Abstract
Hepatitis C virus (HCV) behaves in infected patients as a complex mixture of genetically distinct but closely related variants referred to as a “quasispecies.” By using quasispecies analysis strategies, we showed that HCV nonstructural protein 5A (NS5A) has a quasispecies distribution in infected humans and that NS5A quasispecies undergo significant genetic evolution over time, as a result of random accumulation of nucleotide mutations during replication. Genetic evolution of the NS5A quasispecies results in sporadic amino acid changes in the protein sequence. By using the functional in vitro model of HCV NS5A transcriptional activation in Saccharomyces cerevisiae, we showed that natural NS5A quasispecies variants induce different levels of transcriptional activation, according to the charge of the residues (and possibly minor conformational changes) in the quasispecies variant sequence. These findings show that the accumulation of mutations on HCV genomes during replication randomly generates variant proteins with quantitatively different functional properties. The fact that each new variant protein is initially produced in a single infected hepatocyte and may or may not subsequently spread throughout the liver (depending on the replication capacities of the variant virus) points to cellular compartmentalization of virus-host interactions during chronic infection. This feature of quasispecies-distributed viruses could play an important role in various aspects of the viral life cycle and related disease.
Hepatitis C virus (HCV) infects 170 million individuals worldwide. Acute HCV infection spontaneously resolves in 15 to 40% of cases. The remaining patients, i.e., the vast majority, develop chronic infection and are thus at risk of chronic hepatitis and its complications, namely, cirrhosis and hepatocellular carcinoma (31). An estimated 10 to 15% of HCV-infected patients develop cirrhosis after 20 years of infection, while the annual incidence of hepatocellular carcinoma (primary liver cancer) in patients with HCV-related cirrhosis is about 4 to 5% (31). Prevention of these complications relies on antiviral treatment. The present standard of treatment is the combination of pegylated alpha interferon (IFN-α) and ribavirin (31). This treatment permanently clears the infection in approximately 80% of patients infected by HCV genotypes 2 and 3 and 40% of patients infected by HCV genotype 1 (the most frequent HCV genotype in industrialized countries) (9, 25).
Hepatocellular carcinoma generally occurs against a background of HCV-related cirrhosis, in the form of a single tumor or multiple tumoral foci. Tumor growth results from clonal proliferation of a single transformed hepatocyte and is probably facilitated by the underlying liver disease, i.e., cirrhosis. The role of viral proteins in transforming infected cells is largely unknown. Several viral proteins, including the core protein, nonstructural protein 3 (NS3), and NS5A, have carcinogenic properties in transfected cells in vitro and also in transgenic mice (6, 18, 21, 22, 30, 39, 41, 47, 48). However, it is not known whether these proteins are expressed at high enough levels in vivo to exert similar actions.
HCV behaves in infected patients as a complex mixture of genetically distinct but closely related variants referred to as a “quasispecies” (27, 46). New variants are continuously generated during viral replication, as a result of errors made by the viral RNA-dependent RNA polymerase, which lacks proofreading activity. The fittest of these newly generated variants are continuously selected in the environment in which the virus replicates, on the basis of their own replication capacities and numerous selection pressures, particularly the host immune responses. At a given time point during infection, the HCV quasispecies distribution reflects the balance among continuous generation of new variants, the need to conserve essential viral functions, and positive selection pressures exerted by the replicative environment (reviewed in references 7 and 32).
The quasispecies distribution of HCV implies that novel viral RNAs continuously emerge in infected cells (8). Most such variants are defective: they may be encapsidated and enveloped but fail to propagate in the host liver. In contrast, some give rise to large viral subpopulations that are eventually released into the peripheral circulation if the selective environment is favorable. The simultaneous presence, in a given individual, of different quasispecies variant populations raises the question of the functional capacities of the corresponding viral structures and proteins. It is conceivable that viral proteins with different sequences have different functional properties. If so, quasispecies regulation of protein functions could play a role in viral variant fitness in a given environment but also in virus-host interactions in the cells in which these proteins are produced.
NS5A is a phosphorylated protein integrated into the replication complex. It plays an important (although unclear) role as a direct modulator of RNA-dependent RNA polymerase activity (43). Other properties attributed to NS5A include attenuation of IFN induction of intracellular pathways and interaction with the regulation of cell growth and cellular signaling pathways (reviewed in reference 36). The 146 C-terminal amino acids of NS5A protein have transcriptional activation properties in vitro (10, 20, 44). These properties are supported by a sequence that alternates two acidic regions followed by a proline-rich region, a pattern typical of the acidic class of transcriptional activators (10, 20, 44). Indirect arguments suggest that NS5A transcriptional activation is functional in infected cells in vivo: (i) the NS5A acidic transcriptional activation domain is conserved among HCV strains and genotypes; (ii) NS5A appears to be naturally cleaved by a caspase-like protease in mammalian cells, at a site located upstream of the transcriptional activation domain (42); (iii) the natural C-terminal cleavage product has transcriptional activation properties in vitro (42); (iv) the NS5A sequence contains a nuclear localization signal just downstream of the transcriptional activation domain, capable of addressing the cleaved fragment to the nucleus (16, 37, 42); and (v) Polyak et al. recently demonstrated that natural NS5A sequences activate interleukin 8 gene transcription in mammalian cells; this is the first biological effect shown to result from the transcriptional activity of NS5A (37). The cellular genes physiologically activated by NS5A, the cellular partners involved, and the exact role of NS5A transcriptional activation in the pathogenesis of HCV infection are currently unknown.
Using a functional model of NS5A transcriptional activation in vitro, we show here that continuous accumulation of mutations on HCV genomes during replication randomly generates variant proteins with quantitatively different functional properties and that these functional differences are related to the physicochemical properties of the amino acid residues. These results support the hypothesis that virus-host interactions are compartmentalized at the cellular level, a feature that could play an important role in viral persistence and HCV disease pathogenesis.
MATERIALS AND METHODS
Materials.
