Abstract
A safe and effective dengue vaccine is still not available. Passive immunization with monoclonal antibodies from humans or nonhuman primates represents an attractive alternative for the prevention of dengue virus infection. Fab monoclonal antibodies to dengue type 4 virus (DENV-4) were recovered by repertoire cloning of bone marrow mRNAs from an immune chimpanzee and analyzed for antigen binding specificity, VH and VL sequences, and neutralizing activity against DENV-4 in vitro. Fabs 5A7, 3C1, 3E4, and 7G4 were isolated from a library constructed from a chimpanzee following intrahepatic transfection with infectious DENV-4 RNA. Fabs 5H2 and 5D9, which had nearly identical VH sequences but varied in their VL sequences, were recovered from a library constructed from the same chimpanzee after superinfection with a mixture of DENV-1, DENV-2, and DENV-3. In radioimmunoprecipitation, Fab 5A7 precipitated only DENV-4 prM, and Fabs 3E4, 7G4, 5D9, and 5H2 precipitated DENV-4 E but little or no prM. Fab 3E4 and Fab 7G4 competed with each other for binding to DENV-4 in an enzyme-linked immunosorbent assay, as did Fab 3C1 and Fab 5A7. Fab 5H2 recognized an epitope on DENV-4 that was separate from the epitope(s) recognized by other Fabs. Both Fab 5H2 and Fab 5D9 neutralized DENV-4 efficiently with a titer of 0.24 to 0.58 μg/ml by plaque reduction neutralization test (PRNT), whereas DENV-4-neutralizing activity of other Fabs was low or not detected. Fab 5H2 was converted to full-length immunoglobulin G1 (IgG1) by combining it with human sequences. The humanized chimpanzee antibody IgG1 5H2 produced in CHO cells neutralized DENV-4 strains from different geographical origins at a similar 50% plaque reduction (PRNT50) titer of 0.03 to 0.05 μg/ml. The DENV-4 binding affinities were 0.42 nM for Fab 5H2 and 0.24 nM for full-length IgG1 5H2. Monoclonal antibody IgG1 5H2 may prove valuable for passive immunoprophylaxis against dengue virus in humans.
Among the arthropod-borne flaviviruses, the four dengue virus serotypes (dengue type 1 virus [DENV-1], DENV-2, DENV-3, and DENV-4) that constitute a serologically distinct subgroup are most important in terms of human morbidity and geographic distribution. Dengue viruses cause dengue outbreaks and major epidemics in most tropical and subtropical areas where Aedes albopictus and Aedes aegypti mosquitos are abundant. Dengue virus infection produces fever, rash, and joint pain in humans. A more severe and life-threatening form of dengue, characterized by hemorrhagic fever and hemorrhagic shock, has occurred with increasing frequency in Southeast Asia and Central and South America, where all four dengue virus serotypes circulate. The underlying cause of severe dengue remains controversial (23, 53). An association of severe dengue with increased viral replication has been reported recently (61). A safe and effective vaccine against dengue is currently not available.
The dengue virus contains a positive-strand RNA genome coding for a polyprotein that is cleaved co- and posttranslationally by a combination of cellular and viral proteases to generate the individual viral proteins (9, 19, 40). Dengue virus prM and E structural proteins and nonstructural NS1 protein are glycosylated. The prM glycoprotein is further cleaved by the cellular enzyme furin following viral assembly, generating M, which is present in the mature virus (58). Flavivirus prM and E form heterodimers, which are assembled into viral particles during infection (62). In this manner, the prM serves to protect the functional integrity of E from acid-induced conformational change (26, 31). The E glycoprotein is responsible for cell attachment, possibly mediated by a receptor, and for fusion with the cell membranes following viral entry.
Mouse monoclonal antibodies against the dengue viruses have been valuable for dengue virus serotype determination (20, 27). Studies in which monoclonal antibodies were used against dengue virus and other flaviviruses have also provided valuable information concerning the antigenic structure of the major viral antigen E (24, 25, 29, 39, 52). The three-dimensional structure of the E glycoprotein has been determined at 2-Å resolution for tick-borne encephalitis virus and recently for DENV-2 (45, 51). These studies showed that the monomeric E polypeptide is folded into three distinct domains and that the E glycoprotein consists of a flat, elongated dimer structure with an interdomain ligand-binding pocket.
Monoclonal antibodies reactive to flavivirus envelope proteins have been shown to mediate protection against homologous virus challenge in animal models (6, 22, 34, 35, 42). In most cases, protection by passive immunization has been correlated with the ability of these antibodies to neutralize the virus in vitro. Protection against dengue virus challenge was also demonstrated in mice following passive immunization with monoclonal or polyclonal antibodies specific to prM (7, 34) or NS1 (18, 28).
Most research efforts directed to the development of an attenuated live dengue vaccine have not yielded a satisfactory result. Recently, a clinical evaluation was conducted on a genetically engineered DENV-4 mutant containing a 30-nucleotide deletion in the 3′ noncoding region that exhibited reduced replicative capacity in simian cell culture and in primates (14, 44). Following a single-dose inoculation, a total of 20 volunteers remained afebrile and exhibited very few clinical signs of infection. Each of the vaccinees developed a high titer of DENV-4-neutralizing antibodies 4 to 6 weeks after immunization. However, five vaccinees showed an elevation of serum levels of the liver enzyme alanine transaminase (ALT). The ALT elevations were mostly transient and eventually subsided, but there remains a concern about the safety of a live dengue virus vaccine. Passive immunization with clinically acceptable dengue virus-neutralizing antibodies provides an attractive alternative for the prevention of dengue virus infection. Highly efficient neutralizing antibodies might also be useful for consideration as a possible therapy for severe dengue virus infection. Recently, a phage display of combinatorial antibody libraries has allowed for the isolation of antibodies against important viral pathogens from humans or nonhuman primates (8, 12, 41, 49, 56, 63). In the present study, we employed this technique to identify a panel of chimpanzee Fab antibodies against DENV-4. One of these Fab antibodies neutralized DENV-4 efficiently by an in vitro assay and was combined with human sequences to convert it to the whole immunoglobulin G1 (IgG1) antibody. The humanized chimpanzee IgG1 antibody produced in CHO cells neutralized DENV-4 efficiently.
MATERIALS AND METHODS
Preparation of serotypes DENV-1 to DENV-4.
Mosquito C6/36 cells were grown in minimum essential medium (MEM) supplemented with 10% fetal calf serum. Confluent C6/36 cells were infected with DENV-4 at 0.1 multiplicity of infection (MOI) in MEM containing 2% fetal calf serum and incubated at 28°C. The medium from the infected cells was harvested at 7 days and again at 10 days. It was clarified by centrifugation at 3,000 rpm in a JA10 rotor (1,000 × g) and was then centrifuged at 9,000 rpm in a JA10 rotor (15,000 × g) overnight. The DENV-4 pellet was resuspended in phosphate-buffered saline (PBS) for phage panning and for enzyme-linked immunosorbent assay (ELISA). In addition, DENV-4 grown in C6/36 cells in serum-free medium (VP-SFM; Gibco) was directly used for panning and for ELISA. DENV-1 (Western Pacific strain), DENV-2 (prototype New Guinea C strain) and DENV-3 (strain H87) were prepared in serum-free medium from infected simian Vero cells.
Inoculation of chimpanzees with infectious DENV-4 RNA and with DENV-1, DENV-2, and DENV-3.
