
- 15.Kent, K. A., L. Gritz, G. Stallard, M. P. Cranage, C. Collignon, C. Thiriart, T. Corcoran, P. Silvera, and E. J. Stott. 1991. Production of and monoclonal antibodies to simian immunodeficiency virus envelope glycoproteins. AIDS 5:829-836. [DOI] [PubMed] [Google Scholar]
- 16.Kim, M., B. Chen, R. E. Hussey, Y. Chishti, D. Montefiori, J. A. Hoxie, O. Byron, G. Campbell, S. C. Harrison, and E. L. Reinherz. 2001. The stoichiometry of trimeric SIV glycoprotein interaction with CD4 differs from that of anti-envelope antibody Fab fragments. J. Biol. Chem. 276:42667-42676. [DOI] [PubMed] [Google Scholar]
- 17.Krishna, T. S., X. P. Kong, S. Gary, P. M. Burgers, and J. Kuriyan. 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79:1233-1243. [DOI] [PubMed] [Google Scholar]
- 18.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 95:9134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, K. A., R. Wyatt, M. Farzan, H. Choe, L. Marcon, E. Desjardins, J. Robinson, J. Sodroski, C. Gerard, and N. P. Gerard. 1997. CD4-independent binding of SIV gp120 to rhesus CCR5. Science 278:1470-1473. [DOI] [PubMed] [Google Scholar]
- 22.Moore, J. P., J. A. McKeating, Y. X. Huang, A. Ashkenazi, and D. D. Ho. 1992. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J. Virol. 66:235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore, J. P., Q. J. Sattentau, and P. R. Clapham. 1990. Enhancement of soluble CD4-mediated HIV neutralization and gp 120 binding by CD4 autoantibodies and monoclonal antibodies. AIDS Res. Hum. Retrovir. 6:1273-1279. [DOI] [PubMed] [Google Scholar]
- 24.Morikawa, Y., E. Barsov, and I. Jones. 1993. Legitimate and illegitimate cleavage of human immunodeficiency virus glycoproteins by furin. J. Virol. 67:3601-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sattentau, Q. J., and J. P. Moore. 1993. The role of CD4 in HIV binding and entry. Philos. Trans. R. Soc. Lond. B 342:59-66. [DOI] [PubMed] [Google Scholar]
- 30.Sattentau, Q. J., J. P. Moore, F. Vignaux, F. Traincard, and P. Poignard. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza haemaglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 33.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 34.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 35.Veronese, F. D., A. L. DeVico, T. D. Copeland, S. Oroszlan, R. C. Gallo, and M. G. Sarngadharan. 1985. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science 229:1402-1405. [DOI] [PubMed] [Google Scholar]
- 36.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 37.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 39.Wu, C. W., and G. G. Hammes. 1973. Relaxation spectra of aspartate transcarbamylase. Interaction of the native enzyme with an adenosine 5′-triphosphate analog. Biochemistry 12:1400-1408. [DOI] [PubMed] [Google Scholar]
- 40.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 41.Yamshchikov, G. V., G. D. Ritter, M. Vey, and R. W. Compans. 1995. Assembly of SIV virus-like particles containing envelope proteins using a baculovirus expression system. Virology 214:50-58. [DOI] [PubMed] [Google Scholar]
- 42.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, Z. N., T. C. Mueser, J. Kaufman, S. J. Stahl, P. T. Wingfield, and C. C. Hyde. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J. Struct. Biol. 126:131-144. [DOI] [PubMed] [Google Scholar]