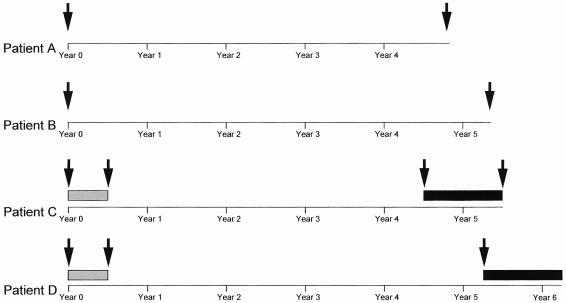
Serum samples from four patients with chronic HCV genotype 1b infection monitored for at least 5 years were used to generate natural HCV NS5A quasispecies sequences and to study their genetic variability and evolution and their transcriptional activation properties in vitro. Two patients (A and B) were monitored for up to 58 and 64 months, respectively, and received no antiviral treatment. These patients' NS5A quasispecies were studied at the beginning and end of follow-up (Fig. 1). The other two patients (C and D) received 3 million units of IFN-α (Roferon-A; Hoffmann-LaRoche, Basel, Switzerland) subcutaneously, three times a week for 6 months. Patient C did not respond (HCV RNA remained detectable throughout therapy), while patient D initially cleared HCV RNA but relapsed before the end of treatment. Both patients were treated again, 54 and 63 months after the end of the first treatment, respectively, with a combination of 3 million units of IFN-α three times a week and 1 g of oral ribavirin/day. Patient C did not respond to this combination therapy, while patient D achieved sustained HCV RNA clearance, i.e., HCV RNA disappeared during treatment and remained undetectable after its withdrawal, a result almost always associated with permanent HCV clearance. In both patients treated, we studied serum samples taken before the first course of treatment, at the end of the first course of treatment (6 months later), and before the second course of treatment. We also studied a sample taken at the end of the second treatment from patient C, who did not respond (Fig. 1). Overall, 11 serum samples from the four patients were studied.
FIG. 1.
Sampling protocol in the four patients with chronic HCV infection. The sampling dates are indicated by black arrows. The gray boxes indicate IFN monotherapy, and the black boxes indicate IFN-ribavirin combination therapy.
Generation of NS5A quasispecies sequences.
RNA was extracted from 200 μl of serum with the High Pure viral RNA kit (Roche Applied Science, Indianapolis, Ind.). RNA was reverse transcribed at 42°C for 90 min with 10 pmol of the antisense primer Y3EAS (5′-TCCTTGAGCACGTCCCGGTA-3′, nucleotides [nt] 7776 to 7795) in the presence of 8 U of avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.). The transcriptional activation domain of NS5A (amino acid positions 2135 to 2332 in the HCV-J genotype 1b prototype sequence) was amplified by nested PCR, with outer primers Y507S (sense) (5′-TTCCCCATCAACGCATACACCAC-3′, nt 6507 to 6529) and ASPR (antisense) (5′-AGCTCCGCCAAGGCAGAAGACAC-3′, nt 7347 to 7369) and inner primers S6745 (sense) (5′-CCGGCGTGCAAACCTCTCCT-3′, nt 6731 to 6750) and MK1b (antisense) (5′-GATTCTGTCAGGACAACCGT-3′, nt 7323 to 7342). The two nested PCR rounds were run in the presence of 0.5 U of high-fidelity Pwo DNA polymerase (Roche Applied Science). The error rate of this enzyme reported by the manufacturer is 3.2 × 10−6 errors/base, ensuring that the vast majority of mutations seen in the NS5A-encoding region were not introduced during the in vitro amplification. The first round consisted of 5 min of denaturation at 94°C, followed by 30 PCR cycles with denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min, and a final extension step at 72°C for 5 min. One microliter of first-round PCR product was used as template for the second round, which consisted of 5 min of denaturation at 94°C, followed by 25 PCR cycles with denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 45 s, and a final extension step at 72°C for 5 min. Amplification products were analyzed by electrophoresis through a 2% agarose gel and staining with ethidium bromide.
PCR amplicons were purified on Microcon columns (Millipore, Bedford, Mass.). Purified products were quantified by ethidium bromide staining with DNA standards as controls, and 50 ng was directly ligated into pCR4.0 plasmid (Invitrogen, Carlsbad, Calif.). Transformation of recombinant plasmids into Escherichia coli competent cells was performed as recommended by the manufacturer. Transformants were selected on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-ampicillin plates overnight, and 30 clones per serum sample were amplified by PCR with the M13 universal (−20) and M13 reverse primers. A total of 330 NS5A clones were generated. The sequencing reactions were based on the Sanger method, implemented with the Thermo Sequenase cycle sequencing kit (Visible Genetics Inc., Toronto, Ontario, Canada). The DNA templates were sequenced in the forward and reverse directions by using Cy5-labeled M13 universal (−20) and Cy5.5-labeled M13 reverse primers, respectively.
Genetic analysis of NS5A quasispecies diversity and evolution.
To calculate genetic distances, nucleotide and amino acid sequences were aligned using the CLUSTAL W program (45). Distances between pairs of nucleotide and amino acid sequences were calculated by using the DNADIST and PROTDIST modules in the PHYLIP package version 3.572. Calculation was based on a Kimura two-parameter distance matrix with a transition-transversion ratio of 4.0. The mean ± standard error of the mean (SEM) within-sample genetic distances were calculated at each time point. The numbers of synonymous and nonsynonymous substitutions per synonymous and nonsynonymous site, respectively, were calculated at each time point in each patient, with the Jukes-Cantor correction for multiple substitutions, by using the MEGA program. In addition, the mean ± SEM between-sample genetic distances were calculated on the basis of distances between pairs of pretreatment (month 0)-posttreatment (month 6) sequences, as were the numbers of synonymous and nonsynonymous substitutions per synonymous and nonsynonymous site. Statistical comparisons were made using a t test. The PHYLIP program version 3.572 was used to construct phylogenetic trees by means of the neighbor-joining method with a sequence matrix determined by Kimura's two-parameter method. Patient phylogenetic trees were constructed with both nucleotide and amino acid sequences.
NS5A protein sequence analysis.