Two dengue virus-seronegative chimpanzees, numbers 1616 and 1618, were intrahepatically inoculated with infectious RNA transcripts made from the full-length cDNA clone of DENV-4 strain 814669 (36). A blood sample was collected weekly from each animal for analysis of the serum ALT levels and for analysis of antibodies to DENV-4. Eleven weeks after DENV-4 RNA inoculation, bone marrow was aspirated from the iliac crest of each chimpanzee, and a combinatorial antibody library (designated library D4) was constructed. Nine and one-half months after inoculation with DENV-4 RNA, each of the chimpanzees was inoculated subcutaneously with a mixture of DENV-1, DENV-2, and DENV-3, each at 106 PFU, in 1 ml of MEM (Gibco) plus 0.25% human serum albumin. Six weeks after inoculation with the dengue virus mixture, serum samples were collected for analysis of antibody response. Twelve weeks after inoculation with DENV-1, DENV-2, and DENV-3, bone marrow was aspirated again, and a second antibody library (designated library D1-4) was constructed. Both libraries were prepared from bone marrow of chimpanzee number 1618, which developed slightly higher antibody titers against DENV-1, DENV-2, and DENV-3 than did chimpanzee number 1616.
Construction of γ1/κ chimpanzee Fab antibody libraries.
The lymphocytes from bone marrow were separated on a Ficoll-Paque gradient by centrifugation, and aliquots of approximately 107 cells/ml in MEM containing 10% dimethyl sulfoxide and 10% fetal calf serum were stored over liquid nitrogen. Total RNA was extracted from 3 × 107 lymphocytes with an RNA extraction kit (Stratagene), and mRNA was reverse transcribed by using oligo(dT) as a primer (ThermoScript RT-PCR system; Invitrogen). The κ light-chain DNA was amplified from the cDNA product by PCR with seven pairs of human κ light-chain family-specific 5′ primers and a 3′ primer in the constant domain (4, 21, 49, 56). The γ1 heavy-chain Fd cDNA was amplified by using nine human γ1 heavy-chain-family-specific 5′ primers plus a chimpanzee γ1-specific 3′ primer (21, 56). A 30-cycle PCR at 94°C for 15 s, 52°C for 50 s, and 68°C for 90 s was performed with AmpliTaq DNA polymerase (Perkin-Elmer).
Cloning of the chimpanzee κ light-chain and γ1 heavy-chain DNA fragments into the pComb 3H phage display vector was performed as described previously (4, 63). Briefly, amplified κ light-chain DNA fragments were pooled and digested with SacI and XbaI and then cloned into pComb 3H (49) by transformation of electrocompetent Escherichia coli XL-1 Blue (Stratagene). A plasmid containing the γ1 light-chain DNA inserts was prepared from E. coli transformants and then cleaved with SpeI and XhoI for insertion with amplified γ1 heavy-chain DNA fragments cleaved with the same enzymes. The plasmid containing both the heavy-chain and the light-chain DNA inserts was used for the transformation of E. coli XL-1 Blue by electroporation. In both electroporation steps, the ligated DNA mixture yielded a library size of 1 × 108 to 3 × 108 E. coli colonies.
Panning of phage library and isolation of DENV-4-specific soluble Fabs.
The construction of phage display libraries, recovery and transfer of Fab sequences, and identification of E. coli transformants expressing DENV-4-specific soluble Fabs were carried out as described previously (21, 56). Briefly, approximately 108 transformants were grown in 2YT broth containing 1% glucose, 10 μg of tetracycline/ml, and 100 μg of ampicillin/ml for 3 h at 37°C. The bacterial culture was then infected with helper phage VSC M13 (Stratagene) at 50 MOI to generate the phage library. The phage library D4 was panned by affinity binding on DENV-4 virions used to coat an ELISA plate that was blocked with 3% nonfat powdered milk in PBS to reduce nonspecific binding. The phage library D1-4 was panned by affinity binding on DENV-4 virions captured by a chimpanzee serum immobilized on an ELISA plate to minimize conformational changes of the DENV-4 antigenic structure. Following three cycles of panning, the selected phage mixture was used to infect E. coli XL-1 Blue, and replicative-form DNA (phagemid) was prepared. The phagemid was cleaved with NheI and SpeI and recircularized to remove the phage gene III portion of the fused Fab sequence. E. coli XL-1 Blue was transformed with the circularized DNA, and colonies that yielded soluble Fab fragments reactive to DENV-4 virus were screened by ELISA.
DNA sequencing of DENV-4-specific Fab clones.
Plasmid from the selected E. coli transformants was initially analyzed by BstN1 digestion to identify Fab clones with distinct patterns. Sequence analysis of the Fab VH and VL DNA segments was performed on an automated DNA sequencer with a fluorescence dideoxynucleotide terminator cycle sequencing kit with TaqDNA polymerase (Perkin-Elmer). The following primers were used: 5′ ACAGCTATCGCGATTGCAGTG (LC-1) and 5′ CACCTGATCCTCAGATGGCGG (LC-4) for sequencing the VL segment and 5′ ATTGCCTACGGCAGCCGCTGG (HC-1) and 5′ GGAAGTAGTCCTTGACCAGGC (HC-4) for sequencing both DNA strands of the VH segment (21, 56). Software Vector NTI (InforMax) was used for sequence analysis. The DNAPLOT software program (MRC Center for Protein Engineering) was used to search for human Ig homologues in the database.
Production and purification of Fab antibodies.
Selected E. coli colonies were grown in 1 liter of L broth containing 1% glucose and 100 μg of ampicillin/ml and 10 μg of tetracycline/ml to an early exponential growth phase (optical density at 600 nm [OD600] of approximately 0.2) at 30°C. The bacteria were then transferred to 2 liters of L broth containing 100 μg of ampicillin/ml and 10 μg of tetracycline/ml and grown at 30°C in the presence of 0.1 mM inducer IPTG (isopropyl-β-d-thiogalactopyranoside) for 5 h. The bacteria were pelleted and resuspended in 20 ml of extraction buffer containing 50 mM sodium phosphate, 10 mM Tris-HCl (pH 8.0), 100 mM NaCl (Clontech), and 0.1 mM protease inhibitor AEBSF [4-(2-aminoethyl)-benzene sulfonyl fluoride]. After three cycles of freezing and thawing to release the soluble Fab product from the bacterial periplasm, the preparation was clarified by centrifugation at 10,000 rpm in a JA20 rotor (10,000 × g) for 60 min. The histidine-tagged Fab in the supernatant was purified through a column containing a 1-ml bed volume of TALON metal affinity resin (Clontech) by the pH elution procedure as suggested by the manufacturer. The Fab purity was verified by polyacrylamide gel electrophoresis with purified human IgG F(ab′)2 (Cappel) as a marker. The Fab concentration was determined colorimetrically with a BCA protein assay kit (Pierce).
Biotinylation of purified Fab fragments and competition ELISA.
Purified Fabs were biotinylated with EZ-Link NHS-LC-biotin (Pierce) according to the procedure suggested by the supplier. After extensive dialysis against PBS, the biotin-labeled Fab was tested for binding to DENV-4 that was used to coat wells of a microtiter plate. For competition ELISA, a fixed concentration of biotinylated Fab was mixed with a competing Fab in serial dilution and the mixture was added to the DENV-4-coated wells. Streptavidin-alkaline phosphatase was used for the detection of biotinylated Fab bound to DENV-4.
Radiolabeling of DENV-4 antigens and radioimmunoprecipitation.