All analyses were performed using our HCV database website facilities (HCVDB, http://hepatitis.ibcp.fr), which contain all previously reported HCV sequences, downloaded from the EMBL database. The NS5A sequence 2332 to 2141 of the HCV-J strain (EMBL accession number D90208) (19) was used to retrieve all reported isolates from the EMBL database by using the FASTA homology search program (34). A final set of 215 genotype 1b sequences were used to analyze the polymorphism of natural NS5A variants, as shown in Fig. 2A. Multiple sequence alignment was carried out with the CLUSTAL W program (45). The MPSA program was used to visualize sequence alignments and to plot the most frequently represented amino acid residues at each position (3). At each amino acid position, the residue types and their respective frequencies were computed with a program developed at the Institut de Biologie et Chimie des Protéines (C. Combet, unpublished data, August 2003).
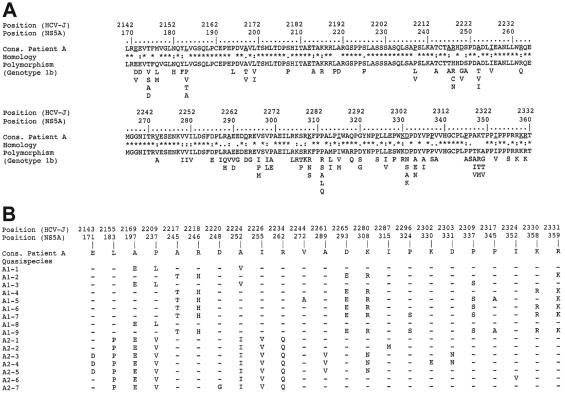
FIG. 2.
Natural variability of previously reported and patient A NS5A sequences in the transcriptional activation region. Amino acids are denoted by their single-letter code, and positions in the sequences are numbered according to both the HCV polyprotein and the NS5A protein in the prototype HCV genotype 1b HCV-J strain. (A) Polymorphism of previously reported NS5A sequences relative to patient A consensus sequence. “Cons.” represents the consensus amino acid sequence of patient A, derived from the alignment of this patient's 16 quasispecies variants with the CLUSTAL W program (45). Homology refers to the polymorphism observed in the 215 natural HCV genotype 1b NS5A variants recovered from EMBL and accessed through our database (HCVDB; http://hepatitis.ibcp.fr). Fully conserved, conserved, and similar residues at each position are symbolized by an asterisk, a colon, and a dot, respectively. The polymorphism of the 215 NS5A variants is presented as the repertoire of amino acids at each position, in decreasing order of observed frequency, from top to bottom (35). The least frequently observed residues (<1%) are not shown, because they could have been due to PCR and/or sequencing errors and/or sequencing of defective viruses. (B) Summary of patient A's quasispecies variant variability. Only the amino acid positions with one or several observed substitutions are shown. “Cons.” refers to patient A's consensus sequence. The sequences of the nine variants isolated at the start of follow-up are designated A1-i, where i varies from 1 to 9, while the seven sequences isolated at the end of follow-up are designated A2-j, where j varies from 1 to 7. Amino acids identical to those in the consensus sequence are represented by a hyphen.
Yeast plasmid constructs and transformation.
Each NS5A sequence generated was inserted into the pGBT9 yeast expression vector to generate a fused protein consisting of the NS5A transcriptional activation domain and the GAL4 DNA-binding domain (Clontech, Palo Alto, Calif.). For this purpose, the cloned NS5A sequences were amplified by PCR with sense primer ND6745-EcoRI (5′-GATGAATTCCCGGCGTGCAAACCTCTCCT-3′; nt 6731 to 6750) and antisense primer AS7296-BamHI (5′-CCGCTGGATCCCGTCCTYTTYCTCCGTGG-3′; nt 7282 to 7301) containing restriction sites for EcoRI and BamHI, respectively (underlined). The PCR products and the pGBT9 vector were digested by EcoRI and BamHI (New England Biolabs, Beverly, Mass.), the vector was dephosphorylated with calf intestinal phosphatase (Amersham Biosciences, Piscataway, N.J.), and DNA was purified by isopropanol precipitation. Ligation was performed overnight at 4°C, and the reaction was stopped by being heated for 20 min at 65°C. For plasmid amplification, XL-2 blue E. coli competent cells (Stratagene, La Jolla, Calif.) were transformed and grown overnight on ampicillin plates. Recombinants were screened by means of PCR with amplimer primers annealing to pGBT9 (Clontech). Positive plasmids were extracted with the Qiagen plasmid minikit (Qiagen Sciences, Valencia, Calif.) and were controlled by EcoRI/BamHI digestion and electrophoresis.
The yeast reporter was Saccharomyces cerevisiae strain Y187 (MATα gal4Δ gal80Δ trp1-901 leu2-3) (Clontech). Y187 contains an integrated lacZ reporter gene regulated by the wild-type GAL1 promoter. Transformation was performed by the lithium acetate method according to the protocol provided with the YeastMaker yeast transformation kit (Clontech). Plasmid pCL1 (Clontech), encoding the full-length wild-type GAL4, was used as a positive control. Transformants were grown on tryptophan-deficient (Trp−) synthetic dropout plates for 3 days at 30°C.
Measurement of NS5A transcriptional activation in vitro.
A quantitative luminescent β-galactosidase assay based on the Galacto Star kit (Tropix, Bedford, Mass.) was used to measure NS5A transcriptional activation in vitro. Well-individualized colonies were grown overnight in Trp− synthetic dropout medium for plasmid maintenance. Two milliliters of this culture was then inoculated into 8 ml of yeast extract-peptone-dextrose medium and grown at 30°C up to mid-log phase (optical density at 600 nm of 0.4 to 0.6), and 1.5 ml was used to prepare the cell extract: it was spun down and resuspended in 300 μl of the Z lysis buffer provided with the kit. One hundred microliters was subjected to two freeze-thaw cycles, and 20 μl was mixed with 300 μl of a reaction buffer containing the substrate. Light emission was measured after incubation for 20 min at room temperature. β-Galactosidase activity obtained with the pGBT9 vector alone was used as a negative control. The results were expressed as a ratio, by the use of β-galactosidase activity obtained with vector pGBT9 alone as the determinant. All assays were repeated twice, with three independent transformants for each construct. The reported results are the means ± SEMs of the six determinations performed for each construct.
Nucleotide sequence accession numbers.
Sequence data from this article have been deposited in GenBank under accession numbers AY379551 to AY379601.
RESULTS
NS5A has a quasispecies distribution in vivo.
We generated a total of 330 NS5A sequences spanning the full transcriptional activation domain of the NS5A protein (amino acid positions 169 to 260 of the protein, i.e., 2141 to 2332 in the HCV-J genotype 1b prototype sequence [19]). Phylogenetic analysis of all viral sequences generated in this study showed distinct clusters of viral sequences corresponding to each patient (data not shown), indicating the absence of PCR cross-contamination.
In each of the 11 samples, taken at various time points from the four patients, NS5A exhibited a typical quasispecies distribution, characterized by a mixture of genetically distinct but closely related sequences. The case of patient A is illustrated in Fig. 2. The NS5A variants differed from one another by a small number of residues (Fig. 2B) distributed throughout the protein sequence (underlined residues in Fig. 2A); no obvious hot spots of amino acid substitution were found. Interestingly, all the amino acid variations observed in the natural quasispecies variants from the four patients had already been described in the 215 previously reported HCV genotype 1b NS5A sequences available in HCVDB (Fig. 2A).
NS5A quasispecies undergo significant genetic evolution over time in vivo.
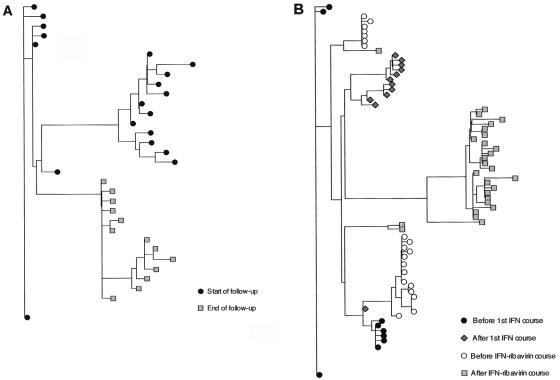
Significant genetic evolution of NS5A quasispecies over time was observed in both the IFN-α-treated and the untreated patients. Indeed, the average between-sample genetic distances (calculated by pairwise comparison of quasispecies nucleotide sequences isolated from two successive samples in a given patient) were always significantly higher than the average within-sample genetic distances (calculated by pairwise comparison of quasispecies sequences isolated from the first of these two samples) (Table 1). Phylogenetic analyses confirmed this finding, consistently showing distinctive clustering of NS5A nucleotide sequences according to the sampling time (examples of patients A and C are in Fig. 3).
TABLE 1.
Mean (± SEM) within-sample and between-sample genetic distances in the four patients
| Patient | Samplea | Mean (SEM) distance within sample | Samplesa | Mean (SEM) distance between samples | P |
|---|---|---|---|---|---|
| A | M0 | 0.0182 (0.0008) | M58 vs M0 | 0.0354 (0.0007) | <0.0001 |
| M58 | 0.0080 (0.0005) | ||||
| B | M0 | 0.0105 (0.0003) | M64 vs M0 | 0.0138 (0.0002) | <0.0001 |
| M64 | 0.0125 (0.0003) | ||||
| C | M0 | 0.0091 (0.0008) | M6 vs M0 | 0.0256 (0.0003) | <0.0001 |
| M6 | 0.0164 (0.0012) | M54 vs M6 | 0.0327 (0.0006) | <0.0001 | |
| M54 | 0.0236 (0.0028) | M69 vs M54 | 0.0336 (0.0006) | <0.0001 | |
| M69 | 0.0247 (0.0061) | ||||
| D | M0 | 0.0109 (0.0007) | M6 vs M0 | 0.0121 (0.0004) | <0.0001 |
| M6 | 0.0087 (0.0006) | M63 vs M6 | 0.0324 (0.0004) | <0.0001 | |
| M63 | 0.0097 (0.0005) |
M, month.
FIG. 3.
Examples of phylogenetic trees plotted with the quasispecies variant nucleotide sequences isolated at the various points of follow-up. (A) Phylogenetic tree plotted with patient A's quasispecies sequences isolated at the start and end of follow-up, respectively. (B) Phylogenetic tree plotted with patient C's quasispecies sequences isolated before and after IFN monotherapy and before and after IFN-ribavirin combination therapy.
NS5A quasispecies genetic evolution in vivo results in sporadic amino acid changes.
In all four patients, pairwise comparisons of NS5A quasispecies sequences isolated from two successive samples showed that the ratio of accumulation of nonsynonymous mutations per nonsynonymous site was significantly lower than the ratio of accumulation of synonymous mutations per synonymous site (data not shown). This was the case not only in the two untreated patients but also in the two patients treated with IFN-α and with the combination of IFN-α and ribavirin. These treatments induce profound metabolic alterations likely to exert strong evolutionary pressure on HCV proteins, like those previously observed in other genomic regions (33, 38). This finding suggests that, despite the continuous random accumulation of nucleotide mutations on the HCV genome during ongoing replication, the amino acid sequence of NS5A is subject to strong conservative constraints. As a result, only a few amino acid substitutions occurred during follow-up, at positions already known to tolerate some variability (Fig. 2). Examination of the physicochemical properties of the amino acids revealed that the variable positions were exclusively occupied by residues bearing the same hydrophobic or hydrophilic properties or by residues exhibiting close structural properties (e.g., Ser and Pro at positions 324 and 337 in patient A [Fig. 2B]). Calculation of hydrophobicity indices for the variant sequences, by using the Black and Mould hydrophobicity scale (2), indeed yielded very close values. In addition, secondary structure predictions by various methods did not reveal significant putative conformational differences among the variants (data not shown). These findings suggest, despite the observed amino acid variations, that the three-dimensional structure of NS5A protein is strictly conserved and that most of these variations should have little if any effect on local conformations and, thus, have at most a mild effect on NS5A functions. In contrast, the observed amino acid substitutions could affect the charge of the residues at hydrophilic positions (i.e., positions thought to be exposed at the surface of NS5A), such variations being more likely to affect NS5A interactions and functions, as shown by functional analyses (see below).
Different NS5A quasispecies variants have different transcriptional activation efficiencies.