Infection with DENV-4 or recombinant vaccinia virus and subsequent radiolabeling of infected cells were performed as described previously (18). Confluent Vero cells in a T-25 flask were infected with DENV-4 strain 814669 at 1 MOI and incubated for 4 days at 37°C. Infected cells were rinsed once, starved for methionine in methionine-free MEM for 30 min, and then labeled with [35S]methionine at 150 μCi/ml (specific activity, 3,000 Ci/mM). After a 6-h labeling period, cells were rinsed with cold PBS and lysed in 2 ml of radioimmunoprecipitation assay (RIPA) buffer containing 1% sodium deoxycholate, 1% NP-40, 0.1% sodium dodecyl sulfate, 0.15 M NaCl, and 0.1 M Tris (pH 7.5). Confluent CV-1 cells were infected with 5 MOI of recombinant vaccinia virus DENV-4 (vDENV-4) PrM (7) or vDENV-4 E (43) containing the full-length PrM or E coding sequence, respectively, for 15 h at 37°C. Infected cells were rinsed and starved for methionine in methionine-free MEM, placed in the labeling medium for 2 h, and then lysed in RIPA buffer as described above. A 20-μl labeled lysate of DENV-4- or recombinant vaccinia virus-infected cells was mixed with 10 μl of the Fab fragment to be tested and 70 μl of RIPA buffer, incubated at 4°C overnight, and then mixed with 2 μl of goat anti-human IgG F(ab′)2 antibody for 2 h. A 10-μl suspension of protein A-Sepharose beads was added to bind the radioimmune complexes. The Sepharose beads were collected by centrifugation and washed three times with RIPA buffer prior to separation by sodium dodecyl sulfate-12% polyacrylamide gel (acrylamide/bisacrylamide ratio of 37.5:1) electrophoresis. Radiolabeled protein bands on the dried gel were visualized by exposure to an X-ray film.
Construction of DNA recombinants and expression of full-length IgG1 in Chinese hamster ovary (CHO) cells.
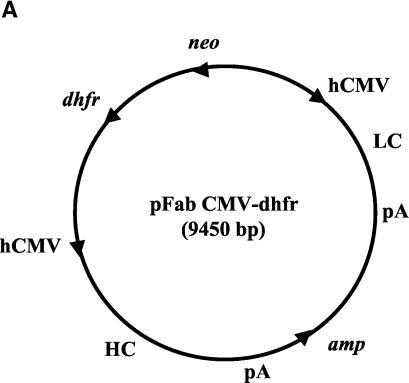
The expression vector pFab cytomegalovirus (CMV), kindly supplied by P. Sanna (Scripps Research Institute), was reengineered for IgG1 production (Fig. 1). The vector contained a neomycin phosphotransferase gene (neo), located between the two human CMV (hCMV) promoters, and a β-lactamase gene (amp), located between the two poly(A) sites as mapped by restriction digestion and by sequencing. The neo and amp locations differed from those of the published map (54). A dihydrofolate reductase (dhfr) gene together with the transcription signals was inserted at the unique NotI site in the original vector as the selecting marker and for gene amplification (64). The dhfr gene insert was the 1.4-kb DNA fragment from PvuII/AfeI cleavage of pCDHC68B, kindly provided by K. Deen (2). The original plasmid vector contained an A at the last nucleotide position of the intron that precedes the CH3 exon. This variant nucleotide was converted to G to allow proper RNA splicing for full-length IgG1 expression. The Fab 5H2 κ light-chain DNA segment cleaved by SacI and XbaI was first inserted into the expression vector. The resulting recombinant was then added with the γ1 heavy-chain DNA segment cleaved by XhoI and SpeI which was regenerated by PCR with the Fab 5H2 DNA template and appropriate primers. The chimpanzee-specific sequence in the hinge region together with the variant sequences introduced during plasmid construction were converted to the human hinge sequence by using positive-strand primer 5′ GACAAAACTCACACATGTCCACCGTGCCCA, which introduced a PciI site (underlined) with silent mutations (15, 59). Accordingly, the IgG1 antibody product would contain the chimpanzee VH and CH1 sequences and the entire human hinge CH2 and CH3 sequences.
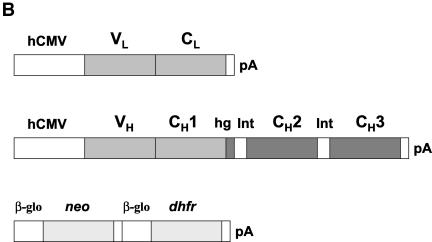
FIG. 1.
Map of pFab CMV dhfr vector for expression of full-length IgG1 in CHO cells and structure of the IgG1 light-chain and heavy-chain DNA inserts. (A) Locations of the various genes present in the expression vector. LC, light-chain DNA; pA, poly(A) addition signal; HC, heavy-chain DNA. The arrows indicate transcription direction. (B) Structure of the humanized IgG1 light-chain and heavy-chain genes under the control of an hCMV early promoter. VL and CL are the light-chain hypervariable region and constant region, respectively. VH, heavy-chain hypervariable region; CH1, constant region 1; hg, hinge; int-1, intron 1 (118 nucleotides); CH2, constant region 2; int-2, intron 2 (97 nucleotides); CH3, constant region 3. The dark-shaded regions are human IgG1 sequences, and the medium-shaded regions represent chimpanzee IgG1 sequences. The selectable neo and dhfr genes (light shaded) are flanked by a β-globin promoter (β-glo) and a poly(A) addition site (pA).
CHO dhfr− (duk−) cells were purchased from the American Type Culture Collection. The production of the whole IgG1 in CHO dhfr- cells was carried out by transfection with RsrII-cleaved recombinant plasmid in the presence of Lipofectamine (Gibco). Two days after transfection, cells in a T-25 flask were replated in Iscove's modified Dulbecco medium (Gibco) supplemented with 10% fetal bovine serum plus 10−7 M methotrexate in the absence of hypoxanthine-thymidine as selecting medium (13, 64). Transformed CHO cells resistant to 10−7 M methotrexate appeared approximately 2 weeks after transfection. Transformed CHO cells producing IgG1 in the medium were identified by ELISA and by plaque reduction neutralization test (PRNT) following subcloning in a 96- or 24-well plate. Gene amplification was carried out step-wise by increasing the methotrexate concentration to 2 × 10−7 M for the selecting medium. CHO cells that produced IgG1 at a high level were selected. The selected CHO cells were adapted to growth in suspension for IgG1 production in serum-free CD CHO medium (Gibco). Medium fluid was concentrated, and the IgG1 product was purified through a protein A affinity column. The full-length IgG1 5H2 antibody was compared with the Fab 5H2 fragment for DENV-4-binding affinity by ELISA. The equilibrium affinity constant (Kd) was calculated as the antibody concentration that gave 50% maximum binding (38, 50).
Determination of DENV-4-neutralizing activity of Fab and whole IgG1 antibodies.
Affinity-purified Fab or full-length IgG1 antibodies were analyzed for DENV-4-neutralizing activity by a modification of PRNT, as described previously (47). Briefly, approximately 50 focus-forming units of DENV-4 were mixed with a serial dilution of Fab or IgG1 antibodies in 250 μl of MEM. The mixture was incubated at 37°C for 30 min and then used for the infection of Vero cell monolayers in a 24-well plate. The cells were overlaid with a semisolid medium containing 1% tragacanth gum (Sigma) and incubated at 37°C for 4 days. Foci of DENV-4-infected cells were visualized following immunostaining with hyperimmune mouse ascites fluid (HMAF) and anti-mouse horseradish peroxidase conjugate (Pierce). The Fab or IgG1 concentration that produced 50% focus reduction was calculated. The neutralizing activity of the IgG1 antibody was tested against DENV-4 strain H241 isolated from the Philippines and two Caribbean DENV-4 isolates, i.e., strain 814669 and strain 341750.
RESULTS
Chimpanzee antibody response to intrahepatic transfection with DENV-4 RNA and to subsequent inoculation with a mixture of DENV-1, DENV-2, and DENV-3.