In order to identify any functional consequences of HCV NS5A quasispecies variability, we used an in vitro yeast assay of transcriptional activation to compare the different NS5A variants isolated at each time point from the four patients. One NS5A clone was used for each of the 98 different NS5A sequences identified among the 330 clones generated. As described in Materials and Methods, the NS5A fragments were fused to the DNA-binding domain of GAL4 and the fusion proteins were transfected into S. cerevisiae reporter strain Y187, which contains an integrated lacZ reporter construct regulated by the wild-type GAL1 promoter. In vitro transcriptional activation was measured by means of a β-galactosidase luminescence assay.
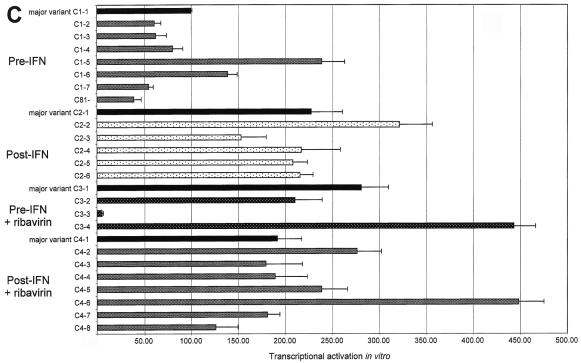
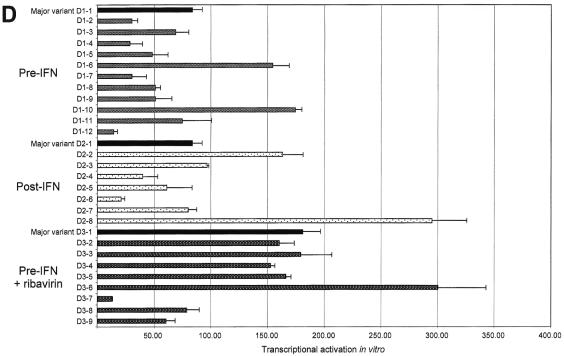
Transcriptional activation efficiency differed significantly among the four patients and between variant sequences isolated from a given patient both at the same time point and at different time points (Fig. 4). In each patient, transcriptional activation by the most frequent variant sequence isolated from the first serum sample was used as a reference (patient A, RA; patient B, RB; patient C, RC; and patient D, RD). Using RC as a reference, we defined transcriptional activation relative units (RU) as 1 RU = RC/100. Marked quantitative differences were found among variants isolated from the first sample of each patient (Fig. 4). Indeed, transcriptional activation efficiency ranged from 60 to 270 RU (median, 110) in patient A, 7 to 56 RU (median, 9) in patient B, 40 to 246 RU (median, 74) in patient C, and 14 to 180 RU (median, 53) in patient D (Fig. 4).
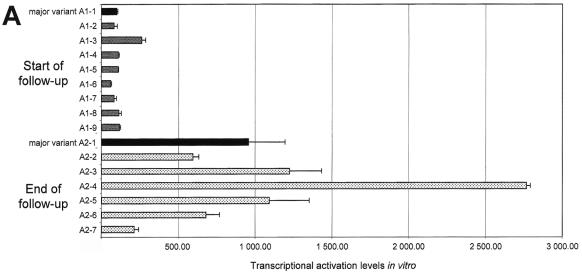
FIG. 4.
Transcriptional activation by HCV NS5A variants, as measured in an in vitro yeast assay. Panels A to D show data for patients A to D, respectively. The quantitative results are shown in RU, where 1 RU = RC/100 (RC is the transcriptional activation level obtained with the major variant in patient C's initial sample). At each time point, the most frequent quasispecies variant (major variant) is shown in black and is followed from top to bottom by the other variants isolated in the same quasispecies mixture. Each bar represents the mean ± standard deviation of three experiments performed in duplicate. Note that the same unit (RU) has been used in all four panels, but the scales of the panels differ due to different average levels of transcriptional activation in the four patients.
NS5A amino acid substitutions resulting from viral evolution in vivo had functional consequences, as the variants isolated at the different time points always had different transcriptional activation efficiencies (Fig. 4). Quantitative functional changes were particularly striking in patient A, in whom transcriptional activation ranged from 60 to 270 RU (median, 110) at the first time point and from 220 to 2,850 RU (median, 990) at the end of follow-up, 58 months later. This represented approximately a 10-fold increase in amplitude (Fig. 4A). In contrast, NS5A quasispecies evolution was not associated with marked functional changes in patient B (Fig. 4B). This was in keeping with slow genetic drift in this patient, as suggested by phylogenetic analysis. Transcriptional activation also changed over time in the two treated patients. No significant relationship was found between treatment outcome and NS5A transcriptional activation efficiency. In patient C, overall NS5A transcriptional activation was increased after the first course of IFN-α and thereafter (Fig. 4C). In patient D, transcriptional activation was stable during the first course of treatment and slightly increased (twofold on average) before the second treatment course (Fig. 4D). This patient cleared HCV RNA after IFN-α-ribavirin combination therapy.
NS5A transcriptional activation efficiency is dependent on the charge of constituent amino acid residues.
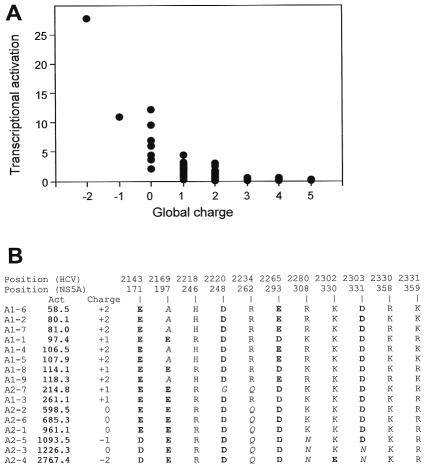
In order to understand how HCV NS5A quasispecies variability modulates NS5A transcriptional activation efficiency, we examined the characteristics of variant sequences associated with stronger or weaker transcriptional activation. The 98 distinct NS5A sequences were aligned in order of increasing activity. The characteristics of the amino acids were studied, especially at variable amino acid positions. Overall, the differences in transcriptional activation appeared to be mostly related to differences in charged amino acids. Indeed, changes from basic to neutral residues and, to a lesser extent, from neutral to acidic residues were always associated with increased transcriptional activation efficiency. A significant relationship was found between the number of basic and acidic residues in the sequence and the level of transcriptional activation in the model (Fig. 5A). Taking patient A as an example, the marked differences in transcriptional activation among the variants isolated at different time points were mainly related to a few mutations decreasing the globally basic or increasing the globally acidic nature of the sequences. Analysis of the impact of individual substitutions on transcriptional activation efficiency in this patient showed that each substitution had measurable consequences (Fig. 5B). For instance, the A197E substitution moderately increased the efficiency, whereas the R262Q substitution had a stronger effect. Activity was further increased when the K308N substitution was also present. Finally, the K330E substitution yielded a further increase.
FIG. 5.
Correlation between the global charge of NS5A region 2141 to 2332 (169 to 360) and in vitro transcriptional activation efficiency. The global charge of each NS5A sequence was calculated by attributing a “+1” charge for each basic residue (R, K, and H) and a “−1” charge for each acidic residue (D and E). (A) Plot of the global charge of all of the quasispecies variants isolated from the four patients versus their transcriptional activation efficiency. (B) Summary of charge variations in patient A's quasispecies. The sequences of the nine variants isolated at the start of follow-up are designated A1-i, where i varies from 1 to 9, while the seven sequences isolated at the end of follow-up are designated A2-j, where j varies from 1 to 7. The “Act.” column displays in vitro transcriptional activation efficiency in RU, where 1 RU = RC/100 (RC is the transcriptional activation level obtained with the major variant in patient C's initial sample). The variants are ordered by increasing transcriptional activation efficiency, from top to bottom. The global charge of the corresponding variant is shown in the “Charge” column. The composition in charged residues at basic and acidic positions is shown.
Although the global charge of the variant sequence appeared to be the main determinant of transcriptional activation efficiency, other features of individual residues were also sometimes involved. This is suggested for instance by the fact that the D331N substitution, which decreased the global acidic character of the relevant sequences, was present in those variant sequences with the highest transcriptional activation efficiency. Furthermore, sequences with the same overall pattern of charged residues exhibited slightly different activation efficiencies in vitro, which could have been due to substitutions placing uncharged residues at various positions. Pro-to-Ser replacements in the proline-rich region of the transcriptional activation domain appeared to play a significant role, highlighting the potential importance of polar Ser residues in transcriptional activation efficiency. For instance, the functional difference between variant sequences A1-6 and A1-7 (Fig. 2B) was related to a P324S substitution, which also appeared to be responsible for the difference between variant sequences A1-5 and A1-9 (Fig. 2B). A P337S substitution appeared to be involved in the difference between variant sequences A1-1 and A1-3 (Fig. 2B). Substitutions generating hydrophobic residues (such as I315M in variant sequences A2-1 and A2-2 and I352V in variant sequences A2-1 and A2-6) also appeared to play a moderate role, suggesting that minor local conformational changes may also modulate NS5A transcriptional activation. Interestingly, most of the substitutions found to have a functional influence that was unrelated to the charges of the residues were located within the proline-rich region, in keeping with an important role of this region in transcriptional activation.
DISCUSSION
This study shows that NS5A quasispecies undergo significant genetic evolution over time in vivo, as a result of random accumulation of mutations on the genome during replication. Most mutations that accumulate are silent. Some result in sporadic amino acid changes that appear to be tolerated at certain positions. All the mutated positions observed in quasispecies variants in this study were already identified as polymorphic positions in our database. This suggests that only these positions can tolerate amino acid changes, because they do not disrupt the global structure of the protein, in a context of strong conservative constraints on NS5A protein function(s).
A key finding in this study was that different NS5A variants from the same quasispecies mixture, or from quasispecies mixtures isolated from a given patient at different time points, induced different levels of transcriptional activation in vitro. These functional differences were principally dependent on the charge of the substituted residues and probably, to a lesser extent, on minor changes in the protein conformation (the latter could only be predicted, as the three-dimensional structure of NS5A is unknown). This implies that a nucleotide substitution randomly occurring during negative- or positive-stranded viral RNA synthesis can lead to the production of a viral protein with modified physicochemical and/or conformational properties and, possibly, enhanced or diminished functional activity. The random occurrence of such nucleotide substitutions during replication in a given cell can thus yield a new viral protein. This new protein may interact with one or several important cellular functions and subsequently induce profound changes in host cell metabolism and behavior. The fact that such events occur randomly in an individual cell and that the resulting variant does not need to propagate to other host cells in order to modify the metabolism of the initial cell points to cellular compartmentalization of virus-host interactions during HCV infection. In other words, HCV quasispecies variability leads to constant generation of new viral proteins with unique biological properties at the level of the individual cell.
It is conceivable that the random emergence and production of NS5A proteins with potent transcriptional activation properties in an infected cell could activate genes involved in the pathogenesis of HCV infection (37). Such genes might be involved in hepatocarcinogenesis (strong activation potentially leading to cell transformation), apoptosis (activation possibly prolonging or shortening the half-life of the infected cell), and/or IFN resistance. Our findings, based on an in vitro model of NS5A transcriptional activation, probably apply to many other HCV proteins and functions, as well as to other viruses with quasispecies behavior. Indeed, we recently showed that the efficiency of HCV internal ribosome entry site-driven translation in vitro and its sensitivity to the regulatory action of the X region of the HCV 3′ noncoding region vary widely among natural variants isolated from a given patient (Y. Michel, C. Malnou, M. Soler, N. Fanzone, A. M. Borman, J. M. Pawlotsky, and K. M. Kean, abstract from the 52nd Annual Meeting of the American Association for the Study of Liver Diseases, 2001, Hepatology 34:435A, 2001). In addition, Marcus et al. reported different levels of IFN induction by individual components of vesicular stomatitis virus quasispecies (26). Our results could therefore point to a general mechanism used by quasispecies-generating viruses to modulate their interaction with host cells. This would permit not only continuous selection of the fittest viruses but also adaptation and continuous selection of the “fittest” host cells in terms of chronic viral replication. This cellular compartmentalization of HCV-host cell interactions would lead to the continuous presence of multiple pools of infected cells, thereby guaranteeing ongoing virus production in a changing host environment. Microdissection experiments should throw further light on the cellular compartmentalization of HCV-host interactions.