Two chimpanzees (numbers 1616 and 1618) were intrahepatically transfected with the full-length RNA transcripts of cloned DENV-4 cDNA (36). Four weeks after inoculation, these chimpanzees showed transient mild serum ALT elevations and became seropositive for DENV-4, indicating that both animals were infected (data not shown). At 9 weeks, the antibodies against DENV-4 reached a 50% plaque reduction (PRNT50) titer of 1/992 and 1/1,065, respectively. This level of neutralizing antibodies was comparable to that in rhesus monkeys infected with DENV-4 by a subcutaneous route (44). To increase the repertoire of dengue virus-specific antibodies, both chimpanzees were inoculated with a mixture of DENV-1, DENV-2, and DENV-3, each at 106 PFU/dose, 9.5 months after DENV-4 RNA transfection. Both chimpanzees developed moderate to high PRNT50 titers of antibodies against DENV-1, DENV-2, and DENV-3 (Table 1), indicating that the chimpanzees were infected with each of these viruses. Meanwhile, the PRNT50 antibody titer against DENV-4 increased approximately twofold following infection with DENV-1, DENV-2, and DENV-3.
TABLE 1.
Serum neutralizing antibody titers of chimpanzeesa
| Chimpanzee | Infection with DENV-1 to DENV-3 | Neutralizing antibody titer against
|
|||
|---|---|---|---|---|---|
| DENV-4 | DENV-1 | DENV-2 | DENV-3 | ||
| 1616 | Preinfection | 1,031 | <10 | 34 | 80 |
| Postinfection | 2,380 | 327 | 880 | 610 | |
| 1618 | Preinfection | 1,110 | 23 | 69 | 156 |
| Postinfection | 1,654 | 730 | 1,787 | 1,271 | |
Chimpanzees were previously inoculated with DENV-4 RNA intrahepatically and then infected with a mixture of DENV-1, DENV-2, and DENV-3 9 months later. Chimpanzees were infected with a mixture of DENV-1, DENV-2, and DENV-3 at a dose of 106 PFU for each virus. The neutralizing antibody titer was the reciprocal of the serum dilution that yielded a 50% plaque reduction.
Chimpanzee γ1/κ combinatorial Fab antibody libraries.
Two phagemid libraries were constructed from bone marrow mRNA of chimpanzee number 1618 as follows: (i) library D4 was prepared from the chimpanzee after intrahepatic inoculation with DENV-4 RNA and (ii) library D1-4 was prepared from the same animal after infection with a mixture of the other three dengue serotype viruses. Phage library D4 was panned for three successive rounds against DENV-4 virions immobilized directly on an ELISA plate. After the third panning cycle, plasmid was isolated and cleaved with SpeI and NheI for the expression of soluble Fabs. Library D1-4 was panned for three successive rounds against DENV-4 virions captured by chimpanzee antibodies which were used to coat an ELISA plate. In this manner, possible conformational distortions of the DENV-4 virion surface due to direct coating on a solid phase might be minimized. Similarly, after the third panning, plasmid was isolated and cleaved with SpeI and NheI for the expression of soluble Fabs.
Identification and characterization of chimpanzee Fabs specific to DENV-4.
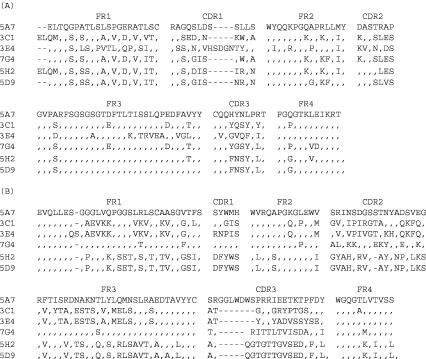
E. coli transformants were screened for the production of soluble Fabs capable of binding to DENV-4. Plasmid containing the Fab insert was analyzed by digestion with BstNI in order to select distinct clones. Sequence analysis of the VH and VL DNA inserts identified Fabs 5A7, 3C1, 3E4, and 7G4 in library D4. Fabs 5H2 and 5D9, which varied in the VL sequences but had nearly identical VH sequences (a single amino acid difference in the FR3 region), were recovered from library D1-4 (Fig. 2). The sequences in the heavy-chain complementarity-determining region 3 (CDR3) (65), critical for antigen binding, showed a greater diversity than the sequences in other regions among these Fabs. A sequence similarity search of the available human Ig genes was conducted to determine the specific germ line origin of these chimpanzee Fab fragments. The chimpanzee VH and VL sequences and their most related human Ig genes of the germ line VH or Vκ families are shown (Table 2). These chimpanzee VH or VL sequences and their human homologues had 88 to 95% identity, excluding the CDR3 region.
FIG. 2.
Alignment of amino acid sequences among DENV-4-specific and cross-reactive Fab monoclonal antibodies. The amino acid sequences of the six chimpanzee Fab monoclonal antibodies recovered by repertoire cloning were compared. (A) Sequences of VL light-chain segments; (B) VH heavy-chain segments. The framework regions (FR1 to FR4) and complementarity-determining regions (CDR1 to CDR3) are shown. A dash indicates where an amino acid deletion occurred, and an identical amino acid is represented by a comma.
TABLE 2.
Sequence similarities between chimpanzee Fab antibodies and their most related human germ line Ig genesa
| Chimpanzee Fab | VH homologue
|
VL homologue
|
||||
|---|---|---|---|---|---|---|
| Family (gene) | % Identity | Reference cited | Family (gene) | % Identity | Reference cited | |
| 5A7 | VH3 (COS-6) | 95 | 60 | Vκ3 (DPK-23) | 90 | 11 |
| 3C1 | VH1 (DP-10) | 88 | 60 | Vκ1 (L12a) | 92 | 32 |
| 3E4 | VH1 (DP-10) | 88 | 60 | Vκ2 (DPK-8) | 88 | 11 |
| 7G4 | VH3 (DP-54) | 92 | 60 | Vκ1 (L12a) | 95 | 32 |
| 5H2 | VH4 (DP-71) | 89 | 60 | Vκ1 (Va) | 94 | 48 |
| 5D9 | VH4 (DP-71) | 88 | 60 | Vκ1 (Va) | 93 | 48 |
The DNAPLOT program was used to search for the most homologous sequence of human IgG molecules in the database. The percent identity in the VH or VL region excluding CDR3 is indicated.
Antigenic specificity of chimpanzee Fab monoclonal antibodies.
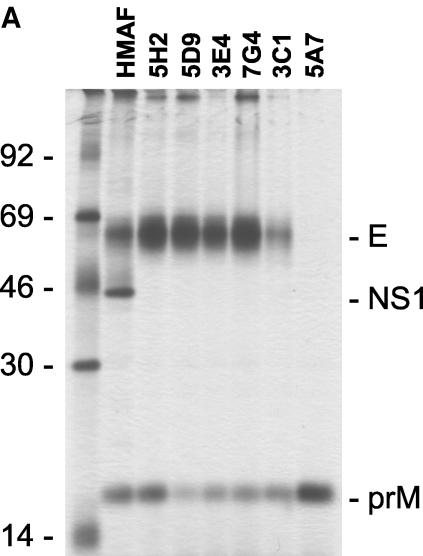
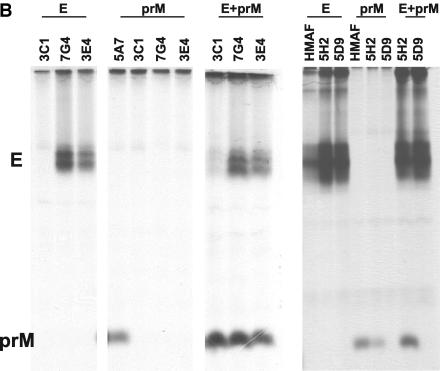
First, the binding activity of the Fab antibodies to DENV-4 was analyzed by ELISA. All six selected Fabs showed strong binding to DENV-4 virions (Table 3). Chimpanzee Fab 1F2, which was selected from library D4 for its ability to bind anti-human F(ab′)2 but not DENV-4, was used as the control. A cross-reactivity to DENV-1, DENV-2, or DENV-3 was detected for Fabs 3E4, 7G4, and 5A7. Fab 3C1 also cross-reacted with DENV-2 at a low titer. Fabs 5H2 and 5D9 showed no detectible cross-reactivity to DENV-1, DENV-2, or DENV-3. Radioimmunoprecipitation with a lysate of DENV-4-infected Vero cells was then performed to determine the antigen-binding specificity (Fig. 3A), Fab 5A7 selectively precipitated prM. All other Fabs precipitated both E and prM. The amount of prM relative to the E precipitated varied, depending on the Fab. Radioimmunoprecipitation was again performed by using labeled E or prM prepared individually in recombinant vaccinia virus-infected cells (Fig. 3B). Fabs 3E4 and 7G4 precipitated E but not prM. Fab 3C1 precipitated neither E nor prM. Fab 5D9 precipitated E but not prM, whereas Fab 5H2 precipitated E and a trace of prM. When the labeled antigens were mixed, coprecipitation of prM and E was again detected for Fabs 3E4, 7G4, 3C1, and 5H2.