We used the yeast model of NS5A transcriptional activation to study HCV quasispecies functional variability and compartmentalization. Transcriptional activators fall into several broad classes, including proline-rich, glutamine-rich, and acidic species (reviewed in reference 13). The NS5A C-terminal domain belongs to the latter category, and indirect arguments (listed in the introduction) suggest that NS5A transcriptional activation is functional in HCV-infected cells. Other viral proteins possessing transactivating properties important for the virus life cycle include HBV X protein, which activates a variety of viral and cellular promoters and enhancers via a protein-protein interaction and plays a role in HBV-induced hepatocarcinogenesis (reviewed in reference 15); papillomavirus E2 protein, which regulates viral transcription and DNA replication through interactions with cellular and viral proteins (14); and acidic transcriptional activators, such as herpes simplex virus VP16 and varicella-zoster virus open reading frame 10 proteins, which stimulate expression of viral immediate-early genes (1, 12, 17, 28, 29, 40). Given its global charge, it cannot be excluded that NS5A, after cleavage and migration to the nucleus, could directly bind to host DNA (5). However, NS5A does not bear known specific DNA-binding domains, and it is likely that cellular partners (such as transcription factors [11]) are involved in the interaction. We found here that negative charges (and potentially minor conformational changes) were responsible for the observed differences in NS5A transcriptional activation efficiency. This is in keeping with results obtained for other acidic transcriptional activators (12). It is possible that fusion with the GAL4 DNA-binding domain in the experimental model used here slightly altered the physiological properties of the intact protein and that the levels of activation in vitro did not perfectly reflect the situation in vivo, as already suggested for other activators (12).
The cellular genes putatively targeted by NS5A transcriptional activation during natural human infection are unknown. To date, the only gene that has been shown in vitro to be potentially activated by NS5A is that for interleukin 8; it has been postulated that such induction could play a role in HCV resistance to IFN (37). We studied four patients at various time points, in two cases before and after treatment with IFN and the IFN-ribavirin combination. No common pattern of change in transcriptional activation efficiency was found during unsuccessful treatment. Alternatively, whether NS5A transcriptional activation influences HCV replication efficiency deserves investigation. In this respect, it has been shown that HCV subgenomic replicons replicating in Huh7 cell lines select adaptive mutations that are frequently localized within the NS5A protein sequence (4, 23, 24). However, these adaptive mutations were recently suggested not to increase replication efficiency per se but to be specific for Huh7 cells (23). None of these amino acid substitutions were found in any of the four patients' quasispecies sequences, and the relationship between NS5A transcriptional activation and replication efficiency during natural human infection remains to be studied. Further studies are thus needed to identify the structural determinants and pathophysiological role of HCV NS5A transcriptional activation.
In conclusion, this study shows that accumulation of genomic mutations during HCV replication randomly generates variant viral proteins with quantitatively different functional properties in individual infected host cells. This points to cellular compartmentalization of virus-host interactions, a feature of quasispecies-distributed viruses that could have an important role in the life cycle and pathogenicity of HCV.
.
Acknowledgments
This work was supported by a grant from the Agence Nationale de Recherches sur le SIDA et l'Hépatite C (ANRS). M.P. is the recipient of a predoctoral grant from the Ligue contre le Cancer.
REFERENCES
- 1.Babb, R., C. C. Huang, D. J. Aufiero, and W. Herr. 2001. DNA recognition by the herpes simplex virus transactivator VP16: a novel DNA-binding structure. Mol. Cell. Biol. 21:4700-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, S. D., and D. R. Mould. 1991. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal. Biochem. 193:72-82. [DOI] [PubMed] [Google Scholar]
- 3.Blanchet, C., C. Combet, C. Geourjon, and G. Deleage. 2000. MPSA: integrated system for multiple protein sequence analysis with client-server capabilities. Bioinformatics 16:286-287. [DOI] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 6.Chang, J., S. H. Yang, Y. G. Cho, S. B. Hwang, Y. S. Hahn, and Y. C. Sung. 1998. Hepatitis C virus core from two different genotypes has an oncogenic potential but is not sufficient for transforming primary rat embryo fibroblasts in cooperation with the H-ras oncogene. J. Virol. 72:3060-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo, E., E. Baranowski, C. M. Ruiz-Jarabo, A. M. Martin-Hernandez, J. C. Saiz, and C. Escarmis. 1998. Quasispecies structure and persistence of RNA viruses. Emerg. Infect. Dis. 4:521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duarte, E. A., I. S. Novella, S. C. Weaver, E. Domingo, S. Wain-Hobson, D. K. Clarke, A. Moya, S. F. Elena, J. C. de la Torre, and J. J. Holland. 1994. RNA virus quasispecies: significance for viral disease and epidemiology. Infect. Agents Dis. 3:201-214. [PubMed] [Google Scholar]
- 9.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 10.Fukuma, T., N. Enomoto, F. Marumo, and C. Sato. 1998. Mutations in the interferon-sensitivity determining region of hepatitis C virus and transcriptional activity of the nonstructural region 5A protein. Hepatology 28:1147-1153. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh, A. K., M. Majumder, R. Steele, P. Yaciuk, J. Chrivia, R. Ray, and R. B. Ray. 2000. Hepatitis C virus NS5A protein modulates transcription through a novel cellular transcription factor SRCAP. J. Biol. Chem. 275:7184-7188. [DOI] [PubMed] [Google Scholar]
- 12.Grapes, M., and P. O'Hare. 2000. Differences in determinants required for complex formation and transactivation in related VP16 proteins. J. Virol. 74:10112-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, S. 1993. Structure and function of acidic transcription activators. Cell 72:481-483. [DOI] [PubMed] [Google Scholar]