TABLE 3.
Binding activities of Fab monoclonal antibodies to DENV-4 and other dengue virus serotypes as determined by ELISAa
| Fab | ELISA titer of Fab binding to
|
|||
|---|---|---|---|---|
| DENV-4 | DENV-1 | DENV-2 | DENV-3 | |
| 5A7 | 3.41 | 2.51 | 2.51 | 2.51 |
| 3C1 | 3.71 | 1.30 | 3.11 | 1.30 |
| 3E4 | 4.61 | 4.61 | 4.31 | 4.61 |
| 7G4 | 4.01 | 4.01 | 4.01 | 4.01 |
| 5D9 | 3.41 | <1.0 | <1.0 | <1.0 |
| 5H2 | 4.01 | <1.0 | <1.0 | <1.0 |
| 1F2b | 1.30 | <1.0 | <1.0 | <1.0 |
Microtiter plates were coated with DENV-1, DENV-2, DENV-3, or DENV-4 virions. The starting amount of each Fab in ELISA was approximately 300 μg/ml. Data are presented as log10 of the reciprocal dilution that gave an OD reading of twofold greater or higher than the background.
Chimpanzee Fab from library D4 was used as the negative control for binding to DENV-4 and other dengue virus serotypes.
FIG. 3.
Analysis of antigenic specificity by radioimmunoprecipitation. (A) [35S]methionine-labeled lysates of DENV-4-infected Vero cells were precipitated with the various Fab preparations indicated. (B) [35S]methionine-labeled lysates were prepared from CV-1 cells infected with vaccinia virus recombinant containing the full-length coding sequence of vDEN-4 prM or E, respectively. E+prM, precipitations with a mixture of both lysates; HMAF, precipitation using HMAF raised against DENV-4.
Mapping Fab antibody-binding sites on DENV-4 virions by competition ELISA.
Biotinylated Fabs 3C1, 3E4, 7G4, and 5H2 were each tested for binding to DENV-4 in the presence of an unlabeled, competing Fab. Chimpanzee Fab 1F2, which did not bind DENV-4, was analyzed in parallel. Fab 5D9, which was nearly identical to Fab 5H2, was not tested. The result (Fig. 4) showed that the binding of Fab 3C1 to DENV-4 was competed for with Fab 5A7, but not with Fabs 3E4, 7G4, 5H2, or 1F2. Thus, the binding site on PrM for Fab 3C1 overlapped with that for Fab 5A7. Fab 3E4 and Fab 7G4 also competed with each other for binding to DENV-4, indicating that their binding sites on E overlapped. The binding site on E for Fab 5H2 was unique, as binding competition with other Fabs was not observed.
FIG. 4.
Epitope analysis of chimpanzee Fab antibodies against DENV-4 by competition ELISA. Selected Fabs were affinity purified, biotinylated, and used for the analysis of binding reactivity to DENV-4 virions by competition ELISA in the presence of competing, unlabeled Fabs. (A) Biotinylated Fab 3C1; (B) biotinylated Fab 3E4; (C) biotinylated Fab 7G4; (D) biotinylated Fab 5H2. Chimpanzee Fab 1F2, which did not bind to DENV-4, was used as a negative control. The numbers on the y axis are OD readings, and the x coordinate represents reciprocal dilutions of the competing Fabs.
DENV-4 and cross-serotype neutralizing activity of Fab antibodies.
Affinity-purified Fabs were used for PRNT50 determination (Table 4). Similar to the Fab 1F2 control, prM-specific Fab 5A7 or Fab 3C1 did not neutralize DENV-4. Fabs 3E4 and 7G4 exhibited a low DENV-4-neutralizing activity with a PRNT50 titer at 91 μg/ml or greater. Fab 3E4, which was most cross-reactive to DENV-1, DENV-2, or DENV-3, was used in a cross-serotype neutralization assay. The cross-neutralizing activity against DENV-1, DENV-2, or DENV-3 was lower than that detected for DENV-4 (data not shown). Importantly, Fab 5H2 and Fab 5D9 neutralized DENV-4 efficiently, with a PRNT50 titer of 0.24 and 0.58 μg/ml, respectively.
TABLE 4.
DENV-4-neutralizing titer of chimpanzee Fab antibodiesa
| Fab | Phage library | PRNT50 titer (μg/ml) |
|---|---|---|
| 5A7 | D4 | >200 |
| 3C1 | D4 | >200 |
| 7G4 | D4 | 121 |
| 3E4 | D4 | 91 |
| 5D9 | D1-4 | 0.58 |
| 5H2 | D1-4 | 0.24 |
| 1F2b | D4 | >200 |
Affinity-purified chimpanzee Fabs were tested for DENV-4 neutralization by PRNT, and the PRNT50 titer was calculated.
Chimpanzee Fab that did not bind to DENV-4.
Humanized chimpanzee full-length IgG1 antibodies produced in CHO cells.
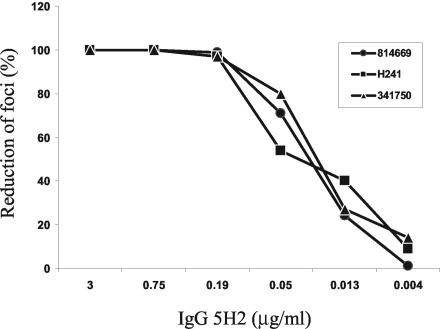
Production of full-length antibodies from the Fab γ1/κ sequences was achieved with expression vector pFab CMV dhfr, which provides a portion of the hinge and the entire CH2 and CH3 sequences of the human γ1 heavy chain (Fig. 1). A dhfr gene was inserted into the vector for the selection of antibody-producing CHO cells with methotrexate and for gene copy amplification. Other modifications of the expression vector included a conversion of the chimpanzee-specific hinge sequence to the human counterpart and an A-to-G substitution at the last nucleotide of the intron between the CH2 and CH3 exons of the heavy-chain sequence (see Materials and Methods). Thus, the product was a full-length, chimeric human-chimpanzee (humanized) IgG1 antibody. Fab 5H2 was chosen for conversion to the whole IgG1 antibody. The full-length IgG1 5H2 was secreted into the culture medium of the transformed CHO cells, and the yield of the affinity-purified product was approximately 1.8 mg per liter. Affinity-purified IgG1 5H2 was compared with Fab 5H2 for binding affinity to DENV-4 by ELISA. The IgG1 5H2 and Fab 5H2 had Kds of 0.24 nM and 0.42 nM, respectively. IgG1 5H2 neutralized three DENV-4 strains from two geographic regions in vitro at a similar high PRNT50 titer of 0.03 to 0.05 μg/ml (Fig. 5). Humanized IgG1 5H2 represents the first DENV-4-neutralizing monoclonal antibody of primate origin.
FIG. 5.
In vitro neutralization of DENV-4 strains by humanized chimpanzee antibody IgG1 5H2. Full-length antibody IgG1 5H2 was concentrated from the culture medium of transformed CHO cells selected with 2 × 10−7 M methotrexate and then affinity purified through a protein A column. The neutralizing activity of the antibody preparation was tested by PRNT against DENV-4 strain H241, isolated in the Philippines, and DENV-4 814669 and DENV-4 341750, isolated in the Caribbean.
DISCUSSION
The last few decades have seen the isolation and characterization of a large number of murine monoclonal antibodies against the four dengue virus types and other arthropod-borne flaviviruses. The clinical utility of these murine monoclonal antibodies is limited because of their propensity to induce an antibody response in humans. To develop a strategy of passive immunization against dengue, we turned to antibodies from chimpanzees, which are closely related to humans and can be experimentally infected with dengue viruses. The present study represents the first successful recovery of chimpanzee Fab monoclonal antibodies against the dengue virus by combinatorial cloning.
An analysis of the series of Fab antibodies against DENV-4 recovered by combinatorial cloning suggested a pattern of chimpanzee antibody response to intrahepatic infection with the infectious DENV-4 RNA transcripts. As in dengue virus infection of mice, both PrM-specific (Fab 5A7) and E-specific (Fabs 3E4 and 7G4) antibodies were identified in the chimpanzee. Interestingly, both Fab 3E4 and Fab 7G4 antibodies coprecipitated prM and E when the two antigens were mixed. Fab 3C1 also coprecipitated prM and E but did not precipitate either antigen when they were present individually. These results suggest that these Fabs recognized either PrM or E in the prM-E heterodimer. These chimpanzee Fab antibodies may be useful for the analysis of PrM-E interactions and the antigenic structure of dengue virus. Nevertheless, their DENV-4-neutralizing activity was low or not detected, and they are not likely to be effective against the virus.
Our goal of recovering antibodies highly efficient for neutralizing DENV-4 was achieved by repertoire cloning of chimpanzee bone marrow following multiple dengue virus infections. In this case, DENV-4 virions captured by polyclonal antibodies immobilized on plates were used for phage panning. The panning modification might better preserve the native conformation of the DENV-4 antigenic structure. This experiment yielded Fabs 5H2 and 5D9 that neutralized DENV-4 efficiently at a PRNT50 titer in the range of 0.2 to 0.6 μg/ml. Both Fabs had nearly identical VH sequences but varied in the CDR1 and CDR2 and other regions of their VL sequences. These differences in the VL and VH sequences might explain the observation that Fab 5H2 coprecipitated E and prM, whereas Fab 5D9 precipitated only E. Importantly, both Fabs neutralized DENV-4 efficiently at a PRNT50 titer in the range of that of human Fab antibodies against the respiratory syncytial virus (12), Ebola virus (41), or human immunodeficiency virus (5) selected by phage display.
Fab antibody fragments have a high clearance rate in humans and, therefore, are not directly useful clinically. The conversion of the Fab fragments to their whole IgG1 antibody molecules was achieved by using expression vector pFab CMV dhfr, which provided a portion of the hinge and the complete CH2 and CH3 heavy-chain sequences from a human germ line DNA segment. A dihydrofolate reductase gene was inserted in the expression vector to increase the IgG1 antibody production. The chimpanzee-specific sequence found in the hinge region was also converted to the human sequence. The humanized IgG1 5H2 had at least equal binding affinity for DENV-4, if not higher than that of Fab 5H2, as measured by equilibrium affinity constants. Importantly, the humanized antibody IgG1 5H2 exhibited a PRNT50 titer of 0.03 to 0.05 μg/ml, which is approximately eightfold more efficient than that of the Fab fragment against DENV-4.
Polyclonal antibody preparations against Caribbean DENV-4 isolates, including strain 814669, have been shown to neutralize DENV-4 strain H241 isolated from the Philippines less efficiently than the homologous DENV-4 strains, suggesting that there are antigenic variations among DENV-4 strains with different geographical origins (30). Sequence analysis also indicates that there is a significant genetic variation among DENV-4 isolates from different geographic regions (37). Thus, it is significant that IgG1 5H2 was able to neutralize geographically diverse DENV-4 isolates at a similar high titer. The DENV-4-neutralizing activity of IgG1 5H2 was approximately 6- to 10-fold higher than the IgG monoclonal antibody against the Ebola virus (41) and 40- to 60-fold higher than the humanized mouse antibody (MEDI-493) against respiratory syncytial virus (33). The CHO cell line obtained in this study produced the humanized chimpanzee antibody IgG1 5H2 at approximately 1.8 mg per liter. An increased production of this antibody in other mammalian cell systems should be possible.
A computer search revealed that the amino acid sequences of VH and VL segments of these Fab antibodies had a strong homology, ranging from 88 to 95%, with the sequences of their human Ig homologues. In particular, the Fab 5H2 γ1 heavy-chain and κ light-chain sequences had 89 and 94% sequence similarity to the human germ line IgG gene homologues, excluding the CD3 region (48, 60). Furthermore, in the CH1 or CL region, there was only one amino acid (or approximately 1.0%) difference between chimpanzee and human sequences (data not shown) (16, 59). The high level of antibody sequence similarity and a number of other observations addressing this issue suggest the possibility that chimpanzee antibodies may be administered directly to humans without further modifications to humanize these reagents (16, 17). Experimental data available indicate that little immunogenicity is seen when components of human antibodies are introduced into chimpanzees (46).
The cause of severe dengue virus infection sometimes associated with secondary dengue virus infection remains controversial. According to one hypothesis, in a secondary infection, dengue virus forms a complex with a subneutralizing level of cross-reactive antibodies produced during the primary infection, leading to an enhanced uptake and replication in susceptible mononuclear cells via their Fc receptors (23). Several classes of FcR receptors have been identified on the cell surface, and their interacting amino acids in the respective IgG have been carefully mapped (1, 10). It is now possible to eliminate the FcR receptor binding sequences in the antibody molecules and to test their activity for enhancing dengue virus replication in vitro (3, 55, 57). Humanized chimpanzee IgG1 monoclonal antibodies lacking the FcR1 binding sequences should permit a critical test of the hypothesis. For clinical application, it is important that these humanized neutralizing antibodies do not enhance dengue virus replication in human monocytes or other FcR receptor-bearing cells. As specific treatment for severe dengue virus infection is still not available, an early intervention during the viremic phase by passive transfer with highly efficient neutralizing antibodies may prove beneficial.
Since there are four dengue virus serotypes, monoclonal antibodies against each of the remaining three dengue virus serotypes will be required for the effective prevention of dengue virus infection with this approach. It should be similarly possible to identify Fab antibodies from the infected chimpanzees that efficiently neutralize each of the other three dengue virus serotypes. Accordingly, a panel of humanized chimpanzee monoclonal antibodies that most efficiently neutralize these dengue viruses will be prepared and evaluated for protection against dengue virus infection in animal models and ultimately of humans.
Acknowledgments
We thank Max Shapiro and other members of Bioqual (Rockville, Md.) for providing animal care. We also thank Ron Engle for performing the serological assays, Lynn Rasmussen for nucleotide sequencing, and Chris Kemp for full-length antibody production and purification.