- 14.Harris, S. F., and M. R. Botchan. 1999. Crystal structure of the human papillomavirus type 18 E2 activation domain. Science 284:1673-1677. [DOI] [PubMed] [Google Scholar]
- 15.Henkler, F. F., and R. Koshy. 1996. Hepatitis B virus transcriptional activators: mechanisms and possible role in oncogenesis. J. Viral Hepat. 3:109-121. [DOI] [PubMed] [Google Scholar]
- 16.Ide, Y., L. Zhang, M. Chen, G. Inchauspe, C. Bahl, Y. Sasaguri, and R. Padmanabhan. 1996. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene 182:203-211. [DOI] [PubMed] [Google Scholar]
- 17.Immaneni, A., P. Lawinger, Z. Zhao, W. Lu, L. Rastelli, J. H. Morris, and S. Majumder. 2000. REST-VP16 activates multiple neuronal differentiation genes in human NT2 cells. Nucleic Acids Res. 28:3403-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, D. Y., H. L. Wang, Y. Zhou, A. C. Chun, K. V. Kibler, Y. D. Hou, H. Kung, and K. T. Jeang. 2000. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 19:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, N., K. H. Lan, S. K. Ono-Nita, Y. Shiratori, and M. Omata. 1997. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J. Virol. 71:8856-8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan, K. H., M. L. Sheu, S. J. Hwang, S. H. Yen, S. Y. Chen, J. C. Wu, Y. J. Wang, N. Kato, M. Omata, F. Y. Chang, and S. D. Lee. 2002. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene 21:4801-4811. [DOI] [PubMed] [Google Scholar]
- 22.Lerat, H., M. Honda, M. R. Beard, K. Loesch, J. Sun, Y. Yang, M. Okuda, R. Gosert, S. Y. Xiao, S. A. Weinman, and S. M. Lemon. 2002. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology 122:352-365. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 26.Marcus, P. I., L. L. Rodriguez, and M. J. Sekellick. 1998. Interferon induction as a quasispecies marker of vesicular stomatitis virus populations. J. Virol. 72:542-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martell, M., J. I. Esteban, J. Quer, J. Genesca, A. Weiner, R. Esteban, J. Guardia, and J. Gomez. 1992. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J. Virol. 66:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matis, C., P. Chomez, J. Picard, and R. Rezsohazy. 2001. Differential and opposed transcriptional effects of protein fusions containing the VP16 activation domain. FEBS Lett. 499:92-96. [DOI] [PubMed] [Google Scholar]
- 29.Moriuchi, H., M. Moriuchi, and J. I. Cohen. 1995. Proteins and cis-acting elements associated with transactivation of the varicella-zoster virus (VZV) immediate-early gene 62 promoter by VZV open reading frame 10 protein. J. Virol. 69:4693-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moriya, K., H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, and K. Koike. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065-1067. [DOI] [PubMed] [Google Scholar]
- 31.National Institutes of Health. 2002. National Institutes of Health consensus development conference statement: management of hepatitis C: 2002—June 10-12, 2002. Hepatology 36:S3-S20. [DOI] [PubMed] [Google Scholar]
- 32.Pawlotsky, J. M. 2003. Hepatitis C virus genetic variability: pathogenic and clinical implications. Clin. Liver Dis. 7:45-66. [DOI] [PubMed] [Google Scholar]
- 33.Pawlotsky, J. M., G. Germanidis, P. O. Frainais, M. Bouvier, A. Soulier, M. Pellerin, and D. Dhumeaux. 1999. Evolution of the hepatitis C virus second envelope protein hypervariable region in chronically infected patients receiving alpha interferon therapy. J. Virol. 73:6490-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penin, F., C. Combet, G. Germanidis, P. O. Frainais, G. Deleage, and J. M. Pawlotsky. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polyak, S. J. 2003. Hepatitis C virus-cell interactions and their role in pathogenesis. Clin. Liver Dis. 7:67-88. [DOI] [PubMed] [Google Scholar]
- 37.Polyak, S. J., K. S. Khabar, D. M. Paschal, H. J. Ezelle, G. Duverlie, G. N. Barber, D. E. Levy, N. Mukaida, and D. R. Gretch. 2001. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J. Virol. 75:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polyak, S. J., S. McArdle, S. L. Liu, D. G. Sullivan, M. Chung, W. T. Hofgartner, R. L. Carithers, Jr., B. J. McMahon, J. I. Mullins, L. Corey, and D. R. Gretch. 1998. Evolution of hepatitis C virus quasispecies in hypervariable region 1 and the putative interferon sensitivity-determining region during interferon therapy and natural infection. J. Virol. 72:4288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray, R. B., L. M. Lagging, K. Meyer, and R. Ray. 1996. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J. Virol. 70:4438-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regier, J. L., F. Shen, and S. J. Triezenberg. 1993. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 90:883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakamuro, D., T. Furukawa, and T. Takegami. 1995. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J. Virol. 69:3893-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satoh, S., M. Hirota, T. Noguchi, M. Hijikata, H. Handa, and K. Shimotohno. 2000. Cleavage of hepatitis C virus nonstructural protein 5A by a caspase-like protease(s) in mammalian cells. Virology 270:476-487. [DOI] [PubMed] [Google Scholar]
- 43.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 44.Tanimoto, A., Y. Ide, N. Arima, Y. Sasaguri, and R. Padmanabhan. 1997. The amino terminal deletion mutants of hepatitis C virus nonstructural protein NS5A function as transcriptional activators in yeast. Biochem. Biophys. Res. Commun. 236:360-364. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiner, A. J., M. J. Brauer, J. Rosenblatt, K. H. Richman, J. Tung, K. Crawford, F. Bonino, G. Saracco, Q. L. Choo, and M. Houghton. 1991. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology 180:842-848. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, T., T. Hanada, T. Tokuhisa, K. Kosai, M. Sata, M. Kohara, and A. Yoshimura. 2002. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J. Exp. Med. 196:641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zemel, R., S. Gerechet, H. Greif, L. Bachmatove, Y. Birk, A. Golan-Goldhirsh, M. Kunin, Y. Berdichevsky, I. Benhar, and R. Tur-Kaspa. 2001. Cell transformation induced by hepatitis C virus NS3 serine protease. J. Viral Hepat. 8:96-102. [DOI] [PubMed] [Google Scholar]