REFERENCES
- 1.Allen, J. M., and B. Seed. 1989. Isolation and expression of functional high-affinity Fc receptor complementary DNAs. Science 243:378-381. [DOI] [PubMed] [Google Scholar]
- 2.Ames, R. S., M. A. Tornetta, K. Deen, C. S. Jones, A. M. Swift, and S. Ganguly. 1995. Conversion of murine Fabs isolated from a combinatorial phage display library to full length immunoglobulins. J. Immunol. Methods 184:177-180. [DOI] [PubMed] [Google Scholar]
- 3.Armour, K. L., M. R. Clark, A. G. Hadley, and L. M. Williamson. 1999. Recombinant human IgG molecules lacking Fcγ receptor I binding and monocyte triggering activities. Eur. J. Immunol. 29:2613-2624. [DOI] [PubMed] [Google Scholar]
- 4.Barbas, C. F., A. S. Kang, R. A. Lerner, and S. J. Benkovic. 1991. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. USA 88:7978-7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbas, C. F., D. Hu, N. Dunlop, L. Sawyer, D. Cababa, R. M. Hendry, P. L. Nara, and D. R. Burton. 1994. In vitro evolution of a neutralizing human antibody to human immunodeficiency virus type 1 to enhance affinity and broaden strain cross reactivity. Proc. Natl. Acad. Sci. USA 91:3809-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandriss, M. W., J. J. Schlesinger, E. E. Walsh, and M. Briselli. 1986. Lethal 17D yellow fever encephalitis in mice: Passive protection by monoclonal antibodies to the envelope proteins of 17D yellow fever and dengue 2 viruses. J. Gen. Virol. 67:229-234. [DOI] [PubMed] [Google Scholar]
- 7.Bray, M., and C. J. Lai. 1991. Dengue virus premembrane and membrane proteins elicit a protective immune response. Virology 185:505-508. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., J. Pyati, R. Kudori, S. J. Sharp, G. B. Thomton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garraty, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas. 1994. Efficient neutralization of primary isolates of HIV by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 9.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 10.Chappel, M. S., D. E. Isenman, M. Everett, Y.-Y. Xu, K. J. Dorrington, and M. H. Klein. 1991. Identification of the Fcγ receptor class I binding site in human IgG through the use of recombinant IgG1/IgG2 hybrid and point-mutated antibodies. Proc. Natl. Acad. Sci. USA 88:9036-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, J. P., I. M. Tomlinson, and G. Winter. 1994. A directory of human germ-line V kappa segments reveals a strong bias in their usage. Eur. J. Immunol. 24:827-836. [DOI] [PubMed] [Google Scholar]
- 12.Crowe, J. E., Jr., B. R. Murphy, R. M. Chanock, R. A. Williamson, C. F. Barbas, and D. R. Burton. 1994. Recombinant human respiratory syncytial virus (RSV) monoclonal antibody Fab is effective therapeutically when introduced directly into the lungs of RSV-infected mice. Proc. Natl. Acad. Sci. USA 91:1386-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorai, H., and G. P. Moore. 1987. The effect of dihydrofolate reductase-mediated gene amplification on the expression of transfected immunoglobulin genes. J. Immunol. 139:4232-4241. [PubMed] [Google Scholar]
- 14.Durbin, A. P., R. A. Karron, W. Sun, D. W. Vaughn, M. J. Reynolds, J. R. Perreault, B. Thumar, R. Men, C. J. Lai, W. R. Elkins, R. M. Chanock, B. R. Murphy, and S. S. Whitehead. 2001. Attenuation and immunogenicity in humans of a live dengue virus type 4 vaccine candidate with a 30 nucleotide deletion in the 3′ untranslated region. Am. J. Trop. Med. Hyg. 65:405-413. [DOI] [PubMed] [Google Scholar]
- 15.Ehrlich, P. H., Z. A. Moustafa, and L. Oestberg. 1991. Nucleotide sequence of chimpanzee Fc and hinge regions. Mol. Immunol. 28:319-322. [DOI] [PubMed] [Google Scholar]
- 16.Ehrlich, P. H., Z. A. Moustafa, K. E. Harfeldt, C. Isaacson, and L. Ostberg. 1990. Potential of primate monoclonal antibodies to substitute for human antibodies: nucleotide sequence of chimpanzee Fab fragments. Hum. Antib. Hybrid. 1:23-26. [PubMed] [Google Scholar]
- 17.Ehrlich, P. H., Z. A. Moustafa, J. C. Justice, K. E. Harfeldt, and L. Ostberg. 1990. Further characterization of the fate of human monoclonal antibodies in rhesus monkeys. Hybridoma 7:385-395. [DOI] [PubMed] [Google Scholar]
- 18.Falgout, B., M. Bray, J. J. Schlesinger, and C. J. Lai. 1990. Immunization of mice with recombinant vaccinia virus expressing authentic dengue virus nonstructural protein NS1 protects against lethal dengue virus encephalitis. J. Virol. 64:4356-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falgout, B., M. Pethel, Y. M. Zhang, and C. J. Lai. 1991. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus non-structural proteins. J. Virol. 65:2467-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentry, M. K., E. A. Henchal, J. M. McCown, W. E. Brandt, and J. M. Dalrymple. 1982. Identification of distinct antigenic determinants on dengue 2 virus using monoclonal antibodies. Am. J. Trop. Med. Hyg. 31:548-555. [DOI] [PubMed] [Google Scholar]
- 21.Glamann, J., D. R. Burton, P. W. H. I. Parren, H. J. Ditzel, K. A. Kent, C. Arnold, D. Montefori, and V. M. Hirsch. 1998. Simian immunodeficiency virus (SIV) envelope-specific Fabs with high-level homologous neutralizing activity: recovery from a long-term-nonprogressor SIV-infected macaque. J. Virol. 72:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gould, E. A., A. Buckley, A. D. Barrett, and N. Cammack. 1986. Neutralizing (54K) and non-neutralizing (54K and 48K) monoclonal antibodies against structural and non-structural yellow fever virus proteins confer immunity in mice. J. Gen. Virol. 67:591-595. [DOI] [PubMed] [Google Scholar]
- 23.Halstead, S. 1982. Immune enhancement of viral infection. Prog. Allergy 31:301-364. [PubMed] [Google Scholar]
- 24.Heinz, F. X., R. Berger, W. Tuma, and C. Kunz. 1983. A topological and functional model of epitopes on the structural glycoprotein of tick-borne encephalitis virus defined by monoclonal antibodies. Virology 126:525-537. [DOI] [PubMed] [Google Scholar]
- 25.Heinz, F. X. 1986. Epitope mapping of flavivirus glycoproteins. Adv. Virus Res. 31:103-168. [DOI] [PubMed] [Google Scholar]
- 26.Heinz, F. X., K. Stiasny, G. Pueschner-Auer, H. Holzmann, S. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 27.Henchal, E. A., M. K. Gentry, J. M. McCown, and W. E. Brandt. 1982. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 31:830-836. [DOI] [PubMed] [Google Scholar]
- 28.Henchal, E. A., L. S. Henchal, and J. J. Schlesinger. 1988. Synergistic interactions of anti-NS1 monoclonal antibodies protect passively immunized mice from lethal challenge with dengue 2 virus. J. Gen. Virol. 69:2101-2107. [DOI] [PubMed] [Google Scholar]
- 29.Henchal, E. A., J. M. McCown, D. S. Burke, M. C. Seguin, and W. E. Brandt. 1985. Epitope analysis of antigenic determinants on the surface of dengue 2 virion using monoclonal antibodies. Am. J. Trop. Med. Hyg. 34:162-169. [DOI] [PubMed] [Google Scholar]
- 30.Henchal, E. A., P. M. Repik, J. M. McCown, and W. E. Brandt. 1986. Identification of an antigenic and genetic variant of dengue-4 virus from the Caribbean. Am. J. Trop. Med. Hyg. 35:393-400. [DOI] [PubMed] [Google Scholar]
- 31.Holzmann, H., K. Stiasny, H. York, F. Dorner, C. Kunz, and F. X. Heinz. 1995. Tick-borne encephalitis virus envelope protein E-specific monoclonal antibodies for the study of low pH-induced conformational changes and immature virions. Arch. Virol. 140:213-221. [DOI] [PubMed] [Google Scholar]
- 32.Huber, C., K. F. Schable, E. Huber, R. Klein, A. Meindl, R. Thiebe, R. Lamm, and H. G. Zachau. 1993. The V kappa genes of the L regions and the repertoire of V kappa gene sequences in the human germ line. Eur. J. Immunol. 23:2868-2875. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, S., C. Oliver, G. A. Prince, V. G. Hemming, D. S. Pfarr, S.-C. Wang, M. Dormitzer, J. O'Grady, S. Koenig, J. K. Tamura, R. Woods, G. Bansal, D. Couchenour, E. Tsao, W. C. Hall, and J. F. Young. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 176:1215-1224. [DOI] [PubMed] [Google Scholar]
- 34.Kaufman, B. M., P. L. Summers, D. R. Dubois, and K. H. Eckels. 1987. Monoclonal antibodies against dengue 2 virus E-glycoprotein protect mice against lethal dengue infection. Am. J. Trop. Med. Hyg. 36:427-434. [DOI] [PubMed] [Google Scholar]
- 35.Kimura-Kuroda, J., and K. Yasui. 1988. Protection of mice against Japanese encephalitis virus by passive administration with monoclonal antibodies. J. Virol. 141:3606-3610. [PubMed] [Google Scholar]
- 36.Lai, C. J., B. Zhao, H. Hori, and M. Bray. 1991. Infectious RNA transcribed from stably cloned full-length cDNA of dengue type 4 virus. Proc. Natl. Acad. Sci. USA 88:5139-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanciotti, R. S., D. J. Gubler, and D. W. Trent. 1997. Molecular evolution and phylogeny of dengue type 4 virus. J. Gen. Virol. 78:2279-2284. [DOI] [PubMed] [Google Scholar]
- 38.Lin, C.-W., and S.-C. Wu. 2003. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing-antibody combining sites. J. Virol. 77:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandl, C. W., F. Guirakhoo, H. Holzmann, F. X. Heinz, and C. Kunz. 1989. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J. Virol. 63:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markoff, L. 1989. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J. Virol. 63:3345-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruyama, T., L. L. Rodriguez, P. B. Jahrling, A. Sanchez, A. S. Khan, S. T. Nichol, C. J. Peter, P. W. Parren, and D. R. Burton. 1999. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J. Virol. 73:6024-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathews, J. H., and J. T. Roehrig. 1984. Elucidation of the topography and determination of the protective epitopes on the E glycoprotein of Saint Louis encephalitis virus by passive transfer with monoclonal antibodies. J. Immunol. 132:1533-1537. [PubMed] [Google Scholar]
- 43.Men, R., M. Bray, and C.-J. Lai. 1991. Carboxy-terminally truncated dengue virus envelope glycoproteins expressed on the cell surface and secreted extracellularly exhibited increased immunogenicity in mice. J. Virol. 65:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Men, R., M. Bray, D. Clark, R. M. Chanock, and C.-J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogata, N., L. Östberg, P. H. Ehrlich, D. C. Wong, R. H. Miller, and R. H. Purcell. 1993. Markedly prolonged incubation period of hepatitis B in a chimpanzee passively immunized with a human monoclonal antibody to the a determinant of hepatitis B surface antigen. Proc. Natl. Acad. Sci. USA 90:3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okuno, Y., T. Fukunaga, M. Tadano, Y. Ohnishi, and M. Tagaki. 1985. Rapid focus reduction neutralization test of Japanese encephalitis virus in microtiter system. Arch. Virol. 86:129-135. [DOI] [PubMed] [Google Scholar]
- 48.Pech, M., H. Smola, H. D. Pohlenz, B. Straubinger, R. Gerl, and H. G. Zachau. 1985. A large section of the gene locus encoding human immunoglobulin variable regions of the kappa type is duplicated. J. Mol. Biol. 183:291-299. [DOI] [PubMed] [Google Scholar]
- 49.Persson, M. A., R. H. Caothien, and D. R. Burton. 1991. Generation of diverse high-affinity human monoclonal antibodies by repertoire cloning. Proc. Natl. Acad. Sci. USA 88:2432-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raffai, R., K. H. Weisgraber, R. MacKenzie, B. Rupp, E. Rassart, T. Hirama, T. L. Innerarity, and R. Milne. 2000. Binding of an antibody mimetic of the human low density lipoprotein receptor to apolipoprotein E is governed through electrostatic forces. J. Biol. Chem. 275:7109-7116. [DOI] [PubMed] [Google Scholar]
- 51.Rey, P. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 52.Roehrig, J. T., R. A. Bolin, and R. G. Kelly. 1998. Monoclonal antibody mapping of envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246:317-328. [DOI] [PubMed] [Google Scholar]
- 53.Rosen, L. 1986. Dengue in Greece in 1927 and 1928 and the pathogenesis of dengue hemorrhagic fever: new data and different conclusion. Am. J. Trop. Med. Hyg. 35:642-653. [DOI] [PubMed] [Google Scholar]
- 54.Sanna, P. P., M. E. Samson, J. S. Moon, R. Rozenshteyn, A. De Logu, R. A. Williamson, and D. R. Burton. 1999. pFab-CMV, a single vector system for the rapid conversion of recombinant Fabs into whole IgG1 antibodies. Immunotechnology 4:185-188. [DOI] [PubMed] [Google Scholar]
- 55.Schlesinger, J. J., and S. Chapman. 1999. Influence of the human high-affinity IgG receptor Fcγ1 (CD64) on residual infectivity of neutralized dengue virus. Virology 260:84-88. [DOI] [PubMed] [Google Scholar]
- 56.Schofield, D. J., J. Glamann, S. U. Emerson, and R. H. Purcell. 2000. Identification by phage display and characterization of two neutralizing chimpanzee monoclonal antibodies to the hepatitis E virus capsid protein. J. Virol. 74:5548-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shields, R. L., A. K. Namenuk, K. Hong, Y. G. Meng, J. Rae, J. Briggs, D. Xie, J. Lai, A. Stadlen, B. Li, J. A. Fox, and L. G. Presta. 2001. High resolution mapping of the binding site on human IgG1 for FcγRI, FcγRII, FcγRIII, and FcγRn and design of IgG1 variants with improved binding to the FcγR. J. Biol. Chem. 276:6591-6604. [DOI] [PubMed] [Google Scholar]
- 58.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takahashi, N., S. Ueda, M. Obata, T. Nikaido, S. Nakai, and T. Honjo. 1982. Structure of human immunoglobulin gamma genes: implication for evolution of a gene family. Cell 29:671-679. [DOI] [PubMed] [Google Scholar]
- 60.Tomlinson, I. M., G. Walter, J. D. Marks, M. B. Llewelyn, and G. Winter. 1992. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J. Mol. Biol. 227:776-798. [DOI] [PubMed] [Google Scholar]
- 61.Wang, W.-K., S.-R. Lin, C.-M. Lee, C.-C. King, and S.-C. Chang. 2002. Dengue type 3 virus in plasma is a population of closely related genomes: quasispecies. J. Virol. 76:4662-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wengler, G., and G. Wengler. 1989. Cell-associated West Nile flavivirus is covered with E + pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J. Virol. 63:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williamson, R. A., R. Burioni, P. P. Sanna, L. J. Patridge, C. F. Barbas III, and D. R. Burton. 1994. 1993. Human monoclonal antibodies against a plethora of viral pathogens from single combinatorial libraries. Proc. Natl. Acad. Sci. USA 90:4141-4145. (Erratum, 91:1193.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood, C. R., A. J. Dorner, G. E. Morris, E. M. Alderman, D. Wilson, R. M. O'Hara, Jr., and R. J. Kaufman. 1990. High level synthesis of immunoglobulins in Chinese hamster ovary cells. J. Immunol. 145:3011-3016. [PubMed] [Google Scholar]
- 65.Wu, T. T., G. Johnson, and E. A. Kabat. 1993. Length distribution of CDRH3 in antibodies. Proteins Struct. Funct. Genet. 16:1-7. [DOI] [PubMed] [Google Scholar]